Abstract
Objective
Influenza season occurs every year in China, but its presentation was unusual in the period from December 2017 to early 2018. During this period, influenza activity was increasing across the country and was much greater than during the same period in previous years, with great harm to people’s health.
Methods
In this study, we isolated two human influenza virus strains—A/Hebei/F076/2018(H1N1) and B/Hebei/16275B/2018—from patients with severe influenza in Hebei, China, during the flu season in January 2018, and explored their genetic characteristics, pathogenicity, and transmissibility.
Results
A/Hebei/F076/2018(H1N1) belongs to the human-like H1N1 influenza virus lineage, whereas B/Hebei/16275B/2018 belongs to the Victoria lineage and is closely related to the World Health Organization reference strain B/Brisbane/60/2008. Pathogenicity tests revealed that A/Hebei/F076/2018(H1N1) replicated much more strongly in mice, with mice exhibiting 40% mortality, whereas B/Hebei/16275B/2018 was not lethal. Both viruses could be transmitted through direct contact and by the aerosol route between guinea pigs, but the H1N1 strain exhibited higher airborne transmissibility.
Conclusions
These results may contribute to the monitoring of influenza mutation and the prevention of an influenza outbreak.
Keywords: Influenza A virus, H1N1pdm09, influenza B virus, pathogenicity, transmissibility, genetic characteristics
Introduction
Influenza viruses can cause seasonal or pandemic influenza in humans and animals. According to the different antigenicity of their nuclear protein (NP) and matrix protein (M1), they can be separated into four types: A, B, C, and D.1,2 Of these, influenza A virus (IAV) is widespread and can infect humans and a variety of animals. It is the most common type of influenza in human and livestock.3 Influenza B virus (IBV) has strong host specificity and is currently reported to infect humans and seals.4,5 Influenza C viruses mainly infect humans, pigs, and cattle,6 and influenza D viruses mainly infect pigs and cattle.7
Influenza A viruses are currently categorized into 18 hemagglutinin (HA) and 11 neuraminidase (NA) subtypes, but only viruses with HA subtypes H1, H2, and H3, and NA subtypes N1 and N2 are known to cause influenza pandemics in humans.8–10 Since the beginning of the 20th century, there have been five major outbreaks of pandemic influenza worldwide.11 H1N1pdm09 was a recent human influenza pandemic that caused outbreaks in 214 countries (or regions) and the deaths of at least 18,000 people worldwide.12 At present, the H1N1pdm09 influenza virus has replaced the H1N1 subtype seasonal influenza virus that dominated before 2009, with H1N1pdm09 spreading widely among the population in the form of new viruses causing seasonal influenza outbreaks every year.13–15 Sequencing and analysis of the genome of the H1N1pdm09 influenza virus showed that it is a triple reassortment virus that carried genes from human, avian, and swine influenza viruses.16,17 Genetic evolution analysis revealed that the PB2 and PA genes of the H1N1pdm09 influenza virus are derived from avian influenza virus, the PB1 gene is derived from H3N2 subtype human influenza virus, and the HA, NP, NA, M, and NS genes are derived from swine influenza viruses.18,19
IBV was first obtained from sick children in 1940 and successfully isolated from seals in 2000.4,20 According to differences in the nucleotide sequence of the HA fragment of the virus, IBV is divided into two major lineages; namely, the B-Victoria branch, which is represented by the B/Victoria/2/87 strain, and the B-Yamagata branch, which is represented by the B/Yamagata/16/88 strain.21 IBV does not pose a pandemic threat, but localized outbreaks cannot be ignored.
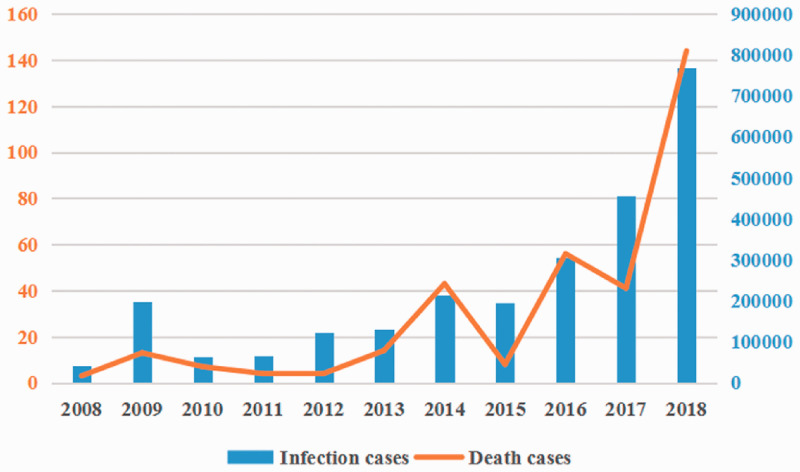
Influenza outbreaks occur almost every year, causing respiratory diseases of varying severity in the population and posing a serious threat to global public health.22–24 Virus monitoring is an effective early warning method against influenza virus pandemics. From December 2017 to early 2018, the morbidity and mortality of influenza were much higher than those in previous years (Figure 1), sounding the alarm for a potential epidemic. To understand the characteristics of the strains involved in this outbreak, our laboratory collected 1570 nasopharyngeal swabs from patients with suspected influenza infection in Hebei, China, during the influenza season in January 2018, from which we isolated 17 influenza virus epidemic strains, including 15 IAV (14 H1N1 and 1 H3N2) strains and two IBV strains. In particular, two strains, A/Hebei/F076/2018(H1N1) and B/Hebei/16275B/2018, were isolated from severely ill patients, whereas the others were isolated from mildly symptomatic patients. In this study, we explored the genetic characteristics, pathogenicity, and transmissibility of these two strains and compared them with a previously isolated seasonal influenza A/Hebei/HB17/2017(H1N1) and a H1N1pdm09 strain A/CA/04/2009(H1N1). The results will aid in influenza pandemic preparedness efforts.
