Abstract
Neuro-oncology largely consists of malignancies of the brain and central nervous system including both primary as well as metastatic tumors. Currently, a significant clinical challenge in neuro-oncology is to tailor therapies for patients based on a priori knowledge of their survival outcome or treatment response to conventional or experimental therapies. Radiomics or the quantitative extraction of subvisual data from conventional radiographic imaging has recently emerged as a powerful data-driven approach to offer insights into clinically relevant questions related to diagnosis, prediction, prognosis, as well as assessing treatment response. Furthermore, radiogenomic approaches provide a mechanism to establish statistical correlations of radiomic features with point mutations and next-generation sequencing data to further leverage the potential of routine MRI scans to serve as “virtual biopsy” maps. In this review, we provide an introduction to radiomic and radiogenomic approaches in neuro-oncology, including a brief description of the workflow involving preprocessing, tumor segmentation, and extraction of “hand-crafted” features from the segmented region of interest, as well as identifying radiogenomic associations that could ultimately lead to the development of reliable prognostic and predictive models in neuro-oncology applications. Lastly, we discuss the promise of radiomics and radiogenomic approaches in personalizing treatment decisions in neuro-oncology, as well as the challenges with clinical adoption, which will rely heavily on their demonstrated resilience to nonstandardization in imaging protocols across sites and scanners, as well as in their ability to demonstrate reproducibility across large multi-institutional cohorts.
Keywords: glioblastoma, machine learning, radiogenomics, radiomics
Key Points.
Radiomics has recently emerged as a powerful data-driven approach that can offer insights into clinically relevant questions related to diagnosis, prediction, prognosis, as well as assessing treatment response.
Using radiomics and radiogenomic approaches to personalize treatment decisions in neuro-oncology will rely heavily on resilience to nonstandardization in imaging protocols across sites and scanners, as well as in their ability to demonstrate reproducibility across large multi-institutional cohorts.
The field of neuro-oncology encompasses primary and metastatic malignant tumors of the central nervous system (CNS) including the brain and spinal cord. While the incidence of brain tumors is rare (lifetime likelihood of less than 1%), their mortality and morbidity rate is unusually high. In 2020, around 23 890 cases of malignant brain tumors and 18 020 brain cancer-related deaths are estimated in adults across the globe.1,2 Among the malignant tumors, brainstem gliomas, glioblastomas (GBMs), and anaplastic astrocytomas have the worst outcome.3,4 While the survival rate for patients with a malignant brain or CNS tumor is poor with only 36% living beyond 5 years following diagnosis, it is particularly abysmal in GBM patients (age group of 55–64 years) who have a 5-year survival of 6%. Unfortunately, at this time, there are no widely recommended tests to screen for brain and spinal cord tumors.
Diagnosis, and treatment response assessment, in neuro-oncology is currently investigated using multi-parametric magnetic resonance imaging (MRI) such as T1-weighted imaging both before (T1w) and after administration of gadolinium-based contrast agent (Gd-T1w), T2-weighted imaging (T2w), and T2w-Fluid attenuation recovery (FLAIR) sequences. For instance, Response Assessment in Neuro-Oncology group (RANO criteria) utilizes the increase/decrease in the product of perpendicular diameter on routine MRI to identify patient’s response to treatment. On routine multi-parametric scans, Gd-T1w MRI is useful for elucidating a brain tumor, with large portions of the tumor region (or the entire tumor) typically enhancing in comparison to the brain parenchyma. Meanwhile, T2w and T2w-FLAIR MRI scans are often used to identify heterogeneously enhancing tumors like gliomas especially GBMs which often have peri-tumoral edema or inflammation, in order to differentiate these regions from the enhancing tumor.
In addition to qualitative/semiquantitative assessment of these imaging modalities by trained neuro-radiologists, radiologic imaging also harbors massive amounts of interpretable quantitative information which may not be appreciable via visual inspection or 2-dimensional assessment of a single measure (ie, tumor diameter).5 Recent advances in the high-throughput computational techniques and rapid algorithm developments have facilitated the extraction of meaningful information from radiologic imaging via “Radiomics.” Radiomics is defined as the extraction of “hand-crafted” features from routine radiological scans (X-rays, CT, MRI, and PET) that quantitatively capture the textural and morphological characteristics of a given tumor. For instance, radiomics attempts to comprehensively characterize the pixel-wise tumor characteristics including (1) semantic or qualitative features that include radiologist-derived assessments of the tumor including spiculations, size of the tumor along several axes, (2) shape-based features that quantitatively measure regular or irregular tumor boundary changes based on their 3D topology,6 (3) intratumoral heterogeneity measures including gray-level features, which investigate pixel-level textural differences to characterize how heterogeneous a tumor is,7,8 as well as (4) deformation features that capture the impact of tumor-related mass effect in the tumor micro-environment.9 These various radiomics-based approaches have been widely employed across multiple cancers including brain,10–14 lung,15–18 colorectal,19–21 breast,22–24 and prostate25,26 among others for diagnosis, prognosis, treatment response prediction, as well as measuring early treatment effects. Specifically, in the context of neuro-oncology, these approaches have shown tremendous promise in the development of noninvasive radiomic prognostic and predictive markers, as well as for distinguishing treatment effects from tumor recurrence, using routine MRI scans.10,27–29
In addition to advances in multi-parametric imaging, next-generation sequencing technology, which allows for the high-throughput sequencing of multiple genes, has been a significant propeller of tailoring personalized treatments in neuro-oncology.30,31 Recent investigative studies have identified several driver mutations and chromosomal anomalies as specific targets for personalizing treatment and improving prognosis32,33 in neuro-oncology. A few of these driver mutations have been reported to have prognostic (eg, isocitrate dehydrogenase [IDH], O6-methylguanine-DNA methyltransferase [MGMT] promoter methylation, and epidermal growth factor receptor [EGFR]) and predictive (eg, MGMT) implications in GBM tumors.34,35 However, molecular profiling involves tissue extraction often from stereotactic/needle biopsies that are inherently prone to sampling bias based on the site of biopsy and thus may not capture the spatial heterogeneity extant within the tumor (specifically in highly heterogeneous tumors such as GBM).36
Interestingly, the field of “radiogenomics” has provided a mechanism for establishing statistical associations of tumoral radiomic features with the underlying genetic profile of the tumor, including point mutations, signaling, and pathways of biological significance. By providing an imaging phenotype for the tumor corresponding to a particular genotype, radiogenomics may allow for circumventing the challenges with biopsy sampling. For example, radiogenomic approaches have been used to create “virtual-biopsy” maps to predict specific prognostic point mutations as well as chromosomal alterations37 based on the 2016 update on the WHO classification of diffuse gliomas in neuro-oncology.13,38,39 Identifying such radiogenomics associations may improve our understanding of how the changes in biological processes at the molecular level affect changes at a radiologic scale40 and may ultimately aid in personalizing treatment decisions.
