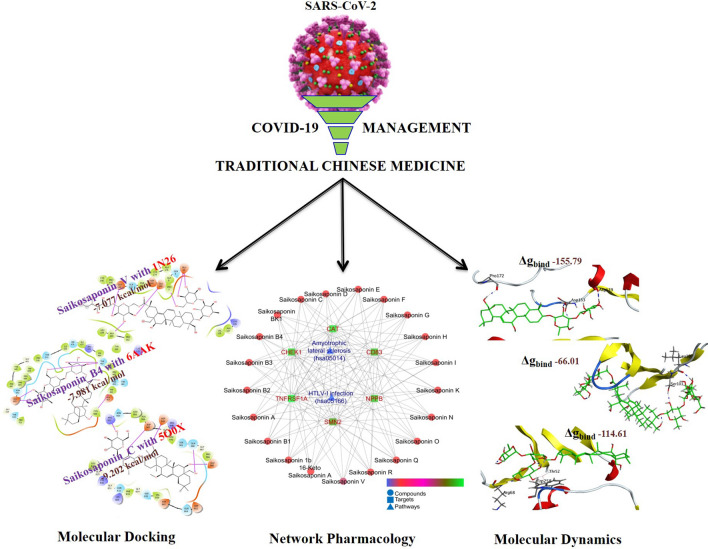
Abstract
Saikosaponins are major biologically active triterpenoids, usually as glucosides, isolated from Traditional Chinese Medicines (TCM) such as Bupleurum spp., Heteromorpha spp., and Scrophularia scorodonia with their antiviral and immunomodulatory potential. This investigation presents molecular docking, molecular dynamics simulation, and free energy calculation studies of saikosaponins as adjuvant therapy in the treatment for COVID19. Molecular docking studies for 23 saikosaponins on the crystal structures of the extracellular domains of human lnterleukin-6 receptor (IL6), human Janus Kinase-3 (JAK3), and dehydrogenase domain of Cylindrospermum stagnale NADPH–oxidase 5 (NOX5) were performed, and selected protein–ligand complexes were subjected to 100 ns molecular dynamics simulations. The molecular dynamics trajectories were subjected to free energy calculation by the MM-GBSA method. Molecular docking and molecular dynamics simulation studies revealed that IL6 in complex with Saikosaponin_U and Saikosaponin_V, JAK3 in complex with Saikosaponin_B4 and Saikosaponin_I, and NOX5 in complex with Saikosaponin_BK1 and Saikosaponin_C have good docking and molecular dynamics profiles. However, the Janus Kinase-3 is the best interacting partner for the saikosaponin compounds. The network pharmacology analysis suggests saikosaponins interact with the proteins CAT Gene CAT (Catalase) and Checkpoint kinase 1 (CHEK1); both of these enzymes play a major role in cell homeostasis and DNA damage during infection, suggesting a possible improvement in immune response toward COVID-19.
Graphic abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11030-021-10183-w.
Keywords: Saikosaponins, SARS-CoV-2, NADPH-Oxidase-5, Janus Kinase-3, Interleukin-6, In silico analysis
Introduction
Since the outbreak of COVID-19 pandemic, constant attempts are being made worldwide to develop a vaccine and utilize repurposing approaches to find out an effective cure and preventive measure [1]. Drug development using antiviral and immunomodulatory botanicals or traditional medicines is no exception to this. In these directions, two types of targets are mainly exploited; first is SARS-CoV-2 proteins to prevent the entry of virus in host cells to reduce viral load, and second is to modulate human proteins to control the replications of virus and/or to counter the immunopathogenicity [2]. A plethora of evidence suggests that viral proteins like spike proteases have been extensively studied within a short period [3]. With continuous research, a lot has been known about the COVID-19 right from SARS-CoV-2 entry in human beings till the severity of pathology it imposes and mechanism involved therein [4].
In the human transmission of COVID-19, the angiotensin-converting enzyme 2 (ACE2) transmembrane protein provides the pathway for SARS-CoV-2 to infect host epithelial cells. ACE2 is highly expressed on lung alveolar epithelial cells; hence, the lungs are particularly vulnerable to injury following SARS-CoV-2 infection [4]. During attachment, SARS-CoV-2 binds to ACE2 via the surface glycoprotein, which is cleaved into S1 and S2. S1 subunit contains a receptor-binding domain and gives easy access to COVID-19 for direct attachment to the peptidase domain of ACE2 [2, 4]. ACE2, being a membrane receptor, needs to be glycosylated to internalize and active.
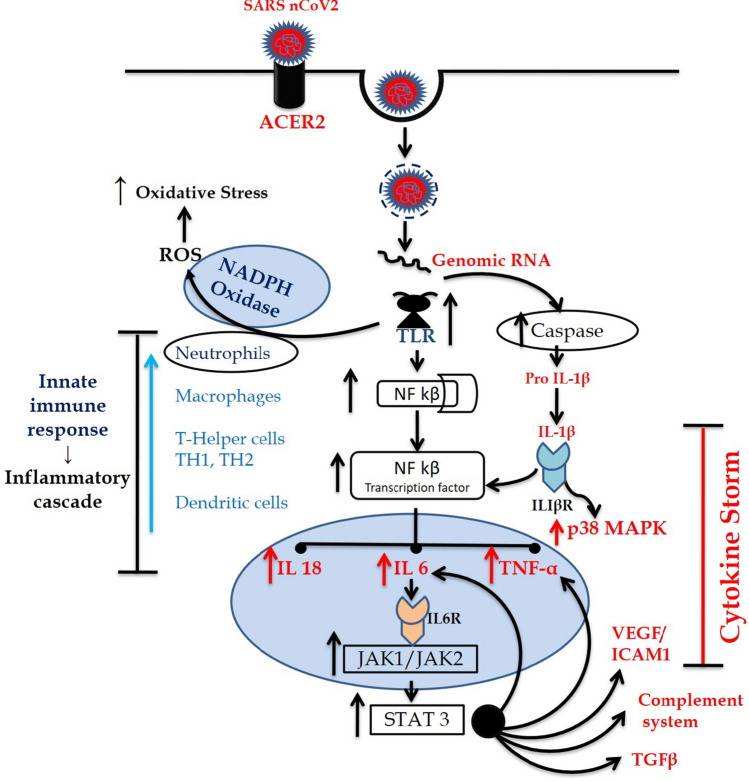
SARS-CoV-2 is a single-stranded RNA virus and its intracellular uptake is mediated by endosomes [1, 2, 4]. After incorporation into endosomes within endothelial cells, a viral protease enzyme 3CLpro of SARS-CoV is entangled in apoptosis of cell causing activation of endosomal NADPH oxidase (a pentameric complex of gp91phox, p67phox, p47phox, p22phox, and Rac) via TLR7. This catalyzes the production of superoxide and hydroxyl radicals increasing the generation of ROS which further leads to activation of NF-κB for inflammation. The viral protease enzyme also activates a NF-kB factor-dependent reporter gene which is linked with ROS generation [5].
On the other hand, 3a protease of SARS-CoV is linked with mitochondrial cell death pathways. The mechanism of this protease depends on the activation of p38 mitogen-activated protein kinase (p38MAPK) which is a key protein kinase to regulate the transcriptional activity of NF-κB and expression of proinflammatory molecules [5]. Thus, SARS-CoV causes the phosphorylation and activation of p38MAPK which then activates NF-κB, a proinflammatory transcription factor. Together with activated p300 (a proinflammatory histone acetyltransferase), the NF-κB subsequently leads to transcription and increased production of different proinflammatory cytokines and chemokines interleukin-2 (IL-2), IL-7, IL-10, granulocyte colony-stimulating factor (G-CSF), tumor necrosis factor α (TNF-α), as well as IL-6–induced genes and TNF-α pathway-related genes. Cytokines are important mediators of the mediator of inflammation. Persistently, high levels of these cytokines, especially IL-6, as observed in severely ill patients, are called cytokine storm which is responsible for the state of hyperinflammation and ultimate lung damage. Janus Kinases are important protein kinases that are known to play an important role in IL6 and other cytokine signaling and hyperimmune activation exacerbating lung damage important features of the acute respiratory distress syndrome.