Figure 1.
Morbidity (blue bars) and mortality (orange curve) of influenza in China 2008 to 2018. Source: National Health Commission of the People’s Republic of China.
Materials and methods
Ethics statement
All animals were adequately cared for, and the animal studies were conducted in strict accordance with the guidelines of animal welfare of the World Organization for Animal Health.25 Experimental protocols involving animals were approved by the Animal Care and Use Committee of Military Veterinary Institute (Changchun, Jilin, China; approval number: SCXK 20160008; approval date: 7 March 2016). All experiments with the influenza A (H1N1) and influenza B viruses were performed in biosafety level three laboratories approved by the Academy of Military Medical Sciences. This study did not involve human participants.
Viruses
Two human influenza viruses, isolated from severely ill influenza patients in 2018, were used in this study. The viruses were A/Hebei/F076/2018(H1N1) (hereafter F076; GenBank accession numbers MH748636 and MH748645) and B/Hebei/16275B/2018 ( hereafter 16275B; GenBank accession number MH748675). Additionally, A/Hebei/HB17/2017(H1N1) (hereafter HB17), isolated from severely ill influenza patients in 2017, and A/CA/04/2009(H1N1) (hereafter CA04), a pandemic flu virus, were used as control group viruses in this study. Viruses were grown in 9-day-old specific-pathogen-free eggs (Merial Vital Laboratory Animal Technology Company, Beijing, China) and stored at −80°C.
Phylogenetic and sequence analyses
Viral RNA was extracted from allantoic fluid using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed into cDNAs using the primer Uni12 (5′-AGCRAAAGCAGG-3′) or Uni9 (5′-AGCAGAAGC-3′). PCR was performed using a previously described method.26,27 The PCR products were subjected to electrophoresis on 1% agarose gels stained with ethidium bromide and visualized with an ultraviolet light transilluminator. The PCR products were purified and sequenced by Comate Bioscience Company Limited (Changchun, Jilin, China). All sequence data were analyzed using the SEQMAN program (DNAStar Inc., Madison, WI, USA). All reference sequences used in this study were obtained from the National Center for Biotechnology Information GenBank database. Phylogenetic analysis was performed by the distance-based neighbor-joining method using MEGA 7 software (DNAStar Inc.).
Receptor-binding specificity assay
The receptor-binding specificities of human influenza viruses were determined by HA assays with 1% chicken and sheep red blood cell (cRBC and sRBC) suspensions. For sialidase treatment, 90 μL of a 10% cRBC suspension was treated with 10 μL of α-2,3-sialidase (50 mU/μL; TaKaRa, Dalian, China) for 10 minutes at 37°C. The sample was then washed twice with PBS, centrifuged at 210 × g for 5 minutes each time, adjusted to a final working concentration (1%) with PBS, and stored at 4°C. For Vibrio cholerae neuraminidase (VCNA; Roche, San Francisco, CA, USA) treatment, 90 μL of a 10% cRBC suspension was treated with 10 μL of VCNA (50 mU/μL) for 1 hour at 37°C, washed twice with PBS, centrifuged at 210 × g for 5 minutes each time, adjusted to a final working concentration (1%) with PBS, and stored at 4°C. For the HA assay, viruses were serially diluted 2-fold with 50 μL of PBS and mixed with 50 μL of a 1% RBC suspension in a 96-well plate. HA titers were read after 20 minutes of reaction at room temperature.
Mouse experiments
Groups of five 6-week-old female BALB/c mice (Merial Vital Laboratory Animal Technology Company) were anesthetized with ether and intranasally inoculated with 50 µL of a 106.0 EID50 (50% embryo infectious dose) solution of strain F076, HB17, CA04, or 16275B.28,29 The mice were monitored for weight loss and mortality daily for 14 days. Mice that lost >30% of their original body weight were humanely euthanized. All animals were euthanized by inhaling excess CO2.28 To assess the growth characteristics of the two viruses and the pathological changes in the lungs of infected mice, four groups of 20 mice were anesthetized with ether and intranasally instilled with 106.0 EID50 of either F076, HB17, CA04, or 16275B, and another 3 mice were intranasally instilled with PBS as controls. Three mice were euthanized at 3, 5, and 7 days post-infection (dpi). The lungs, hearts, livers, spleens, kidneys, and brains of infected mice were removed to determine the virus titers. Briefly, the tissues were weighed, and 0.1 g of each tissue was placed into 1 mL of PBS containing 100 U/mL penicillin, generating 10% (wt/vol) tissue homogenates. The tissue samples were homogenized by Tissue Lyser (Qiagen, Hilden, Germany) and centrifuged at 1680 × g. Then, the supernatants were collected and inoculated into 9-day-old embryonated eggs. After 72 hours of incubation at 37°C or 32°C, HA activity was tested and the EID50 was determined by the Reed and Muench method. The lungs of infected mice euthanized at 3 dpi were fixed in formalin, and the fixed tissues were embedded in paraffin and stained with hematoxylin and eosin for pathological examination.
