Abstract
Abstract
Cyclophilin A (CypA), a key member of the immunophilin family, is the most abundantly expressed isozyme of the 18 known human cyclophilins. Besides acting as an intracellular receptor for cyclosporine A, CypA plays a vital role in microorganismal infections, cardiovascular diseases, liver diseases, kidney diseases, neurodegeneration, cancer, rheumatoid arthritis, periodontitis, sepsis, asthma, and aging. This review focuses on the pivotal roles of CypA in the infection of etiological agents, which manifests mainly in promoting or inhibiting viral replication based on the host cell type and viral species. CypA can interact with viral proteins and thus regulate the replication cycle of the virus. CypA is involved in pathogenic bacterial infections by regulating the formation of host actin skeleton or membrane translocation of bacterial toxins, or mediated the adhesion of Mycoplasma genitalium during the infection processes by acting as a cellular receptor of M. genitalium. CypA also plays a critical role in infection or the life cycle of certain parasites or host immune regulation. Moreover, we summarized the current understanding of CypA inhibitors acting as host-targeting antiviral agents, thus opening an avenue for the treatment of multiple viral infections due to their broad antiviral effects and ability to effectively prevent drug resistance. Therefore, the antiviral effect of CypA has the potential to promote CypA inhibitors as host-targeting drugs to CypA-involved etiological agent infections and human diseases.
Key points
• CypA is involved in the replication and infection of several viruses, pathogenic bacteria, mycoplasma, and parasites.
• CypA inhibitors are in a strong position to inhibit the infection of viruses, bacterial, and mycoplasma.
Keywords: Cyclophilin A, Viruses, Bacteria, Mycoplasmas, Parasites, CypA inhibitors
Introduction
Cyclophilins are a family of proteins from vertebrates and other organisms that bind to ciclosporin (cyclosporine A), an immunosuppressant which was used to suppress rejection after organ transplantations. These proteins exhibit peptidyl proline isomerase activity, which catalyzes the trans to cis isomerization of peptide bonds on proline residues and facilitates protein folding (Qian et al. 2010). At present, 17 types of cyclophilins with different structures have been identified in human cells, among which the most abundant one is cyclophilin A (CypA), which accounts for 0.1–0.6% of the total cytoplasmic protein (Harikishore and Yoon 2015). With the increasing research efforts on CypA, its biological role is recognized steadily. It not only participates in many biological processes, such as protein folding, cholesterol metabolism, regulation of immune function, and inflammatory reaction of the body (Qian et al. 2010) but also has an important clinical significance in the development of diseases such as inflammatory diseases, viral infections (de Wilde et al. 2018a), and malignant tumors (Qian et al. 2010). More specifically, during infection, oxidative stress, and hypoxia, CypA is secreted spontaneously into the extracellular space and plays an essential role in some physiological or pathological processes, such as inflammation, periodontitis, etiological agent infection and replication, atherosclerosis, diabetes, cancer proliferation, and metastasis (Nigro et al. 2013). In addition, overexpression of CypA not only promotes the proliferation of tumor cells and arrests hypoxia-induced cell apoptosis but also regulates malignant transformation and neonatal metabolism of tumor cells. However, the specific mechanism requires further research. Furthermore, CypA contains a binding site for caveolin-1and thus can bind to caveolin-1 on the cell membrane, and participates in the synthesis of cytoplasmic microcapsules, which is involved in cholesterol metabolism. CypA can also maintain the balance of extracellular and intracellular cholesterol by mediating the transfer of cholesterol into and out of cells (Qian et al. 2010).
Additionally, accumulating evidences indicated that CypA is a vital host protein for successful infection by microorganisms. The available data showed that CypA may either promote or inhibit virus replication, depending on the host cell type and the viral species. CypA plays an essential role in regulating the life cycle of several etiological agents by interacting with microbial proteins or promoting an antiviral immune response by mediating the release of inflammatory cytokines such as IFN-I. CypA also participated in the pathogenic progress of several bacteria by regulating the formation of host actin skeleton or membrane translocation of bacterial toxin (Dhanda et al. 2018). Furthermore, our previous studies verified that CypA was the receptor of MgPa and thus mediate M. genitalium to adhering and invading human urethral epithelial cells (Deng et al. 2018). The infection or life cycle of various parasites are also affected by CypA in several ways, which demonstrated the unique role of CypA in the replication and infection of several viruses, pathogenic bacteria, mycoplasmas, and parasites.
Moreover, an exciting recent development in CypA is research on CypA inhibitors. The CypA inhibitors attracted the attention of researchers due to their broad antiviral effects and the ability to effectively prevent drug resistance. Advances in CypA inhibitors will provide new methods to combat multiple pathogen infections in the future. This review summarizes the critical role of CypA in the replication of etiological agents, which makes CypA a new therapeutic target for etiological agent infection.
The role of CypA in viral infection
The role of CypA in influenza A virus infection
The influenza A virus is the pathogen of influenza with high morbidity and mortality. Recent studies indicated that CypA inhibits influenza virus replication and infection through the ubiquitination pathway and antiviral immune pathways (Liu et al. 2017; Mahesutihan et al. 2018).
CypA inhibits the influenza A virus through the M1 ubiquitination pathway
M1, a matrix protein of the influenza A virus, plays a vital role in the morphology and pathogenicity of the influenza A virus particle. Studies have shown that CypA interacts with M1 and thus inhibits the replication of the influenza A virus by promoting the degradation of M1, which regulated the assembly and budding of influenza A virus particle (Liu et al. 2012b; Mahesutihan et al. 2018). Liu et al. established a 293T cell line depleted of CypA expression (293T/CypA-) and showed that CypA participated in the ubiquitin-proteasome system (UPS) to downregulate M1 and thus inhibited the replication of the influenza A virus at the posttranslational level (Liu et al. 2012b). Researches on the mechanism of CypA regulation of M1 has resulted in significant progress. By disrupting the interaction between M1 and the E3 ubiquitin ligase atrophin-interacting protein-4 (AIP4), CypA inhibits AIP4-mediated M1 ubiquitination at K102 and K104 sites, thereby inhibiting the nuclear export of M1 and ultimately reducing the replication of the influenza A virus. Meanwhile, CypA and the K102 and K104 M1 mutants could alter the cellular localization of M1 (Mahesutihan et al. 2018). However, the K35 M1 mutant increased M1 ubiquitination, although the exact mechanism remains unclear.
CypA inhibits the influenza A virus through antiviral immune pathways
In addition to promoting the degradation of M1, CypA also inhibits influenza A virus replication by facilitating the retinoic acid-inducible gene-I-mediated (RIG-I-mediated) antiviral immune response. Earlier reports showed that RIG-I recognized RNA viruses such as Newcastle disease virus, Sendai virus, vesicular stomatitis virus, and influenza A virus (Kato et al. 2006; Loo et al. 2008) and stimulated IFN-I production by activating the RIG-I-mediated signaling pathway, inhibiting RNA virus replication. Liu et al. showed that viral infection upregulated CypA, further promoting the production of IFN-I to inhibit the infection of RIG-I-recognized RNA viruses (Liu et al. 2017). In addition, using TMN355, a CypA high-affinity inhibitor, and CsH a CsA analog, Ma et al. demonstrated that CsA inhibited influenza virus replication through a CypA-independent pathway (Ma et al. 2016). Moreover, a previous study by Liu et al. showed that CsA reduced the M1 protein expression in both normal 293T cell and CypA depleted 293T cell line, and the inhibitory effect of the M1 protein expression in the 293T/CypA+ cell line was stronger than that of the CypA depleted 293T cell line, the CST pull-down assay further demonstrated that CsA increased the interaction between CypA and M1, which indicated that CsA treatment could inhibit influenza A virus replication via both CypA-dependent and -independent mechanisms (Liu et al. 2012a). Undoubtedly, CsA shows exciting prospects for development as a drug against influenza virus infection.
In summary, based on the results of current research on the influenza A virus, we can conclude that CypA inhibits the replication and infection of the influenza virus by two mechanisms, one is regulating AIP-4-mediated M1 ubiquitination, leading to inhibition of M1 ubiquitination and M1 nuclear export, and the other is promoting RIG-I-mediated antiviral immune responses (Fig. 1). So far, the role of the ubiquitination mechanism of influenza A virus proteins in the replication and infection of the influenza A virus remains worthy of further investigation.
Fig. 1.