Overview of Radiomic and Radiogenomics Pipeline
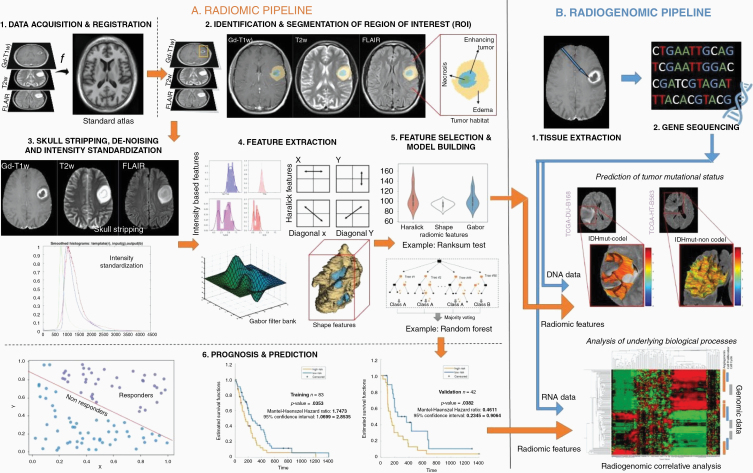
One of the primary advantages of developing and implementing a radiomic/radiogenomics pipeline is that, ideally, by leveraging routine MRI scans, the analysis does not significantly disrupt the existing clinical workflow. A typical pipeline of a radiomic/radiogenomics-based approach consists of the conversion of radiographic images into mineable data and involves the following steps: (1) image acquisition and registration, (2) segmentation of region of interest, (3) preprocessing, (4) feature extraction, (5) feature selection and building machine learning models for predictive and prognostic applications, and lastly (6) radiogenomic associations to either predict a genotype or identify the biological processes that drive the tumor biology. Figure 1 shows the complete radiomic/radiogenomics pipeline.
Figure 1.
Overall workflow of radiomic and radiogenomic pipeline.
Image Acquisition, Tumor Segmentation, and Preprocessing
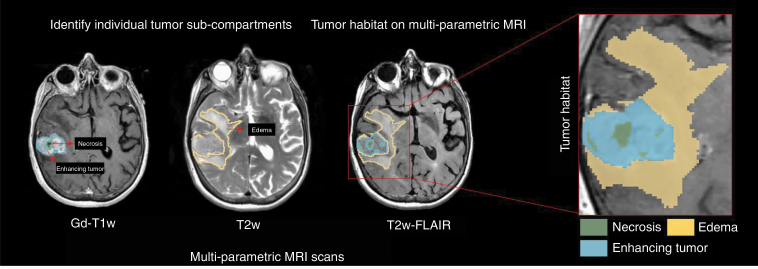
Preprocessing is a key step prior to radiomic feature extraction and involves multi-protocol registration to account for patient movement during image acquisition, as well as accounting for image variations in MRI scans across different manufacturers and multiple participating sites. Next, the region of interest is identified and segmented, either manually or automatically. On Gd-T1w images, necrosis is relatively represented as hypo-intense regions that are commonly located in the central region of the tumor and occasionally have a ring enhancement. Similarly, hyper-intense T2w-FLAIR signals correlate with greater interstitial leakage and low cellular density, reflecting edema. Therefore, T2w and T2w-FLAIR scans are typically used to identify edema and necrosis and enhancing tumor is delineated based on Gd-T1w MRI. The “tumor habitat” consisting of the 3 segmented tumor subcompartments, necrotic core, enhancing, and peri-tumoral edema, can then be interrogated by extracting radiomic features from each of the tumor subcompartments across different MRI protocols. Figure 2 illustrates the tumor habitat in GBM as delineated by an expert radiologist using routine MRI protocols. Currently, manual segmentations are considered the gold standard for radiomic analysis as they ensure high accuracy. However, there exist several automated segmentation approaches using deep learning (DL) architectures including U-Net, Conv-Net, Transfer Learning, and Deep Hourglass approaches as well as semiautomatic seeding-based algorithms that have gained popularity.41–43 These automated tools have shown promise in annotating the tumor subcompartments including enhancement, nonenhancement, necrosis, as well as peri-tumoral edema from the immediate periphery of the tumor.
Figure 2.
Using multi-parametric MRI scans to identify the entire tumor habitat of glioblastoma as delineated by an expert radiologist. Necrotic core (as marked in green) and enhancing tumor (as marked in blue) can be identified on post-contrast T1w MRI scans. Similarly, the peri-tumoral edema (as marked in yellow) can be identified on the T2w/T2w-FLAIR MRI scans.
Further preprocessing techniques such as skull stripping and intensity standardization are executed to account for varying magnetic strengths, and slice thicknesses in the curated dataset.44–47 Intensity standardization algorithms are implemented across multi-parametric MRI protocols to correct for intensity nonstandardness, which refers to the issue of MRI “intensity drift” across different imaging acquisitions. The intensity nonstandardness results in MRI intensities lacking tissue-specific numeric meaning within the same MRI protocol, for the same body region, or for images of the same patient obtained on the same scanner. Therefore, it is important that the radiomic pipelines in neuro-oncology implement the necessary preprocessing techniques to ensure consistency in the dataset and reproducibility of results.48
Radiomic Feature Extraction
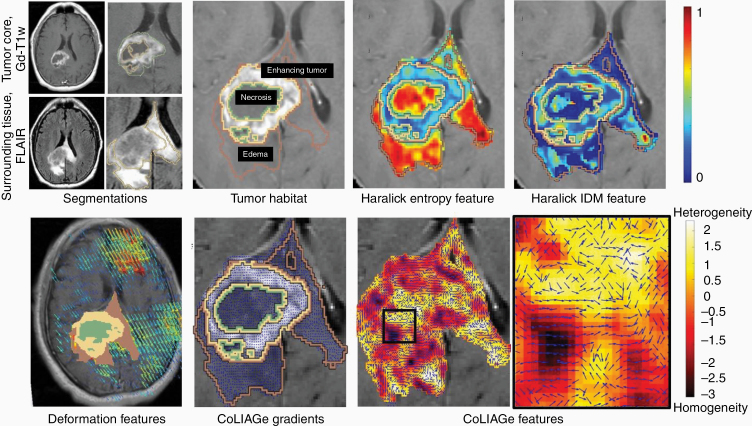
Following preprocessing and lesion segmentation, quantitative features are extracted for radiomic analysis, via algorithms that capture tumor heterogeneity across local pixel neighborhoods within a given radiologic image. Common radiomic features are currently divided into the following classes: semantic, shape, texture/gradient-based, deformation, and wavelet. Figure 3 illustrates an example of a few radiomic features extracted from the multi-parametric MRI scans from across the GBM tumor habitat. The specifics of different feature families are discussed in the subsequent subsections.
Figure 3.
(Top Row) Multi-parametric MRI is used to annotate the tumor subcompartments in GBM patients. The GBM tumor habitat consists of (1) necrotic core, (2) enhancing tumor, and (3) edema. Haralick features such as entropy and IDM are extracted from the GBM tumor habitat. (Bottom Row) Deformation features are calculated from the brain around tumor region. CoLlAGe gradients detect the homogeneous and heterogeneous regions based on the local intensity patterns on MRI scans.