Looking into these pathways of COVID-19 pathologies involving host proteins, the drugs/molecules can be developed into prospective drug candidates for COVID-19 treatment through possible influence on mechanisms, viz. suppression of the glycosylation and membrane attachment of ACE2; inhibition of the phosphorylation of p38 to inhibit the production of different pro-inflammatory cytokines; prevention of the translocation of gp91phox to the membrane to inhibit the activation of NADPH oxidase and reduce oxidative stress and inflammation; and inhibition of JAK-dependent cytokine signaling to prevent hyperinflammation (Fig. 1).
Fig. 1.
Cytokine storm pathway
Currently, chemoinformatics and molecular modeling method approaches are becoming vital components in the drug discovery due to reduction in cost and time. Computational methods signify a competent approach to efficiently alter large and diverse compound libraries to potential candidates for drug development. In this sense, there are many compounds suggested by computational methods that could be evaluated quickly with in vitro techniques [6, 7].
Saikosaponins are major biologically active triterpene glucosides, mostly occurring in Traditional Chinese Medicines (TCM) like Bupleurum spp., Heteromorpha spp., and Scrophularia scorodonia. The traditional uses of these TCM plants and saikosaponins are well known and reported in Chinese Pharmacopeia and Japanese Pharmacopeia. Saikosaponins possess antiviral, anti-hepatitis, antihepatoma, anti-inflammatory, anti-depressant, anti-neurodegenerative, antitumor, nephron-protective, and antibacterial effects, both in vitro and in vivo [8–10]. Saikosaponins (a, b, c, and d) reported to have sedative and analgesic, antiviral, anticancer, anti-inflammatory, and antibacterial properties and protect the liver and kidney [11–13].
Saikosaponins demonstrated immunomodulatory potential with enhanced production of immunosuppressive mediators (Th1/Th2 cells, IL-4, and IL-10) and suppression of pro-immune mediators (IFN-c and TNF-a in splenocytes) [12]. As compared to lamivudine, saikosaponins revealed better HBV-DNA replication and HBsAg secretion, without inhibiting the cell proliferation [14]. Moreover, Saikosaponins (A, B2, C, and D) also inhibited the early stage of viral replication including absorption and penetration of the coronaviruses [1]. In support of established in vitro anticoronaviral potential, our recent in silico studies of 23 saikosaponins on NSP15 endoribonuclease (PDB ID: 6W01) and prefusion 2019-nCoV spike glycoprotein (PDB ID: 6VSB) proteins of 2019-nCoV2 reported the plausible role of Saikosaponin_U and Saikosaponin_V in the treatment for COVID-19 [15]. Recently, in silico studies have reported repurposing of existing drugs and plausible role of natural lead in the treatment for COVID-19 [16–18].
In view of the association of cytokine storm in COVID-19 and the major role of inflammatory responses in the pathogenesis of SARS-CoV-2, targeting the coronavirus alone with antiviral therapy might not be sufficient to reverse highly pathogenic infections. The current prophylactic measures are insufficient, and as of now, there are no vaccines or antiviral drugs declared to be officially useful against the infection.
The present investigation demonstrates molecular docking, simulation, and MM-PBSA studies of TCM as adjuvant therapy in the treatment for COVID-19. Three different proteins targets involved in inflammation and oxidative stress were selected for molecular docking and simulation studies.
Material and methods
Molecular docking
Molecular docking studies of 23 bioactive saikosaponins (Supplementary File) were performed on the crystal structures of extracellular domains of human interleukin-6 receptor alpha chain (PDB ID: 1N26) with 2.40 Å resolution; JAK3 in complex with peficitinib (PDB ID: 6AAK) with 2.67 Å resolution and dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) (PDB ID: 5O0X) with 2.20 Å resolution. Schrödinger maestro 2018-1 MM share version was used for docking simulation.
Molecular dynamics (MD) simulation and molecular mechanics-generalized born solvent accessibility (MM-GBSA) analysis
The AMBER18 software package was used for MD simulations, and ligands were parameterized with ANTECHAMBER. The protein–ligand complexes prepared in xLeap were subjected to 100 ns MD simulations on Nvidia V100-SXM2-16 GB Graphic Processing Unit using the PMEMD.CUDA module. Further, the 100 ns trajectories were subjected to MM-GBSA analysis using Amber18 and Amber18 tools on all the 10,000 frames (Supplementary File).
Enrichment and network analysis
Initially, the Isomeric SMILES of individual bioactives were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/) or ChemSpider (www.chemspider.com) wherever applicable and their protein-based targets were identified at the minimum pharmacological activity > 0.5 using DIGEP-Pred [19]. The compounds with negative Mol Charge and molecular weight 1250 were excluded from the study as the inputs of these compounds are not recognized by the cheminformatic tool. The list of predicted targets was then queried using STRING [20] Version 11.0, and the protein–protein interaction was enriched concerning Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database.
Results and discussion
The human IL-6R is a 468 amino acid protein which comprises a signal peptide (19 residues), an extracellular region (339 residues), a transmembrane domain (28 residues), and a short cytoplasmic domain (82 residues). The N-terminal domain D1 (residues 1–93) is distinctive of the Ig subfamily, and domains D2 (residues 94–194) and D3 (residues 195–299) are similar to cytokine binding domains [21, 22]. SARS-CoV-2 induces fatal acute respiratory disease, caused by combative inflammation initiated by viral replication [23]. Viral import and cell infection causes enormous autophagy of the epithelial and endothelial cells, vascular leakage and initiates the immune response of the host, which produces the ebullient proinflammatory cytokines and chemokines [24, 25]. SARS-CoV-2 patient’s clinical results indicate that IL-6, IL-10, and tumor necrosis factor (TNF)-α increase during sickness and decrease after recovery [26–28]. Based on the docking score (Table 1, Supplementary File), the best five saikosaponins and appropriate reference drug with respect to IL-6 and other proteins have been discussed.
Table 1.
Molecular docking score and binding free energy for the protein–ligand complexes calculated by MM-GBSA analysis, all energies are in kcal/mol with standard deviation in parenthesis
| Compounds | MM-GBSA* | |
|---|---|---|
| Dock Score | ΔGbind | |
| Human interleukin-6 receptor (PDB ID: 1N26) | ||
| Saikosaponin-U | − 6.987 | − 145.65 (18.76) |
| Saikosaponin-V | − 7.077 | − 155.79 (18.08) |
| Janus Kinase-3 (PDB ID: 6AAK) | ||
| Saikosaponin-B4 | − 7.981 | − 66.01 (5.97) |
| Saikosaponin-I | − 7.942 | − 300.07 (56.37) |
| NADPH-Oxidase 5 (NOX5) (PDB ID: 5O0X) | ||
| Saikosaponin-BK1 | − 7.813 | − 176.90 (18.05) |
| Saikosaponin-C | − 9.202 | − 114.61 (12.30) |
ΔGbind = binding free energy; Dock Score = Glide Score
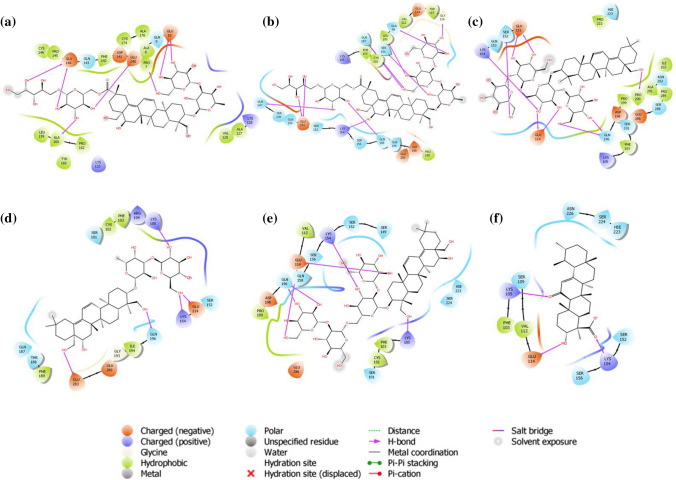
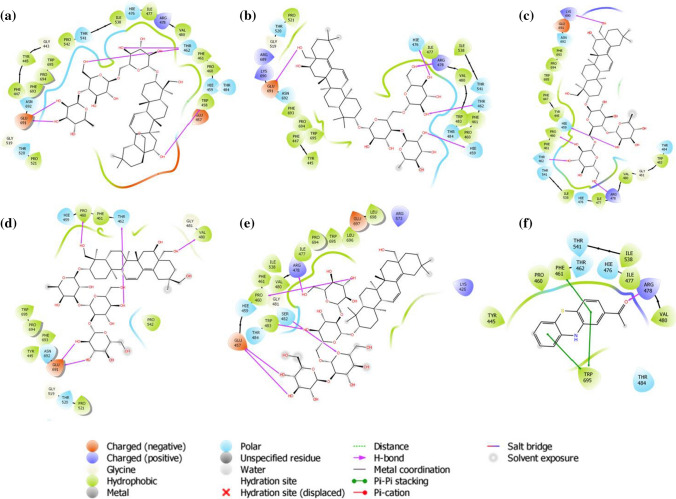
The docking results of extracellular domains of human interleukin-6 receptor alpha chain revealed that Saikosaponin_V was properly positioned into the binding pocket constructed by polar Gln 143, Gln 9, charged Glu 144, Asp 141, Glu 140, Glu 10, Lys 126, Lys 133 hydrophobic Pro 7, Leu 159, Val 128, Ala 176, Cys 146, Cys 174, Ala 8, Ala 127, Ala 160, Pro 162, and Tyr 169 amino acids with docking score − 7.077 kcal/mol. The residues Glu 10, Glu 11, Glu 140, Ala 160 exhibited H-bonding with a hydroxyl group of oxane ring, other hydroxyl groups of Saikosaponin_V showed H-bonding with Glu 144, and hydroxymethyl group of oxane ring of Saikosaponin_V showed H-bonding with Pro 7 (Fig. 2a).