Guinea pig experiments
Hartley strain albino female guinea pigs weighing 300 to 350 g (Merial Vital Laboratory Animal Technology Company) were used in this study. In the transmission studies, three guinea pigs per group were intranasally inoculated with 200 μL of the test viruses at 106 EID50 and housed in a cage placed inside an isolator. The next day, three naïve guinea pigs were cohoused (in the same cage) with the three infected guinea pigs to study direct-contact transmission, and another three naïve guinea pigs per group were housed in a wire-frame cage adjacent to the infected guinea pigs to study aerosol transmission. The distance between the infected and aerosol-contact guinea pig cages was 5 cm. To monitor virus shedding, nasal washes were collected and titrated from all animals at 2, 4, 6, and 8 dpi.
Statistical analysis
Significant differences were identified using one-way analysis of variance with GraphPad Prism software (GraphPad Inc., San Diego, CA, USA). All assays were run in triplicate and are representative of at least 3 separate experiments. The error bars represent the standard deviation.
Results
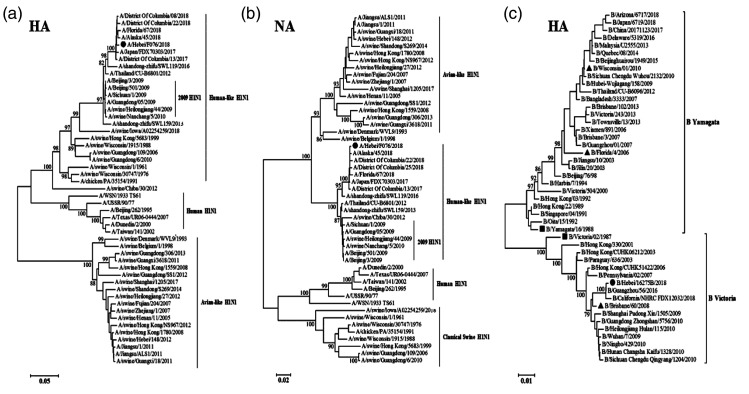
Phylogenetic analysis of surface genes
Full-length sequences of the influenza virus A/Hebei/F076/2018(H1N1) and B/Hebei/16275B/2018 isolates were compared with those of known influenza viruses in the GenBank database. The eight gene segments of F076 and 16275B were found to have the highest nucleotide sequence homologies predominantly with the epidemic strain (H1N1 and influenza B) from recent years, which are available from the influenza sequence database (https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=database; Table 1). Phylogenetic trees based on the HA and NA genes of F076 are shown in Figure 2a and 2b. The H1N1 strain in this study (indicated by solid circles) is divided into the human-like H1N1 influenza virus lineage and is closely related to the epidemic strain of recent years. The HA phylogenetic tree for 16275B is shown in Figure 2c; it consists of epidemic strains and WHO reference epidemic strains (marked with solid triangles), as well as reference strains of the Yamagata and Victoria lineages (marked with solid squares).19 The results showed that B/Hebei/16275B/2018 (marked with solid circles) is a member of the Victoria lineage and is closely related to the reference strain B/Brisbane/60/2008.
Table 1.
Sequence homology of human influenza strains F076 (A/Hebei/F076/2018) and 16275B (B/Hebei/16275B/2018), both associated with severe illness in 2018, with known influenza viruses (https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=database).
| Virus strain | Gene | Virus from the database with the highest homology | % | Accession number |
|---|---|---|---|---|
| A/Hebei/F076/2018(H1N1) | PB2 | A/Baltimore/P0252/2018(H1N1) | 99 | MH637811.1 |
| PB1 | A/Porto Alegre/LACENRS-378/2016(H1N1) | 99 | KY926165.1 | |
| PA | A/Delaware/02/2018(H1N1) | 99 | MH125473.1 | |
| HA | A/Baltimore/P0269/2018(H1N1) | 99 | MH637665.1 | |
| NP | A/Baltimore/P0264/2018(H1N1) | 99 | MH637700.1 | |
| NA | A/Baltimore/P0252/2018(H1N1) | 99 | MH637715.1 | |
| M | A/Baltimore/P0269/2018(H1N1) | 99 | MH637519.1 | |
| NS | A/Baltimore/P0258/2018(H1N1) | 99 | MH637476.1 | |
| B/Hebei/16275B/2018 | PB2 | B/Georgia/22/2016 | 99 | CY215312.1 |
| PB1 | B/Delaware/03/2016 | 100 | KX612876.1 | |
| PA | B/South Carolina/11/2016 | 100 | KX920063.1 | |
| HA | B/Virginia/28/2017 | 99 | CY249484.1 | |
| NP | B/Hawaii/46/2017 | 99 | CY249167.1 | |
| NA | B/Ohio/08/2016 | 100 | KY043629.1 | |
| M | B/South Carolina/09/2016 | 100 | KX920051.1 | |
| NS | B/Pennsylvania/32/2018 | 100 | MH671688.1 |
Figure 2.
Phylogenetic analysis of the hemagglutinin (HA) and neuraminidase (NA) genes of strain F076 (A/Hebei/F076/2018, human, associated with severe illness in 2018; panels a and b, respectively) and the HA genes of strain 16275B (B/Hebei/16275B/2018, human, associated with severe illness in 2018; panel c). Phylogenetic trees of the HA and NA genes were constructed by the distance-based neighbor-joining method with 1000 bootstrap replicates using MEGA 7 software (DNAStar Inc., Madison, WI, USA). Horizontal distances are proportional to genetic distances. Influenza virus isolates in this study are marked with solid circles. The WHO reference strains are marked with solid triangles. Reference strains of the Yamagata and Victoria lineage are marked with solid squares.