“Hand-crafted” radiomic descriptors
Semantic features
In a large National Cancer Institute’s The Cancer Genomic Atlas (TCGA)-based effort, brain tumors (GBMs) were examined by experienced neuro-radiologists to define “semantic” features that capture the visual phenotypic characterization of the tumors. These unique semantic features were based on the 4 commonly analyzed tumor subcompartments (nonenhanced tumor, enhancing tumor, necrosis, and edema) and included features such as location of the lesion, morphology, major axis length, minor axis length margin, and lesion vicinity. This comprehensively characterized feature set by expert radiologists is now known as Visually AcceSAble Rembrandt Images (VASARI) features.49 Multiple studies have demonstrated that prognostic imaging attributes, such as percent of necrosis and edema, can be correlated with VASARI features (ρ = 0.67 [P < .001] and ρ = 0.41 [P < .001]) in GBM tumors.50,51 Another study found a significant correlation between overall survival (OS) in GBMs and VASARI features. These studies have established that VASARI features (such as degree of contrast enhancement as well as the length of the major axis of the lesion) may provide additional prognostic value on top of standard clinical variables (such as Karnofsky Performance Score [KPS]).52 Several radiogenomic-based studies have similarly demonstrated that genetic expression or RNA sequencing of GBMs may be correlated with the VASARI features.53–55 For instance, Diehn et al.55 have presented an extensive study that seemed to suggest that GBM tumor contrast enhancement may be associated with activation of specific hypoxia gene expression.
Shape-based features
Irregular and aggressive tumor infiltration may prompt surface and shape changes in the tumor and peri-tumoral regions. A larger surface-area-to-volume ratio, for example, has been found to be indicative of a more spiculated tumor, with more malignant potential in comparison to a round mass with a smaller ratio.56 Driven by this intuition, shape features may be informative of tumor malignancy. These features are broadly classified into 2 categories—local and global features. Local surface features capture characteristics such as curvature that identifies flat areas of surface from highly curved ones, sharpness that measures how sharp the curvature is, with highly curved masses exhibiting sharper curvatures, as well as shape index that characterizes the shape topology of the tumor.15 Whereas some of the global shape features include the major and minor axis, and elongation (ratio between major and minor axes of the region of interest). Several studies have shown that shape-based features of the tumor subcompartments in GBMs may be used to predict OS.8,57–60 Gevaert et al.51 have shown that high irregularity of enhancing lesion shape (ie, degree of spiculation) was associated with a lower OS. In a radiogenomic setting, Itakura et al.61 have reported that shape-driven features can classify GBMs into 3 distinct clusters, where each cluster is mapped to a unique set of molecular signaling pathways and differential probabilities of survival. For instance, they reported that cluster 2 (characterized by spherical tumors with regular edges and small volumes) was associated with downregulation of VEGFR signaling pathway. Apart from this, shape features have also been shown to be better in evaluating treatment response. Ismail et al.6 have demonstrated that 2 local features (total curvature of the enhancing lesion and curvedness of the T2w/T2w-FLAIR hyper-intense peri-tumoral edema) may be able to differentiate between pseudo-progression and tumor progression in GBMs.
Texture/gradient-based features—
The most commonly used texture features include (1) gray-level co-occurrence matrix (GLCM) or Haralick features that capture the variations in gray-level image characteristics via second-order intensity statistics (eg, entropy, inverse differential moment [IDM], angular second moment, contrast, and differential entropy).62 Haralick features potentially capture the structural heterogeneity within the region of interest. For instance, IDM is a reflection of the presence or absence of uniformity and hence is a measure of local regions of homogeneity. (2) Gray-level run length matrix (GLRLM), which in comparison with GLCM features, investigates the pixel runs instead of pairs of pixels. A pixel run includes the number of pixels of a specific gray value that are in a right direction, in the right sequence. While the rows of the matrix still represent gray levels, the columns represent run lengths. (3) Laws features that define various texture parameters including spot, edge, ripple, and level surfaces present within the tumor63 and (4) Co-occurrence of Local Anisotropic Gradient Orientations (CoLlAGe), which captures local anisotropic differences that exist in micro-structures by measuring entropy (a mathematical construct to measure disorder) of co-occurrences of pixel/voxel-level gradient orientations computed within a local neighborhood.64
Haralick features have been used extensively in the context of neuro-oncology to predict survival.7,12 Kickingereder et al.65 have shown that Haralick features extracted from the T2w-FLAIR MRI sequences can predict survival and stratify patients with newly diagnosed GBM. In 2 independent studies, it has been demonstrated that GLRLM features, extracted from multi-parametric MRI scans, are also prognostic of OS in GBMs.66,67 GLRLM features extracted from 18F-FDG-PET have also shown to distinguish primary CNS lymphoma from GBMs.68 In a cohort of 42 patients, CoLlAGe has been shown to differentiate radiation necrosis, a benign yet confounding effect of radiation treatment, from recurrent tumors, with an accuracy of 83.79% in primary cases, and 88.52% in metastatic brain tumors.64,69
Deformation-based features
Deformation features seek to measure the extent of tissue deformation in the brain parenchyma (ie, brain around tumor [BAT]) due to the mass effect in brain tumors. MRI scans are nonrigidly registered to equivalent healthy, age-matched, and/or gender-matched imaging atlases. Then, the resulting deformation field (represented as a displacement vector at every voxel location) is obtained through a combination of forward as well as inverse mapping between the patient’s 3D volume and the reference atlas. The per-voxel deformation measurements from the BAT region are then used as radiomic features for analysis. Prasanna et al.70 hypothesized that larger variations in deformation within functional areas of the contralateral hemisphere are likely related to decreased survival in GBMs. They demonstrated that decreased OS was found to be associated with increased deformation in areas of language comprehension, visual perception, cognitive, and motor-control functions, particularly in the memory areas in the left hemisphere. In another study, they also showed that combining textural-based radiomic features with deformation values may result in improved prediction of survival in GBMs, for identifying short-term survivors (OS <240 days) from long-term survivors (OS >540 days).9 In a radiogenomic setting, Iyer et al.71 have shown that deformation-based radiomic features can also be used to differentiate the molecular subtypes of medulloblastomas (Sonic Hedgehog [SHH], Wingless [WNT], Group 3, Group 4) in pediatric brain tumors.