Fig. 2.
Binding-interaction analysis of a Saikosaponin-V, b Saikosaponin-U, c Saikosaponin-K, d Saikosaponin-B1, e Saikosaponin-O, f 11-keto-B-boswellic acid with human interleukin-6 receptor alpha chain (PDB ID: 1N26)
Saikosaponin_V binds mainly into the domains D1 and D2 key residues. Saikosaponin_U, K, B1, and O showed a similar interaction (Fig. 2b, c, d, e) with docking scores of − 6.987, − 6.683, − 6.136, and − 6.072 kcal/mol respectively. All the saikosaponins showed better binding modes and affinity to the binding pocket than the standard 11-keto-β-boswellic acid, which was found to interact with the binding pocket assembled by polar, Ser 109, Asn 226, Ser 152, Hie 223, Ser 156, Ser 224 charged Glu 114, Lys 154, Lys 105 hydrophobic Val 112, Phe 103 amino acids with docking score − 3.723 kcal/mol. The residues Lys 154, Lys 105 exhibited H-bonding with carboxyl group, and Glu 114 showed H-bonding with hydroxyl group of 11-keto-β-boswellic acid (Fig. 2f).
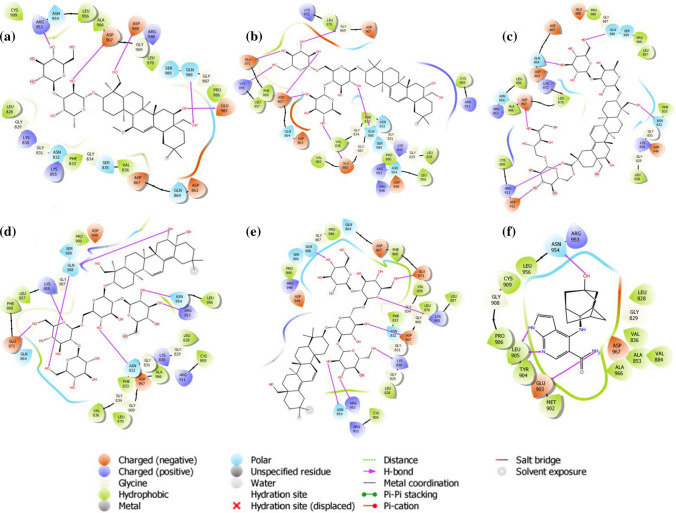
Janus Kinase 3 (JAK3) is a cytosolic tyrosine kinase associated with cytokine receptors and has a pivotal role in several chronic inflammations [29, 30]. Docking studies are found helpful to examine the plausible role of saikosaponins in inhibiting the JAK3. The JAK family comprises JAK1, JAK2, JAK3, and Tyk2 with over 1000 amino acids and between 120 and 140 kDa of molecular weight. There are four domains in each JAK protein: an N-terminal domain, an SH2 domain, a pseudokinase domain, and a tyrosine kinase domain [31]. The ATP-binding pocket in the tyrosine kinase domain from the crystal structure of JAK3 was chosen in docking studies. Saikosaponin_B4 was found to be positioned into the binding pocket assembled by polar Ser 989, Asn 954, Gln 988, Ser 835, Gln 864, Asn 832 charged Asp 967, Glu 985, Asp 949, Asp 867, Asp 863 hydrophobic Leu 828, Pro 986, Leu 956, Val 836, Ala 966, Cys 909, Leu 970, Phe 833 and Gly 987, Gly 969, Gly 834, Gly 831, Gly 829 amino acids with docking score − 7.981 kcal/mol. The nonprotonated Asp 967 and protonated Arg 953 residues exhibited H-bonding with the hydroxyl group of oxane ring, other hydroxyl group showed H-bonding with Glu 985, and hydroxymethyl group depicted H-bonding with Gln 988, Asp 949 (Fig. 3a). The hydroxyl group of oxane ring of Saikosaponin_I exhibited H-bonding with Gln 988, Asp 867, Glu 871, Gly 969, Glu 985 with docking score − 7.942 kcal/mol (Fig. 3b). The carboxyl group of Saikosaponin_V showed H-bonding with Arg 911 and the hydroxyl group gets interacted by H-bonding with Asp 967, Asn 832 amino acids with docking score of − 7.773 kcal/mol (Fig. 3c). Saikosaponin_O and Saikosaponin_K exhibited similar interactions with docking scores of − 7.664 and 7.664 kcal/mol, respectively (Fig. 3d, e). All the saikosaponins showed lower binding affinity to the binding pocket than the cocrystallized ligand peficitinib, which was found to interact with catalytic site assembled by polar Asn 954 charged Glu 903, Asp 967, Arg 953 hydrophobic Leu 828, Leu 956, Leu 905, Val 836, Cys 909, Pro 906, Tyr 904, Met 902, Ala 966, Ala 853, Val 884, and Gly 908, Gly 829 amino acids with docking score of − 9.681 kcal/mol (Fig. 3f).
Fig. 3.
Binding interaction analysis of a Saikosaponin-B4, b Saikosaponin-I, c Saikosaponin-V, d Saikosaponin-O, e Saikosaponin-K, f peficitinib with Janus Kinase-3 (PDB ID: 6AAK)
Superoxide, a reactive oxygen species, is known to cause cellular damages through oxidative stress [32]. The NADPH oxidases (NOXs) are the only enzymes that are devoted exclusively to the production of reactive oxygen species [33]. NOXs are membrane proteins and consist of six transmembrane domains (TM) and a cytosolic C-terminal dehydrogenase (DH) domain. DH includes the FAD and NADPH binding sites, while TM binds with two hemes [34]. For the development of superoxide and reactive oxygen molecules, DH transfers the electron from NADPH to oxygen bound to the moieties of the heme. Among these, Pro 694 was the most significant for function changes and reduction in NOX5 actions [35]. The docking study was performed on the crystal structure of the dehydrogenase domain of NOX5. In NOX5 binding pocket, Saikosaponin_C was found to be the best fit ligand, constructed by polar Thr 541, Hie 476, Asn 692, Thr 520, Thr 462, Hie 459, Thr 484 charged Glu 691, Arg 478, Glu 457 hydrophobic Pro 521, Phe 447, Phe 693, Pro 694, Tyr 445, Trp 695, Trp 458, Pro 542, Pro 460, Ile 538, Ile 477, Val 480, Phe 461 and Gly 519, Gly 443 amino acids with the docking score of − 9.202 kcal/mol. The residues Thr 462, Glu 691 exhibited H-bonding with the hydroxyl group of oxane ring, and hydroxymethyl group of oxane ring showed H-bonding with Thr 462 (Fig. 4a).
Fig. 4.