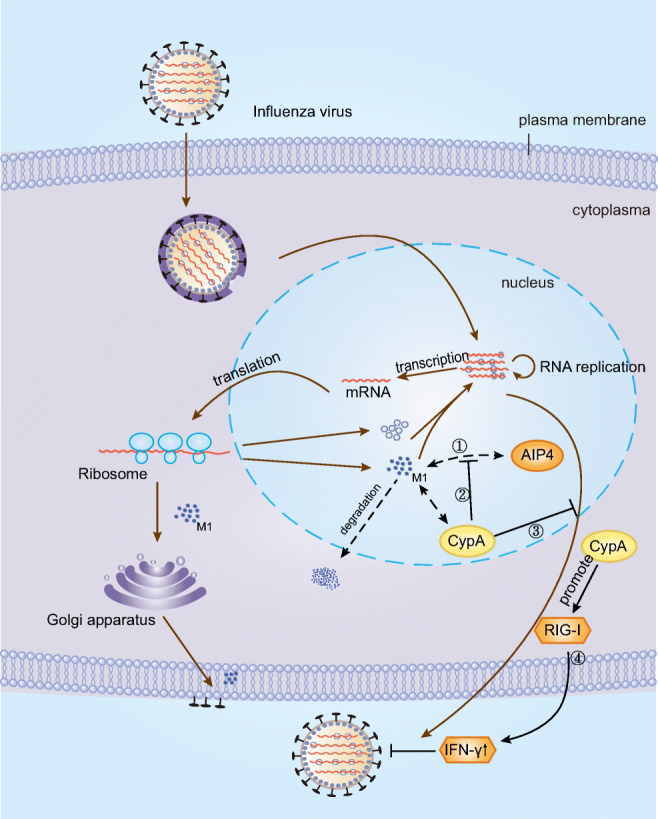
The roles of CypA in the lifecycle of influenza A virus. ①E3 ubiquitin ligase AIP4 can accelerate ubiquitin proteasome-mediated degradation of M1. ②By disrupting the interaction between M1 and the AIP4, CypA inhibits AIP4-mediated M1 ubiquitination. ③CypA inhibits the nuclear export of M1.④CypA promotes the production of type I IFN by triggering the RIG-I-mediated signaling pathway
The role of CypA in human immunodeficiency virus infection
The human immunodeficiency virus (HIV) is the etiological agent of the human acquired immunodeficiency syndrome (AIDS), which destroys the immune system to allow life-threatening opportunistic infections and cancers to thrive. Researchers have achieved significant progress using multiple experimental approaches to understand the pathogenic mechanism of HIV. The accumulating evidences indicated that the HIV-1 capsid plays a vital role in the life cycle of HIV-1 by interacting with cellular proteins. The replication and infection of HIV-1 are determined mainly by the combination of HIV-1 capsid and host factor CypA (Gamble et al. 1996). It has been known that CypA interacts with residues 85-93 of the N-terminal domain of HIV-1’s capsid (Gamble et al. 1996), but the precise mechanism of CypA interaction with the capsid and thus regulates the replication and infection of HIV-1 remains unclear. Current studies show that CypA either inhibits or enhances HIV infection in a cell-specific manner (Burse et al. 2017; De Iaco and Luban 2014; Li et al. 2009). Here, we review recent advances in the study of CypA inhibition and enhancement of HIV infection, as well as antiviral drugs targeting CypA binding sites on HIV.
CypA acts as a positive regulator in HIV infection
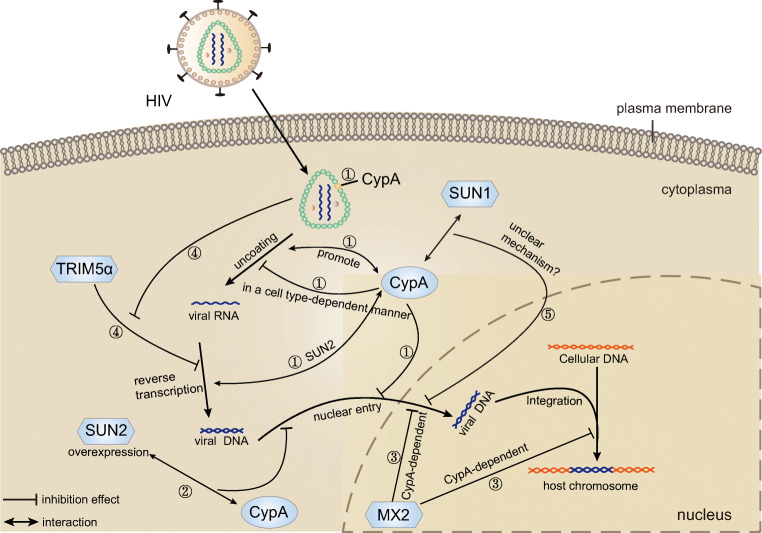
The positive effect of CypA on promoting HIV-1 infection had been investigated for decades. CypA positively regulates HIV infection by stabilizing the capsid, changing uncoating, improving the efficiency of reverse transcription, and nuclear import (Burse et al. 2017). CypA promotes the reverse transcription of HIV-1 under many conditions but inhibits the nuclear entry of HIV-1 in a cell type-dependent manner by delaying uncoating (De Iaco and Luban 2014). For example, the interaction between CypA and the viral capsid facilitates the early stages of the HIV replication cycle in most human cells, such as 293T, Jurkat T cells, and primary human CD4+ T cells. But the replication of HIV-1 in HeLa and H9 T cells is independent of CypA (De Iaco and Luban 2014; Meehan et al. 2014). Moreover, studies showed that the interaction between CypA and HIV-1 capsid could prevent the triggering of the innate immune pathway by evading the detection of viral DNA in the cytosol (Meehan et al. 2014). Stress granules (SGs), translationally silent ribonucleoprotein complexes (RNPs), were shown to be aggregated during viral infection and following physical or environmental trauma. Valiente-Echeverría and colleagues showed that the Gag-CypA association could stabilize the interaction between the capsid domain of Gag and the eukaryotic elongation factor and SG-dependency factor (SGDF), which promotes HIV-1 replication by inducing a blockade of SG assembly (Valiente-Echeverria et al. 2014).
Furthermore, several recent studies have demonstrated that TRIM5α may act as an HIV-1 restriction factor (Jimenez-Guardeño et al. 2019). Tripartite motif-containing proteins (TRIMs) are a large class of proteins that can directly interfere with viral activity by negatively affecting distinct stages of the viral life cycle, while TRIM5α as a CypA-sensitive HIV restriction factor, the restricting effect on HIV-1 is closely related to the interaction of CypA and capsid (Kim et al. 2019). Kyusik Kim et al. disrupted capsid-CypA interactions by using HIV-1-P90A mutants that cannot bind CypA, CSA, or shRNA targeting CypA, the results turned out that only when the capsid-CypA interaction was disrupted could the TRIM5α restrict HIV-1 infection, and the blocking effect occurs early in HIV-1 infection before the reverse transcription. HIV-1 could exploit CypA to escape the potent antiviral activity of human TRIM5α under the premise of successful establishment of capsid-CypA interaction (Kim et al. 2019). Selyutina et al. further demonstrated that CypA positively regulated HIV-1 infection in lymphocytes by blocking the binding of TRIM5α to capsid. They depleted TRIM5α, constructed HIV-1-P90A mutants and HIV-1-A92E/G94D mutants that lost the ability to bind to TRIM5α, which demonstrated that TRIM5a could restrict HIV-1 infection when capsid-CypA interaction was interrupted, while the successful establishment of capsid-CypA interaction could protect HIV-1 against TRIM5a attacks (Selyutina et al. 2020) (Fig. 2).
Fig. 2.
The functions of CypA in the lifecycle of HIV. ①CypA binds directly to the HIV-1 capsid, positively modulates viral infectivity by stabilizing the capsid, changing uncoating, improving the efficiency of reverse transcription and nuclear import, and negatively regulates viral infectivity by delaying capsid core uncoating and inhibiting the nuclear entry of HIV-1 in a cell type-dependent manner. ②CypA can help block the nuclear shape and early events of HIV infection in SUN2-overexpressed cells by interacting with SUN2. And SUN2 is an essential host protein for the positive effects of CypA on HIV-1 reverse transcription and infection. ③MxB protein can inhibit HIV replication and infection by hindering the nuclear import and integration of HIV-1 DNA in a CypA-dependent manner. ④ The successful establishment of capsid-CypA interaction could disrupt the restricting effect of TRIM5α on HIV-1 before the completion of reverse transcription. ⑤SUN1 regulates HIV infection by participating in the HIV-1 nuclear entry process through the mechanism dependent on the interaction between capsid and CypA in an unclear mechanism
CypA acts as a negative regulator in HIV infection
Although the positive effects of CypA in enhancing HIV-1 infection have been investigated for decades, the mechanisms underlying CypA inhibition of HIV-1 replication and infection remain unclear. Researchers are focusing their attention on understanding the mechanisms by which CypA participates in inhibiting HIV replication or infection. Several host proteins (TRIM5α, SUN1, SUN2, CPSF6, NUP153, NUP358/RanBP2, and MxB) can inhibit HIV-1 infection via interaction with the capsid (Burse et al. 2017; Donahue et al. 2016, 2017; Lahaye et al. 2016; Liu et al. 2013, 2015; Valiente-Echeverria et al. 2014). Myxovirus resistance protein-B (MxB, also called Mx2) is an interferon-induced dynamin GTPase that inhibits HIV-1 infection and replication by blocking nuclear import and integration of HIV-1 DNA (Cortines et al. 2015). Certain HIV strains can escape MxB inhibition, provided these HIV-1 strains lose the viral capsid that binds to CypA. This observation suggested that MxB protein can inhibit HIV-1 replication and infection in a CypA-dependent manner (Liu et al. 2013, 2015). The CypA-binding region of HIV-1 capsid also plays a critical role in the replication and infection of HIV-1. Although the experiments showed only a minor effect of the CypA-binding region mutations on CypA binding to the capsid, the mutations in the CypA-binding region can lead to an increase in the stability of the capsid, which hinders macrophage infectivity of HIV. A more stable capsid will lead to a decline in the early steps of HIV replication cycle efficiency (Setiawan et al. 2016). Using proteasome inhibitors MG132, CsA, and others, Setiawan et al. showed the dependence of effective replication of HIV on the levels of CypA present on target cells, and both high- and low-concentrations of CypA levels inhibit the virus replication (Setiawan et al. 2016).
