Abstract
Recent scientific advances have greatly enhanced our understanding of the complex link between the gut microbiome and cancer. Gut dysbiosis is an imbalance between commensal and pathogenic bacteria and the production of microbial antigens and metabolites. The immune system and the gut microbiome interact to maintain homeostasis of the gut, and alterations in the microbiome composition lead to immune dysregulation, promoting chronic inflammation and development of tumors. Gut microorganisms and their toxic metabolites may migrate to other parts of the body via the circulatory system, causing an imbalance in the physiological status of the host and secretion of various neuroactive molecules through the gut-brain axis, gut-hepatic axis, and gut-lung axis to affect inflammation and tumorigenesis in specific organs. Thus, gut microbiota can be used as a tumor marker and may provide new insights into the pathogenesis of malignant tumors.
Keywords: Gut microbiota, Immunity, Metabolism, Gut-brain/liver/lung axis
Background
Human intestines harbor approximately 3.8 × 1013 microorganisms that maintain the physiology and health of the host by influencing basic functions, such as metabolism, nutrition, immunomodulation, and pathogen resistance [1–5]. Recently, gut microbiota have been reported to play key roles in the regulation of several processes related to brain function and mental health [6]. At the same time, gut microbiota play an important role in host disease pathogenesis [7]. Drugs and particular diseases (autoimmune and chronic diseases) may cause intestinal microbial dysfunction [8]. Other factors, such as activation of inflammatory signaling, dietary changes, infection, and lack of nucleotide-binding oligomerization domain 2 (NOD2), can also lead to dysbiosis [9, 10]. Dysbiosis of the microbiome has differential effects on the abundance of certain gut microbiota. It may increase metabolic disorders and the abundance of inflammation-inducing bacteria, which can induce carcinogenesis [11–13]. Gut microbiota regulates cancer at the level of genetic instability, susceptibility to host immune response, progression, and response to therapy [14, 15].
Using animal models, researchers have gained insights into the mechanisms through which microbes trigger carcinogenesis [16–18]. Escherichia coli and Bacteroides fragilis have been shown to potentiate intestinal tumorigenesis in chronic inflammation [19]. Besides, specific microbes and microbial dysbiosis have been shown to induce and even promote carcinogenesis by releasing genotoxins that may damage host DNA [20, 21]. Recent research shows, host innate immune responses against the resident microbiome may lead to tumor growth [16]. Thus, the gut microbiome is an essential factor for consideration in the precise treatment of cancer and can be used as a biomarker for diagnosis and treatment purposes [22]. Moreover, the efficacy of cancer treatments has been shown to be reduced in antibiotic-treated and germ-free mice, suggesting that intact gut microbiota is necessary for optimal treatment response [23].
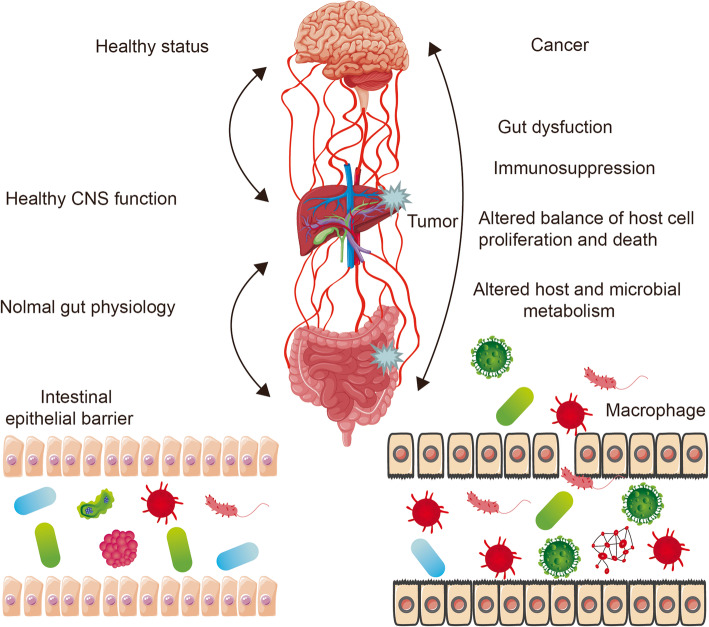
Regardless of health or disease status, gut microbiota affect metabolism, tissue development, inflammation, and immunity in the host [24]. Of course, the large amount of communication in the gut-organ axis cannot be separated from the involvement of the gut microbiota. It has been shown that there are bidirectional interactions within the brain-gut-microbiome axis, involving neural, endocrine, and inflammatory mechanisms [25, 26]. In addition, intestinal flora has been shown to affect liver immune function and bile acid metabolism through the gut-liver axis [27]. Similarly, lung inflammation originating in the gut has been reported in a study of the gut-lung axis [28]. In this review, we summarize how gut dysbiosis and immune dysregulation can lead to the induction and maintenance of tumors. Moreover, alterations in the microbiome may participate in immune modulation to promote cancer through metabolic pathways. We propose that gut microbiota dysbiosis affects cancer development through the gut-organ axis.
Gut microbiota dysbiosis is associated with the occurrence and development of cancer
The human gut is populated by trillions of archaea, bacteria, eukaryotes, viruses, and microbes belonging to four major microbial phyla: Proteobacteria, Firmicutes, Bacteroides, and Actinobacteria; these account for 95% of the gut microbiome [29]. Notably, true oncomicrobes in the intestine account for a very small proportion of all microbial populations. Furthermore, components of the microbiota such as flagellin can alter this balance and promote chronic inflammation, promoting intestinal tumor development [30]. HPV can cause the overexpression of the E6 and E7 genes of the virus, thereby cooperating to make the host cell immortal [31, 32]. Cunningly, microorganisms and their metabolites may migrate to other parts of the body and contribute to tumor development [32]. Disruption of the intestinal barrier function may trigger inflammation and carcinogenesis [12, 32]. Impaired barrier function can cause bacteria to enter the intestinal epithelium, allowing toxins to be transmitted. Bacterial toxins, such as colistin that is produced by Escherichia coli, have been shown to potentiate colorectal cancer in azoxymethane-exposed mice [12]. The toxin produced by the enterotoxigenic Bacteroides fragilis is related to colorectal tumors [33, 34]. In fact, microbes also drive cancerous transformation by affecting genome stability, resistance to cell death, and proliferation. For example, soluble fiber microbial fermentation disorders can induce cholestatic liver cancer [35]. Gut microbiota dysbiosis modulates the responses of CD8+ T cells to influence colitis-associated tumorigenesis [18]. Peptide tyrosine tyrosine expression induced by gut microbiota dysbiosis has been linked to the development of pancreatic cancer [36, 37]. Dysbiosis of gut microbiota has been shown to lead to the progression of chronic inflammation and liver disease, thereby increasing the risk of hepatocellular carcinoma (HCC) [38]. Interventions that regulate intestinal flora and improve immune function may be new regimens for future cancer treatment [29, 30]. Specific microbial changes can cause flora imbalance through signaling pathways and promote related cancer progression. Representative cancers related to gut dysbiosis are listed in Table 1.