Binding-interaction analysis of a Saikosaponin-C, b Saikosaponin-BK1, c Saikosaponin-N, d Saikosaponin-R, e Saikosaponin-K, f 2-acetylphenothiazine with dehydrogenase domain of Cylindrospermum stagnale NADPH-oxidase 5 (NOX5) (PDB ID: 5O0X)
The hydroxyl group of oxane ring of Saikosaponin_BK1 exhibited H-bonding with Thr 462, Hie 459, Hie 476, Arg 478, and Glu 691 with a docking score of − 7.813 kcal/mol (Fig. 4b). The hydroxyl groups substituted on oxane rings of Saikosaponin_N exhibited H-bonding with Hie 459, Pro 460, and Thr 462; while hydroxymethyl groups on oxane ring and the aglycone showed H-bonding with Arg 478 and Lys 690 respectively with docking score of − 7.743 kcal/mol (Fig. 4c). Saikosaponin_R and Saikosaponin_K showed a little bit low binding affinity as compared to the rest of saikosaponins with a docking score of − 7.646 and − 7.278 kcal/mol ,respectively (Fig. 4d, e). In this study, we used 2-acetylphenothiazine as a standard that showed good binding affinity and was stabilized by π–π stacking interaction with Phe 461 and Trp 695 and exhibited docking score of − 8.338 kcal/mol (Fig. 4f). Saikosaponin_C was the only ligand that showed better binding affinity than the standard, but other saikosaponins also exhibited comparable binding energy with the standard 2-acetylphenothiazine.
Molecular dynamics simulations
Molecular dynamics simulations (MDS) were performed to explore the dynamic nature and stability of the protein–ligand complex in water. It helps to understand the interaction between ligand and protein amino acid residues at the atomistic level. In the present work, the integrated workflow of molecular docking, molecular dynamics simulations and free energy calculations was employed to understand the properties of various small molecule inhibitors with the target human proteins [17, 36, 37]. The MDS studies on protein–ligand complexes for 100 ns allow exploring a wide space of conformational optimization and binding affinity. Herein, 100 ns MDS was performed on each of the complexes: human interleukin-6 bound to Saikosaponin_V and Saikosaponin_U; Janus Kinase 3 (JAK3) bound to Saikosaponin_B4 and Saikosaponin_I; and dehydrogenase domain of NOX5 bound to Saikosaponin_C and Saikosaponin_BK1. The selection of these compounds was based on their binding affinity to target receptors compared to the other phytochemicals screened in the present study. Preprocessing of the complex was performed in three stages with two steps of minimization of the waters and completes complex, simulated annealing, NPT, and NVT equilibration of 5 ns each followed by 100 ns of production stage [18]. Trajectory analysis of these complexes gives information on their binding modes, the formation of hydrogen bond, pi–pi interactions and van der Waals interactions. RMSD, RMSF, Ligand-RMSD, and the MMGBSA-binding free energies were reported for individual complexes.
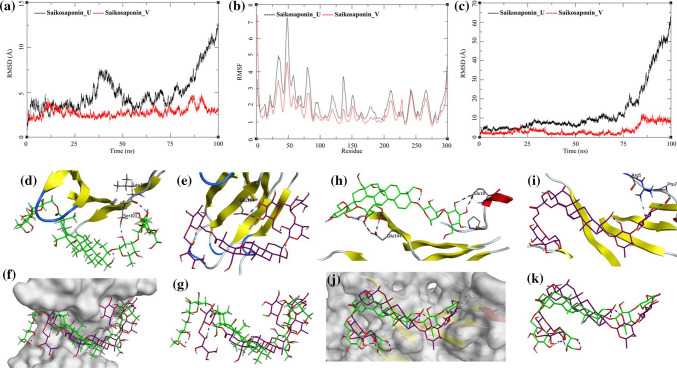
The MDS trajectories of human interleukin-6 with Saikosaponin_U and Saikosaponin_V were analyzed, and the protein RMSD, ligand RMSD, and per amino acid (aa) fluctuations, RMSF were recorded (Fig. 5a, b, c). The RMSD analyses of human interleukin-6 bound to Saikosaponin_U showed an increase in RMSD from the beginning of the MDS with initial equilibration at around 4 Å for the first 25 ns after which it had gradual rise to 7.5 Å between 25 and 50 ns and later frequent changes increasing to 12.5 Å. This could be attributed to the relative motion of the loop region between aa25-aa60 during MDS; this is supported by the RMSF for the aa in this region of the receptor. The RMSF of aa50 is about 7.5 Å in the complex of Saikosaponin_U [Fig. 5b (black)]. These RMSD fluctuations are not observed in the second complex with Saikosaponin_V but the RMSF has similar trends [Fig. 5b (red and black)]. The Lig-RMSD of Saikosaponin_U remains between 5 and 10 Å for about 75 ns and then sharply rises to 60 Å [Fig. 5c (black)]. To investigate this sharp increase in Lig-RMSD, visual inspection of the MDS trajectory was performed (Fig. 5d, e, f, g). Saikosaponin_U is a large molecule, and its binding to the shallow cavity of the receptor allows it to move into a conformationally more stable position and hence even a small change in the ligand conformation is reflected with high RMSD in the plot (Fig. 5c, black). The hydrogen bonds initially present between the residues Leu100 and Ser101 of human interleukin-6 with Saikosaponin_U (Saikosaponin_U-O6-Leu100-O; bond length = 2.69 Å. Saikosaponin_U-O8-Leu101-OG; bond length = 3.17 Å) breaks and new hydrogen bond with aa Glu114 is formed (Saikosaponin_U-O23-Glu114-OE1; Bond length = 2.76 Å) (Fig. 5d, e, f, g). This making and breaking of bond happens during the last 25 ns of the MDS which is reflected in the Lig-RMSD. The ligand superposition also shows some structural conformational changes in the Saikosaponin_U as well but these help in improving the stability of the protein–ligand complex. Several other new hydrogen bond interactions are formed like Saikosaponin_U-O15-Leu156-OG; bond length = 2.64 Å, Saikosaponin_U-O22-Gln99-O; bond length = 2.63 Å and Saikosaponin_U-O12-Gln187-NE2; bond length = 2.23 Å. The Saikosaponin_V also undergoes minor conformational changes and improves its binding with the receptor, at the beginning of the MDS, it had hydrogen bonds with the Pro7, Glu10, and Glu144 (Saikosaponin_V-O19-Pro7-O; bond length = 2.72 Å, Saikosaponin_V-O17-Glu10-OE1; bond length = 2.71 Å and Saikosaponin_V-O22-Gln144-OE1; bond length = 2.62 Å) (Fig. 5h, i, j, k). During the MDS, it forms a new hydrogen bond with the Arg5 while the Glu144 and Pro7 are conserved (Saikosaponin_V-O14-Glu10-N; bond length = 2.74 Å and Saikosaponin_V-O19-Pro7-O; bond length = 2.79 Å) (Fig. 5h, i, j, k). The ligand RMSD is also nominal with fluctuations between 0 and 5 Å for about 90 ns and an increase to 10 Å toward the end of MDS. The ligand superposition does not show a huge change in conformations, and the surface analysis also confirms the stability of the complex. The MDS studies on the protein–ligand complex for the human interleukin-6 bound to Saikosaponin_U and Saikosaponin_V suggest that these complexes are quite stable.
Fig. 5.