SUN1 and SUN2 are inner nuclear membrane proteins that play a critical role in the nuclear-cytoplasmic connection by forming the linker inner part of the nucleo-skeleton and cytoskeleton (LINC) complex. An overexpression of SUN2 inhibits HIV-1 infection by disrupting the nuclear shape and early events of HIV-1 infection. Meanwhile, CypA blocks HIV infection in cells overexpressing SUN2 by interacting with SUN2 or SUN2-interacting cofactor or playing a role in the nuclear import pathway usage. Moreover, SUN2 is an essential host protein for the positive effects of CypA on HIV-1 reverse transcription and infection and is required for the antiviral activity of CypA (Donahue et al. 2016; Lahaye et al. 2016). More recently, the role of SUN2 in HIV-1 infection was investigated in greater detail. Donahue et al. showed that SUN2 plays a critical role in the proliferation, activation, and survival of primary CD4+ T cells. The experimental results showed that the silencing of SUN2 could reduce HIV infection to CD4+ T cells and endogenous SUN2 was not involved in the positive regulation of CypA on HIV infection. All these results showed that the infection of primary human CD4+ T cells with wild-type HIV was not mediated by a combination of both CypA and SUN2-dependent manner, but was affected individually by these proteins. Similarly, the overexpression of SUN1 can reduce the accumulation of 2-long-terminal-repeat (2-LTR) circular DNA and integrated viral DNA, thus blocking the process of nuclear import and inhibiting HIV infection. Results by Luo et al. suggested that SUN1 regulates HIV-1 infection by participating in the HIV-1 nuclear entry process through a mechanism dependent on the interaction between capsid and CypA (Luo et al. 2018). However, the exact mechanism remains unclear and further research is needed (Donahue et al. 2017). Collectively, these data indicated that CypA enhances the restriction ability of various protein and small molecule inhibitors on HIV-1 infection (Fig. 2).
Roles of CypA and CypA inhibitor in anti-HIV infection
Until now, studies showed the effect of CypA on inducing dendritic cell (DC) maturation and a series of innate immune responses, including the activation of the transcription factor IRF3, induction of an antiviral type I interferon response, and stimulation of T cell responses (Manel et al. 2010). Hou et al. demonstrated that the triggering of these immune responses by an interaction of HIV-1 capsids with cellular CypA specifically promoted HIV-1 Gag-specific cellular immunity. Therefore, CypA could be used as Gag antigen-specific gene adjuvant in DNA vaccine formulation (Hou et al. 2016). Moreover, based on the positive and negative effects of CypA on HIV replication and infection, researchers began to focus on the development of antiviral drugs targeted at the CypA or CypA-binding loop. CPI-431-32, a novel cyclophilin inhibitor, inhibited both HCV and HIV-1 infections by blocking the CypA-capsid interaction, thus destabilizing the core to cause premature uncoating and detection of the viral genome by host cell sensors. Compared with CsA and D-Methyl-Ala3-Ethyl-Val4 (DEB025, ALV), CPI-431-32 is better in blocking CypA-HIV-1 capsid interaction and therefore shows a better antiviral effect (Gallay et al. 2015). The antiviral drugs targeting CypA or CypA-binding loop may provide new thoughts to conquer HIV, but it is still a long way to go.
The role of CypA and CypA inhibitors in hepatitis C virus infection
The hepatitis C virus (HCV) is the cause of hepatitis C and certain cancers such as liver cancer and lymphomas in humans (Yi et al. 2019). In the HCV life cycle, after the viral particles were internalized into the cell, the HCV genome is translated to generate a polyprotein of 3011 amino acids. The polyprotein is then proteolytically processed by viral and cellular proteases into four structural proteins (core, E1, E2, and p7) and six nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) viral proteins (Suzuki et al. 2007). CypA acts as an essential factor in the HCV life cycle by interacting with NS5A and NS5B.
Role of CypA in HCV infection
CypA, the primary host cofactor of HCV replication, enhances the replication of HCV by interacting with NS5B or NS5A (Chatterji et al. 2015). NS4B is a small (27 kDa) hydrophobic integral membrane protein with four trans-membrane domains. It causes a morphological change in the endoplasmic reticulum and cooperates with other replicase factors, including NS3, NS4A, NS5A, and NS5B, to form a structure called a membranous web (MW) composed of double-membrane vesicles (DMVs). DMVs are a membrane chamber that guarantees the safety and efficiency of HCV replication, protected by cellular RNA sensors and degradation factors (Chatterji et al. 2016). Chatterji et al. suggested that the expression of the HCV NS3-NS5B polyprotein generates DMVs resembling shapes, numbers, and sizes to those created by full-length replicating HCV. However, the NS3-NS5B polyprotein cannot form DMVs in CypA knockdown cells or mutant cells with CypA isomerase activity abolished, indicating that CypA regulates the formation of DMVs synergistically through its isomerase with NS5A (Chatterji et al. 2015; Liu et al. 2009). Concurrently, based on evidence that both CypA inhibitors and NS5Ai could prevent NS3-NS5B-mediated DMV formation, it led to the conclusion that CypA is essential for DMV formation (Chatterji et al. 2015). Moreover, recent data suggests that NS5A self-interaction, in addition to its role in regulating core and NS5A subcellular localization, drove the interaction between NS5A and CypA. More specifically, CypA interacted with domain-II and domain-III of HCV NS5A to promote NS5A function to promote HCV replication, while mutations in NS5A domain-I will disrupt the interaction of NS5A with CypA (Ngure et al. 2016; Shanmugam et al. 2018). The nuclear magnetic resonance (NMR) spectroscopy and molecular dynamics (MD) simulations on HCV NS5A domain-II showed that the interaction of the short structural motif (PW-turn) of NS5A, embedded in proline-rich sequences, with CypA was directly related to the replication efficiency of HCV RNA. Similar to HIV-1 CA-CypA interactions, CypA fine-tuned the replication efficiency of HCV RNA by allosterically transforming the structural motif of NS5A domain-II in HCV (Dujardin et al. 2019). By using NMR, Dujardin et al. further demonstrated that the interaction of short structural motifs (PW-turn) in NS5A-D2 proline-rich sequences with human proline isomerase cycloproteins was directly related to the HCV RNA replication efficiency, similar to the HIV-1 CA-CypA interactions, CypA the structural motifs in the HCV were allosterically transformed to fine-tune the replication efficiency (Dujardin et al. 2019), but the specific underlying mechanisms require further investigation.
Roles of CypA inhibitors in HCV infection
It should be pointed out that the potential of CypA inhibitors in anti-viral infection has been studied extensively due to the role of CypA as a necessary host factor for the entry and replication of the HCV virus. There are currently two distinct developmental types of hepatitis C antiviral drugs, host-targeting antivirals (HTAs) and direct-acting antivirals (DAAs). HTAs provided a higher barrier to drug resistance for the HCV resistant against anti-viral compounds due to their minimal mutational rate in the host during viral or chemotoxic stress, while HTAs also had an advantage of pan-genotypic coverage to all HCV genotypes and serotypes (Baugh et al. 2013). Previous studies showed that CypA inhibitors played a pivotal role in suppressing HCV replication by blocking de novo formation of DMVs (Madan et al. 2014). Further studies on DMVs showed that CypA and NS5A inhibitors did not block the synthesis of HCV RNA replication complexes (RCs) (as opposed to NS5Bi), but rather prevented the formation of DMVs which was critical for HCV RNA replication (on the contrary, other anti-HCV agents, S5Bi, Mir-122i, and PI4KIII had no inhibitory effect) (Chatterji et al. 2015). Therefore, although the interaction of CypA with NS5A is essential for HCV RNA replication as described in “Role of CypA in HCV infection,” studies have shown inhibition of replication of HCV by NS5Ai-mediated disruption of the interaction between NS5A and phosphatidylinositol-4 kinase III (PI4KIII) (which is critical for HCV replication) (Eyre and Beard 2014; Reghellin et al. 2014). Intriguingly, recent evidence showed a unique regulatory effect of CypA inhibitors on the endoplasmic reticulum (ER) of HCV-infected cells. The CypA inhibitors recombined the ER of HCV-infected cells, and CypA inhibitor-treated HCV-infected cells remain completely insusceptible to HCV reinfection, suggesting that CypA inhibitor-mediated ER recombination made cells resistant to reinfection. Therefore, the accumulating evidence indicated that CypA inhibitors mainly inhibited HCV infection and replication in two ways: (i) inhibiting the formation of DMVs essential for HCV RNA replication by inhibiting the interaction of CypA with NS5A, and (ii) ER recombination to render cells resistant to HCV reinfection (Chatterji et al. 2016).
Application potential of CypA inhibitors in anti-HCV infection
Altogether, CypA inhibitors have received extensive attention, and the anti-HCV CypA inhibitors identified in recent years include ALV, NIM811 (Gallay et al. 2013), SCY-635, CPI-431-32 (Gallay et al. 2015), bis-amide derivative 25, bisamide derivatives 7e (Han et al. 2020), C31 (a small-molecule CypI) (Nevers et al. 2018), and the interferon-induced dynamin GTPase MxB (Yi et al. 2019). Recent studies on MxB further showed an interaction of MxB with the HCV non-structural protein NS5A, to interfere with HCV replication virus by attenuating the interaction between NS5A and CypA and the localization of NS5A in the endoplasmic reticulum. Specifically, MxB may combine with NS5A via domainI, which blocked CypA from accessing domain II of NS5A through steric hindrance (Yi et al. 2019). Moreover, studies showed that MxB also significantly inhibited Japanese encephalitis virus (JEV) and dengue virus (DENV), which belong to two other CypA-dependent viruses of the Flaviviridae family (Yi et al. 2019). Therefore, it is tempting to speculate that MxB may have the ability to inhibit the replication or infection of most CypA-dependent viruses. Currently, a novel CsA analog CPI-431-32 was identified with the ability to inhibit HCV replication by inhibiting the formation of DMVs (Gallay et al. 2015) with a superior anti-HCV effect than D-Methyl-Ala3-Ethyl-Val4 (DEB025, ALV), and was the only compound that entered the late-phase clinical development but was stopped due to severe side effects unrelated to CypA inhibition (Gallay et al. 2013; Zeuzem et al. 2015). In addition, CPI-431-32 also had an anti-HIV function, as described in “The role of CypA in human immunodeficiency virus infection.”
