Abstract
Gastric cancer is the third most common cause of death from cancer in the world and infection with Helicobacter pylori (H. pylori) is the main cause of gastric cancer. In addition to Helicobacter infection, the overall stomach microbiota has recently emerged as a potential factor in gastric cancer progression. Previously we had established that mice deficient in myeloid differentiation primary response gene 88 (MyD88, Myd88−/−) rapidly progressed to neoplasia when infected with H. felis. Thus, in order to assess the role of the microbiota in this fast-progressing gastric cancer model we investigated changes of the gastric microbiome in mice with different genotypic backgrounds: wild type (WT), MyD88-deficient (Myd88−/−), mice deficient in the Toll/interleukin-1 receptor (TIR) domain-containing adaptor-inducing interferon-β (TRIF, TrifLps2), and MyD88- and TRIF-deficient (Myd88−/−/TrifLps2, double knockout (DKO)) mice. We compared changes in alpha diversity, beta diversity, relative abundance, and log-fold differential of relative abundance ratios in uninfected and Helicobacter infected mice and studied their correlations with disease progression to gastric cancer in situ. We observed an overall reduction in microbial diversity post-infection with H. felis across all genotypes. Campylobacterales were observed in all infected mice, with marked reduction in abundance at 3 and 6 months in Myd88−/− mice. A sharp increase in Lactobacillales in infected Myd88−/− and DKO mice at 3 and 6 months was observed as compared to TrifLps2 and WT mice, hinting at a possible role of these bacteria in gastric cancer progression. This was further reinforced upon comparison of Lactobacillales log-fold differentials with histological data, indicating that Lactobacillales are closely associated with Helicobacter infection and gastric cancer progression. Our study suggests that differences in genotypes could influence the stomach microbiome and make it more susceptible to the development of gastric cancer upon Helicobacter infection. Additionally, increase in Lactobacillales could contribute to faster development of gastric cancer and might serve as a potential biomarker for the fast progressing form of gastric cancer.
Keywords: Helicobacter, microbiome, gastric cancer, MyD88, Lactobacillales, TRIF
1. Introduction
Gastric cancer is the sixth most common cancer and third most common cause of cancer mortality in the world [1]. By far the largest risk factor for gastric cancer development is the presence of the carcinogenic microbe Helicobacter pylori [2]. Infection with H. pylori leads to development of premalignant lesions that progress from gastric atrophy to metaplasia, dysplasia, and finally to gastric adenocarcinoma. While H. pylori infects almost 50% of the global population, only 1–3% of the infected individuals develop gastric cancer [3]. Several other factors contribute to gastric cancer progression, such as Helicobacter strains, environmental factors, external factors like alcohol consumption [4] and host immune response. In addition, the microbiota of the stomach may also influence final disease outcome [3]. Recently, studies have revealed that thrombosis also can be one of the reasons for fatality in gastric cancer patients [5].
The stomach had been traditionally considered a sterile organ due to its highly acidic environment and digestive juices. It was only after the discovery of H. pylori in 1982, and its ability to survive in such harsh conditions, that this led to the idea of a gastric microbiome, similar to microbial communities inhabiting other parts of the human host. Advancement in DNA sequencing strategies and computational methods have uncovered a complex microbiome of the stomach [3]. The gastric microbial density is estimated to be 102–104 colony forming units (CFU)/mL, a much lower density than other body sites, such as the colon, which often reaches 1010–1012 CFU/mL [6]. Early characterization of the gastric microbiota relied on culturing techniques but the advancement of sequencing techniques has led to the identification of different species falling into five predominant phyla—Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes and Fusobacteria, indicating a distinct microbiome of the stomach [6,7]. Thus, with the presence of such complex microbial diversity in the stomach, interactions between Helicobacter and other gut bacteria could potentially play a role in deciding the fate of gastric cancer progression, or may act as an aide along with other factors in driving the disease.
H. felis is a close relative of human gastric pathogen H. pylori and studies have shown that H. felis infection leads to the development of high-grade dysplastic lesions in C57BL/6 mice as compared to H. pylori, mimicking gastric carcinogenesis as seen in human [8,9,10,11]. Moreover, we have previously shown that Myeloid differentiation primary response gene 88 (MyD88) regulates Helicobacter induced gastric cancer progression, where MyD88 deficient mice (Myd88−/−) infected with H. felis showed fast progression to gastric cancer in situ as compared to wild-type (WT) mice proving it to be a better Helicobacter model in rodents [8]. Furthermore, we also observed that cells from mice deficient in MyD88 showed increased secretion of interferon (IFN)-α after H. pylori infection suggesting MyD88-independent induction of type I IFNs, suggesting role of Toll/IL-1R (TIR) domain-containing adaptor-inducing interferon-β (TRIF)-dependent pathway [12]. This prompted us to study the microbial diversity of the stomach in mice with different genotypic backgrounds: Wild type (WT), MyD88 deficient (Myd88−/−), TRIF (TrifLps2), and MyD88 and TRIF deficient (Myd88−/−/TrifLps2, double knockout (DKO)) mice.