Trajectory analysis for human interleukin-6 (PDB: 1N26) bound to Saikosaponin_U and Saikosaponin_V; a root-mean-square deviation (RMSD), b root-mean-square fluctuations per amino acid (aa) (RMSF) and c root-mean-square deviation for each ligand (Lig-RMSD). Interaction analysis of the human interleukin-6 (PDB: 1N26) bound to Saikosaponin_U during the molecular dynamics simulation; d equilibrated structure of Saikosaponin_U (green) bound to the human interleukin-6 before MDS production phase; e Saikosaponin_U (purple) bound to the human interleukin-6 post-MDS production phase; f receptor surface analysis; g ligand analysis. Interaction analysis of the human interleukin-6 (PDB: 1N26) bound to Saikosaponin_V during the molecular dynamics simulation; h equilibrated structure of Saikosaponin_V (green) bound to the human interleukin-6 before MDS production phase; i Saikosaponin_V (purple) bound to the human interleukin-6 post-MDS production phase; j receptor surface analysis; k ligand analysis
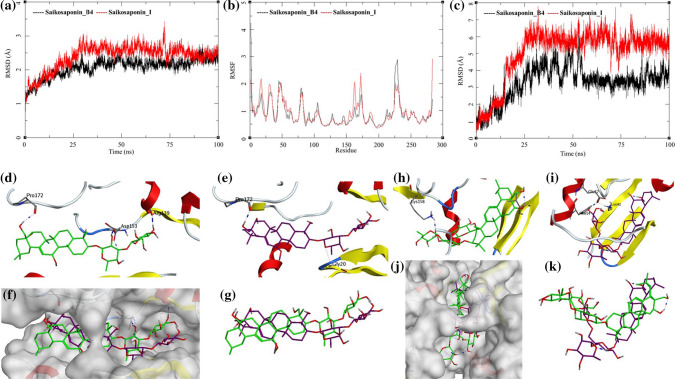
The MDS trajectories of Janus Kinase 3 (JAK3) bound to Saikosaponin_B4 and Saikosaponin_I were analyzed, and the protein RMSD, ligand RMSD, and per amino acid (aa) fluctuations, RMSF were recorded (Fig. 6a, b, c). The protein RMSD for Saikosaponin_B4 rises to about 2 Å during the first 25 ns and then equilibrated at around 2 Å till 85 ns after which it fluctuates slightly toward the end of MDS. The Saikosaponin_I bound to JAK3 protein equilibrates around 30 ns between 2.5 and 3.0 Å with a slight fluctuation between 70 to 75 ns. The long-equilibrated states in RMSD suggest that these two saikosaponins can form stable complexes with the JAK3 (Fig. 6a). This finding also reflects in the RMSF and Ligand RMSD. The aa RMSF for both the complexes has a similar pattern with the fluctuations ranging between 0.5 and 2.5 Å; however, the Saikosaponin_B4 bound to residues aa226, aa227 has slightly higher RMSF compared to Saikosaponin_I bound to JAK3 (Fig. 6b). The ligand RMSD of Saikosaponin_B4 gradually increased to 5 Å till 50 ns of the MDS after which it equilibrated at 3–4 Å for the rest of the MDS. Saikosaponin_I RMSD increases gradually to 7 Å during the first 25 ns; afterward, it equilibrated in the same conformation for the rest of the MDS (Fig. 6c). These observations support the stability of these complexes. The protein–ligand interactions for JAK3 bound to Saikosaponin_B4, and Saikosaponin_I during the MDS are shown in Fig. 6d, e, f, g.
Fig. 6.
Trajectory analysis for Janus kinase 3 (JAK3) (PDB: 6AAK) bound to Saikosaponin-B4 and Saikosaponin-I; a root-mean-square deviation (RMSD), b root-mean-square fluctuations per amino acid (aa) (RMSF) and c root-mean-square deviation for each ligand (Lig-RMSD). Interaction analysis of the Janus kinase 3 (JAK3) (PDB: 6AAK) bound to Saikosaponin-B4 during the molecular dynamics simulation; d equilibrated structure of Saikosaponin_B4 (green) bound to the JAK3 before MDS production phase; e Saikosaponin_B4 (purple) bound to the JAK3 post-MDS production phase; f receptor surface analysis; g ligand analysis. Interaction analysis of the Janus kinase 3 (JAK3) (PDB: 6AAK) bound to Saikosaponin-I during the molecular dynamics simulation; h equilibrated structure of Saikosaponin_I (green) bound to the JAK3 before MDS production phase; i Saikosaponin_I (purple) bound to the JAK3 post-MDS production phase; j receptor surface analysis; k ligand analysis
In the initial conformation of Saikosaponin_B4 bound to JAK3 before MDS, a hydrogen bond between the 4-hydroxymethyl group of tetradecahydropicen-3-yl ring and the Pro172 of the active site was observed (Saikosaponin_B4-O5-Pro172-O; bond length = 3.13 Å) (Fig. 6d). The first oxane ring of Saikosaponin_B4 forms a hydrogen bond with the Arg139 (Saikosaponin_B4-O7-Asp153-OD1; bond length = 2.63 Å) and the second oxane ring hydrogen bonds with Asp153 (Saikosaponin_B4-O7-Asp153-OD1; bond length = 2.90 Å). The post-MDS analysis shows retention of the hydrogen bond with Pro172 (Saikosaponin_B4-O7-Asp153-OD1; bond length = 2.90 Å), but interactions between the oxane rings of Saikosaponin_B4 and the receptor are lost (Fig. 6e). In this process, the ligand stabilizes and gains an energetically lower conformation (Fig. 6f, g). However, the positional conformation is retained by the hydrogen bond with Pro172 and the channel shape of the active site was shown by the white surface in Fig. 5f.
Before performing the MDS ligand–receptor interaction analysis, it shows a hydrogen bond interaction between the hydroxy group of oxane ring in Saikosaponin_I with the Lys158 of JAK3 (Saikosaponin_I-O11-Lys158-NZ; bond length = 2.75 Å) (Fig. 6h). The post-MDS analysis suggests the breaking of this bond. Figure 6c shows a sharp rise in the RMSD around 20 ns which suggests a sudden conformational change in the ligand. This could have caused the breaking of the old hydrogen bond and formation of a new hydrogen bond with Glu57 (Saikosaponin_I-O16-Glu57-OE1; bond length = 2.77 Å) (Fig. 6i). The surface analysis of the superposed MDS trajectory structures shows the shift from the initial structure (Fig. 6j, k). These observations suggest the stabilization of Saikosaponin_I occur during the MDS. It achieves energetically and conformationally stable binding position during the MDS and further stabilizes the protein–ligand complex which is evident from equilibrated protein and ligand RMSDs (Fig. 6a, c).
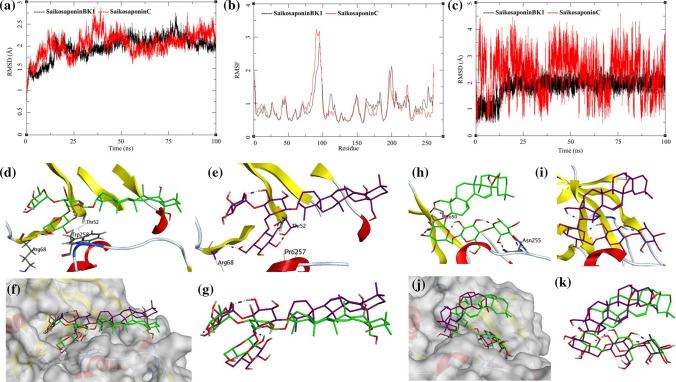
The MDS trajectories of NADPH-Oxidase 5 (NOX5) bound to Saikosaponin_BK1 and Saikosaponin_C were analyzed, and the protein RMSD, ligand RMSD and per amino acid (aa) fluctuations, RMSF were recorded (Fig. 7a, b, c). The Saikosaponin_BK1 bound to NOX5 protein shows convergence after 20 ns of MDS, and the receptor protein is an oxidase domain with more than 260 amino acid residues (Fig. 7a). Initially, the RMSD increases to around 2.25 Å and then 1.75 to 2.5 Å, which is highly acceptable for a protein of this size. It suggests that the binding of Saikosaponin_BK1 stabilizes the complex. The NOX5 bound to Saikosaponin_C shows fluctuations between 1.5 and 2.5 Å throughout the MDS. The RMSF of NOX5 bound to both the ligands was analyzed (Fig. 7b). The RMSF for NOX5 is similar for both the complexes with a high fluctuation for residues between Gly95-Val100 of about 3.25 Å. The ligand RMSD of Saikosaponin_BK1 and Saikosaponin_C shows interesting information (Fig. 7c). The Saikosaponin_BK1 RMSD increases for 1 to 2 Å between 15 and 20 ns of MDS and stabilizes to 2 Å for the rest of the simulation. This suggests a stable complex and strong binding between the protein and ligand. On the other hand, RMSD of Saikosaponin_C fluctuates between 1 and 4 Å, but it has a pattern in fluctuations suggesting a flapping motion at an interval of every 20 ns. It suggests that the NOX5–Saikosaponin_C complex is stable but the ligand achieves various conformations within the active site of the protein which allows for some sort of flexibility to Saikosaponin_C. To understand the ligand interactions, visual assessment of the trajectories was performed.
Fig. 7.