Table 1
| Cancers Associated | microbiota | signaling | reference |
|---|---|---|---|
| With Gut Dysbiosis | |||
| Pancreatic cancer | Proteobacteria, Bacteroidetes, Firmicutes | TLR | [37] |
| Colorectal cancer | Bifidobacteria, Helicobacter, Bacteroides | TGF-β | [39–41] |
| Liver cancer | Fiber-Fermenting Bacteria Proteobacteria | TLR | [35, 42] |
| Lung cancer | Enterococcus,Streptococcus, Prevotella | [43, 44] | |
| Gastrointestinal cancer | H. pylori | STAT3 | [45–47] |
| Breast cancer | Pseudomonas aeruginosa, human papilloma virus | NF-κΒ | [48, 49] |
| Thyroid cancer | Neisseria,Streptococcus | STAT3 | [50] |
| Bladder cancer | Bacteroides fragilis and Clostridium cluster I | IL-6 | [51, 52] |
Gut microbiome and immune dysregulation in cancer
Intriguingly, the gut microbiome can inhibit infection by intestinal pathogens by occupying a niche, adjusting the niche environment, competing for nutrients, releasing bacteriocins, and regulating host immune defense. This process starts during the constitution of the microbiome at birth, affecting the maturation of the immune system, the development of tolerance and containment of the microbiome [53, 54]. In the mucosa, the T and B cells of immune system have the phenotype and function of specific locations affected by the microflora. These cells play a key role in maintaining immune homeostasis by inhibiting the response to harmless antigens and preserving the integrity of the intestinal mucosal barrier function [54].
Indeed, the host intestinal mucosal surface barrier allows microbial symbiosis [45, 55]. Gut microbiota are susceptible to continuous damage caused by the environment and must be repaired quickly to restore homeostasis. Disruption of the gut barrier results in confrontation between the microorganisms and the immune system, which may result in cancer and inflammatory diseases. The immune response in the developing tumor microenvironment, including the triggering of pro-inflammatory or immunosuppressive processes, can be further affected by microorganisms [46].
The central role of immunity in the biology of cancer calls for attention to the exact contribution of microbiota in oncogenesis. For example, gut microbiome dysbiosis promotes inflammation via chemokine C-C chemokine ligand 5 (CCL5), which recruits a non-physiological number of lymphocytes in the intestine, and the resulting inflammatory state promotes epithelial cell proliferation through local activation of the interleukin-6 (IL-6) pathway [56, 57] (Fig. 1). Upregulation of toll-like receptors (TLRs) by lipopolysaccharide (LPS) and other microbial products can activate the nuclear factor (NF)-κB, c-Jun/JNK, and JAK/STAT3 pathways, which have well-defined roles in cell proliferation and immunosuppression [21, 50].
Fig. 1.
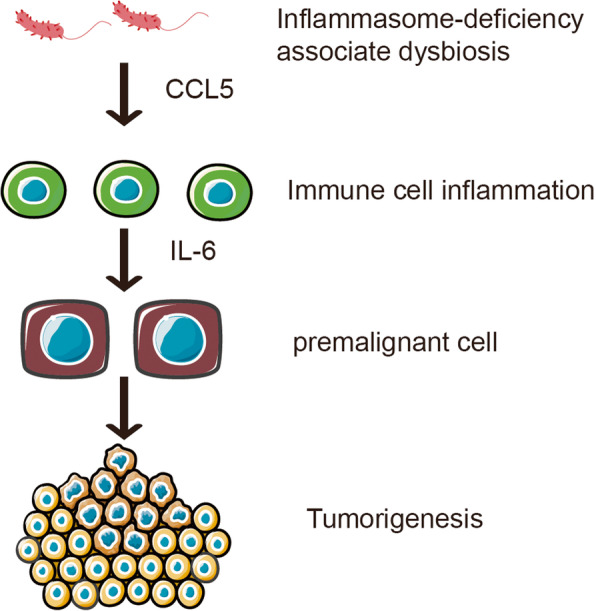
Gut dysbiosis can drive inflammation-induced cancer; it causes epithelial reprogramming and induces CCL5 transcription to induce local inflammation. In turn, it leads to local induction of IL-6 secretion and proliferation of intestinal epithelial cells, ultimately leading to tumor formation
Gut microbiome participates in immune modulation to promote cancer through metabolic pathways
Research on the interaction between the gut microbiome and immunity is an emerging field that examines the role of environmental factors, such as diet, as well as genetic and immune signals in metabolism, immunity, and host response to infection [58]. Studies on immune dysregulation may contribute to our understanding of the effects exerted by the microbiome in cancer development and treatment. In patients with colorectal cancer (CRC), the gut microbiome can directly or indirectly affect CRC by secreting metabolites, invading tissues, and modulating host immune response [39, 40]. Clostridium, Peptostreptococcus, Porphyromonas Genus, Prutella, Bacteroides, and twin cocci are the most significant bacteria associated with CRC [41, 59].