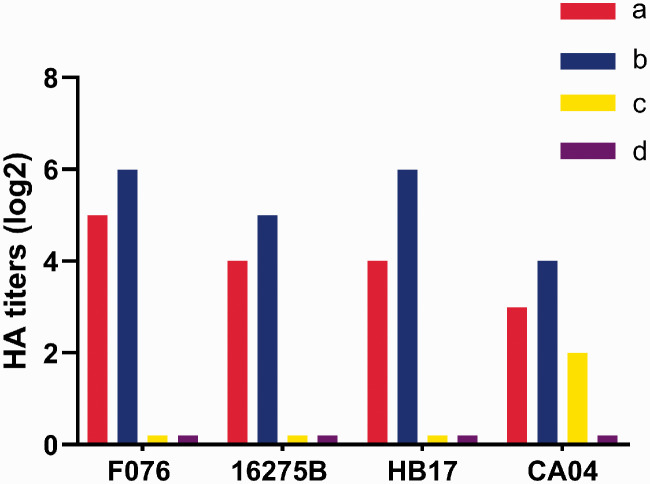
F076 and 16275B exhibited comparable binding affinities for avian and human sialic acid receptors
We evaluated the receptor-binding specificities of F076 and 16275B using an HA assay, and compared them with HB17 and CA04. The surface of cRBCs contains α-2,3-linked and α-2,6-linked sialic acid receptors. cRBCs treated with α-2,3-sialidase contain only α-2,6-linked sialic acid receptors, cRBCs treated with VCNA contain no receptors, and the surface of sRBCs contain only α-2,3-linked sialic acid receptors. As expected, all viruses could agglutinate untreated cRBCs but not VCNA-treated cRBCs or sRBCs, as shown in Figure 3. The HA titers are shown in Figure 3. These results indicated that the F076, HB17, and 16275B strains maintained affinity for human-like (α-2,6) receptors but exhibited no affinity for avian-like (α-2,3) receptors, suggesting that they are less likely to infect avian species.
Figure 3.
Agglutination activity (hemagglutinin [HA] titers) of four influenza strains in red blood cells (RBCs) of different types: (a) chicken RBCs (with α-2,3-linked sialic acid receptors and α-2,6-linked sialic acid receptors); (b) chicken RBCs treated with α-2,3-sialidase (with only α-2,6-linked sialic acid receptors); (c) sheep RBCs (with only α-2,3-linked sialic acid receptors); and (d) chicken RBCs treated with Vibrio cholerae neuraminidase (VCNA; no receptors). Influenza strains: F076 (A/Hebei/F076/2018, human, associated with severe illness in 2018), HB17 (A/Hebei/HB17/2017, human, associated with severe illness in 2017; control), CA04 (A/CA/04/2009, pandemic flu virus; control), and 16275B (B/Hebei/16275B/2018, human, associated with severe illness in 2018).
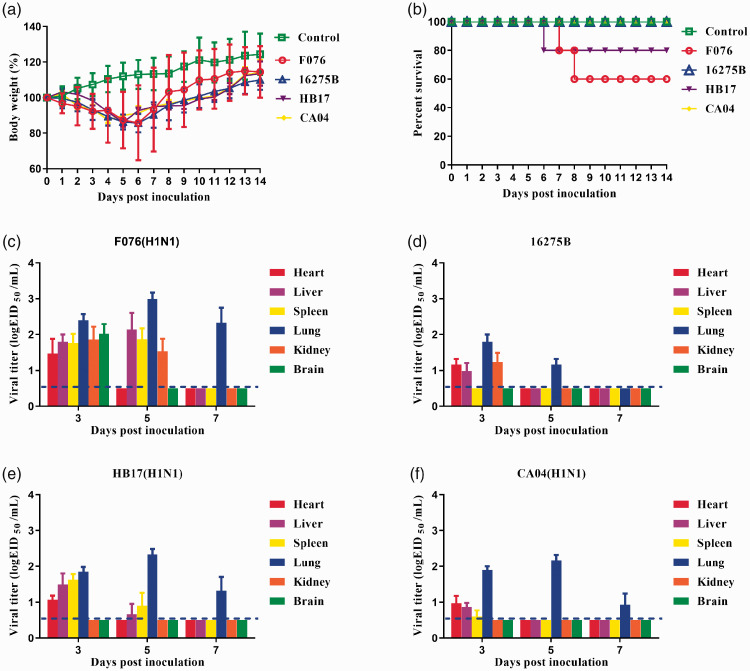
Pathogenicity of F076 and 16275B in mice
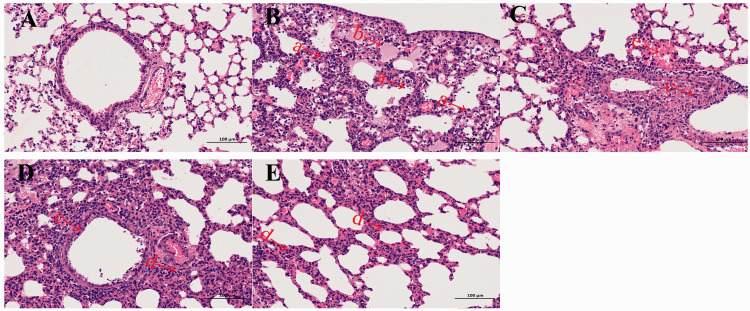
The pathogenicity of F076 and 16275B viruses was evaluated in a mouse model and compared with that of HB17 and CA04. Groups of five 6-week-old female BALB/c mice were used in this experiment. After 24 hours of infection with F076, HB17, CA04, or 16275B, the mice showed clinical manifestations such as coat inversion and loss of appetite. Mice inoculated with viruses rapidly lost approximately 15% of their weight (Figure 4a). Mice infected with F076 exhibited 40% mortality, mice infected with HB17 exhibited 20% mortality, whereas the CA04 and 16275B infections were not lethal (Figure 4b). The change in body weight of mice infected with F076 was more obvious (p < 0.005). Viral titers were investigated in heart, liver, spleen, lungs, kidneys, and brain of mice infected with each of these four viruses at a dose of 106 EID50. The F076 virus could be detected in all six organs at 3 dpi, whereas only liver, spleen, lungs, and kidneys were positive at 5 dpi. At 7 dpi, the virus could be detected only in the lungs, which had the highest viral titer (Figure 4c). The HB17 virus could be detected in heart, liver, spleen, and lungs at 3 dpi, whereas only the liver, spleen, and lungs were positive at 5 dpi. At 7 dpi, the virus could be detected only in the lungs, which had the highest viral titer (Figure 4e). The CA04 virus