Trajectory analysis for NADPH-Oxidase 5 (NOX5) (PDB: 5O0X) bound to Saikosaponin-BK1 and Saikosaponin-C; a root-mean-square deviation (RMSD), b root-mean-square fluctuations per amino acid (aa) (RMSF) and c root-mean-square deviation for each ligand (Lig-RMSD). Interaction analysis of the NADPH-oxidase 5 (NOX5) (PDB: 5O0X) bound to Saikosaponin-BK1 during the molecular dynamics simulation; d equilibrated structure of Saikosaponin-BK1 (green) bound to the NOX5 before MDS production phase; e Saikosaponin-BK1 (purple) bound to the NOX5 post-MDS production phase; f receptor surface analysis; g ligand analysis. Interaction analysis of the NADPH-Oxidase 5 (NOX5) (PDB: 5O0X) bound to Saikosaponin-C during the molecular dynamics simulation; h equilibrated structure of Saikosaponin-C (green) bound to the NOX5 before MDS production phase; i Saikosaponin-C (purple) bound to the NOX5 post-MDS production phase; j receptor surface analysis; k ligand analysis
The visual inspection of the NOX5–Saikosaponin_BK1 complex before performing MDS revealed the presence of a hydrogen bond acceptor donor pair between the backbone of NOX5 and Saikosaponin_BK1 (Saikosaponin_BK1-O16-Arg68-N; bond length = 2.98 Å) (Fig. 7d). This complex also formed a weaker side-chain donor pair (Saikosaponin_BK1-O15-Thr52-OG1; bond length = 3.17 Å). Post-MDS analysis shows the hydrogen bond observed before the simulation breaks and is replaced by the other one (Saikosaponin_BK1-O17-Arg68-N; bond length = 2.22 Å). The Saikosaponin_BK1 also forms other interactions; Saikosaponin_BK1-O14-Thr52-N; bond length = 3.12 Å and Saikosaponin_BK1-O17-Thr52-OG1; bond length = 1.77 Å (Fig. 7e). The surface analysis of the complex before and after MDS suggests a good affinity of ligands to the active site (Fig. 7f, g) and hence low RMSD as discussed previously.
The visual inspection of the NOX5–Saikosaponin_C complex before performing MDS revealed the presence of a hydrogen bond acceptor donor pair between the backbone of NOX5 and Saikosaponin_BK1 (Saikosaponin_C-O7-Pro50-O; bond length = 2.76 Å) (Fig. 7h). This complex also formed other bond acceptor donor pairs (Saikosaponin_C-O16-Asn255-O; bond length = 2.85 Å: Saikosaponin_C-O17-Asn255-N; bond length = 2.93 Å). Post-MDS analysis showed that ligand–receptor complex forms new hydrogen bond interactions, viz., Saikosaponin_C-O7-Pro50-O; bond length = 2.74 Å: Saikosaponin_C-O6-His49-NE2; bond length = 2.94 Å; Saikosaponin_C-O16-Asn255-O; bond length = 2.92 Å: Saikosaponin_C-O17-Asn255-O17; bond length = 2.05 Å (Fig. 7i). The newly formed interactions aid the complex to remain stable and within the active site (Fig. 7j, k), albeit higher fluctuation in the ligand RMSD observed during the MDS analysis.
MM-GBSA analysis was performed on all of the six protein–ligand complexes, viz. human interleukin-6 receptor, Janus Kinase-3, and NADPH-Oxidase 5 (NOX5) bound to various saikosaponins (Table 1 and Supplementary File) to evaluate the affinity of ligands to the target protein receptors. The MM-GBSA-based binding free energy (∆Gbind) calculations were performed on the 100 ns long MDS trajectories. The binding energies measured by this method are more efficient than the GlideScore values for the selection of protein–ligand complexes. The major energy components such as H-bond interaction energy (∆Gbind_H-bond), Coulomb or electrostatics interaction energy (∆Gbind_Coul), covalent interaction energy (∆Gbind_Cov), lipophilic interaction energy (∆Gbind_Lipo), electrostatic solvation free energy (∆Gbind_Solv) and van der Waals interaction energy (∆Gbind_vdW) altogether contribute to the calculation of MM-GBSA-based relative binding affinity. The binding energies and the contributing factors calculated for the MDS trajectories are mentioned in Table 1 and Supplementary File. The human interleukin-6 receptor–Saikosaponin_U complex had binding energy of − 145.65 kcal/mol with the largest contribution from the gas phase interaction energy (ΔGgas) of − 207.22 kcal/mol. The human interleukin-6 receptor–Saikosaponin_V complex had binding energy of − 155.79 kcal/mol, which is higher than the binding energy of Saikosaponin_U. The Human JAK3–Saikosaponin_B4 complex had binding energy of − 66.01 kcal/mol; however the JAK3–Saikosaponin_I complex had a large biding energy of − 300.07 kcal/mol with the largest contribution from the gas phase interaction energy (ΔGgas) of − 375.61 kcal/mol. The NADPH-–oxidase 5 (NOX5)–Saikosaponin_BK1 complex had binding energy of − 176.90 kcal/mol; however, the NADPH-oxidase 5 (NOX5)–Saikosaponin_C complex had lower binding energy of − 114.61 kcal/mol compared to Saikosaponin_BK1. These results indicate that the JAK3 would be the best interacting partner for the saikosaponin compounds.
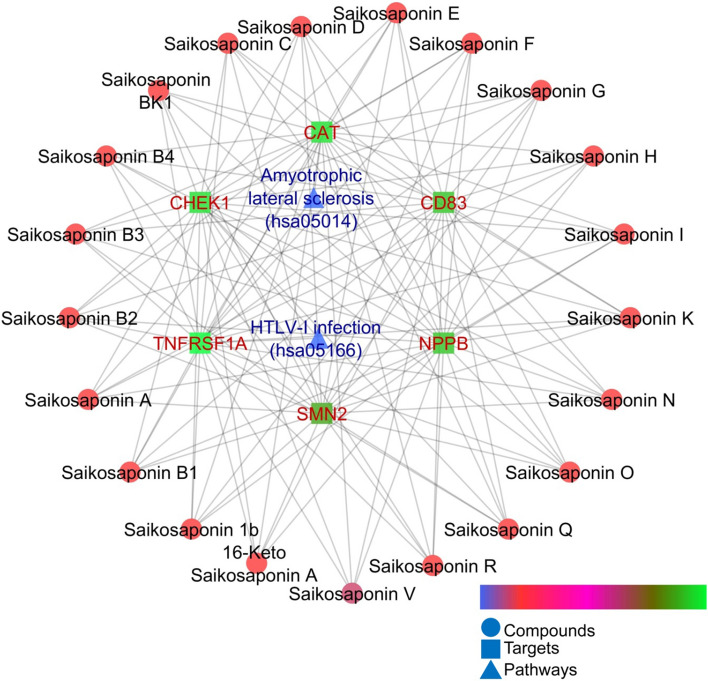
During network pharmacology analysis, out of 23 bioactives, Saikosaponin_U and Saikosaponin_2 could not be predicted for any targets because of molecular weight > 1250 and negative mol charge, respectively. Rest of the compounds collectively modulated six genes, i.e., CAT, CD83, CHEK1, NPPB, SMN2, and TNFRSF1A; where all the proteins except CHEK1 were upregulated. Further, KEGG analysis exhibited the regulation of amyotrophic lateral sclerosis (hsa05014) and HTLV-I infection (hsa05166) pathways via common modulation of TNFRSF1A (Table 2). The interaction of each compound with its targets and respective modulated proteins is presented in Fig. 8.
Table 2.
Enriched proteins with modulated pathways
| Pathway ID | Pathway description | Observed gene count | Background gene count | False discovery rate | Matching proteins in network |
|---|---|---|---|---|---|
| hsa05014 | Amyotrophic lateral sclerosis (ALS) | 2 | 50 | 0.0039 | CAT, TNFRSF1A |
| hsa05166 | HTLV-I infection | 2 | 250 | 0.0455 | CHEK1, TNFRSF1A |
Fig. 8.