In another aspect, to overcome the immunosuppressive activity and severe side effects (including hepatotoxicity, nephrotoxicity, neurotoxicity, hypertension, and dyslipidemia) of CsA, researchers developed a liver-specific, sustained drug delivery system by conjugating the liver-targeting peptide (LTP) to PEGylated CsA-encapsulated poly (lactic-co-glycolic) acid (PLGA) nanoparticles, with high specificity to the liver and a superior anti-HCV effect. However, the nanoparticles still merit a larger trial to confirm their utility as a safe and effective drug delivery system for the treatment of HCV infection (Jyothi et al. 2015). In summary, the application of CypA inhibitors in anti-HCV infection might be forthcoming.
Taken together, although HCV can rapidly mutate the viral targets of DAAs, CypA inhibitors (a kind of HTA) bring promise to multiple viral infections treatment with their broad antiviral effects and ability to effectively prevent drug resistance. Therefore, compared with DAAs, HTAs have considerable advantages as follows: (i) high barrier to resistance, broad antiviral effects, and potential synergistic effects with DAAs and (ii) convenient to perceive the future of these types of drugs, in conjunction with next-generation DAAs, to provide a new approach to combat the most challenging HCV cases.
The role of CypA in hepatitis B virus infection
The hepatitis B virus is the pathogen of hepatitis B, which causes chronic disease and results in a high risk of death from cirrhosis and liver cancer. Studies showed that CypA not only plays an essential role in the life cycle of HCV but also participates in several steps of the HBV life cycle in infected cells. The expression and secretion of HBV small surface protein (SHBs) in human hepatoma cell lines can specifically stimulate CypA secretion, and an increase in secreted CypA is accompanied by a decrease in intracellular CypA (Zhao et al. 2007). Moreover, the secreted CypA has a strong chemotactic effect on inflammatory cells and further causes inflammation of the liver, which may be one of the pathogenic mechanisms of HBV (Tian et al. 2010). In regard to anti-HBV infection, Mohebbi A et al. predicted by using an in silico procedure that HBF-0259, a lead-like compound, can be used as a non-toxic anti-HBV molecule to inhibit HBV by interacting with CypA involved in HBsAg secretion and HBV integration, but the specific mechanism and effect require further investigation. Similarly, Phillips and colleagues showed that CypA inhibition could reduce the recycling of HBV nucleocapsids from the cytoplasm to the nucleus, a key mechanism for complementing viral templates. In addition, CypA is an essential co-factor for lipid and apolipoprotein B (ApoB) transport, and cellular lipids are a portion of HBV envelope proteins. Therefore, the inhibitory effect of CypA on ApoB secretion may hinder the intracellular formation and secretion of lipoprotein in HBV and subviral particles. For these reasons, cyclophilin inhibitors (alisporivir and NIM811) reduce HBV replication and the production and secretion of HBV envelope protein, thereby significantly decreasing HBV and HBsAg levels (Phillips et al. 2015). Finally, the use of cyclophilin inhibitors in combination with direct antiviral drugs targeting HBV polymerase will create a more effective treatment against chronic hepatitis B.
The role of CypA in flavivirus infection
The family of flaviviruses includes the West Nile virus, dengue virus, tick-borne encephalitis virus, Yellow fever virus (YFV), Zika virus, and several other viruses (Shi 2012). Most of these viruses are transmitted to vertebrates or humans through the bite of infected arthropods (mosquitoes or ticks), causing natural epidemic diseases such as Japanese encephalitis, dengue fever, yellow fever, and others. Importantly, the replication of Flaviviruses is highly dependent on host cell factors. Three structural proteins (capsid [C], pre-membrane [prM], or membrane [M], and envelope [E]), and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) form a single polyprotein which is encoded by YFV’s single 11-kb positive-strand RNA genome (Vidotto et al. 2017). Recent studies showed that MxB could inhibit the Japanese encephalitis virus and dengue virus, both of which are CypA-dependent viruses, through proteasomal pathway-induced NS5 degradation, suggesting that the susceptibility of the virus to MxB could be associated with CypA (Yi et al. 2019). Moreover, further research showed that NS4B protein, a nonstructural protein of flavivirus, plays a critical role in the formation of virus replication complexes. CypA has a critical role in the positive regulation of flavivirus replication by interacting with NS4B protein (Vidotto et al. 2017). However, the mechanism underlying the interaction between NS4B and CypA in the replication of flavivirus, and the pathogenesis of flavivirus infection still require further research and NS4B protein could be a target for the development of flavivirus vaccines.
The role of CypA in nidovirus infection
The nidovirus order is comprised of four families of positive-stranded RNA viruses, including well-known human pathogens such as MERS- and SARS-coronavirus (SARS-CoV) and economically important animal nidoviruses that cause porcine reproductive and respiratory syndrome virus (PRRSV), porcine epidemic diarrhea virus (PEDV), equine arteritis virus (EAV), and chicken infectious bronchitis virus (IBV) (de Wilde et al. 2018a). Due to the substantial morbidity and mortality of nidoviruses, considerable research on the virus in recent years, focused mainly on the interaction between nidoviruses and the host proteins, for developing broad-range antiviral agents.
CypA and coronavirus infections
Coronaviruses primarily infect the upper respiratory and gastrointestinal tract of mammals and birds and mainly cause colds and pneumonia in humans or digestive and respiratory diseases in animals. Since the discovery of human coronavirus SARS-CoV that causes severe acute respiratory syndrome (SARS), coronaviruses have attracted extensive attention in the research community. The research progresses on CypA and coronaviruses in recent years are summarized below. Previous research showed that CypA interacts with the N protein of SARS-CoV and gets incorporated into SARS-CoV particles (Neuman et al. 2008). To understand the detailed interaction between viral and the host proteins, and to identify the new targets for viral replication, Pfefferle et al. showed the influence of the N-terminal part of SARS-CoV non-structural protein Nsp1 on nuclear factor of activated T cells (NFAT) activation by interacting with several immunophilins (cyclophilins and FK506-binding proteins) and calcipressin. Using cyclosporin A (cyclophilin inhibitor), a highly active antagonist of NFAT activation, Pfefferle et al. showed that cyclophilins might play a significant role in the replication of all coronavirus prototype members (Pfefferle et al. 2011). Based on earlier studies, Carbajo-Lozoya et al. also showed that CsA inhibits coronavirus replication completely (Carbajo-Lozoya et al. 2012). Furthermore, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), an emerging novel coronavirus that causes coronavirus disease 2019 (COVID-19), could cause respiratory illness ranging from mild disease to severe disease and death. Studies had demonstrated that cyclophilin inhibitor alisporivir (Debio 025), a nonimmunosuppressive analog of CsA, could inhibit the infection of SARS-CoV-2 by inhibiting a post-entry step of the SARS-CoV-2 life cycle (Softic et al. 2020). Moreover, since CypA is one of the crucial elements of human coronavirus NL63 (HCoV-NL63) and HCoV-229E replication (Carbajo-Lozoya et al. 2014; Ma-Lauer et al. 2019), researchers used CypA as an antiviral target for CoV infection to evaluate the inhibitory effects of non-immunosuppressive derivatives alisporivir and NIM811 and immunosuppressive agents CsA and FK506 on HCoV-NL63. All these agents inhibit the replication of HCoV-NL63 and HCoV-229E at low micromolar concentrations (Carbajo-Lozoya et al. 2014; Ma-Lauer et al. 2019). Hence, based on the above knowledge, cyclophilins could serve as a new antiviral target, and non-immunosuppressive derivatives of cyclosporine A could be used as broad-range CoV inhibitors. Besides, other researchers showed that potent inhibition of the replication of EAV in CypA-deficient cells, while MERS-CoV was only slightly affected, and HCoV-229E was unaffected (de Wilde et al. 2018b). Possible explanations include the existence of CypA in the virus particles or different types of viruses, however, the specific underlying mechanisms need further investigation.
Virus replication can be effectively inhibited by antiviral therapies that directly affect the viral components involved in the virus's life cycle but has the disadvantage of not being able to prevent the high mutation frequency of the virus. Therefore, the host-targeting agents (HTAs) are more efficient to inhibit virus replication and infection by targeting host factors involved in virus entry, replication, assembly, and release, or in protein translation, folding, and other functions.
CypA and arterivirus infections
Arteriviruses include equine arteritis virus, porcine reproductive and respiratory syndrome virus, simian hemorrhagic fever virus, and lactate dehydrogenase-elevating virus (LDV) of mice (de Wilde et al. 2018a). The role of CypA in arteriviruses is similar as in coronavirus. Low concentrations of CSA can strongly inhibit viral replication by inhibiting the RNA synthesis of equine arteritis virus (EAV) and porcine reproductive and respiratory syndrome virus (PRRSV). The nonstructural proteins (NSPs) of these viruses can be assembled into membrane-associated replication, and transcription complexes (RTCs) with various host factors and CsA inhibits the synthesis of viral RNA by inhibiting the host factor CypA involved in this process (de Wilde et al. 2013). In addition, CypA effects on the replication of various nidoviruses are different. Studies showed potent inhibition of the replication of EAV in CypA-deficient cells, while MERS-CoV was only slightly affected, and HCoV-229E was unaffected (de Wilde et al. 2018b).
The exact mechanism of CypA in nidovirus replication and infection is still unclear, and the specific mechanism by which CypA interacts with nidovirus proteins is also worthy of further study. Moreover, as the replications of the RNA virus genome have low fidelity, antiviral drugs targeting CypA can solve the problem of the rapid emergence of resistance of RNA virus which will have broad research prospects in the future.