could be detected in heart, liver, spleen, and lungs at 3 dpi, but at 5 and 7 dpi, the virus could be detected only in the lungs, which had the highest viral titer (Figure 4f).The influenza B virus (16275B) could be detected in heart, liver, lungs, and kidneys at 3 dpi, whereas only the lungs were positive at 5 dpi. At 7 dpi, no virus was detected in any of the six organs (Figure 4d). The above results show that the influenza A (F076) virus replicates much more strongly in mice than the influenza B (16275B) virus and could cause death during the infection process. In addition, histopathological analysis of the lungs of the infected mice showed multiple pathological phenomena, such as alveolar wall thickening and lymphocyte infiltration (Figure 5). Histological analysis showed that the mice infected with F076 or 16275B exhibited severe histopathological changes.
Figure 4.
Pathogenicity of the isolated viruses in mice. Five mice per group were intranasally inoculated with the viruses at 106 EID50 (50% embryo infectious dose). (a) Body weights during 14 days of monitoring. Values represent the average score of overall body weight loss compared with the initial body weight ± standard deviation (SD). The change in body weight of mice infected with F076 was more obvious (p < 0.005). (b) Percentage survival of mice infected with the influenza viruses. (c–f) Heart, liver, spleen, lungs, kidneys and brain were collected from the infected mice (n = 3) on the indicated days post-infection (dpi) and viral titers were determined in 9-day-old specific-pathogen-free embryonated eggs. Influenza strains: F076 (A/Hebei/F076/2018, human, associated with severe illness in 2018), HB17 (A/Hebei/HB17/2017, human, associated with severe illness in 2017; control), CA04 (A/CA/04/2009, pandemic flu virus; control), and 16275B (B/Hebei/16275B/2018, human, associated with severe illness in 2018).
Figure 5.
Histopathological analysis. At 3 days post inoculation (dpi), lungs were collected from mice inoculated with 106.0 EID50 (50% embryo infectious dose) of F076, 16275B, HB17, or CA04, and were fixed with formalin, embedded in paraffin, and stained with hematoxylin and eosin. The images were obtained at a magnification of 200×. Panels a–e show pathological changes in the lungs of mice inoculated with PBS, F076, 16275B, HB17, and CA04, respectively. Arrow a indicates alveolar wall thickening, accompanied by lymphocyte infiltration, neutrophil infiltration; arrow b indicates pulmonary edema, a small amount of eosinophilic serous substance exuding in the alveolar cavity; arrow c indicates local bronchus with necrosis and shedding of epithelial cells and nuclear fragmentation; arrow d indicates alveolar wall thickening, accompanied by a large amount of lymphocytes and neutrophil infiltration; arrow e indicates bleeding. The scale bar represents 100 µm. Influenza strains: F076 (A/Hebei/F076/2018, human, associated with severe illness in 2018), HB17 (A/Hebei/HB17/2017, human, associated with severe illness in 2017; control), CA04 (A/CA/04/2009, pandemic flu virus; control), and 16275B (B/Hebei/16275B/2018, human, associated with severe illness in 2018).