The liver is sensitive to intestinal bacterial metabolites, and changes in the intestinal microbiome affect the function of immune cells in the liver. Moreover, commensal microbiota can mediate the metabolism of primary to secondary bile acids. Gut microbiota are thought to be involved in the physiological activities of the host by affecting the bile acid pool, thus regulating hormone secretion and immunity via the resulting metabolites [60, 61]. We hypothesize that the gut microbiome promotes host immunity mainly through anabolic pathways. Ma et al. have found that the immune response to liver cancer has the opposite effect and that reducing the abundance of intestinal Clostridial bacteria through the use of antibiotics can increase the levels of primary bile acids and inhibit liver tumors by increasing the expression of CXC chemokine ligand 16 (CXCL16) in sinusoidal endothelial cells, after primary bile acids are metabolized to secondary bile acids by Clostridium bacteria [27]. Primary bile acids increase CXCL16 expression, whereas secondary bile acids exert the opposite effect. Schramm et al. have reported that the expression of CXCL16 in patients with liver cancer is linked to primary bile acids. However, there are differences in the composition of human and mouse immune systems, intestinal microbiome, and bile acid [62]. Therefore, the clinical significance of the relevant research results is limited.
In mice, gut microbiota and bile acid products play diverse roles in cancer development. For example, elimination of Clostridium XIV, increase in primary bile acids, and reduction in secondary bile acids inhibits the progress of liver cancer. Immune cells, such as dendritic cells, macrophages, and myeloid-derived suppressor cells, can be regulated by bile acids or their receptors, thus promoting anti-cancer immune responses. Bile acid receptors and flora metabolites may be novel targets for cancer treatment in the future [63]. Moreover, microbial pathogen-associated molecular patterns can activate toll-like receptor (TLR) signaling in a variety of cell types, leading to cytokine production and NF-κB-mediated inflammation, which can fuel tumor growth. Gut microbiota exacerbate metabolic inflammation through TLR signaling [64]. For example, the LPS receptor TLR4 has been shown to promote hepatocellular carcinoma, pancreatic cancer, and colon cancer. TLR-induced activation of NF-kB and STAT3 is a key signaling pathway that promotes cancer [32]. NF-κB signaling can stimulate glycolytic energy flux during acute inflammation [65]. (Fig. 2).
Fig. 2.
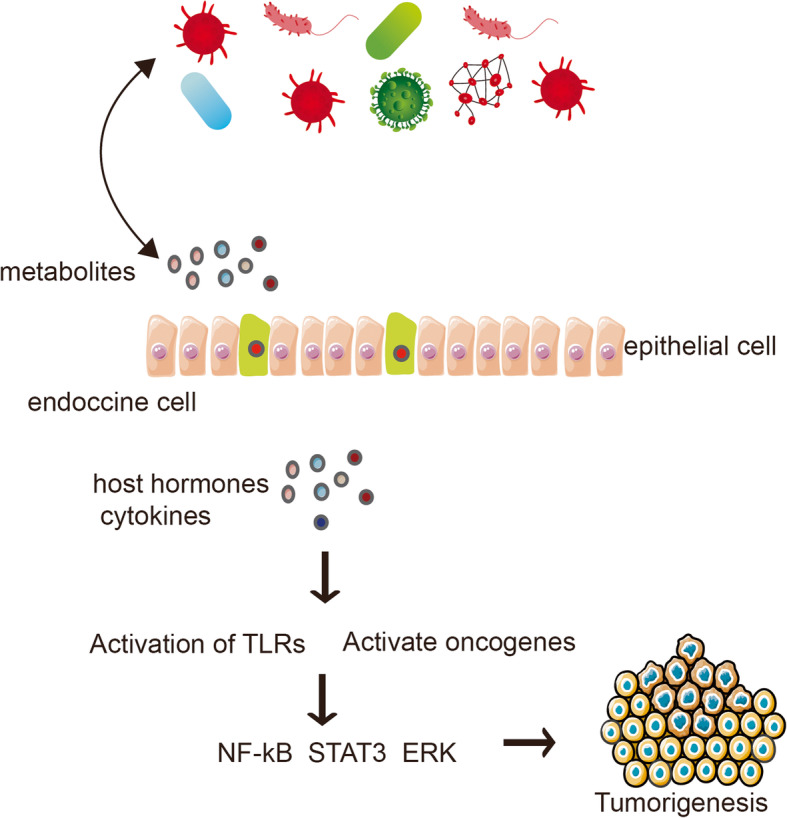
Microbiota metabolites are related to tumor development. Pro-inflammatory pathways are activated when the mucosal barrier is broken. The loss of the boundary between the host and the microorganism is related to the pattern recognition receptor signaling cascade. The feedforward circuit of chronic inflammation mediated by NF-kB and STAT3 signal transduction promotes canceration in transformed cells and non-tumor cells. Microbial PAMP activate TLR signaling in a variety of cell types, leading to cytokine production and NF-kB-mediated inflammation, thereby exacerbating tumor growth
Gut microbiota may affect the efficacy of PD-1 inhibitors
Previous studies in mouse models and humans have shown that regulation of the fecal microbiome significantly affects the outcome of cancer immunotherapy in terms of toxicity and efficacy [66]. Cancer immunotherapy based on the blockade of programmed cell death protein 1 (PD-1) and programmed death-ligand 1 (PD-L1) has become an essential approach for the treatment of various cancers in advanced stages [66]. Fecal bacterial transplantation may alter the gut microbiome of patients with cancer to improve the efficacy of drugs such as anti-PD-1 monoclonal antibody. Routy et al. have found that the response of patients with lung or kidney cancer to PD-1 monoclonal antibody is related to a higher abundance of Akkermansia muciniphila [67].
It is worth noting that the efficacy of tumor immunotherapy is related to the composition of intestinal bacteria [4]. Gopalakrishnan et al. have shown that, in patients with melanoma, the response to treatment with PD-1 monoclonal antibody was related to Faecalibacterium-based flora in patients [68]. There is a link between the efficacy of the cancer immune drug PD-1 blocker and the gut microbiota of patients, as gut microbiota may affect the efficacy of PD-1 inhibitors. Scientists suspect that the cytokine IL-12, which is released in response to A muciniphila, may help recruit T cells to combat cancer [3, 69]. Cancer patients harboring diverse intestinal flora rich in Clostridium lentum and Clostridiaceae have a better response to PD-1 inhibitors, and may show more significant effects of immunotherapy [3, 4].
A study examining the response of patients with different flora to PD-1 monoclonal antibodies has shown that responding patients were found to have a higher diversity of bacteria and a higher abundance of Bifidobacterium longum [69]. Therefore, the composition of the gut microbiome can affect the anti-tumor immunity of cancer patients, and can be used as a biomarker to predict the response of patients to immune checkpoint blockade therapy.