Interaction of bioactives with their targets and regulated pathways
As recorded in KEGG, hsa05014 pathway is networked with 12 different pathways (https://www.genome.jp/dbget-bin/www_bget?hsa05014) in which autophagy, apoptosis, PI3K signaling, oxidative phosphorylation, ubiquitin–proteasome system, cytoskeletal regulation, and RNA processing may play an important concern in the COVID-19 management. In COVID-19 as there is the invasion of the viral infection, it is important to remove the unwanted cellular components which are affected by the coronavirus which could play an important role in its management. Since corona virus is reported to cell necrosis by triggering inflammatory responses and activating caspase-8 [38], triggering of apoptosis via hsa05014 pathway could play a tricky game as its regulation may inhibit the excessive cell necrosis. Similarly, it has been a great concern with the upregulation of immunity against COVID-19 and multiple approaches have been made to identify the lead hits against COVID-19 via this approach [39, 40]. Since PI3K signaling, oxidative phosphorylation, ubiquitin–proteasome system, cytoskeletal regulation, and RNA processing directly concerned with immune system regulation [41–45] are networked with hsa05014, modulation of this pathway would add a beneficial effect in COVID-19 management by upregulating the immune system.
Additionally, hsa05166 is concerned with multiple pathways that are associated with the pathways that are involved in the regulation of viral infection, i.e., PI3K signaling (nt06114), TGFB signaling (nt06118), JAK-STAT signaling (nt06119), calcium signaling (nt06120), TNF signaling (nt06123), MHC presentation (nt06129), cell cycle (nt06130), cytokine-cytokine receptor interaction (nt06150) and human T-cell leukemia virus 1 (nt06160) (https://www.genome.jp/dbget-bin/www_bget?hsa05166). Since hsa05166 was affected by the saikosaponins, they may affect the multiple signaling pathways concerned with coronavirus. Additionally, PI3K signaling (nt06214), TGFB signaling (nt06218), JAK-STAT signaling (nt06219), calcium signaling (nt06220), TNF signaling (nt06223), and MHC presentation (nt06229) are associated with regulation of immune system [46–51] and are concerned with hsa05166, regulated by saikosaponins. Hence, saikosaponins could upregulate the immune system and add a beneficial effect to the immune compromised subjects.
Gene CAT codes catalase, an enzymatic anti-oxidant biomarker [52], that scavenges free radicals in multiple infectious and non-infectious pathogenesis which in turn minimizes the cell necrosis and inflammation. Since inflammation is one of the major causes of progressive necrosis and cell damage, activation of this enzyme could contribute to minimizing the above conditions via scavenging free radicals. CD83 plays a prime role important role in boosting the immune system through activation of lymphocytes and scavenging of exogenic components [53, 54]. This helps the immune system to fight back with infection which is beneficial to immune-compromised subjects suffering from multiple infectious and noninfectious diseases. CHEK1 is involved in the cell damage [55] and its downregulation could minimize necrosis and attenuate the cell damage. NPPB plays a key role in cardiovascular homeostasis through natriuresis, diuresis, vasorelaxation, and inhibition of renin and aldosterone secretion (https://www.prospecbio.com). Activation of this protein could minimize the fluid retention, thereby facilitating O2 and CO2 exchange in COVID-19-infected subjects. SMN2 plays an important role in the splicing of cellular pre-mRNAs [56]. Upregulation of this protein could provide a more favorable condition for the synthesis of homeostatic proteins and boosting of the immune system. TNFRSF1A contributes to inducing noncytocidal effects including antiviral state and activation of the acid sphingomyelinase (https://www.prospecbio.com).
CAT (Identifier: ENSP00000241052, CAT; organism: Homo sapiens) codes the protein catalase and STRING database links it with ten different genes, i.e., HAO1, SCP2, DAO, ACOX1, HSD17B4, SOD2, AKT1, SOD1, SOD3 and GSR. The catalase promotes growth of cells including T-cells, B-cells, myeloid leukemia cells, melanoma cells, mastocytoma cells, and normal and transformed fibroblast cells which are concerned with the immune system regulation [57]. Additionally, catalase also acts as an antioxidant [58] which would play an important role in scavenging hydroxyl free radicals produced during cell necrosis due to corona virus infection. Further, STRING networks CD83 (Identifier: ENSP00000368450, CD83; Organism: Homo sapiens) with CD86, TNF, CD40, CD80, IL10, CD1A, CD1B, CD1C, CD1D, and CD1E and encodes CD83 antigen which could play a significant role in antigen presentation or the cellular interactions followed by the activation of lymphocytes and T cell response which are associated to the symptoms of COVID-19 [59]. Likewise, CHEK1 and encodes serine/threonine-protein kinase Chk1; links with ATM, EXO1, CDC45, CDC6, CDC25A, CDC25C, RAD51, TOPBP1, ATR, and CLSPN and is required for checkpoint-mediated cell cycle arrest and is also involved in immune regulation [60] which could add a beneficial effect to the immune compromised subjects.
Conclusion
In the present work, the role of saikosaponins as adjuvants in the treatment for COVID-19 is analyzed through in silico methods. The molecular docking studies and the dynamic simulations projected the plausible potential of Saikosaponins_U and Saikosaponin_V as promising adjuvants in the COVID-19 treatment. The present study could be the starting point for the future ligands from natural sources in 2019-nCoV as adjuvant treatment.
Supplementary Information
Below is the link to the electronic supplementary material.
Authors’ contributions
SSG and MMW contributed to conceptualization, supervision, investigation. RVC, SKS, and RBP conducted molecular docking and dynamics studies. PK, NSG, and YDD performed network pharmacology and analysis. SKS, SKP, and MA performed methodology, software data analysis. SSG and NSG contributed to writing—original draft preparation. SKP and MA were involved in formal analysis, visualization, reviewing and editing.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rupesh Chikhale, Saurabh Sinha and Manish Wanjari have contributed equally to this work and are treated as first authors.
Contributor Information
Rajesh B. Patil, Email: rajshama1@yahoo.com
Shailendra S. Gurav, Email: shailendra.gurav@nic.in
References
- 1.Stower H. Spread of SARS-CoV-2. Nat Med. 2020;26:465–465. doi: 10.1038/s41591-020-0850-3. [DOI] [PubMed] [Google Scholar]
- 2.Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santibáñez-Morán MG, López-López E, Prieto-Martínez FD, Sánchez-Cruz N, Medina-Franco JL. Consensus virtual screening of dark chemical matter and food chemicals uncover potential inhibitors of SARS-CoV-2 main protease. RSC Adv. 2020;10(42):25089–25099. doi: 10.1039/d0ra04922k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng H, Wang Y, Wang G. Organ-protective effect of angiotensin-converting enzyme 2 and its effect on the prognosis of COVID-19. J Med Virol. 2020;92:726–730. doi: 10.1002/jmv.25785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe T, Jono H, Han J, et al. Synergistic activation of NF-B by nontypeable Haemophilus influenzae and tumor necrosis factor. Proc Natl Acad Sci. 2004;101:3563–3568. doi: 10.1073/pnas.0400557101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.López-López E, Barrientos-Salcedo C, Prieto-Martínez FD, Medina-Franco JL. In silico tools to study molecular targets of neglected diseases: inhibition of TcSir2rp3, an epigenetic enzyme of Trypanosoma cruzi. Adv Protein Chem Struct Biol. 2020 doi: 10.1016/bs.apcsb.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Prieto-Martínez FD, López-López E, Eurídice Juárez-Mercado K, Medina-Franco JL (2019) Computational drug design methods—current and future perspectives. In: In silico drug design, pp 19–44. 10.1016/b978-0-12-816125-8.00002-x
- 8.Li X, Li X, Huang N, et al. A comprehensive review and perspectives on pharmacology and toxicology of saikosaponins. Phytomedicine. 2018;50:73–87. doi: 10.1016/j.phymed.2018.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X-Q, Song Y-N, Wang S-J, et al. Saikosaponins: a review of pharmacological effects. J Asian Nat Prod Res. 2018;20:399–411. doi: 10.1080/10286020.2018.1465937. [DOI] [PubMed] [Google Scholar]