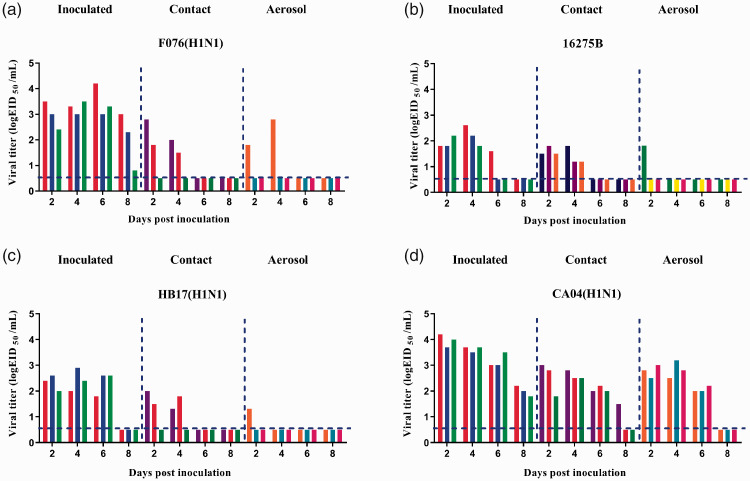
Transmissibility of F076 and 16275B in guinea pigs
Hartley strain albino female guinea pigs weighing 300 to 350 g were used to investigate the contact or airborne transmissibility of the two viruses compared with control strains. Three guinea pigs were intranasally inoculated with 200 μL of the test virus at 106 EID50/mL and housed in a cage placed inside an isolator. Infection was detected by a nasal wash of the influenza A virus (F076) infection group, which had the highest viral titer of 104.2 EID50/mL. F076 was transmitted to two guinea pigs via the direct contact route and to one guinea pig via the aerosol route (three animals were examined in each group; Figure 6a). The highest viral titers in the F076 contact and aerosol infection groups were 102.8 EID50/mL. HB17 was transmitted to two guinea pigs via the direct contact route and to one guinea pig via the airborne route (three animals were examined in each group; Figure 6c). The highest viral titers in the HB17 contact and aerosol infection group were 102.0 EID50/mL and 101.3 EID50/mL, respectively. CA04 was transmitted to three guinea pigs via the direct contact route and to three guinea pigs via the airborne route (three animals were examined in each group; Figure 6d). The highest viral titers in the CA04 contact and aerosol infection group were 103.0 EID50/mL. 16275B was transmitted to three guinea pigs via the direct contact route and to one guinea pig via the airborne route (three animals were examined in each group; Figure 6b). The highest viral titers in the 16275B contact and aerosol infection group were 101.8 EID50/mL. These results suggest that both influenza A and B viruses included in this study could be transmitted via direct contact and by the airborne route.
Figure 6.
Horizontal transmission of influenza viruses between guinea pigs. Groups of three guinea pigs were inoculated with the indicated viruses at 106 EID50 (50% embryo infectious dose): (a) F076, (b) 16275B, (c) HB17, and (d) CA04. The next day, the inoculated animals were paired by cohousing with direct-contact guinea pigs; aerosol-transmission animals were also housed in a wire-frame cage adjacent to the infected guinea pigs. Nasal washes were collected from all animals for detection of virus shedding every other day beginning on day 2 after initial infection. Each color bar represents the virus titer in an individual animal. Dashed lines indicate the lower limit of virus detection. Influenza strains: F076 (A/Hebei/F076/2018, human, associated with severe illness in 2018), HB17 (A/Hebei/HB17/2017, human, associated with severe illness in 2017; control), CA04 (A/CA/04/2009, pandemic flu virus; control), and 16275B (B/Hebei/16275B/2018, human, associated with severe illness in 2018).
Discussion
Influenza epidemics and pandemics pose a severe threat to public health and the global economy. Long-term viral surveillance can reflect an outbreak situation in a timely manner and provide early warning of an epidemic or pandemic. In 2018, the proportion of outpatient and emergency influenza-like cases and the positive rate of influenza virus tests reported by the influenza surveillance sentinel hospital in China were higher than those reported during the same period in the previous 3 years. The number of severe cases was also higher.30–32 Therefore, there was an urgent need to study the pathogenicity and transmissibility of the major influenza viruses circulating in 2018.
In this study, we isolated two influenza strains from severely ill patients in Hebei, China, during the 2018 flu season. Phylogenetic analyses revealed that F076 belongs to the human-like H1N1 influenza virus lineage and 16275B belongs to the Victoria lineage. Both are closely related to the epidemic strain present in recent years. In addition, 16275B is closely related to the WHO reference epidemic strain B/Brisbane/60/2008, which in 2009 was recommended by the WHO for inclusion as a vaccine strain.33 We compared the gene sequences of influenza epidemic strains in recent years and found that F076 has the PB1 D701N mutation. Asparagine (N) at residue 701 improves the binding of PB2 to mammalian importin-a isoforms,34,35 and for H5N1 avian influenza viruses, it is associated with increased viral replication in mammalian cell lines, enhanced virulence in the mouse model, and more efficient transmission in the guinea pig model.36,37 These changes may increase the pathogenicity and transmissibility of F076. Interestingly, at the same time, we compared other H1N1 influenza viruses isolated in the laboratory and found that they did not have this mutation. This might explain why patients infected with the F076 strain develop severe illness. Future studies in our laboratory will explore in depth the molecular mechanisms of F076. Therefore, monitoring of seasonal influenza is very necessary, and more attention should be paid to strains isolated from severely ill patients with influenza.33
The infection caused by influenza viruses begins with the HA glycoprotein binding to sialic acid receptors on the surface of host cells.38,39 Therefore, the receptor-binding specificity of influenza viruses is a key determinant of cross-species transmission.39,40 Avian influenza viruses preferentially bind to α-2,3 sialic acid receptors, whereas mammalian influenza viruses preferentially bind to α-2,6 sialic acid receptors. In this study, both the F076 strain and 16275B strain showed specificity only for α-2,6 sialic acid receptor binding. Therefore, these 2 viruses preferentially infect mammals and are less likely to infect poultry.