Prediction of the efficacy of PD-1 through microbial signals may require combining RNA sequencing, metabolomics, cancer process management, and intestinal fungal/viral analysis through clinical trials to fully understand the relationship between microbiota and the efficacy of tumor immunotherapy [70, 71]. Gut microbiota can regulate anti-PD-1 efficacy by interacting with the host immune system and thus represent a new therapeutic target [68, 72].
Patients with cancer should use antibiotics judiciously
Cancer treatment with microbial preparations or their products has the potential to shrink tumors [73]. Because flora can affect cancer progression, it may also affect the efficacy of chemotherapy and immunotherapy. Intervention with fecal flora affects the toxicity and effectiveness of immunotherapy. Gut microbes have been considered as key modulators of host immunity, raising the possibility that they could influence the outcome of cancer immunotherapy [74]. For example, it has been suggested that the use of antibiotics early in the treatment reduces the survival rate of patients with renal cell carcinoma and non-small cell lung cancer [66]. Antibiotics have saved countless lives, but they have also many side effects, such as Clostridium difficile infection, antibiotic resistance, and flora changes [75]. A study by Wu et al. has found that the use of antibiotics increases the risk of colon cancer, but reduces the risk of rectal cancer [76]. Antibiotic exposure is the main reason for the emergence of drug resistance, leading to the accumulation of drug-resistant genes in specific locations; the abundance and diversity of drug-resistant genes in the intestinal flora are high, which may be closely related to the use of antibiotics [77].
Gut microbiota dysbiosis affects cancer through gut-organ axis
The gut-organ axis establishes links or a two-way or multi-directional communication between organs through nerve, endocrine, immune, humoral, and metabolic pathways [78]. Intestinal flora and their secretions can be involved in the occurrence and development of tumors through the gut-organ axis [79, 80]. The bidirectional relationship between the gut and vital human organs (such as the lung, brain, and liver) is discussed below.
Gut microbiota dysbiosis affects cancer through the gut-brain axis
The gut-brain axis plays an important role in tumor proliferation, invasion, apoptosis, autophagy, and metastasis [47, 81]. Ruty et al. have proposed that gut microbiota can follow many routes to the brain and impact brain tumor therapeutic interventions [82]. In support, gut microbiota has been shown to have significant associations with cancer treatment-related psychoneurological symptoms [83].
The gut microbiota-brain axis includes gut microbiota and their metabolic products, the enteric nervous system (ENS), sympathetic and parasympathetic branches within the autonomic nervous system, neural-immune system, neuroendocrine system, and central nervous system (CNS) [84]. The gut microbiome produces most neurotransmitters found in the human brain [85, 86]. Recent studies have shown that the CNS and ENS can interact with gut microbiota to regulate nutrient metabolism. The vagal nerve system facilitates communication between the CNS and ENS to control gastrointestinal tract functions and feeding behavior. Vagal afferent neurons also express receptors for gut peptides that are secreted from enteroendocrine cells such as cholecystokinin (CCK), ghrelin, leptin, peptide tyrosine tyrosine (PYY), glucagon-like peptide-1 (GLP-1), and 5-hydroxytryptamine (5-HT; serotonin). The gut microbiome can regulate the levels of these peptides to influence the vagal afferent pathway and thus regulate intestinal metabolism via the microbiota-gut-brain axis. Serotonin functions as a key neurotransmitter at both terminals of this network. Local alterations in serotonin concentrations with subsequent relay of signals along the brain-gut axis influence CNS neurotransmission and regulate the function of neural processes in the gastrointestinal tract [87]. Gut microbiota dysbiosis can result in changes in serotonin levels. In triple-negative breast cancer, it has been shown that serotonin promotes cancer progression through autocrine serotonin signaling [48, 49, 88]. Dysbacteriosis-mediated expression of the glucagon-like peptide-1 has been shown to affect autophagy in endometrial cancer and is related to the occurrence of pancreatic cancer [89, 90].
Gut microbiota dysbiosis affects cancer through the gut-liver axis
The gut microbiome may contribute to cancer pathogenesis and progression through the gut-liver axis [91]. The two-way relationship between the intestine, microbiota, and liver integrates signals generated by dietary, genetic, and environmental factors [92]. This reciprocal interaction is facilitated by the portal vein [93]. Dysbiosis of gut microbiota leads to the progression of chronic inflammation and liver disease, thereby increasing the risk of HCC [35, 42]. Of note, bile acid and LPS provide an important link between the liver, bacterial microbiota, and the intestine. Intestinal microbiome-mediated bile acid metabolism regulates liver cancer through natural killer cells [27]. In a mouse model with dysbiotic intestinal flora, the synthesis of long-chain fatty acids is reduced, accompanied by oxidative stress and inflammation [94]. Bile acid, short-chain fatty acids (SCFAs), trimethylamine-N-oxide (TMAO), and immunoglobulin A (IgA) can also exert metabolic control through the microbiota-gut-liver axis. Further research should focus on the role of gut microbiota in the neuroendocrine regulation of nutrient metabolism via the microbiota-gut-brain-liver axis [95–97]. Intestinal microbes can promote cancer progression by changing the balance between host cell proliferation and death, and by affecting the immune system (Fig. 3).
Fig. 3.
The dysbacteriosis leads to continuous destruction of the barrier and delayed restoration of homeostasis. In these cases, the microbiota can affect canceration by changing the proliferation and apoptosis of host cells, disrupting the function of the immune system, and affecting the metabolism of the host
Gut microbiota dysbiosis affects cancer through the gut-lung axis
The human respiratory tract is the main and continuous entrance for many microorganisms and particles (such as viruses and bacteria), and the lung is an environment rich in flora [98, 99]. The intestinal flora may also have an impact on lung health. Changes in the microbial composition and function of the intestine are related to the development of lung diseases [100]. Metabolites such as SCFAs are produced by the microbiota and may regulate inflammation in the lungs [101, 102]. It appears that chronic lung diseases, such as cancer, are linked to dysbiotic airway microbiota and commonly occur alongside GI disorders [103, 104].