Wavelet-based features—
Wavelet features utilize different wavelengths, amplitudes, and frequencies to recognize textural attributes across a wide range of scales within the image. For instance, Gabor wavelet features are the steerable class of gradients that attempts to match localized frequency characteristics.72 A Gabor filter can be defined as the modulation of a complex sinusoid by a Gaussian function. Each descriptor quantifies response to a given Gabor filter at a specific frequency (f = 0, 4, or 16) and orientation (µ = 45°, 90°, 135°, 180°) and attempts to capture the prominent direction in which intensity changes occur. Tixier et al.73 have reported that GBM patients with large negative skewness of Gabor wavelets had a significantly longer median OS of 22.7 months (P = .004). In another study, a wavelet scattering-based noise robust radiomic method was implemented to predict the gliomas grade in brain tumors.74
Feature Selection and Building Prognostic and Predictive Models
Building radiomic-based machine learning classifiers involves reducing the high-dimensional radiomic feature set by selecting the most discriminative features using a feature selection scheme, in order to reduce the curse of dimensionality.75 Feature selection can either be using univariate or multivariate statistical models. Univariate methods (also known as filter methods) only depend on feature association, while ignoring redundancy, whereas multivariate methods (wrapper methods) inspect inter-actions within different features and account for both association and redundancy. Fisher score, Chi-squared test, and Wilcoxon are the most common filter methods that are used for feature selection.16 Wrapper models are computationally expensive as their objective is to find a subset of features that result in the best performing model. Notable examples for wrapper methods include forward feature selection, backward feature elimination, exhaustive feature selection (greedy algorithm), or bidirectional search.16,76
After identifying a subset of features following feature selection, machine learning models are developed by categorizing and classifying various datasets according to defined labels (ie, GBM vs low-grade glioma [LGG], poor survival vs improved survival, radiation effects vs tumor recurrence). Generally, classifiers can be divided into supervised and unsupervised approaches. While supervised methods utilize a predefined set of known labels to identify features that best represent the outcomes of interest on the radiologic images, an unsupervised approach (such as clustering) can be employed when the target labels are unknown. A range of classifiers including random forest, support vector machines (SVMs), and generalized linear models have been used for diagnosis and treatment response evaluation applications.6,12,76 For instance, previous studies have used SVM classifier to predict the histopathological grade (LGGs vs GBMs) of a given primary brain tumor using MRI scans.76–78 Research groups have employed least absolute shrinkage and selection operator (LASSO) logistic regression and SVM models79,80 to differentiate pseudo-progression from early tumor progression in GBM patients. Prasanna et al.7 have shown that radiomic features extracted from the peri-tumoral edema on multi-parametric MRI were able to distinguish short-term survivors (OS <7 months) and long-term survivors (OS >18 months) in 65 GBMs studies. Recently, DL-based architectures such as convolutional neural networks and auto-encoders have also been used to extract features from the radiologic imaging data.27,81 While DL is capable of apprehending more abstract and higher-dimensional relationships between imaging features and the clinical end point, these approaches are still largely considered black-box.82
Survival Analysis
In most of the survival-based (ie, prognostic) radiomic/radiogenomic studies, Cox proportional hazards model are used, where the event of disease diagnosis is chosen as the time of origin. The commonly used end points for survival analysis are either OS (time from disease diagnosis to death due to the cancer in question) or time duration from a given treatment to response or outcome (progression-free survival [PFS]).83 In a Cox proportional hazard regression model, hazard rate (HR) quantifies the effect of individual feature on survival and measures the risk of failure (ie, the risk or probability of suffering the event of interest), given that the patient has survived up to a specific time. Risk parameters yielding negative regression coefficients (ie, low feature values correlated with long-term survival) produce a HR between 0 and 1; features yielding positive regression coefficients (ie, low feature values correlated with short-term survival) produce a HR between 1 and infinity.40,84,85
Kaplan–Meier (KM) curves, a nonparametric survival analysis method, have also been well accepted for evaluation of the “time-to-event” data.86 KM curves evaluate the probability of survival over time (dependent variable), under different conditions (independent variable), where the horizontal axis represents the time and the vertical axis shows the probability of survival. After the KM curves have been plotted, the log-rank test is used to compare the 2 curves (high risk vs low risk). The log-rank test calculates the chi-square (χ 2) for each event time across all groups and sums up the results. χ 2 from the log-rank test concludes whether 2 curves from the groups are statistically different.87
Radiogenomic Studies
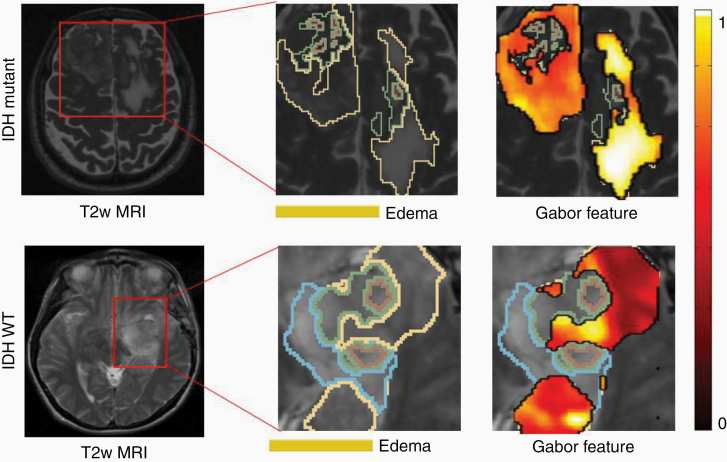
Multiple radiomic features have been shown, via radiogenomic analysis, to capture genomic alterations within tumor DNA, on routine MRI scans. These radiomic features are shown to identify the presence of specific mutations that have implications in the management and outcome of neuro-oncology patients.54,88,89 For instance, several groups have demonstrated that the IDH mutations in diffuse gliomas may be predicted via radiomic signatures on pretreatment MRI (Figure 4).32,38,90,91 In a recent study, Shboul et al.92 have demonstrated that radiomic features can discriminate IDH mutation from IDH wild type in LGG with an AUC of 0.84. They presented that higher values of the size ratio between the enhancing tumor and the necrosis, and higher values of the vertical orientation of edema major axis were significantly associated (ANOVA test, P value < .005) with IDH wild-type status. Similarly, radiogenomic models have also been developed to predict other imaging biomarkers for MGMT,13,33,73 EGFR,93 TERT,94,95 PTEN,96 and ATRX97 mutations in neuro-oncology.
Figure 4.
Isocitrate dehydrogenase (IDH) is an independent prognostic factor in gliomas, with mutated IDH1 and IDH2 having improved prognosis compared to gliomas with wild-type IDH. Gradient and intensity statistics texture features within edema in our preliminary work were found to discriminate IDH1 mutation versus wild-type gliomas on n = 78 studies.90
Furthermore, radiogenomic analysis could be leveraged to elucidate the underlying biological basis of the prognostic radiomic features, by identifying signaling pathways that drive tumor biology between the radiomics-driven survival groups.40,98,99 These radiogenomic approaches first identify the prognostic radiomic features that can identify low-risk from high-risk groups based on patient outcome (OS or PFS). Next, bioinformatics tools such as Gene Ontology (GO)100,101 and single-sample gene set enrichment analysis (ssGSEA)102 are employed to detect associated downstream signaling pathways of biological significance across the low-risk and the high-risk categories. For a predefined set of genes, ssGSEA captures the significantly enriched or depleted biological processes and calculates an enrichment score for every patient in the cohort. GO is a knowledge base of the functions of genes and thus forms a foundation for computational analysis of large-scale molecular biology and genetics experiments. GO also highlights the most overrepresented genes and finds the systematic linkages between those genes and biological processes.