The role of CypA in other viral infections
Furthermore, CypA also promotes or inhibits the replication and infection of other viruses such as tombusvirus (TBSV), enterovirus 71 (EV71), rotavirus, human cytomegalovirus (HCMV), vaccinia virus (VV), and vesicular stomatitis virus (VSV) by promoting the secretion of host cytokines (IFN-β, etc.) or interacting with viral proteins. Using a Saccharomyces cerevisiae model based on genome-wide screening and global proteomics, Kovalev and Nagy showed that CypA binds to the RNA-binding region of TBSV replication proteins and inhibits the recruitment of viral RNA and blocks the assembly of viral replicase complexes (VRCs) to inhibit the replication of TBSV (Kovalev and Nagy 2013). CypA regulates the uncoating process of the EV71 virus by interacting with the H-I loop of VP1 protein in the capsid of EV71 in a pH-dependent manner (Qing et al. 2014). And CypA is incorporated selectively into vaccinia virus particles and VSV virions (Bose et al. 2003) to participate in their replication cycle. The stable expression of CypA plays a vital role in HCMV replication, production of HCMV virions and reactivation of HCMV in infected cells (Keyes et al. 2012). In contrast, human rotavirus (HRV) infection activates the PI3K/Akt pathway in host cells, further promoting the expression of transcription factor hypoxia-inducible factor-1 (HIF-1), and HIF-1 expression significantly promotes the transcription of CypA. He et al. used proteomic methods to identify CypA, a host protein upregulated in HRV infection, and demonstrated that CypA could interact with HRV structural protein VP2, thus inhibiting viral protein expression from blocking the replication of HRV (He et al. 2013). CypA also promotes IFN-I reaction to reduce host susceptibility to HRV and further affects the replication and infection of HRV (He et al. 2012).
The role of CypA in bacterial infection
The accumulating evidence indicates that CypA also played a pivotal role in the pathogenesis of several bacteria. In view of the biological function of CypA to promote protein folding and transport, it has been shown to be involved in biological processes requiring actin, such as general cytoskeletal remodeling and cell migration (Saleh et al. 2016). And the actin cytoskeleton is the common host subcellular target of bacteria such as Listeria monocytogenes, Salmonella enterica serovar Typhimurium, enterohemorrhagic E. coli (EHEC), and enteropathogenic Escherichia coli (EPEC), which could assist these bacteria to attach or invade host cells, and help bacteria disseminated from one cell to another (Dhanda et al. 2018). More specifically, CypA is involved in the formation of actin-rich L monocytogenes-generated membrane protrusions, which is a prerequisite for the effective disseminated of Listeria among cells. And CypA acts as a component of actin-rich structures so as to play a role in bacterial infection or transmission present not only in Listeria infection but also in S. Typhimurium infection. The membrane ruffles, a CypA-rich structures produced by S. Typhimurium-induced actin polymerization, which could encapsulate nearby S. Typhimurium and engulf them into host cells, while the enrichment of CypA in membrane ruffles also inhibited bacterial internalization and thus limiting S. Typhimurium invasion into host cells (Dhanda et al. 2018). Furthermore, EPEC and EHEC could regulate their attachment and invasion to host cells by hijacking the actin cytoskeleton of the host cells to form a CypA-rich structure called pedestals (Dhanda et al. 2018). The host actin cytoskeleton is the main attack target of many pathogenic enterobacteria, which gives us a hint that CypA may also play an essential role in the infection of other pathogens that rely on the host actin skeleton to disease development. Moreover, CypA could also promote the pathogenicity of some bacterial toxins by promoting their membrane translocation. LFNDTA, a fusion protein of the N-terminal domain of lethal factor (LF) of Bacillus anthracis fused to the catalytic subunit of diphtheria toxin, could interact with CypA in vitro to promote LFNDTA membrane transport in vitro, and CypA was further proved to be necessary for the efficient translocation of LFNDTA from endosome to cytoplasm through in vitro translocation experiments (Dmochewitz et al. 2011). Analogous phenomena are also found in Clostridium difficile, CypA is essential for the membrane translocation of the binary actin-ADP-ribosylating toxin C. difficile transferase (CDT). By using CypA inhibitors CSA, Kaiser et al. reversely confirmed that CypA contribute to CDT membrane ectopic and cell invasion (Kaiser et al. 2011). But no systematic study concerning the role of CypA in the pathogenesis of bacteria has been published yet, further effort is required to understand the importance of CypA in the infection of pathogens that rely on the host actin skeleton or bacterial toxin membrane translocation to disease development.
The role of CypA in mycoplasma infection
Some pathogenic mycoplasmas can be parasitic in the human respiratory and genitourinary tract through unique attachment organelles, leading to atypical pneumonia and other respiratory diseases, pelvic inflammatory diseases, fetal respiratory distress syndrome in premature infants, bronchopulmonary dysplasia, and intraventricular hemorrhage (Browning and Citti 2014). Several mycoplasmas act as cofactors to activate AIDS (Mahlangu et al. 2019). Because the interactions between the pathogens and the host cells are essential for the pathogens to infect the host cell successfully, our group identified the receptor of the M. genitalium protein of adhesion (MgPa) and proposed for the first time that CypA may be the receptor protein of MgPa which can partially inhibit M. genitalium to adhere and even invade into human urethral epithelial cells (Deng et al. 2018). And MgPa can stimulate the secretion of extracellular CypA in SV-HUC-1 cells and thus induce the expression of proinflammatory cytokines in SV-HUC-1 cells via the CypA-CD147-ERK-NF-κB signal transduction pathway (Li et al. 2020). Our findings laid the foundation for further study of the possible mechanisms by which M. genitalium adheres and infects host cells (Deng et al. 2018). Similarly, adhesins (P1 and P30) and adhesin-related accessory proteins (HMW1-4) of M. pneumoniae is a proline-rich component that mediates the successful adhesion of M. pneumoniae to host cells. And cyclophilins can activate proline-rich proteins to regulate cell adhesion, virulence, and growth cycle events of M. pneumonia. This effect could be inhibited by CsA, further causing the cytadherence defects of M. pneumonia (Reddy et al. 1996). However, research on the role of CypA in pathogenic mycoplasma remains at a very shallow level and requires further efforts.
The role of CypA in parasites infection
In recent years, the studies on immunophilins of parasites mainly focused on the identification of immunophilin genes and biochemical characterization of their products. A combination with immunosuppressant CsA may have several anti-parasitic roles (except Leishmania donovani) (Dutta et al. 2001). However, the detailed mechanisms involved in the immunophilin role in the life cycle of parasites are still unclear.
Clonorchis sinensis is the most prevalent human trematode in Asia and can cause clonorchiasis. Previous studies have shown that CypA aggravates sepsis by promoting intracellular communication, endothelial cell apoptosis, and chemotaxis to inflammatory cells (Song et al. 2015). Against this mechanism, researchers have found that in patients infected with clonorchiasis, CypA of Clonorchis sinensis (CsCypA) produces specific anti-CsCypA antibodies, and this antibody can neutralize mouse CypA (MuCypA), which may effectively protect patients with sepsis caused by C. sinensis (Song et al. 2015). Later, by using the cecal ligation and puncture (CLP) model, Jiang et al. further showed that cross-reactivity between anti-CsCyPAs and MuCyPA could offer a protective effect for improving organ injury, reducing the expression of inflammatory factors and chemokines, diminishing the bacterial burden of peripheral blood, and decreasing the ROS generation in peritoneal macrophages induced by LPS (Jiang et al. 2018).
In addition, trypanosome CypA is involved in the process of African trypanosome host infection, which is localized in the cytosol and flagellum of African trypanosomes and plays an essential role in the immune regulation of susceptible animals (Pellé et al. 2002). The deprivation of host-cell CypA in macrophages caused a delay of the replication of Leishmania major to achieve the effect of the anti-Leishmania (Hoerauf et al. 1997). All the above-described studies confirmed that host-cell CypA and parasite CypA play a vital role in the infection or life cycle of various parasites in several ways. As an immunosuppressant with a high affinity for CypA, CsA inhibits the infection or reproduction of parasites, such as Plasmodium falciparum, Plasmodium chabaudi, Plasmodium berghei, Toxoplasma gondii, Trypanosoma cruzi, Eimeria tenella, Eimeria vermiformis, Eimeria mitis, Caenorhabditis elegans, and Cryptosporidium parvum in several ways (Krucken et al. 2009; Potenza et al. 2006). The identification of potential receptor molecules for CsA antiparasitic activity will aid the development of anti-parasitic drugs.
Conclusions and perspectives
The roles of CypA in etiological agent infection and replication are under investigation for decades. In this review, we described the current understanding of the function of CypA in the life cycle of several viruses, bacterial, mycoplasmas, and parasites as well as the mechanisms by which CypA inhibitors are emerging as novel antiviral drugs. Further studies are warranted in influenza viruses, to understand the role of the ubiquitination mechanism of influenza virus proteins in the replication and infection of the influenza virus. The specific underlying mechanisms on how CypA assists several host proteins such as SUN1/2, MxB, and others in inhibiting HIV-1 or HCV infection needs further investigation. More specifically, since CypA interacts with a variety of microbial proteins (e.g., HCV NS5A, HIV CA, influenza virus M1, flavivirus NS4B, and SARS-CoV Nsp1), further researches are required directed towards exploring the crystal structure of microbial proteins and to construct the interaction model through biological methods such as spectrometry-based protein foot-printing approach. While the studies involved the role of CypA in pathogenic bacterial infections by regulating the formation of host actin skeleton or membrane translocation of bacterial toxin provided numerous insights and hinted that the prospect of understanding the interaction of between CypA and these pathogens. Furthermore, research on the role of CypA in pathogenic mycoplasmas is scanty, and the mechanism of M. genitalium adherence to host cells by interacting with CypA needs further investigation. In addition, it may be meaningful to explore whether the mechanisms underlying CypA activity in various microbial co-infections are different from a single infection. Finally, the development of CypA inhibitors may open new avenues to the development of novel anti-pathogenic therapeutics. It is necessary to understand the changes in cell structure after the introduction of CypA inhibitors into infected cells and to elucidate a precise mechanism of interaction.