There is increasing evidence of a close relationship between the gastrointestinal tract and the respiratory tract. The exacerbation of chronic intestinal and lung diseases has key conceptual features related to the disorder and imbalance of the microbial ecosystem [105]. The surviving bacteria, cell wall fragments, or protein fragments of dead bacteria escape along with the cytokines and chemokines produced in the intestine, and then enter the general circulation. Entering the pulmonary circulation may lead to the activation of dendritic cells and macrophages and differentiation of T cells [106]. The concentration of circulating SCFAs in the intestine affects IL-6 and IL-8 in lung cancer and is related to the occurrence and development of lung cancer [107, 108]. In addition, patients with non-small-cell lung cancer experience gut butyrate-producing bacterial dysbiosis [109]. A significant relationship has also been found between Mycobacterium tuberculosis (TB) and lung cancer [43, 44]. A possible reason is that persistent tuberculosis infection can cause the production of tumor necrosis factor and cause lung inflammation. In addition, pulmonary fibrosis caused by TB leads to the synthesis of extracellular matrix, which is involved in the development of lung cancer [110, 111].
Conclusion and perspectives
Dysregulation of the gut microbiota and its interaction with the host may be important in tumorigenesis. First, we need to identify relevant bacteria in humans, study their abundance and the impact of their products on cancer progression, and elucidate their interactions with the human immune system as well as their ultimate impact on the mechanism of tumor occurrence and development. We then need to identify novel therapeutic microbial interventions and combine them with conventional therapies to treat tumors and other multifactorial human diseases.
Acknowledgments
Not applicable.
Abbreviations
- PD-1
Programmed cell death protein 1
- PD-L1
Programmed death-ligand 1
- NOD2
Nucleotide-binding oligomerization domain 2
- HPV
Human papillomavirus
- SYDS
Pleen-yang-deficiency syndrome
- CCL5
C-C chemokine ligand 5
- CXCL16
CXC chemokine ligand 16
- PAMPs
Pathogen-associated molecular patterns
- IL-6
Interleukin-6
- IL-12
Interleukin-12
- CNS
Central nervous system
- ENS
Enteric nervous system
- EECs
Enteroendocrine cells
- CCK
Cholecystokinin
- PYY
Peptide tyrosine tyrosine
- GLP-1
Glucagon-like peptide-1
- 5-HT
5-hydroxytryptamine
- HCC
Hepatocellular carcinoma
- SCFAs
Short-chain fatty acids
- TMAO
Trimethylamine-N-oxide
- IgA
Immunoglobulin A
- SCC
Small cell carcinoma
- AC
Adenocarcinoma
- NSCLC
Non-small-cell lung cancer
- TB
Tuberculosis
- TNF
Tumor necrosis factor
- ECM
Extracellular matrix
- SARS-Cov-2
Severe acute respiratory syndrome coronavirus-2
- COVID-19
Coronavirus disease 2019
- ACE2
Angiotensin-converting enzyme 2
- LPS
Lipopolysaccharide
Authors’ contributions
WMH, GYS, WXH, GYL, YJT, and ALY collected the related paper and finished the manuscript and figures. WMH, YJ, and KSMJ provided constructive guidance and made critical revisions. GYS and WMH participated in the design of this review. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Science Foundation of China under Grant number 81272297. Hunan Province Key Research and Development Project, (2020SK2053).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar RD. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal Cancer. Gastroenterology. 2020;158:322–340. doi: 10.1053/j.gastro.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hampton T. Gut microbes may shape response to Cancer immunotherapy. JAMA. 2018;319:430–431. doi: 10.1001/jama.2017.12857. [DOI] [PubMed] [Google Scholar]
- 4.Kaiser J. Gut microbes shape response to cancer immunotherapy. Science. 2017;358:573. doi: 10.1126/science.358.6363.573. [DOI] [PubMed] [Google Scholar]
- 5.Yang JY, Kim MS, Kim E, Cheon JH, Lee YS, Kim Y, Lee SH, Seo SU, Shin SH, Choi SS, et al. Enteric viruses ameliorate gut inflammation via toll-like receptor 3 and toll-like receptor 7-mediated interferon-beta production. Immunity. 2016;44:889–900. doi: 10.1016/j.immuni.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Cenit MC, Sanz Y, Codoner-Franch P. Influence of gut microbiota on neuropsychiatric disorders. World J Gastroenterol. 2017;23:5486–5498. doi: 10.3748/wjg.v23.i30.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen TD. Diet and gut microbiota in health and disease. Nestle Nutr Inst Workshop Ser. 2017;88:117–126. doi: 10.1159/000455220. [DOI] [PubMed] [Google Scholar]
- 8.Drago L, Valentina C, Fabio P. Gut microbiota, dysbiosis and colon lavage. Dig Liver Dis. 2019;51:1209–1213. doi: 10.1016/j.dld.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16:559–564. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Tajik N, Frech M, Schulz O, Schalter F, Lucas S, Azizov V, Durholz K, Steffen F, Omata Y, Rings A, et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat Commun. 2020;11:1995. doi: 10.1038/s41467-020-15831-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schott EM, Farnsworth CW, Grier A, Lillis JA, Soniwala S, Dadourian GH, Bell RD, Doolittle ML, Villani DA, Awad H, et al. Targeting the gut microbiome to treat the osteoarthritis of obesity. JCI Insight. 2018;3. [DOI] [PMC free article] [PubMed]
- 13.Chen X, Li P, Liu M, Zheng H, He Y, Chen MX, Tang W, Yue X, Huang Y, Zhuang L, et al. Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut. 2020;69:513–522. doi: 10.1136/gutjnl-2019-319101. [DOI] [PubMed] [Google Scholar]
- 14.Panebianco C, Andriulli A, Pazienza V. Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome. 2018;6:92. doi: 10.1186/s40168-018-0483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzutsev A, Badger JH, Perez-Chanona E, Roy S, Salcedo R, Smith CK, Trinchieri G. Microbes and Cancer. Annu Rev Immunol. 2017;35:199–228. doi: 10.1146/annurev-immunol-051116-052133. [DOI] [PubMed] [Google Scholar]