Using a radiogenomic approach, Liu et al.11 demonstrated that PFS in LGG may be noninvasively predicted using radiomic features from T2w MRI scans, and then by using GO analysis, revealed that their identified prognostic radiomic features were associated with biology processes of programmed cell death and cell proliferation. Another radiogenomic study developed and independently evaluated a LASSO-based Radiomic Risk Score (RRS), using radiomic features from the tumor habitat, to stratify 203 GBM patients into low-risk and high-risk groups based on PFS. The RRS consisted of a total of 25 radiomic features including textural features belonging to Laws energy and Gabor wavelet families, and shape features from the peri-tumoral edema region, on Gd-T1w MRI. The study then identified significant radiogenomic correlations (P < .05) between the prognostic radiomic features with molecular signaling pathways, such as cell differentiation, cell adhesion, and angiogenesis, using GO and ssGSEA.40 Thus, by establishing multi-scale associations of radiomic phenotypes with corresponding transcriptomic data, radiogenomics approaches may allow for an improved understanding of the underlying disease biology as well as potentially aid in build patient-centric treatment plans.
DL-Based Approaches
Unlike traditional radiomic-based approaches that rely on accurate segmentation and selection of hand-crafted engineered features, DL methods are domain-agnostic. Most DL approaches do not require labor-intensive segmentations and have the ability to capture complex hidden visual representations of radiologic data.82,103–105 Convolutional neural networks (CNNs) are the most common DL models used in medical image analysis.106 A CNN has an input layer, an output layer, and the parallel computations are performed within multiple hidden layers, which include convolutional and pooling layers. Convolutional layers are the building blocks of CNNs from which features are extracted from the input layer (to which small patches of image data are fed). Pooling layers transfer the maximum or average convolved values and consequently reduce the size of the features extracted. Repeated mathematical operations of convolution and pooling result in altered representations of the imaging data and capture several features including (but not limited to) edge detection, color variations, sharpening, blurring, and focusing. During the training phase, guided by a loss function (which estimates the difference between the labels and predictions), the CNN determines the weights of these convolution filters in recognizing subtle visual signatures hidden in images.
In the context of brain tumors, Lao et al.81 developed a DL-based radiomics model using Gd-T1, T2w, T2w-FLAIR MRI protocols and clinical data (age and KPS) from 112 GBM patients and obtained a C-index of 0.71 in predicting OS. In a recent study, Bae et al.107 demonstrated that a deep neural nets can distinguish GBMs from metastasis with an accuracy of 0.95 (95% CI, 0.92–0.99). In another study by Han et al.,108 a bi-directional recurrent CNN on 260 TCGA-GBM patients was developed to obtain an accuracy of 62% in predicting MGMT gene status on an independent test data using multi-parametric MRI.
Limitations
While highly promising, a key challenge in enabling clinical utility of radiomic/radiogenomic approaches is to demonstrate their generalizability to variations in image acquisition protocols across scanners and sites. Sources of variations in MRI acquisition often include differences in image contrast, voxel resolutions, slice thicknesses, image reconstruction methods, magnetic field strengths, echo times, and repetition times. This issue of reproducibility of radiomic features has become even more pertinent in retrospective studies that involve publicly available repositories such as TCGA-GBM and Ivy GAP, where image scans are pooled-in from different institutions.
Several efforts are currently underway to fill these technical gaps, including attempting to standardize the image acquisition, preprocessing, segmentation guidelines, and radiomic features extraction pipelines across multiple sites.109 Most recently, the image biomarker standardization initiative has provided best-practice guidelines for standardizing feature extraction pipelines from MRI scans from different sites and scanners.110 Additionally, open-source software platforms such as Cancer Imaging Phenomics Toolkit, which is specifically developed for neuro-oncology applications111 as well as a more generalized Pyradiomics112 platform, also aim to provide the medical community with standardized set of pipeline for radiomic feature analysis. Additionally, a few recent studies113–115 have explored the issue of repeatability, which refers to the variability in radiomic features across scans obtained at 2 different times on the same scanner, as well as reproducibility, which is defined as the variations in radiomic features on account of differences in image acquisition across sites and scanners. These repeatability and reproducibility radiomic studies have been conducted by leveraging test–retest datasets or phantom studies with varying imaging acquisition protocols. However, rigorous analysis is warranted to improve the generalizability of radiomic features, in terms of repeatability, reproducibility as well as efficacy, across large multi-site cohorts (preferably using retrospective clinical trial datasets). Another important aspect of the radiomic pipeline is the segmentation of the tumor habitat. Manual tumor segmentation is not only labor intensive, but is also affected by inter-observer variability.5,16 While some radiomic studies use automatic and semiautomatic methods for segmentation, the existing segmentation algorithms are not consistent among different research groups, and may further have an impact on the radiomic analysis as well as downstream prognostic and predictive analysis.
Similarly, while the advent of DL networks has opened new avenues of research in GBM analysis, it comes with its own set of limitations. Unlike the radiomic features that often provide at least some degree of interpretability, DL features are considered more of a “black-box”.116 These deep features are limited in their explanatory capacity with neither a set of diagnostic rules nor an insight into the results. Additionally, DL models are limited by the relative sparsity of training samples, which is even more relevant in rare cancers such as GBMs, where obtaining very large data cohorts may not be feasible. Table 1 briefly contrasts the advantages and disadvantages of expert-based, radiomics and DL-based approaches in the context of brain tumor characterization.
Table 1.
Comparison of Radiomics and Deep Learning-Based Approaches
| Expert-Based Evaluation | Radiomics | Deep Learning | |||
|---|---|---|---|---|---|
| Advantages | Limitations | Advantages | Limitations | Advantages | Limitations |
| Observation-driven | Qualitative/semiquantitative | Hand-crafted engineered features | Impacted by variance in image acquisition parameters introduced across sites and scanners | Domain agnostic data-driven | Known as “black-box” due to limited biological interpretability offered the deep features |
| Experience-driven | Labor intensive | Often dependent on segmentation of the tumor habitat | |||
| Low computational costs | Intra- and inter-observer variability | Hand-crafted engineered features | Often used for small retrospective data and may not be generalizable | Does not require segmentation of tumor habitat | Limited by relative sparsity of training samples, not always suited for applications with limited availability of well-curated samples |
| Abundant historical literature | Poor reproducibility |
Conclusion and Future Scope
Radiomic, radiogenomic, and DL studies have made notable progress in the last few years and have demonstrated potential in the field of neuro-oncology, including aiding in diagnosis, outcome prediction, as well as evaluating response to both conventional and experimental treatments.80,89,97 However, for clinical deployment of these approaches, a few important considerations need to be accounted for. First, it may be important to establish causal inference of the radiogenomic associations between radiomic features and the underlying tumor biology, either through controlled preclinical models or spatially co-localized imaging and -omics datasets.117 Second, rigorous repeatability, reproducibility, and efficacy analysis of these approaches will need to be conducted across large multi-site retrospective cohorts. These large multi-institutional retrospective studies could then pave the way for prospective randomized trials to evaluate the efficacy of radiomic and radiogenomic markers in predicting response to treatment and guiding treatment decisions. Lastly, for translation of radiomic/radiogenomic/DL models as decision support in a clinical setting, careful planning will be needed in order to integrate the human and machine interpretations together and improve diagnostic and prognostic reads. Hence, unified efforts from all stakeholders including neuro-radiologists, neuro-oncologists, neuro-surgeons, along with data scientists, and machine learning researchers will be required toward development of these models from conception to deployment. Such cross-disciplinary collaboration will ensure that the tools being developed are tailored and aligned to benefit the patient in a variety of clinical settings including cancer screening, diagnosis, prediction of prognosis, and evaluating treatment response in neuro-oncology.