Author contributions
YL wrote the manuscript. YZ conceived and provided the main direction of this manuscript and guided the writing and modification of the manuscript. DL and KP assisted in the preparation of references. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (no: 81871256).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yating Liao, Email: liaoyatingsix@126.com.
Dan Luo, Email: 2239269298@qq.com.
Kailan Peng, Email: pkl970910@163.com.
Yanhua Zeng, Email: zengyihua21cn@126.com.
References
- Baugh JM, Garcia-Rivera JA, Gallay PA (2013) Host-targeting agents in the treatment of hepatitis C: a beginning and an end? Antivir Res 100(2):555–561. 10.1016/j.antiviral.2013.09.020 [DOI] [PMC free article] [PubMed]
- Bose S, Mathur M, Bates P, Joshi N, Banerjee AK (2003) Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J Gen Virol 84(Pt 7):1687–1699. 10.1099/vir.0.19074-0 [DOI] [PubMed]
- Browning GF, Noormohammadi AH, Markham PF (2014) Identification and Characterization of Virulence Genes in Mycoplasmas. In: Browning GF, Citti C (eds) Mollicutes: molecular biology and pathogenesis. Norfolk, UK, pp 77–90
- Burse M, Shi J, Aiken C (2017) Cyclophilin A potentiates TRIM5α inhibition of HIV-1 nuclear import without promoting TRIM5α binding to the viral capsid. PLoS One 12(8):e0182298. 10.1371/journal.pone.0182298 [DOI] [PMC free article] [PubMed]
- Carbajo-Lozoya J, Muller MA, Kallies S, Thiel V, Drosten C, von Brunn A (2012) Replication of human coronaviruses SARS-CoV, HCoV-NL63 and HCoV-229E is inhibited by the drug FK506. Virus Res 165(1):112–117. 10.1016/j.virusres.2012.02.002 [DOI] [PMC free article] [PubMed]
- Carbajo-Lozoya J, Ma-Lauer Y, Malesevic M, Theuerkorn M, Kahlert V, Prell E, von Brunn B, Muth D, Baumert TF, Drosten C, Fischer G, von Brunn A (2014) Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including Alisporivir. Virus Res 184:44–53. 10.1016/j.virusres.2014.02.010 [DOI] [PMC free article] [PubMed]
- Chatterji U, Bobardt M, Tai A, Wood M, Gallay PA (2015) Cyclophilin and NS5A inhibitors, but not other anti-hepatitis C virus (HCV) agents, preclude HCV-mediated formation of double-membrane-vesicle viral factories. Antimicrob Agents Chemother 59(5):2496–2507. 10.1128/aac.04958-14 [DOI] [PMC free article] [PubMed]
- Chatterji U, Bobardt M, Schaffer L, Wood M, Gallay PA (2016) Cyclophilin Inhibitors Remodel the Endoplasmic Reticulum of HCV-Infected Cells in a Unique Pattern Rendering Cells Impervious to a Reinfection. PLoS One 11(7):e0159511. 10.1371/journal.pone.0159511 [DOI] [PMC free article] [PubMed]
- Cortines JR, Lima LM, Mohana-Borges R, Millen Tde A, Gaspar LP, Lanman JK, Prevelige PE Jr, Silva JL (2015) Structural insights into the stabilization of the human immunodeficiency virus type 1 capsid protein by the cyclophilin-binding domain and implications on the virus cycle. Biochim Biophys Acta 1854(5):341–348. 10.1016/j.bbapap.2014.12.008 [DOI] [PubMed]
- De Iaco A, Luban J (2014) Cyclophilin A promotes HIV-1 reverse transcription but its effect on transduction correlates best with its effect on nuclear entry of viral cDNA. Retrovirology 11:11. 10.1186/1742-4690-11-11 [DOI] [PMC free article] [PubMed]
- de Wilde AH, Li Y, van der Meer Y, Vuagniaux G, Lysek R, Fang Y, Snijder EJ, van Hemert MJ (2013) Cyclophilin inhibitors block arterivirus replication by interfering with viral RNA synthesis. J Virol 87(3):1454–1464. 10.1128/JVI.02078-12 [DOI] [PMC free article] [PubMed]
- de Wilde AH, Pham U, Posthuma CC, Snijder EJ (2018a) Cyclophilins and cyclophilin inhibitors in nidovirus replication. Virology 522:46–55. 10.1016/j.virol.2018.06.011 [DOI] [PMC free article] [PubMed]
- de Wilde AH, Zevenhoven-Dobbe JC, Beugeling C, Chatterji U, de Jong D, Gallay P, Szuhai K, Posthuma CC, Snijder EJ (2018b) Coronaviruses and arteriviruses display striking differences in their cyclophilin A-dependence during replication in cell culture. Virology 517:148–156. 10.1016/j.virol.2017.11.022 [DOI] [PMC free article] [PubMed]
- Deng XY, Dai P, Yu MJ, Chen LS, Zhu CM, You XX, Li LL, Zeng YH (2018) Cyclophilin A is the potential receptor of the Mycoplasma genitalium adhesion protein. Int J Med Microbiol 308(3):405–412. 10.1016/j.ijmm.2018.03.001 [DOI] [PubMed]
- Dhanda AS, Warren KE, Chiu RH, Guttman JA (2018) Cyclophilin A Controls Salmonella Internalization Levels and is Present at E. coli Actin-Rich Pedestals. Anat Rec (Hoboken) 301(12):2086–2094. 10.1002/ar.23957 [DOI] [PubMed]
- Dmochewitz L, Lillich M, Kaiser E, Jennings LD, Lang AE, Buchner J, Fischer G, Aktories K, Collier RJ, Barth H (2011) Role of CypA and Hsp90 in membrane translocation mediated by anthrax protective antigen. Cell Microbiol 13(3):359–373. 10.1111/j.1462-5822.2010.01539.x [DOI] [PMC free article] [PubMed]
- Donahue DA, Amraoui S, di Nunzio F, Kieffer C, Porrot F, Opp S, Diaz-Griffero F, Casartelli N, Schwartz O (2016) SUN2 Overexpression Deforms Nuclear Shape and Inhibits HIV. J Virol 90(8):4199–4214. 10.1128/JVI.03202-15 [DOI] [PMC free article] [PubMed]
- Donahue DA, Porrot F, Couespel N, Schwartz O (2017) SUN2 Silencing Impairs CD4 T Cell Proliferation and Alters Sensitivity to HIV-1 Infection Independently of Cyclophilin A. J Virol 91(6). 10.1128/jvi.02303-16 [DOI] [PMC free article] [PubMed]
- Dujardin M, Madan V, Gandhi NS, Cantrelle FX, Launay H, Huvent I, Bartenschlager R, Lippens G, Hanoulle X (2019) Cyclophilin A allows the allosteric regulation of a structural motif in the disordered domain 2 of NS5A and thereby fine-tunes HCV RNA replication. J Biol Chem 294(35):13171–13185. 10.1074/jbc.RA119.009537 [DOI] [PMC free article] [PubMed]
- Dutta M, Delhi P, Sinha KM, Banerjee R, Datta AK (2001) Lack of abundance of cytoplasmic cyclosporin A-binding protein renders free-living Leishmania donovani resistant to cyclosporin A. J Biol Chem 276(22):19294–19300. 10.1074/jbc.M009379200 [DOI] [PubMed]
- Eyre NS, Beard MR (2014) HCV NS5A inhibitors disrupt replication factory formation: a novel mechanism of antiviral action. Gastroenterology 147(5):959–962. 10.1053/j.gastro.2014.09.024 [DOI] [PubMed]
- Gallay PA, Ptak RG, Bobardt MD, Dumont JM, Vuagniaux G, Rosenwirth B (2013) Correlation of naturally occurring HIV-1 resistance to DEB025 with capsid amino acid polymorphisms. Viruses 5(3):981–997. 10.3390/v5030981 [DOI] [PMC free article] [PubMed]
- Gallay PA, Bobardt MD, Chatterji U, Trepanier DJ, Ure D, Ordonez C, Foster R (2015) The Novel Cyclophilin Inhibitor CPI-431-32 Concurrently Blocks HCV and HIV-1 Infections via a Similar Mechanism of Action. PLoS One 10(8):e0134707. 10.1371/journal.pone.0134707 [DOI] [PMC free article] [PubMed]
- Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP (1996) Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87(7):1285–1294. 10.1016/s0092-8674(00)81823-1 [DOI] [PubMed]
- Han J, Lee HW, Jin Y, Khadka DB, Yang S, Li X, Kim M, Cho WJ (2020) Molecular design, synthesis, and biological evaluation of bisamide derivatives as cyclophilin A inhibitors for HCV treatment. Eur J Med Chem 188:112031. 10.1016/j.ejmech.2019.112031 [DOI] [PubMed]
- Harikishore A, Yoon HS (2015) Immunophilins: Structures, Mechanisms and Ligands. Curr Mol Pharmacol 9(1):37–47. 10.2174/1874467208666150519113427 [DOI] [PubMed]
- He H, Zhou D, Fan W, Fu X, Zhang J, Shen Z, Li J, Li J, Wu Y (2012) Cyclophilin A inhibits rotavirus replication by facilitating host IFN-I production. Biochem Biophys Res Commun 422(4):664–669. 10.1016/j.bbrc.2012.05.050 [DOI] [PubMed]