- 16.Chen GY, Shaw MH, Redondo G, Nunez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 2008;68:10060–10067. doi: 10.1158/0008-5472.CAN-08-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu AI, Zhao L, Eaton KA, Ho S, Chen J, Poe S, Becker J, Gonzalez A, McKinstry D, Hasso M, et al. Gut microbiota modulate CD8 T cell responses to influence colitis-associated tumorigenesis. Cell Rep. 2020;31:107471. doi: 10.1016/j.celrep.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park CH, Eun CS, Han DS. Intestinal microbiota, chronic inflammation, and colorectal cancer. Intest Res. 2018;16:338–345. doi: 10.5217/ir.2018.16.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodwin AC, Destefano SC, Wu S, Huso DL, Wu X, Murray-Stewart TR, Hacker-Prietz A, Rabizadeh S, Woster PM, Sears CL, Casero RJ. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cammarota G, Ianiro G. Gut microbiota and Cancer patients: a broad-ranging relationship. Mayo Clin Proc. 2017;92:1605–1607. doi: 10.1016/j.mayocp.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osadchiy V, Martin CR, Mayer EA. The gut-brain Axis and the microbiome: mechanisms and clinical implications. Clin Gastroenterol Hepatol. 2019;17:322–332. doi: 10.1016/j.cgh.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin CR, Osadchiy V, Kalani A, Mayer EA. The brain-gut-microbiome Axis. Cell Mol Gastroenterol Hepatol. 2018;6:133–148. doi: 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360. [DOI] [PMC free article] [PubMed]
- 28.Mjosberg J, Rao A. Lung inflammation originating in the gut. Science. 2018;359:36–37. doi: 10.1126/science.aar4301. [DOI] [PubMed] [Google Scholar]
- 29.Biragyn A, Ferrucci L. Gut dysbiosis: a potential link between increased cancer risk in ageing and inflammaging. Lancet Oncol. 2018;19:e295–e304. doi: 10.1016/S1470-2045(18)30095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gagliani N, Hu B, Huber S, Elinav E, Flavell RA. The fire within: microbes inflame tumors. Cell. 2014;157:776–783. doi: 10.1016/j.cell.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Pagano JS, Blaser M, Buendia MA, Damania B, Khalili K, Raab-Traub N, Roizman B. Infectious agents and cancer: criteria for a causal relation. Semin Cancer Biol. 2004;14:453–471. doi: 10.1016/j.semcancer.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Rajagopala SV, Vashee S, Oldfield LM, Suzuki Y, Venter JC, Telenti A, Nelson KE. The human microbiome and Cancer. Cancer Prev Res (Phila) 2017;10:226–234. doi: 10.1158/1940-6207.CAPR-16-0249. [DOI] [PubMed] [Google Scholar]
- 33.Prindiville TP, Sheikh RA, Cohen SH, Tang YJ, Cantrell MC, Silva JJ. Bacteroides fragilis enterotoxin gene sequences in patients with inflammatory bowel disease. Emerg Infect Dis. 2000;6:171–174. doi: 10.3201/eid0602.000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60:208–215. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh V, Yeoh BS, Chassaing B, Xiao X, Saha P, Aguilera OR, Lapek JJ, Zhang L, Wang WB, Hao S, et al. Dysregulated microbial fermentation of soluble Fiber induces Cholestatic liver Cancer. Cell. 2018;175:679–694. doi: 10.1016/j.cell.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Wang Z, Dong L, Jiang J, Xu X, Zhou L, Wan Y. Peptide tyrosine-tyrosine combined with its receptors exhibits an anti-cancer potential in pancreatic cancer MiaPaCa-2 cell. Chin Med J. 2014;127:4235–4242. [PubMed] [Google Scholar]
- 37.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, et al. The pancreatic Cancer microbiome promotes Oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8:403–416. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keku TO. Dulal S, Deveaux A, Jovov B, Han X. The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver Physiol. 2015;308:G351–G363. doi: 10.1152/ajpgi.00360.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, Moriyama EH, Copeland JK, Surendra A, Kumar S, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 41.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16:690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 42.Jia B, Wang R, Zhang J, Chi Y. Commentary: Dysregulated microbial fermentation of soluble Fiber induces Cholestatic liver Cancer. Front Cell Infect Microbiol. 2019;9:155. doi: 10.3389/fcimb.2019.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuang H, Cheng L, Wang Y, Zhang YK, Zhao MF, Liang GD, Zhang MC, Li YG, Zhao JB, Gao YN, et al. Dysbiosis of the gut microbiome in lung Cancer. Front Cell Infect Microbiol. 2019;9:112. doi: 10.3389/fcimb.2019.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu F, Li J, Guan Y, Lou Y, Chen H, Xu M, Deng D, Chen J, Ni B, Zhao L, et al. Dysbiosis of the gut microbiome is associated with tumor biomarkers in lung Cancer. Int J Biol Sci. 2019;15:2381–2392. doi: 10.7150/ijbs.35980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung J, Yu J. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67:1024–1032. doi: 10.1136/gutjnl-2017-314281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsuei J, Chau T, Mills D, Wan YJ. Bile acid dysregulation, gut dysbiosis, and gastrointestinal cancer. Exp Biol Med (Maywood) 2014;239:1489–1504. doi: 10.1177/1535370214538743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng C, Bai C, Brown TD, Hood LE, Tian Q. Human gut microbiota and gastrointestinal Cancer. Genomics Proteomics Bioinformatics. 2018;16:33–49. doi: 10.1016/j.gpb.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plaza-Diaz J, Alvarez-Mercado AI, Ruiz-Marin CM, Reina-Perez I, Perez-Alonso AJ, Sanchez-Andujar MB, Torne P, Gallart-Aragon T, Sanchez-Barron MT, Reyes LS, et al. Association of breast and gut microbiota dysbiosis and the risk of breast cancer: a case-control clinical study. BMC Cancer. 2019;19:495. doi: 10.1186/s12885-019-5660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J, Douglass J, Prasath V, Neace M, Atrchian S, Manjili MH, Shokouhi S, Habibi M. The microbiome and breast cancer: a review. Breast Cancer Res Treat. 2019;178:493–496. doi: 10.1007/s10549-019-05407-5. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Zhang F, Zhao C, Xu Q, Liang C, Yang Y, Wang H, Shang Y, Wang Y, Mu X, et al. Dysbiosis of the gut microbiome is associated with thyroid cancer and thyroid nodules and correlated with clinical index of thyroid function. Endocrine. 2019;64:564–574. doi: 10.1007/s12020-018-1831-x. [DOI] [PubMed] [Google Scholar]