In our mouse study, F076 exhibited much higher growth properties and pathogenicity than 16275B, which might be caused by the weak binding affinity for the sialic acid receptor of IBV compared with IAV.41–46 In addition, F076 caused lethal pneumonia in mice without prior adaptation (fatality rate of 40%), demonstrating higher pathogenicity than previous seasonal influenza viruses (Figure 4),47 which might be associated with the PB1 D701N mutation. F076 virus was detected in other tissues in addition to the respiratory tract, which indicated that it was a highly pathogenic virus. Highly pathogenic avian influenza viruses, such as H5N1, H5N6, and H7N9, can also replicate in various tissues.48–50
Airborne transmission capacity is an important indicator of whether an influenza strain can cause an epidemic or a pandemic.51,52 The H5N1, H5N6, and H7N9 strains can infect humans but show limited airborne transmissibility.53,54 Therefore, they are less likely to cause an epidemic. In this study, both F076 and 16275B were transmissible by airborne or direct contact in guinea pigs, but their transmission was not as extensive as that of H1N1pdm09 (Figure 6), indicating that they represent a potential localized outbreak risk.
This study has two limitations. First, the two strains isolated in this study do not and cannot reflect the overall situation of this epidemic. More strains should be characterized in future studies. Second, variations in the virus as they spread from person to person were not considered in this study.
Overall, we assessed the genetic characteristics, pathogenicity, and transmissibility of two influenza strains isolated from severely ill patients during the 2018 flu season. Our results increase our knowledge of current circulating strains and contribute to the prevention and control of influenza epidemic. Continuous monitoring and assessment are needed in the future.
Footnotes
Authors’ contributions: ZDG, CZ, JXL, and LGC conceived and designed the experiments; HC, CZ, JXL, SSD, CMZ, and JXL performed the experiments; and ZDG, ZYW, and CZ wrote the manuscript. All authors approved the content of the final article.
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: This work was supported by National Key Research and Development Program of China (grant no. 2016YFD0501000). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ORCID iDs: Cheng Zhang https://orcid.org/0000-0003-3049-1587
Zhongyi Wang https://orcid.org/0000-0002-8982-2806
References
- 1.Hause BM, Collin EA, Liu R, et al. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the Orthomyxoviridae family. mBio 2014; 5: e00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith DB, Gaunt ER, Digard P, et al. Detection of influenza C virus but not influenza D virus in Scottish respiratory samples. J Clin Virol 2016; 74: 50–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnur JM, El Moussi A, Pozo F, et al. Virological surveillance of influenza viruses during the 2008–09, 2009–10 and 2010–11 seasons in Tunisia. PLoS One 2013; 8: e74064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osterhaus AD, Rimmelzwaan GF, Martina BE, et al. Influenza B virus in seals. Science 2000; 288: 1051–1053. [DOI] [PubMed] [Google Scholar]
- 5.Ramis AJ, Van Riel D, Van De Bildt MW, et al. Influenza A and B virus attachment to respiratory tract in marine mammals. Emerg Infect Dis 2012; 18: 817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Porter E, Lohman M, et al. Influenza C virus in cattle with respiratory disease, United States, 2016-2018. Emerg Infect Dis 2018; 24: 1926–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCullers JA, Hayden FG. Fatal influenza B infections: Time to reexamine influenza research priorities. J Infect Dis 2012; 205: 870–872. [DOI] [PubMed] [Google Scholar]
- 8.Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis 2006; 12: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong S, Zhu X, Li Y, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog 2013; 9: e1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon SW, Webby RJ, Webster RG. Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol 2014; 385: 359–375. [DOI] [PubMed] [Google Scholar]
- 11.Gao GF. From “A”IV to “Z”IKV: Attacks from emerging and re-emerging pathogens. Cell 2018; 172: 1157–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan SJ, Jacobson RM, Dowdle WR, et al. 2009 H1N1 influenza. Mayo Clin Proc 2010; 85: 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb SAR, Pettila V, Seppelt I, et al. Critical care services and 2009 H1N1 influenza in Australia and New Zealand. New Engl J Med 2009; 361: 1925–1934. [DOI] [PubMed] [Google Scholar]
- 14.Novel Swine-Origin Influenza AVIT, Dawood FS, Jain S, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 2009; 360: 2605–2615. [DOI] [PubMed] [Google Scholar]
- 15.Ambrose CS, Luke C, Coelingh K. Current status of live attenuated influenza vaccine in the United States for seasonal and pandemic influenza. Influenza Other Respir Viruses 2008; 2: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducatez MF, Hause B, Stigger-Rosser E, et al. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg Infect Dis 2011; 17: 1624–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith G, Dhanasekaran V, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 Influenza A epidemic. Nature 2009; 459: 1122–1125. [DOI] [PubMed] [Google Scholar]
- 18.Tscherne DM, Garcia-Sastre A. Virulence determinants of pandemic influenza viruses. J Clin Invest 2011; 121: 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarocostas J. Too early to declare H1N1 swine flu pandemic over, says WHO chief. BMJ 2009; 339: b5681. [DOI] [PubMed] [Google Scholar]
- 20.Glezen WP, Schmier JK, Kuehn CM, et al. The burden of influenza B: a structured literature review. Am J Public Health 2013; 103: E43–E51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutsakos M, Nguyen TH, Barclay WS, et al. Knowns and unknowns of influenza B viruses. Future Microbiol 2016; 11: 119–135. [DOI] [PubMed] [Google Scholar]
- 22.Meseko CA, Odaibo GN, Olaleye DO. Detection and isolation of 2009 pandemic influenza A/H1N1 virus in commercial piggery, Lagos Nigeria. Vet Microbiol 2014; 168: 197–201. [DOI] [PubMed] [Google Scholar]
- 23.Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci USA 1999; 96: 13910–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Lau YC, Wu P, et al. Variation in influenza B virus epidemiology by lineage, China. Emerg Infect Dis 2018; 24: 1536–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Escobar LS, Hidalgo Jara W, Nizam QNH, et al. The perspective of the World Organisation for Animal Health. In: Mench JA (ed.) Advances in Agricultural Animal Welfare: Science and Practice. Royston Rd, Cambridge, Duxford, UK: Woodhead Publ Ltd, Officers Mess Business Centre, 2018, pp.169–182.