- He H, Mou Z, Li W, Fei L, Tang Y, Zhang J, Yan P, Chen Z, Yang X, Shen Z, Li J, Wu Y (2013) Proteomic methods reveal cyclophilin A function as a host restriction factor against rotavirus infection. Proteomics 13(7):1121–1132. 10.1002/pmic.201100579 [DOI] [PubMed]
- Hoerauf A, Rascher C, Bang R, Pahl A, Solbach W, Brune K, Röllinghoff M, Bang H (1997) Host-cell cyclophilin is important for the intracellular replication of Leishmania major. Mol Microbiol 24(2):421–429. 10.1046/j.1365-2958.1997.3401716.x [DOI] [PubMed]
- Hou J, Zhang Q, Liu Z, Wang S, Li D, Liu C, Liu Y, Shao Y (2016) Cyclophilin A as a potential genetic adjuvant to improve HIV-1 Gag DNA vaccine immunogenicity by eliciting broad and long-term Gag-specific cellular immunity in mice. Hum Vaccin Immunother 12(2):545–553. 10.1080/21645515.2015.1082692 [DOI] [PMC free article] [PubMed]
- Jiang J, Yin HL, Sun Y, Huang HQ, Hu XC (2018) Clonorchis sinensis cyclophilin A immunization protected mice from CLP-induced sepsis. Int Immunopharmacol 59:347–353. 10.1016/j.intimp.2018.03.039 [DOI] [PubMed]
- Jimenez-Guardeño JM, Apolonia L, Betancor G, Malim MH (2019) Immunoproteasome activation enables human TRIM5α restriction of HIV-1. Nat Microbiol 4(6):933–940. 10.1038/s41564-019-0402-0 [DOI] [PMC free article] [PubMed]
- Jyothi KR, Beloor J, Jo A, Nguyen MN, Choi TG, Kim JH, Akter S, Lee SK, Maeng CH, Baik HH, Kang I, Ha J, Kim SS (2015) Liver-targeted cyclosporine A-encapsulated poly (lactic-co-glycolic) acid nanoparticles inhibit hepatitis C virus replication. Int J Nanomedicine 10:903–921. 10.2147/IJN.S74723 [DOI] [PMC free article] [PubMed]
- Kaiser E, Kroll C, Ernst K, Schwan C, Popoff M, Fischer G, Buchner J, Aktories K, Barth H (2011) Membrane translocation of binary actin-ADP-ribosylating toxins from Clostridium difficile and Clostridium perfringens is facilitated by cyclophilin A and Hsp90. Infect Immun 79(10):3913–3921. 10.1128/iai.05372-11 [DOI] [PMC free article] [PubMed]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441(7089):101–105. 10.1038/nature04734 [DOI] [PubMed]
- Keyes LR, Bego MG, Soland M, St Jeor S (2012) Cyclophilin A is required for efficient human cytomegalovirus DNA replication and reactivation. J Gen Virol 93(Pt 4):722–732. 10.1099/vir.0.037309-0 [DOI] [PMC free article] [PubMed]
- Kim K, Dauphin A, Komurlu S, McCauley SM, Yurkovetskiy L, Carbone C, Diehl WE, Strambio-De-Castillia C, Campbell EM, Luban J (2019) Cyclophilin A protects HIV-1 from restriction by human TRIM5α. Nat Microbiol 4(12):2044–2051. 10.1038/s41564-019-0592-5 [DOI] [PMC free article] [PubMed]
- Kovalev N, Nagy PD (2013) Cyclophilin A Binds to the Viral RNA and Replication Proteins, Resulting in Inhibition of Tombusviral Replicase Assembly. J Virol 87(24):13330–13342. 10.1128/Jvi.02101-13 [DOI] [PMC free article] [PubMed]
- Krucken J, Greif G, von Samson-Himmelstjerna G (2009) In silico analysis of the cyclophilin repertoire of apicomplexan parasites. Parasit Vectors 2(1):27. 10.1186/1756-3305-2-27 [DOI] [PMC free article] [PubMed]
- Lahaye X, Satoh T, Gentili M, Cerboni S, Silvin A, Conrad C, Ahmed-Belkacem A, Rodriguez EC, Guichou JF, Bosquet N, Piel M, Le Grand R, King MC, Pawlotsky JM, Manel N (2016) Nuclear Envelope Protein SUN2 Promotes Cyclophilin-A-Dependent Steps of HIV Replication. Cell Rep 15(4):879–892. 10.1016/j.celrep.2016.03.074 [DOI] [PMC free article] [PubMed]
- Li Y, Kar AK, Sodroski J (2009) Target cell type-dependent modulation of human immunodeficiency virus type 1 capsid disassembly by cyclophilin A. J Virol 83(21):10951–10962. 10.1128/JVI.00682-09 [DOI] [PMC free article] [PubMed]
- Li L, Luo D, Liao Y, Peng K, Zeng Y (2020) Mycoplasma genitalium Protein of Adhesion Induces Inflammatory Cytokines via Cyclophilin A-CD147 Activating the ERK-NF-κB Pathway in Human Urothelial Cells. Front Immunol 11:2052. 10.3389/fimmu.2020.02052 [DOI] [PMC free article] [PubMed]
- Liu Z, Yang F, Robotham JM, Tang H (2009) Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J Virol 83(13):6554–6565. 10.1128/JVI.02550-08 [DOI] [PMC free article] [PubMed]
- Liu X, Zhao Z, Li Z, Xu C, Sun L, Chen J, Liu W (2012a) Cyclosporin A inhibits the influenza virus replication through cyclophilin A-dependent and -independent pathways. PLoS One 7(5):e37277. 10.1371/journal.pone.0037277 [DOI] [PMC free article] [PubMed]
- Liu X, Zhao Z, Xu C, Sun L, Chen J, Zhang L, Liu W (2012b) Cyclophilin A restricts influenza A virus replication through degradation of the M1 protein. PLoS One 7(2):e31063. 10.1371/journal.pone.0031063 [DOI] [PMC free article] [PubMed]
- Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, Cen S, Guo F, Liang C (2013) The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14(4):398–410. 10.1016/j.chom.2013.08.015 [DOI] [PubMed]
- Liu Z, Pan Q, Liang Z, Qiao W, Cen S, Liang C (2015) The highly polymorphic cyclophilin A-binding loop in HIV-1 capsid modulates viral resistance to MxB. Retrovirology 12:1. 10.1186/s12977-014-0129-1 [DOI] [PMC free article] [PubMed]
- Liu W, Li J, Zheng W, Shang Y, Zhao Z, Wang S, Bi Y, Zhang S, Xu C, Duan Z (2017) Cyclophilin A-regulated ubiquitination is critical for RIG-I-mediated antiviral immune responses. Elife 6:e24425 [DOI] [PMC free article] [PubMed]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M Jr (2008) Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 82(1):335–345. 10.1128/JVI.01080-07 [DOI] [PMC free article] [PubMed]
- Luo X, Yang W, Gao G (2018) SUN1 Regulates HIV-1 Nuclear Import in a Manner Dependent on the Interaction between the Viral Capsid and Cellular Cyclophilin A. J Virol 92(13):e00229–e00218. 10.1128/jvi.00229-18 [DOI] [PMC free article] [PubMed]
- Ma C, Li F, Musharrafieh RG, Wang J (2016) Discovery of cyclosporine A and its analogs as broad-spectrum anti-influenza drugs with a high in vitro genetic barrier of drug resistance. Antivir Res 133:62–72. 10.1016/j.antiviral.2016.07.019 [DOI] [PMC free article] [PubMed]
- Madan V, Paul D, Lohmann V, Bartenschlager R (2014) Inhibition of HCV replication by cyclophilin antagonists is linked to replication fitness and occurs by inhibition of membranous web formation. Gastroenterology 146(5):1361–1372.e1361-1369. 10.1053/j.gastro.2014.01.055 [DOI] [PubMed]
- Mahesutihan M, Zheng W, Cui L, Li Y, Jiao P, Yang W, Liu W, Li J, Fan W, Yang L, Liu W (2018) CypA Regulates AIP4-Mediated M1 Ubiquitination of Influenza A Virus. 33(5):440–448. 10.1007/s12250-018-0058-6 [DOI] [PMC free article] [PubMed]
- Mahlangu MP, Müller EE, Venter JME, Maseko DV, Kularatne RS (2019) The Prevalence of Mycoplasma genitalium and Association With Human Immunodeficiency Virus Infection in Symptomatic Patients, Johannesburg, South Africa, 2007-2014. Sex Transm Dis 46(6):395–399. 10.1097/olq.0000000000000984 [DOI] [PMC free article] [PubMed]
- Ma-Lauer Y, Zheng Y, Malešević M, von Brunn B, Fischer G, von Brunn A, Center G (2019) Interaction of cyclophilin A and human coronavirus 229E N protein is essential for virus replication. 10.1016/j.antiviral.2019.104620 [DOI] [PMC free article] [PubMed]
- Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, Littman DR (2010) A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467(7312):214–217. 10.1038/nature09337 [DOI] [PMC free article] [PubMed]
- Meehan AM, Saenz DT, Guevera R, Morrison JH, Peretz M, Fadel HJ, Hamada M, van Deursen J, Poeschla EM (2014) A cyclophilin homology domain-independent role for Nup358 in HIV-1 infection. PLoS Pathog 10(2):e1003969. 10.1371/journal.ppat.1003969 [DOI] [PMC free article] [PubMed]