Given the potential connection between the gastric microbiome and cancer progression, we hypothesized that the fast-progressing Myd88−/− cancer model would show distinct gastric microbiome changes compared to the other genotypes. Gastric microbiome diversity and composition was, therefore, studied both in uninfected and H. felis infected mice of all four genotypes. In addition, we also investigated whether the gastric microbiome changed over time in response to infection and studied the correlation of these changes with gastric cancer progression. Therefore, this study would aid in identifying microbial species whose abundance or scarcity may contribute to progression of Helicobacter-induced lesions and towards adenocarcinoma. Ultimately, identification of other contributing factors would open new avenues of research in gastric cancer disease progression.
2. Material and Methods
2.1. Animals
Six- to 10-week-old wild-type (WT) (n = 42), MyD88 deficient (Myd88−/−) (n = 47), TRIF deficient (TrifLps2) (n = 46), and double knockout (Myd88−/−/TrifLps2, DKO) (n = 37) mice in the C57BL/6 background were used in this study. WT mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Myd88−/−, TrifLps2, and DKO mice were from our breeding colony originally provided by Dr. Akira (Osaka University, Japan) and backcrossed 10 times onto a C57BL/6 background, bred, and maintained at University of California, San Diego (UCSD). All mice were housed together before infection with H. felis and for the duration of the study for each genotype. The institutional Animal Care and Use Committee at the University of California, San Diego, approved all animal procedures and they were performed using accepted veterinary standards.
2.2. Bacterial Growth Conditions
Helicobacter felis, strain CS1 (ATCC 49179) was purchased from the American Type Culture Collection (Manassas, VA, USA). H. felis was routinely maintained on solid medium, Columbia agar (Becton Dickinson, MD, USA) supplemented with 5% laked blood under microaerophilic conditions (5% O2, 10% CO2, 85% N2) at 37 °C and passaged every 2–3 days as described previously [8,12,13]. Prior to mouse infections, H. felis was cultured in liquid medium, brain heart infusion broth (BHI, Becton Dickinson, MD, USA) supplemented with 10% fetal calf serum and incubated at 37 °C under microaerophilic conditions for 48 h. Spiral bacteria were enumerated using a Petroff-Hausser chamber before infections.
2.3. Mouse Infections
This study used a well-characterized cancer mouse model, which involves infecting C57BL/6 mice with H. felis (strain CS1), a close relative of the human gastric pathogen H. pylori. Mice were inoculated with 109 organisms in 300 μL of BHI by oral gavage three times at 2-day intervals as previously described [8,12]. Control mice received BHI only. At 1 month, 3 months, and 6 months post-infection, mice were euthanized, and the stomachs removed under aseptic conditions. The stomach was cut longitudinally and tissue sections were processed for DNA extraction and histopathology.
2.4. Histology
Longitudinal sections of stomach tissue from each mouse were fixed in neutral buffered 10% formalin and embedded in paraffin, and 5 µm sections were stained with hematoxylin and eosin (H&E). Gastric histopathology mucous metaplasia was scored by a blinded comparative pathologist (Rickman) using criteria developed by Rogers et al. [14]. Scores ranging from 0 (no lesions) to 4 (severe lesions) were measured in increments of 0.5, as previously described (9).
2.5. DNA Extraction
DNA was extracted from gastric tissue obtained from H. felis-infected and uninfected WT, Myd88−/−, TrifLps2, and DKO mice. Stomach tissue sections were analyzed at different time points of 1 month, 3 months, and 6 months. DNA was extracted using DNAeasy Blood and Tissue kit (Qiagen, MD, USA) following manufacturer’s instructions. The DNA concentration was quantified using NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) before proceeding to DNA Sequencing.