- 51.Goulet CR, Champagne A, Bernard G, Vandal D, Chabaud S, Pouliot F, Bolduc S. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of bladder cancer cells through paracrine IL-6 signalling. BMC Cancer. 2019;19:137. doi: 10.1186/s12885-019-5353-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He C, Huang L, Lei P, Liu X, Li B, Shan Y. Sulforaphane normalizes intestinal Flora and Enhances gut barrier in mice with BBN-induced bladder Cancer. Mol Nutr Food Res. 2018;62:e1800427. doi: 10.1002/mnfr.201800427. [DOI] [PubMed] [Google Scholar]
- 53.Schnupf P, Gaboriau-Routhiau V, Cerf-Bensussan N. Modulation of the gut microbiota to improve innate resistance. Curr Opin Immunol. 2018;54:137–144. doi: 10.1016/j.coi.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 55.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, Jin C, Wunderlich C, Wunderlich T, Eisenbarth SC, Flavell RA. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci U S A. 2013;110:9862–9867. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 59.Ternes D, Karta J, Tsenkova M, Wilmes P, Haan S, Letellier E. Microbiome in colorectal Cancer: how to get from meta-omics to mechanism? Trends Microbiol. 2020;28:401–423. doi: 10.1016/j.tim.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 60.Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, Geva-Zatorsky N, Jupp R, Mathis D, Benoist C, Kasper DL. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature. 2020;577:410–415. doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chevre R, Silvestre-Roig C, Soehnlein O. Nutritional modulation of innate immunity: the fat-bile-gut connection. Trends Endocrinol Metab. 2018;29:686–698. doi: 10.1016/j.tem.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 62.Schramm C. Bile acids, the microbiome, immunity, and liver tumors. N Engl J Med. 2018;379:888–890. doi: 10.1056/NEJMcibr1807106. [DOI] [PubMed] [Google Scholar]
- 63.Sipe LM, Chaib M, Pingili AK, Pierre JF, Makowski L. Microbiome, bile acids, and obesity: how microbially modified metabolites shape anti-tumor immunity. Immunol Rev. 2020;295:220–239. doi: 10.1111/imr.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Backhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kauppinen A, Suuronen T, Ojala J, Kaarniranta K, Salminen A. Antagonistic crosstalk between NF-kappaB and SIRT1 in the regulation of inflammation and metabolic disorders. Cell Signal. 2013;25:1939–1948. doi: 10.1016/j.cellsig.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 66.Luke JJ, Pal SK. Further evidence to support judicious use of antibiotics in patients with cancer. Ann Oncol. 2018;29:1349–1351. doi: 10.1093/annonc/mdy153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Routy B, Le Chatelier E, Derosa L, Duong C, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 68.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Owens B. Gut bacteria link to immunotherapy sparks interest. Nat Biotechnol. 2018;36:121–123. doi: 10.1038/nbt0218-121. [DOI] [PubMed] [Google Scholar]
- 70.Gharaibeh RZ, Jobin C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut. 2019;68:385–388. doi: 10.1136/gutjnl-2018-317220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peters BA, Wilson M, Moran U, Pavlick A, Izsak A, Wechter T, Weber JS, Osman I, Ahn J. Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients. Genome Med. 2019;11:61. doi: 10.1186/s13073-019-0672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zitvogel L, Daillere R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. 2017;15:465–478. doi: 10.1038/nrmicro.2017.44. [DOI] [PubMed] [Google Scholar]
- 74.West NR, Powrie F. Immunotherapy not working? Check your microbiota. Cancer Cell. 2015;28:687–689. doi: 10.1016/j.ccell.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 75.Tamma PD, Miller MA, Cosgrove SE. Rethinking how antibiotics are prescribed: incorporating the 4 moments of antibiotic decision making into clinical practice. JAMA. 2019;321:139–140. doi: 10.1001/jama.2018.19509. [DOI] [PubMed] [Google Scholar]
- 76.Wan QY, Zhao R, Wang Y, Wu Y, Wu XT. Antibiotic use and risk of colorectal cancer: a meta-analysis of 412 450 participants. Gut. 2020;69:2059–2060. doi: 10.1136/gutjnl-2020-320826. [DOI] [PubMed] [Google Scholar]
- 77.Chewapreecha C. Your gut microbiota are what you eat. Nat Rev Microbiol. 2014;12:8. doi: 10.1038/nrmicro3186. [DOI] [PubMed] [Google Scholar]
- 78.Ahlawat S. Asha, Sharma KK: gut-organ axis: a microbial outreach and networking. Lett Appl Microbiol. 2020. [DOI] [PubMed]
- 79.Tsang SW, Auyeung KK, Bian ZX, Ko JK. Pathogenesis, experimental models and contemporary pharmacotherapy of irritable bowel syndrome: story about the brain-gut Axis. Curr Neuropharmacol. 2016;14:842–856. doi: 10.2174/1570159X14666160324144154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ondicova K, Mravec B. Role of nervous system in cancer aetiopathogenesis. Lancet Oncol. 2010;11:596–601. doi: 10.1016/S1470-2045(09)70337-7. [DOI] [PubMed] [Google Scholar]
- 81.Di YZ, Han BS, Di JM, Liu WY, Tang Q. Role of the brain-gut axis in gastrointestinal cancer. World J Clin Cases. 2019;7:1554–1570. doi: 10.12998/wjcc.v7.i13.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mehrian-Shai R, Reichardt J, Harris CC, Toren A. The gut-brain Axis, paving the way to brain Cancer. Trends Cancer. 2019;5:200–207. doi: 10.1016/j.trecan.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song BC, Bai J. Microbiome-gut-brain axis in cancer treatment-related psychoneurological toxicities and symptoms: a systematic review. Support Care Cancer. 2020. [DOI] [PMC free article] [PubMed]
- 84.Wang HX, Wang YP. Gut microbiota-brain Axis. Chin Med J. 2016;129:2373–2380. doi: 10.4103/0366-6999.190667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morris G, Berk M, Carvalho A, Caso JR, Sanz Y, Walder K, Maes M. The role of the microbial metabolites including tryptophan Catabolites and short chain fatty acids in the pathophysiology of immune-inflammatory and Neuroimmune disease. Mol Neurobiol. 2017;54:4432–4451. doi: 10.1007/s12035-016-0004-2. [DOI] [PubMed] [Google Scholar]