- Neuman BW, Joseph JS, Saikatendu KS, Serrano P, Chatterjee A, Johnson MA, Liao L, Klaus JP, Yates JR 3rd, Wuthrich K, Stevens RC, Buchmeier MJ, Kuhn P (2008) Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol 82(11):5279–5294. 10.1128/JVI.02631-07 [DOI] [PMC free article] [PubMed]
- Nevers Q, Ruiz I, Ahnou N, Donati F, Brillet R, Softic L, Chazal M, Jouvenet N, Fourati S, Baudesson C, Bruscella P, Gelin M, Guichou JF, Pawlotsky JM, Ahmed-Belkacem A (2018) Characterization of the Anti-Hepatitis C Virus Activity of New Nonpeptidic Small-Molecule Cyclophilin Inhibitors with the Potential for Broad Anti-Flaviviridae Activity. Antimicrob Agents Chemother 62(7):e00126–e00118. 10.1128/aac.00126-18 [DOI] [PMC free article] [PubMed]
- Ngure M, Issur M, Shkriabai N, Liu HW, Cosa G, Kvaratskhelia M, Götte M (2016) Interactions of the Disordered Domain II of Hepatitis C Virus NS5A with Cyclophilin A, NS5B, and Viral RNA Show Extensive Overlap. ACS Infect Dis 2(11):839–851. 10.1021/acsinfecdis.6b00143 [DOI] [PubMed]
- Nigro P, Pompilio G, Capogrossi MC (2013) Cyclophilin A: a key player for human disease. Cell Death Dis 4(10):e888. 10.1038/cddis.2013.410 [DOI] [PMC free article] [PubMed]
- Pellé R, McOdimba F, Chuma F, Wasawo D, Pearson TW, Murphy NB (2002) The African trypanosome cyclophilin A homologue contains unusual conserved central and N-terminal domains and is developmentally regulated. Gene 290(1-2):181–191. 10.1016/s0378-1119(02)00559-0 [DOI] [PubMed]
- Pfefferle S, Schöpf J, Kögl M, Friedel CC, Müller MA, Carbajo-Lozoya J, Stellberger T, von Dall'Armi E, Herzog P, Kallies S, Niemeyer D, Ditt V, Kuri T, Züst R, Pumpor K, Hilgenfeld R, Schwarz F, Zimmer R, Steffen I, Weber F, Thiel V, Herrler G, Thiel HJ, Schwegmann-Wessels C, Pöhlmann S, Haas J, Drosten C, von Brunn A (2011) The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog 7(10):e1002331. 10.1371/journal.ppat.1002331 [DOI] [PMC free article] [PubMed]
- Phillips S, Chokshi S, Chatterji U, Riva A, Bobardt M, Williams R, Gallay P, Naoumov NV (2015) Alisporivir inhibition of hepatocyte cyclophilins reduces HBV replication and hepatitis B surface antigen production. Gastroenterology 148(2):403–414.e407. 10.1053/j.gastro.2014.10.004 [DOI] [PMC free article] [PubMed]
- Potenza M, Galat A, Minning TA, Ruiz AM, Duran R, Tarleton RL, Marin M, Fichera LE, Bua J (2006) Analysis of the Trypanosoma cruzi cyclophilin gene family and identification of Cyclosporin A binding proteins. Parasitology 132(Pt 6):867–882. 10.1017/S0031182005009558 [DOI] [PubMed]
- Qian Z, Yue W, Li B (2010) Progress of CypA and lung cancer-related research. Zhongguo Fei Ai Za Zhi 13(8):827–831. 10.3779/j.issn.1009-3419.2010.08.15 [DOI] [PMC free article] [PubMed]
- Qing J, Wang Y, Sun Y, Huang J, Yan W, Wang J, Su D, Ni C, Li J, Rao Z, Liu L, Lou Z (2014) Cyclophilin A associates with enterovirus-71 virus capsid and plays an essential role in viral infection as an uncoating regulator. PLoS Pathog 10(10):e1004422. 10.1371/journal.ppat.1004422 [DOI] [PMC free article] [PubMed]
- Reddy SP, Rasmussen WG, Baseman JB (1996) Correlations between Mycoplasma pneumoniae sensitivity to cyclosporin A and cyclophilin-mediated regulation of mycoplasma cytadherence. Microb Pathog 20(3):155–169. 10.1006/mpat.1996.0014 [DOI] [PubMed]
- Reghellin V, Donnici L, Fenu S, Berno V, Calabrese V, Pagani M, Abrignani S, Peri F, De Francesco R, Neddermann P (2014) NS5A inhibitors impair NS5A-phosphatidylinositol 4-kinase IIIalpha complex formation and cause a decrease of phosphatidylinositol 4-phosphate and cholesterol levels in hepatitis C virus-associated membranes. Antimicrob Agents Chemother 58(12):7128–7140. 10.1128/aac.03293-14 [DOI] [PMC free article] [PubMed]
- Saleh T, Jankowski W, Sriram G, Rossi P, Shah S, Lee KB, Cruz LA, Rodriguez AJ, Birge RB, Kalodimos CG (2016) Cyclophilin A promotes cell migration via the Abl-Crk signaling pathway. Nat Chem Biol 12(2):117–123. 10.1038/nchembio.1981 [DOI] [PMC free article] [PubMed]
- Selyutina A, Persaud M, Simons LM, Bulnes-Ramos A, Buffone C, Martinez-Lopez A, Scoca V, Di Nunzio F, Hiatt J, Marson A, Krogan NJ, Hultquist JF, Diaz-Griffero F (2020) Cyclophilin A Prevents HIV-1 Restriction in Lymphocytes by Blocking Human TRIM5α Binding to the Viral Core. Cell Rep 30(11):3766–3777.e3766. 10.1016/j.celrep.2020.02.100 [DOI] [PMC free article] [PubMed]
- Setiawan LC, van Dort KA, Rits MA, Kootstra NA (2016) Mutations in CypA Binding Region of HIV-1 Capsid Affect Capsid Stability and Viral Replication in Primary Macrophages. AIDS Res Hum Retrovir 32(4):390–398. 10.1089/AID.2014.0361 [DOI] [PubMed]
- Shanmugam S, Nichols AK, Saravanabalaji D, Welsch C, Yi M (2018) HCV NS5A dimer interface residues regulate HCV replication by controlling its self-interaction, hyperphosphorylation, subcellular localization and interaction with cyclophilin A. PLoS Pathog 14(7):e1007177. 10.1371/journal.ppat.1007177 [DOI] [PMC free article] [PubMed]
- Shi P-Y. Molecular virology and control of flaviviruses. Norfolk: Caister Academic Press; 2012. [Google Scholar]
- Softic L, Brillet R, Berry F, Ahnou N, Nevers Q, Morin-Dewaele M, Hamadat S, Bruscella P, Fourati S, Pawlotsky JM, Ahmed-Belkacem A (2020) Inhibition of SARS-CoV-2 Infection by the Cyclophilin Inhibitor Alisporivir (Debio 025). Antimicrob Agents Chemother 64(7). 10.1128/AAC.00876-20 [DOI] [PMC free article] [PubMed]
- Song T, Yang M, Chen J, Huang L, Yin H, He T, Huang H, Hu X (2015) Prognosis of sepsis induced by cecal ligation and puncture in mice improved by anti-Clonorchis Sinensis cyclopholin a antibodies. Parasit Vectors 8:502. 10.1186/s13071-015-1111-z [DOI] [PMC free article] [PubMed]
- Suzuki T, Aizaki H, Murakami K, Shoji I, Wakita T (2007) Molecular biology of hepatitis C virus. J Gastroenterol 42(6):411–423. 10.1007/s00535-007-2030-3 [DOI] [PubMed]
- Tian X, Zhao C, Zhu H, She W, Zhang J, Liu J, Li L, Zheng S, Wen YM, Xie Y (2010) Hepatitis B virus (HBV) surface antigen interacts with and promotes cyclophilin a secretion: possible link to pathogenesis of HBV infection. J Virol 84(7):3373–3381. 10.1128/JVI.02555-09 [DOI] [PMC free article] [PubMed]
- Valiente-Echeverria F, Melnychuk L, Vyboh K, Ajamian L, Gallouzi IE, Bernard N, Mouland AJ (2014) eEF2 and Ras-GAP SH3 domain-binding protein (G3BP1) modulate stress granule assembly during HIV-1 infection. Nat Commun 5:4819. 10.1038/ncomms5819 [DOI] [PMC free article] [PubMed]
- Vidotto A, Morais AT, Ribeiro MR, Pacca CC, Terzian AC, Gil LH, Mohana-Borges R, Gallay P, Nogueira ML (2017) Systems Biology Reveals NS4B-Cyclophilin A Interaction: A New Target to Inhibit YFV Replication. J Proteome Res 16(4):1542–1555. 10.1021/acs.jproteome.6b00933 [DOI] [PubMed]
- Yi DR, An N, Liu ZL, Xu FW, Raniga K, Li QJ, Zhou R, Wang J, Zhang YX, Zhou JM, Zhang LL, An J, Qin CF, Guo F, Li XY, Liang C, Cen S (2019) Human MxB Inhibits the Replication of Hepatitis C Virus. J Virol 93(1). 10.1128/JVI.01285-18 [DOI] [PMC free article] [PubMed]
- Zeuzem S, Flisiak R, Vierling JM, Mazur W, Mazzella G, Thongsawat S, Abdurakhmanov D, Van Kinh N, Calistru P, Heo J, Stanciu C, Gould M, Makara M, Hsu SJ, Buggisch P, Samuel D, Mutimer D, Nault B, Merz M, Bao W, Griffel LH, Brass C, Naoumov NV, Grp EIS (2015) Randomised clinical trial: alisporivir combined with peginterferon and ribavirin in treatment-naive patients with chronic HCV genotype 1 infection (ESSENTIAL II). Aliment Pharmacol Ther 42(7):829–844. 10.1111/apt.13342 [DOI] [PubMed]
- Zhao C, Fang CY, Tian XC, Wang L, Yang PY, Wen YM (2007) Proteomic analysis of hepatitis B surface antigen positive transgenic mouse liver and decrease of cyclophilin A. J Med Virol 79(10):1478–1484. 10.1002/jmv.20945 [DOI] [PubMed]