- 26.Hoffmann E, Stech J, Guan Y, et al. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 2001; 146: 2275–2289. [DOI] [PubMed] [Google Scholar]
- 27.Zou S, Stansfield C, Bridge J. Identification of new influenza B virus variants by multiplex reverse transcription-PCR and the heteroduplex mobility assay. J Clin Microbiol 1998; 36: 1544–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J, Deng G, Kong H, et al. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res 2017; 27: 1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Z, Cheng K, Sun W, et al. Multiple adaptive amino acid substitutions increase the virulence of a wild waterfowl-origin reassortant H5N8 avian influenza virus in mice. Virus Res 2018; 244: 13–20. [DOI] [PubMed] [Google Scholar]
- 30.Mao H, Sun Y, Chen Y, et al. Co-circulation of influenza A(H1N1), A(H3N2), B(Yamagata) and B(Victoria) during the 2017-2018 influenza season in Zhejiang Province, China. Epidemiol Infect 2020: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu XF, Zhou YQ, Wu J, et al. Clinical characteristics and outcomes during a severe influenza season in China during 2017-2018. BMC Infect Dis 2019; 19: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye CC, Zhu WP, Yu JX, et al. Understanding the complex seasonality of seasonal influenza A and B virus transmission: Evidence from six years of surveillance data in Shanghai, China. Int J Infect Dis 2019; 81: 57–65. [DOI] [PubMed] [Google Scholar]
- 33.Horm SV, Mardy S, Rith S, et al. Epidemiological and virological characteristics of influenza viruses circulating in Cambodia from 2009 to 2011. PLoS One 2014; 9: e110713. DOI: 10.1371/journal.pone.0110713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabriel G, Herwig A, Klenk HD, et al. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog 2008; 4: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Chen H, Jiao P, et al. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol 2005; 79: 12058–12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Y, Zhang Y, Shinya K, et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog 2009; 5: e1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou B, Pearce MB, Yi L, et al. Asparagine substitution at PB2 residue 701 enhances the replication, pathogenicity, and transmission of the 2009 pandemic H1N1 influenza A virus. PLoS One 2013; 8: e67616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol 2012; 2: 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorrell EM, Schrauwen EJA, Linster M, et al. Predicting ‘airborne’ influenza viruses: (trans-) mission impossible? Curr Opin Virol 2011; 1: 635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki Y, Ito T, Suzuki T, et al. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol 2000; 74: 11825–11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parrish CR, Kawaoka Y. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol 2005; 59: 553–586. [DOI] [PubMed] [Google Scholar]
- 42.Tewawong N, Suwannakarn K, Prachayangprecha S, et al. Molecular epidemiology and phylogenetic analyses of influenza B virus in Thailand during 2010 to 2014. PLoS One 2015; 10: e0116302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harvala H, Smith D, Salvatierra K, et al. Burden of influenza B virus infections in Scotland in 2012/13 and epidemiological investigations between 2000 and 2012. Euro Surveill 2014; 19: 6–12. [DOI] [PubMed] [Google Scholar]
- 44.Sam IC, Su YCF, Chan YF, et al. Evolution of influenza B virus in Kuala Lumpur, Malaysia, between 1995 and 2008. J Virol 2015; 89: 9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pica N, Chou YY, Bouvier NM, et al. Transmission of influenza B viruses in the guinea pig. J Virol 2012; 86: 4279–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCullers JA, Wang GC, He S, et al. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J Virol 1999; 73: 7343–7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walther T, Karamanska R, Chan RW, et al. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog 2013; 9: e1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun R, Luo J, Gao Y, et al. Different infection routes of avian influenza A (H5N1) virus in mice. Integr Zool 2010; 4: 402–408. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Fu Y, Yang J, et al. Genetic and biological characterization of two novel reassortant H5N6 swine influenza viruses in mice and chickens. Infect Genet Evol 2015; 36: 462–466. [DOI] [PubMed] [Google Scholar]
- 50.Belser JA, Brock N, Sun X, et al. Mammalian pathogenesis and transmission of avian influenza A(H7N9) viruses, Tennessee, USA, 2017. Emerg Infect Dis 2018; 24: 149–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pulit-Penaloza JA, Belser JA, Tumpey TM, et al . Sowing the seeds of a pandemic? Mammalian pathogenicity and transmissibility of H1 variant influenza viruses from the swine reservoir. Trop Med Infect Dis 2019; 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo ZD, Wang ZY, Zhang SF, et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg Infect Dis 2020; 26: 1586–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu ZQ, Zhang Y, Zhao N, et al. Comparative epidemiology of human fatal infections with novel, high (H5N6 and H5N1) and low (H7N9 and H9N2) pathogenicity avian influenza A viruses. Int J Env Res Pub He 2017; 14: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bi Y, Chen Q, Wang Q, et al. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe 2016; 20: 810–821. [DOI] [PubMed] [Google Scholar]