Funding
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number NCI 1U01CA248226-01, the DOD Peer Reviewed Cancer Research Program W81XWH-18-1-0404, Dana Foundation David Mahoney Neuroimaging Program, the V Foundation Translational Research Award, the Ohio Third Frontier Technology Validation Fund, and the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering.
Conflict of interest statement. N.B. is an employee of Tempus Labs, Inc. K.B. is a resident at Maimonides Medical Center, Brooklyn, New York. No potential conflicts of interest were disclosed by the other authors.
References
- 1. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol. 2017;19(suppl 5):v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. American Cancer Society Cancer Facts & Figures 2020. Atlanta: American Cancer Society; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fisher JL, Schwartzbaum JA, Wrensch M, Wiemels JL. Epidemiology of brain tumors. Neurol Clin. 2007;25(4):867–890, vii. [DOI] [PubMed] [Google Scholar]
- 4. DeAngelis LM. Brain tumors. N Engl J Med. 2001;344(2):114–123. [DOI] [PubMed] [Google Scholar]
- 5. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ismail M, Hill V, Statsevych V, et al. Shape features of the lesion habitat to differentiate brain tumor progression from pseudoprogression on routine multiparametric MRI: a multisite study. AJNR Am J Neuroradiol. 2018;39(12):2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prasanna P, Patel J, Partovi S, Madabhushi A, Tiwari P. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: preliminary findings. Eur Radiol. 2017;27(10):4188–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rathore S, Akbari H, Doshi J, et al. Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: implications for personalized radiotherapy planning. J Med Imaging (Bellingham). 2018;5(2):021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prasanna P, Mitra J, Beig N, et al. Radiographic-deformation and textural heterogeneity (r-DepTH): an integrated descriptor for brain tumor prognosis. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S, eds. Medical Image Computing and Computer-Assisted Intervention − MICCAI 2017. Quebec City, QC, Canada: Springer International Publishing; 2017:459–467. [Google Scholar]
- 10. Zhou M, Scott J, Chaudhury B, et al. Radiomics in brain tumor: image assessment, quantitative feature descriptors, and machine-learning approaches. AJNR Am J Neuroradiol. 2018;39(2):208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu X, Li Y, Qian Z, et al. A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. Neuroimage Clin. 2018;20:1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beig N, Patel J, Prasanna P, et al. Radiogenomic analysis of hypoxia pathway is predictive of overall survival in Glioblastoma. Sci Rep. 2018;8(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xi Y, Guo F, Xu Z, et al. Radiomics signature: a potential biomarker for the prediction of MGMT promoter methylation in glioblastoma: GBM radiomics features reflect MGMT. J Magn Reson Imaging. 2018;47(5):1380–1387. [DOI] [PubMed] [Google Scholar]
- 14. Bakas S, Shukla G, Akbari H, et al. Overall survival prediction in glioblastoma patients using structural magnetic resonance imaging (MRI): advanced radiomic features may compensate for lack of advanced MRI modalities. J Med Imaging (Bellingham). 2020;7(3):031505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Orooji M, Alilou M, Rakshit S, et al. Combination of computer extracted shape and texture features enables discrimination of granulomas from adenocarcinoma on chest computed tomography. J Med Imaging (Bellingham). 2018;5(2):024501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thawani R, McLane M, Beig N, et al. Radiomics and radiogenomics in lung cancer: a review for the clinician. Lung Cancer. 2018;115:34–41. [DOI] [PubMed] [Google Scholar]
- 17. Kalpathy-Cramer J, Mamomov A, Zhao B, et al. Radiomics of lung nodules: a multi-institutional study of robustness and agreement of quantitative imaging features. Tomography. 2016;2(4):430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilson R, Devaraj A. Radiomics of pulmonary nodules and lung cancer. Transl Lung Cancer Res. 2017;6(1):86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Antunes JT, Ofshteyn A, Bera K, et al. Radiomic features of primary rectal cancers on baseline T2‐weighted MRI are associated with pathologic complete response to neoadjuvant chemoradiation: a multisite study. J Magn Reson Imaging. 2020;52:1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horvat N, Veeraraghavan H, Khan M, et al. MR Imaging of rectal cancer: radiomics analysis to assess treatment response after neoadjuvant therapy. Radiology. 2018;287(3):833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma X, Shen F, Jia Y, Xia Y, Li Q, Lu J. MRI-based radiomics of rectal cancer: preoperative assessment of the pathological features. BMC Med Imaging. 2019;19(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perou CM, Fan C, Morris E, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016;2:16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu Y, Li H, Guo W, et al. Deciphering genomic underpinnings of quantitative MRI-based radiomic phenotypes of invasive breast Carcinoma. Sci Rep. 2015;5:17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shiradkar R, Ghose S, Jambor I, et al. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: preliminary findings. J Magn Reson Imaging. 2018;48(6):1626–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Penzias G, Singanamalli A, Elliott R, et al. Identifying the morphologic basis for radiomic features in distinguishing different Gleason grades of prostate cancer on MRI: preliminary findings. PLoS One. 2018;13(8):e0200730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park JE, Kickingereder P, Kim HS. Radiomics and deep learning from research to clinical workflow: neuro-oncologic imaging. Korean J Radiol. 2020;21(10):1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kickingereder P, Isensee F, Tursunova I, et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 2019;20(5):728–740. [DOI] [PubMed] [Google Scholar]
- 29. Lohmann P, Kocher M, Ruge MI, et al. PET/MRI Radiomics in patients with brain metastases. Front Neurol. 2020;11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cancer Genome Atlas Research Network, Brat DJ, Verhaak RGW, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29(34):4482–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Butler M, Pongor L, Su YT, et al. MGMT Status as a clinical biomarker in Glioblastoma. Trends Cancer. 2020;6(5):380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Westphal M, Maire CL, Lamszus K. EGFR as a target for glioblastoma treatment: an unfulfilled promise. CNS Drugs. 2017;31(9):723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taghizadeh H, Müllauer L, Furtner J, et al. Applied precision cancer medicine in neuro-oncology. Sci Rep. 2019;9(1):20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Louis DN, Perry A, Reifenberger G, et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 38. Park YW, Han K, Ahn SS, et al. Prediction of IDH1-mutation and 1p/19q-codeletion status using preoperative MR imaging phenotypes in lower grade Gliomas. AJNR Am J Neuroradiol. 