- 86.Aktar R, Parkar N, Stentz R, Baumard L, Parker A, Goldson A, Brion A, Carding S, Blackshaw A, Peiris M. Human resident gut microbe Bacteroides thetaiotaomicron regulates colonic neuronal innervation and neurogenic function. Gut Microbes. 2020;11:1745–1757. doi: 10.1080/19490976.2020.1766936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.O'Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res. 2015;277:32–48. doi: 10.1016/j.bbr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 88.Gautam J, Banskota S, Regmi SC, Ahn S, Jeon YH, Jeong H, Kim SJ, Nam TG, Jeong BS, Kim JA. Tryptophan hydroxylase 1 and 5-HT7 receptor preferentially expressed in triple-negative breast cancer promote cancer progression through autocrine serotonin signaling. Mol Cancer. 2016;15:75. doi: 10.1186/s12943-016-0559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanda R, Hiraike H, Wada-Hiraike O, Ichinose T, Nagasaka K, Sasajima Y, Ryo E, Fujii T, Osuga Y, Ayabe T. Expression of the glucagon-like peptide-1 receptor and its role in regulating autophagy in endometrial cancer. BMC Cancer. 2018;18:657. doi: 10.1186/s12885-018-4570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Monami M, Nreu B, Scatena A, Cresci B, Andreozzi F, Sesti G, Mannucci E. Safety issues with glucagon-like peptide-1 receptor agonists (pancreatitis, pancreatic cancer and cholelithiasis): data from randomized controlled trials. Diabetes Obes Metab. 2017;19:1233–1241. doi: 10.1111/dom.12926. [DOI] [PubMed] [Google Scholar]
- 91.Schwabe RF, Greten TF. Gut microbiome in HCC - mechanisms, diagnosis and therapy. J Hepatol. 2020;72:230–238. doi: 10.1016/j.jhep.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 92.Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol. 2020;72:558–577. doi: 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 93.Arab JP, Martin-Mateos RM, Shah VH. Gut-liver axis, cirrhosis and portal hypertension: the chicken and the egg. Hepatol Int. 2018;12:24–33. doi: 10.1007/s12072-017-9798-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen P, Torralba M, Tan J, Embree M, Zengler K, Starkel P, van Pijkeren JP, DePew J, Loomba R, Ho SB, et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology. 2015;148:203–214. doi: 10.1053/j.gastro.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang SZ, Yu YJ, Adeli K. Role of gut microbiota in neuroendocrine regulation of carbohydrate and lipid metabolism via the microbiota-gut-brain-liver Axis. Microorganisms. 2020;8. [DOI] [PMC free article] [PubMed]
- 96.Holzer P, Farzi A. Neuropeptides and the microbiota-gut-brain axis. Adv Exp Med Biol. 2014;817:195–219. doi: 10.1007/978-1-4939-0897-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bauer PV, Hamr SC, Duca FA. Regulation of energy balance by a gut-brain axis and involvement of the gut microbiota. Cell Mol Life Sci. 2016;73:737–755. doi: 10.1007/s00018-015-2083-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sze MA, Tsuruta M, Yang SW, Oh Y, Man SF, Hogg JC, Sin DD. Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS One. 2014;9:e111228. doi: 10.1371/journal.pone.0111228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, Aitken ML, Fligner CL, Singh PK. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci U S A. 2012;109:13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, Hansbro PM. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 101.Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol. 2019;20:1279–1290. doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- 102.Chakradhar S. A curious connection: teasing apart the link between gut microbes and lung disease. Nat Med. 2017;23:402–404. doi: 10.1038/nm0417-402. [DOI] [PubMed] [Google Scholar]
- 103.Wang H, Liu JS, Peng SH, Deng XY, Zhu DM, Javidiparsijani S, Wang GR, Li DQ, Li LX, Wang YC, Luo JM. Gut-lung crosstalk in pulmonary involvement with inflammatory bowel diseases. World J Gastroenterol. 2013;19:6794–6804. doi: 10.3748/wjg.v19.i40.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jess T, Horvath-Puho E, Fallingborg J, Rasmussen HH, Jacobsen BA. Cancer risk in inflammatory bowel disease according to patient phenotype and treatment: a Danish population-based cohort study. Am J Gastroenterol. 2013;108:1869–1876. doi: 10.1038/ajg.2013.249. [DOI] [PubMed] [Google Scholar]
- 105.He Y, Wen Q, Yao F, Xu D, Huang Y, Wang J. Gut-lung axis: the microbial contributions and clinical implications. Crit Rev Microbiol. 2017;43:81–95. doi: 10.1080/1040841X.2016.1176988. [DOI] [PubMed] [Google Scholar]
- 106.Keely S, Talley NJ, Hansbro PM. Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol. 2012;5:7–18. doi: 10.1038/mi.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sanmamed MF, Carranza-Rua O, Alfaro C, Onate C, Martin-Algarra S, Perez G, Landazuri SF, Gonzalez A, Gross S, Rodriguez I, et al. Serum interleukin-8 reflects tumor burden and treatment response across malignancies of multiple tissue origins. Clin Cancer Res. 2014;20:5697–5707. doi: 10.1158/1078-0432.CCR-13-3203. [DOI] [PubMed] [Google Scholar]
- 108.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkila J, Monti D, Satokari R, Franceschi C, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bingula R, Filaire M, Radosevic-Robin N, Berthon JY, Bernalier-Donadille A, Vasson MP, Thivat E, Kwiatkowski F, Filaire E. Characterisation of gut, lung, and upper airways microbiota in patients with non-small cell lung carcinoma: study protocol for case-control observational trial. Medicine (Baltimore) 2018;97:e13676. doi: 10.1097/MD.0000000000013676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pilaniya V, Gera K, Kunal S, Shah A. Pulmonary tuberculosis masquerading as metastatic lung disease. Eur Respir Rev. 2016;25:97–98. doi: 10.1183/16000617.00002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liang HY, Li XL, Yu XS, Guan P, Yin ZH, He QC, Zhou BS. Facts and fiction of the relationship between preexisting tuberculosis and lung cancer risk: a systematic review. Int J Cancer. 2009;125:2936–2944. doi: 10.1002/ijc.24636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.