- 10.Cheng P-W, Ng L-T, Chiang L-C, Lin C-C. Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin Exp Pharmacol Physiol. 2006;33:612–616. doi: 10.1111/j.1440-1681.2006.04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashour ML, Wink M. Genus Bupleurum: a review of its phytochemistry, pharmacology and modes of action. J Pharm Pharmacol. 2011;63:305–321. doi: 10.1111/j.2042-7158.2010.01170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pistelli L, Bilia AR, Marsili A, et al. Triterpenoid saponins from Bupleurum fruticosum. J Nat Prod. 1993;56:240–244. doi: 10.1021/np50092a009. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Duan M, Zhao Y, et al. Saikosaponin A inhibits influenza A virus replication and lung immunopathology. Oncotarget. 2015;6:42541–42556. doi: 10.18632/oncotarget.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C-W, Tsai F-J, Tsai C-H, et al. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Res. 2005;68:36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha SK, Prasad SK, Islam MA, et al. Identification of bioactive compounds from Glycyrrhiza glabra as possible inhibitor of SARS-CoV-2 spike glycoprotein and non-structural protein-15: a pharmacoinformatics study. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1779132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha SK, Shakya A, Prasad SK, et al. An in silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1762741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chikhale RV, Sinha SK, Patil RB, Prasad SK, Shakya A, Gurav N, et al. In silico investigation of phytochemicals from Asparagus racemosus as plausible antiviral agent in COVID-19. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1784289. [DOI] [PubMed] [Google Scholar]
- 18.Chikhale RV, Gurav SS, Patil RB, Sinha SK, Prasad SK, Shakya A, et al. Sars-CoV-2 host entry and replication inhibitors from Indian ginseng: an in silico approach. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1778539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagunin A, Ivanov S, Rudik A, et al. DIGEP-Pred: web service for in silico prediction of drug-induced gene expression profiles based on structural formula. Bioinformatics. 2013;29:2062–2063. doi: 10.1093/bioinformatics/btt322. [DOI] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varghese JN, Moritz RL, Lou M-Z, et al. Structure of the extracellular domains of the human interleukin-6 receptor α-chain. Proc Natl Acad Sci. 2002;99:15959–15964. doi: 10.1073/pnas.232432399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hui DS, Wong GW. Advancements in the battle against severe acute respiratory syndrome. Expert Opin Pharmacother. 2004;5:1687–1693. doi: 10.1517/14656566.5.8.1687. [DOI] [PubMed] [Google Scholar]
- 24.Fu Y, Cheng Y, Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol Sin. 2020;35:266–271. doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Y, Yang H, Ji W, et al. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 2020;12:372. doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conti P, Ronconi G, Caraffa A, Gallenga CE, Ross R, Frydas I, Kritas SK. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J Biol Regul Homeost Agents. 2020;34:327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 28.Siddiqi HK, Mehra MR. COVID-19 illness in native and immunosuppressed states: a clinical–therapeutic staging proposal. J Heart Lung Transplant. 2020;39:405–407. doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Shea JJ, Schwartz DM, Villarino AV, et al. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–328. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benveniste EN, Liu Y, McFarland BC, Qin H. Involvement of the janus kinase/signal transducer and activator of transcription signaling pathway in multiple sclerosis and the animal model of experimental autoimmune encephalomyelitis. J Interferon Cytokine Res. 2014;34:577–588. doi: 10.1089/jir.2014.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xin P, Xu X, Deng C, et al. The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int Immunopharmacol. 2020;80:106210. doi: 10.1016/j.intimp.2020.106210. [DOI] [PubMed] [Google Scholar]
- 32.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drummond GR, Selemidis S, Griendling KK, Sobey CG. Combating oxidative stress in vascular disease: NADPH oxidases as therapeutic targets. Nat Rev Drug Discov. 2011;10:453–471. doi: 10.1038/nrd3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finegold AA, Shatwell KP, Segal AW, et al. Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J Biol Chem. 1996;271:31021–31024. doi: 10.1074/jbc.271.49.31021. [DOI] [PubMed] [Google Scholar]
- 35.Magnani F, Nenci S, Millana Fananas E, et al. Crystal structures and atomic model of NADPH oxidase. Proc Natl Acad Sci. 2017 doi: 10.1073/pnas.1702293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerzare D, Chikhale R, Bansode R, et al. Design, synthesis, pharmacological evaluation and molecular docking studies of substituted oxadiazolyl-2-oxoindolinylidene propane hydrazide derivatives. J Braz Chem Soc. 2016 doi: 10.5935/0103-5053.20160090. [DOI] [Google Scholar]
- 37.Pant AM, Chikhale RV, Menghani SS, Khedekar PB. LEDGF/p75 IN interaction inhibitors: in silico studies of an old target with novel approach. BMC Infect Dis. 2014 doi: 10.1186/1471-2334-14-s3-p18. [DOI] [Google Scholar]
- 38.Li S, Zhang Y, Guan Z, et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Sig Transduct Target Ther. 2020;5:235. doi: 10.1038/s41392-020-00334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khanal P, Duyu T, Patil BM, Dey YN, Pasha I, Wanjari M, Gurav SS, Maity A. Network pharmacology of AYUSH recommended immune-boosting medicinal plants against COVID-19. J Ayurveda Integr Med. 2020 doi: 10.1016/j.jaim.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khanal P, Patil BM, Chand J, et al. Anthraquinone derivatives as an immune booster and their therapeutic option against COVID-19. Nat Prod Bioprospect. 2020;10:325–335. doi: 10.1007/s13659-020-00260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saleh J, Peyssonnaux C, Singh KK, Edeas M. Mitochondria and microbiota dysfunction in COVID-19 pathogenesis. Mitochondrion. 2020 doi: 10.1016/j.mito.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickramarachchi DC, Theofilopoulos AN, Kono DH. Immune pathology associated with altered actin cytoskeleton regulation. Autoimmunity. 2010;43(1):64–75. doi: 10.3109/08916930903374634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ansel KM. RNA regulation of the immune system. Immunol Rev. 2013;253:5–11. doi: 10.1111/imr.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weichhart T, Säemann MD. The PI3K/Akt/mTOR pathway in innate immune cells: emerging therapeutic applications. Ann Rheum Dis. 2008;67:iii70–iii74. doi: 10.1136/ard.2008.098459. [DOI] [PubMed] [Google Scholar]
- 45.Reinstein E. Immunologic aspects of protein degradation by the ubiquitin-proteasome system. Isr Med Assoc J. 2004;6(7):420–424. [PubMed] [Google Scholar]
- 46.Han JM, Patterson SJ, Levings MK. The role of the PI3K signaling pathway in CD4+ T cell differentiation and function. Front Immunol. 2012;3:245. doi: 10.3389/fimmu.2012.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50(4):924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shuai K, Liu B. Regulation of JAK–STAT signalling in the immune system. Nature Rev Immunol. 2003;3(11):900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 49.Vig M, Kinet J-P. Calcium signaling in immune cells. Nature Immunol. 2009;10(1):21–27. doi: 10.1038/ni.f.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Šedý J, Bekiaris V, Ware CF. Tumor necrosis factor superfamily in innate immunity and inflammation. Cold Spring Harbor Perspect Biol. 2014;7(4):a016279. doi: 10.1101/cshperspect.a016279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wieczorek M, Abualrous ET, Sticht J, Álvaro-Benito M, Stolzenberg S, Noé F, Freund C. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gil D, Rodriguez J, Ward B, et al. Antioxidant activity of SOD and catalase conjugated with nanocrystalline ceria. Bioeng Basel Switz. 2017 doi: 10.3390/bioengineering4010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Ju X, Silveira PA, et al. CD83: Activation marker for antigen presenting cells and its therapeutic potential. Front Immunol. 2019;10:1312. doi: 10.3389/fimmu.2019.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grosche L, Knippertz I, König C, et al. The CD83 molecule—an important immune checkpoint. Front Immunol. 2020 doi: 10.3389/fimmu.2020.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patil M, Pabla N, Dong Z. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell Mol Life Sci. 2013;70:4009–4021. doi: 10.1007/s00018-013-1307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J, Zheng X, Shen H. Targeting RNA-splicing for SMA treatment. Mol Cells. 2012;33:223–228. doi: 10.1007/s10059-012-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cano RLE, Lopera HDE, et al. Introduction to T and B lymphocytes. In: Anaya JM, Shoenfeld Y, Rojas-Villarraga A, et al., editors. Autoimmunity: from bench to bedside. Bogota: El Rosario University Press; 2013. [PubMed] [Google Scholar]
- 58.Nandi A, Yan L-J, Jana CK, Das N. Role of catalase in oxidative stress- and age-associated degenerative diseases. Oxid Med Cellul Longev. 2019;2019:1–19. doi: 10.1155/2019/9613090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Z, John Wherry E. T cell responses in patients with COVID-19. Nature Rev Immunol. 2020 doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guerra B, Issinger O-G, Wang JY. Modulation of human checkpoint kinase Chk1 by the regulatory β-subunit of protein kinase CK2. Oncogene. 2003;22(32):4933–4942. doi: 10.1038/sj.onc.1206721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.