2018;39(1):37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rivera AL, Pelloski CE, Gilbert MR, et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12(2):116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beig N, Bera K, Prasanna P, et al. Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI is associated with biological processes in Glioblastoma. Clin Cancer Res. 2020;26(8): 1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bakas S, Zeng K, Sotiras A, et al. GLISTRboost: combining multimodal MRI segmentation, registration, and biophysical tumor growth modeling with gradient boosting machines for glioma segmentation. Brainlesion (2015). 2016;9556:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hesamian MH, Jia W, He X, Kennedy P. Deep learning techniques for medical image segmentation: achievements and challenges. J Digit Imaging. 2019;32(4):582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Malathi M, Sinthia P. Brain tumour segmentation using convolutional neural network with tensor flow. Asian Pac J Cancer Prev. 2019;20(7):2095–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tao X, Chang M-C. A skull stripping method using deformable surface and tissue classification. In: Dawant BM, Haynor DR, eds. San Diego, CA; 2010. [cited 2018 Oct 24]. p. 76233L. http://proceedings.spiedigitallibrary.org/proceeding.aspx? [Google Scholar]
- 45. Madabhushi A, Udupa JK. New methods of MR image intensity standardization via generalized scale. In: Fitzpatrick JM, Reinhardt JM, eds. San Diego, CA; 2005. [cited 2018 Dec 25]. p. 1143. http://proceedings.spiedigitallibrary.org/proceeding.aspx? [DOI] [PubMed] [Google Scholar]
- 46. Smith SM, Brady JM. SUSAN—a new approach to low level image processing. Int J Comput Vis. 1997;23(1):45–78. [Google Scholar]
- 47. Bakas S, Akbari H, Sotiras A, et al. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data. 2017;4:170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Um H, Tixier F, Bermudez D, Deasy JO, Young RJ, Veeraraghavan H. Impact of image preprocessing on the scanner dependence of multi-parametric MRI radiomic features and covariate shift in multi-institutional glioblastoma datasets. Phys Med Biol. 2019;64(16):165011. [DOI] [PubMed] [Google Scholar]
- 49. VASARI Research Project—The Cancer Imaging Archive (TCIA) Public Access—Cancer Imaging Archive Wiki [Internet]. [cited 2020 Sep 5]. https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project
- 50. Rios Velazquez E, Meier R, Dunn WD Jr, et al. Fully automatic GBM segmentation in the TCGA-GBM dataset: prognosis and correlation with VASARI features. Sci Rep. 2015;5:16822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gevaert O, Mitchell LA, Achrol AS, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273(1):168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gutman DA, Cooper LA, Hwang SN, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267(2):560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Colen R, Foster I, Gatenby R, et al. NCI Workshop report: clinical and computational requirements for correlating imaging phenotypes with genomics signatures. Transl Oncol. 2014;7(5):556–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jamshidi N, Diehn M, Bredel M, Kuo MD. Illuminating radiogenomic characteristics of glioblastoma multiforme through integration of MR imaging, messenger RNA expression, and DNA copy number variation. Radiology. 2014;270(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008;105(13):5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee G, Lee HY, Park H, et al. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: state of the art. Eur J Radiol. 2017;86:297–307. [DOI] [PubMed] [Google Scholar]
- 57. Chaddad A, Desrosiers C, Hassan L, Tanougast C. A quantitative study of shape descriptors from glioblastoma multiforme phenotypes for predicting survival outcome. Br J Radiol. 2016;89(1068):20160575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Henker C, Kriesen T, Glass Ä, Schneider B, Piek J. Volumetric quantification of glioblastoma: experiences with different measurement techniques and impact on survival. J Neurooncol. 2017;135(2):391–402. [DOI] [PubMed] [Google Scholar]
- 59. Sanghani P, Ti AB, Kam King NK, Ren H. Evaluation of tumor shape features for overall survival prognosis in glioblastoma multiforme patients. Surg Oncol. 2019;29:178–183. [DOI] [PubMed] [Google Scholar]
- 60. Czarnek N, Clark K, Peters KB, Mazurowski MA. Algorithmic three-dimensional analysis of tumor shape in MRI improves prognosis of survival in glioblastoma: a multi-institutional study. J Neurooncol. 2017;132(1):55–62. [DOI] [PubMed] [Google Scholar]
- 61. Itakura H, Achrol AS, Mitchell LA, et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med. 2015;7(303):303ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;3(6):610–621. [Google Scholar]
- 63. Laws KI. Textured Image Segmentation. University of Southern California Los Angeles Image Processing Inst, University of Southern California Los Angeles Image Processing Inst; 1980. Jan. Report No.: USCIPI-940. http://www.dtic.mil/docs/citations/ADA083283 [Google Scholar]
- 64. Prasanna P, Tiwari P, Madabhushi A. Co-occurrence of Local Anisotropic Gradient Orientations (CoLlAGe): a new radiomics descriptor. Sci Rep. 2016;6:37241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kickingereder P, Burth S, Wick A, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280(3):880–889. [DOI] [PubMed] [Google Scholar]
- 66. Bae S, Choi YS, Ahn SS, et al. Radiomic MRI phenotyping of glioblastoma: improving survival prediction. Radiology. 2018;289(3): 797–806. [DOI] [PubMed] [Google Scholar]
- 67. Li Q, Bai H, Chen Y, et al. A fully-automatic multiparametric radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in glioblastoma multiforme. Sci Rep. 2017;7(1):14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kong Z, Jiang C, Zhu R, et al. 18F-FDG-PET-based radiomics features to distinguish primary central nervous system lymphoma from glioblastoma. Neuroimage Clin. 2019;23:101912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Prasanna P, Rogers L, Lam TC, et al. Disorder in pixel-level edge directions on T1WI is associated with the degree of radiation necrosis in primary and metastatic brain tumors: preliminary findings. AJNR Am J Neuroradiol. 2019;40(3):412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Prasanna P, Mitra J, Beig N, et al. Mass effect deformation heterogeneity (MEDH) on gadolinium-contrast T1-weighted MRI is associated with decreased survival in patients with right cerebral hemisphere glioblastoma: a feasibility study. Sci Rep. 2019;9(1):1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Iyer S, Ismail M, Tamrazi B, et al. Deformation heterogeneity radiomics to predict molecular subtypes of pediatric medulloblastoma on routine MRI. Medical Imaging 2019: Computer-Aided Diagnosis. International Society for Optics and Photonics; 2019. [cited 2020 Sep 5]. p. 109501E. https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10950/109501E/Deformation-heterogeneity-radiomics-to-predict--molecular-subtypes-of-pediatric/10.1117/12.2513567.short
- 72. Jain AK, Farrokhnia F. Unsupervised texture segmentation using Gabor filters. 1990 IEEE International Conference on Systems, Man, and Cybernetics Conference Proceedings; 1990:14–19.
- 73. Tixier F, Um H, Bermudez D, et al. Preoperative MRI-radiomics features improve prediction of survival in glioblastoma patients over MGMT methylation status alone. Oncotarget. 2019;10(6):660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen Q, Wang L, Wang L, Deng Z, Zhang J, Zhu Y. Glioma grade prediction using wavelet scattering-based radiomics. IEEE Access. 2020;8:106564–106575. [Google Scholar]
- 75. Friedman JH. On bias, variance, 0/1—loss, and the curse-of-dimensionality. Data Min Knowl Discov. 1997;1(1):55–77. [Google Scholar]
- 76. Lohmann P, Galldiks N, Kocher M, et al. Radiomics in neuro-oncology: basics, workflow, and applications. Methods. 2020:S1046–2023(19)30317-2. [DOI] [PubMed] [Google Scholar]
- 77. Tian Q, Yan LF, Zhang X, et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J Magn Reson Imaging. 2018;48(6):1518–1528. [DOI] [PubMed] [Google Scholar]
- 78. Vamvakas A, Williams SC, Theodorou K, et al. Imaging biomarker analysis of advanced multiparametric MRI for glioma grading. Phys Med. 2019;60:188–198. [DOI] [PubMed] [Google Scholar]
- 79. Kim JY, Park JE, Jo Y, et al. Incorporating diffusion- and perfusion-weighted MRI into a radiomics model improves diagnostic performance for pseudoprogression in glioblastoma patients. Neuro Oncol. 2019;21(3):404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Akbari H, Rathore S, Bakas S, et al. Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma. Cancer. 2020;126(11):2625–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lao J, Chen Y, Li ZC, et al. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep. 2017;7(1):10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shen D, Wu G, Suk H-I. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cox DR. Regression models and life‐tables. J R Stat Soc Ser B (Methodological). 1972;34:187–202. [Google Scholar]
- 84. McGarry SD, Hurrell SL, Kaczmarowski AL, et al. Magnetic resonance imaging-based radiomic profiles predict patient prognosis in newly diagnosed glioblastoma before therapy. Tomography. 2016;2(3):223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Weninger L, Haarburger C, Merhof D. Robustness of radiomics for survival prediction of brain tumor patients depending on resection status. Front Comput Neurosci. 2019;13:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53(282):457–481. [Google Scholar]
- 87. Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res. 2010;1(4):274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tejada Neyra MA, Neuberger U, Reinhardt A, et al. Voxel-wise radiogenomic mapping of tumor location with key molecular alterations in patients with glioma. Neuro Oncol. 2018;20(11):1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ellingson BM. Radiogenomics and imaging phenotypes in glioblastoma: novel observations and correlation with molecular characteristics. Curr Neurol Neurosci Rep. 2015;15(1):506. [DOI] [PubMed] [Google Scholar]
- 90. Beig N, Ramon C, Prateek P, et al. Predicting IDH mutation status on routine treatment-naïve MRI using radiogenomic features from peritumoral brain parenchyma. Neuro Oncol. 2016;18:124. [Google Scholar]
- 91. Jakola AS, Zhang YH, Skjulsvik AJ, et al. Quantitative texture analysis in the prediction of IDH status in low-grade gliomas. Clin Neurol Neurosurg. 2018;164:114–120. [DOI] [PubMed] [Google Scholar]
- 92. Shboul ZA, Chen J, M Iftekharuddin K. Prediction of molecular mutations in diffuse low-grade gliomas using MR imaging features. Sci Rep. 2020;10(1):3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bakas S, Akbari H, Pisapia J, et al. In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ-Index. Clin Cancer Res. 2017;23(16):4724–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fukuma R, Yanagisawa T, Kinoshita M, et al. Prediction of IDH and TERT promoter mutations in low-grade glioma from magnetic resonance images using a convolutional neural network. Sci Rep. 2019;9(1):20311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ivanidze J, Lum M, Pisapia D, et al. MRI features associated with TERT promoter mutation status in Glioblastoma. J Neuroimaging. 2019;29(3):357–363. [DOI] [PubMed] [Google Scholar]
- 96. McCann SM, Jiang Y, Fan X, et al. Quantitative multiparametric MRI features and PTEN expression of peripheral zone prostate cancer: a pilot study. AJR Am J Roentgenol. 2016;206(3): 559–565. [DOI] [PubMed] [Google Scholar]
- 97. Hong EK, Choi SH, Shin DJ, et al. Radiogenomics correlation between MR imaging features and major genetic profiles in glioblastoma. Eur Radiol. 2018;28(10):4350–4361. [DOI] [PubMed] [Google Scholar]
- 98. Colen RR, Wang J, Singh SK, Gutman DA, Zinn PO. Glioblastoma: imaging genomic mapping reveals sex-specific oncogenic associations of cell death. Radiology. 2015;275(1):215–227. [DOI] [PubMed] [Google Scholar]
- 99. Zinn PO, Mahajan B, Majadan B, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One. 2011;6(10):e25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. The Gene Ontology Consortium. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45(D1):D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pinho MC, Bera K, Beig N, Tiwari P. MRI Morphometry in brain tumors: challenges and opportunities in expert, radiomic, and deep-learning-based analyses. In: Seano G, ed. Brain Tumors. New York, NY: Springer US; 2021. [cited 2020 Sep 24]. p. 323–368. [Google Scholar]
- 104. Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019;29(2):102–127. [DOI] [PubMed] [Google Scholar]
- 105. Choy G, Khalilzadeh O, Michalski M, et al. Current applications and future impact of machine learning in radiology. Radiology. 2018;288(2):318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. LeCun Y, Boser BE, Denker JS, et al. Handwritten digit recognition with a back-propagation network. In: Touretzky DS, ed. Advances in Neural Information Processing Systems 2. Morgan-Kaufmann; 1990. [cited 2020 Sep 24]. p. 396–404. http://papers.nips.cc/paper/293-handwritten-digit-recognition-with-a-back-propagation-network.pdf [Google Scholar]
- 107. Bae S, An C, Ahn SS, et al. Robust performance of deep learning for distinguishing glioblastoma from single brain metastasis using radiomic features: model development and validation. Sci Rep. 2020;10(1):12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Han L, Kamdar MR. MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Pac Symp Biocomput. 2018;23:331–342. [PMC free article] [PubMed] [Google Scholar]
- 109. Davatzikos C, Barnholtz-Sloan JS, Bakas S, et al. AI-based prognostic imaging biomarkers for precision neuro-oncology: the ReSPOND consortium. Neuro Oncol. 2020;22(6):886–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Davatzikos C, Rathore S, Bakas S, et al. Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J Med Imaging (Bellingham). 2018;5(1):011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chirra P, Leo P, Yim M, et al. Empirical evaluation of cross-site reproducibility in radiomic features for characterizing prostate MRI. Medical Imaging 2018: Computer-Aided Diagnosis. International Society for Optics and Photonics; 2018. [cited 2018 Oct 24]. p. 105750B. https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10575/105750B/Empirical-evaluation-of-cross-site-reproducibility-in-radiomic-features-for/10.1117/12.2293992.short
- 114. Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys. 2018;102(4):1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Baeßler B, Weiss K, Pinto Dos Santos D. Robustness and reproducibility of radiomics in magnetic resonance imaging: a phantom study. Invest Radiol. 2019;54(4):221–228. [DOI] [PubMed] [Google Scholar]
- 116. Zlochower A, Chow DS, Chang P, Khatri D, Boockvar JA, Filippi CG. Deep learning AI applications in the imaging of Glioma. Top Magn Reson Imaging. 2020;29(2):115–110. [DOI] [PubMed] [Google Scholar]
- 117. Zinn PO, Singh SK, Kotrotsou A, et al. A coclinical radiogenomic validation study: conserved magnetic resonance radiomic appearance of periostin-expressing glioblastoma in patients and xenograft models. Clin Cancer Res. 2018;24(24):6288–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]