2.6. 16. S rRNA Gene Sequencing
Purified DNA was amplified and processed according to Earth Microbiome Project (EMP) standard protocols (https://www.earthmicrobiome.org/protocols-and-standards/16s/) Amplicon polymerase chain reaction (PCR) was performed on the V4 region of the 16S rRNA gene using the primer pair 515F/806R with Golay error-correcting barcodes on the reverse primer. 240 nanograms of each amplicon was pooled and purified with the MoBio UltraClean PCR cleanup kit (Qiagen, MD, USA) and sequenced on the Illumina MiSeq sequencing platform. Demultiplexed FASTQ files were processed using Quantitative Insights into Microbial Ecology (QIIME2) version 2019.10 (https://qiime2.org) [15]. Sequences were denoised using Deblur (default settings, --p-trim-length 150) [16]. Taxonomy was assigned using SATé-enabled phylogenetic placement (SEPP) fragment insertion with a classifier trained on the Greengenes13_8 99% operational taxonomic units (OTUs) dataset, with sequences trimmed to contain 150 bases from the region amplified in sequencing [17]. All reads assigned to the phylum Cyanobacteria were filtered from the counts table before analysis, as this is considered a food contaminant in gut microbiome sequencing. A pseudocount of 1 was added to the final counts table in Songbird analysis to prevent samples dropping out of the log fold-differential analysis due to inability to calculate the logarithm of 0. FASTQ files can be found through the NCBI BioSample database (accession number PRJNA685500). Code used to process sequencing and conduct bioinformatics processing can be found on Github (https://github.com/jkccoker/Murine_gastric_microbiome).
2.7. Bioinformatics Processing
Taxonomy abundance plots were generated using the PhyloSeq package [18]. Taxonomy was collapsed to the order of interest in PhyloSeq. Alpha diversity plots were generated in R using data exported from the QIIME2 analysis (--p-sampling-depth 2300). Beta diversity principal component analysis plots were generated in QIIME2 with the DEICODE plug-in [19]. Taxonomy abundance log-fold differentials were calculated through QIIME2 using the Songbird plug-in (--p-formula “Genotype + Infection + Time”) [20], with visualization through Qurro (intercept = “Infection: Yes”) [21]. Receiver operating characteristic (ROC) curves were generated in Prism 7 (GraphPad Software, San Diego, CA, USA) using log-fold differential values from the Songbird analysis.
2.8. Statistics
Differences in alpha diversity and log-fold differentials were calculated in R using Student’s t-test, analysis of variance (ANOVA) or the Kruskal-Wallis H test, where appropriate. Data were checked for normal distribution before the Student’s t-test and ANOVA. Differences in beta diversity were assessed with permutational multivariate analysis of variance (PERMANOVA) in QIIME2 with Benjamini-Hochberg false discovery rate (FDR) correction. Ordinal logistic regression was conducted in R using the polr command (formula = histology_score ~ <log-fold differential>, Hess = T).
3. Results
3.1. Gastric Mucosal Microbial Diversity Varies with Infection Status and Genotype
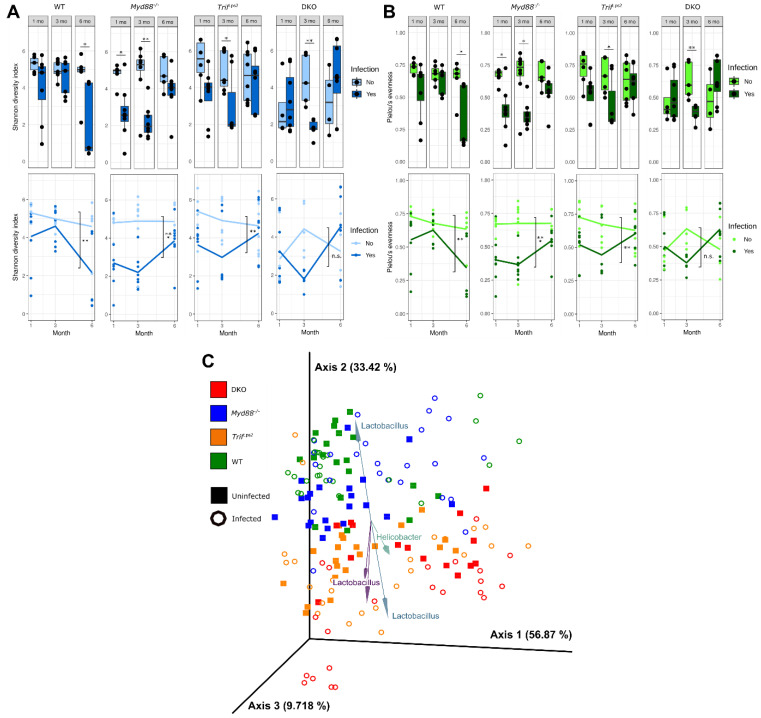
To assess the diversity of the gastric microbial community in each mouse genotype, we first determined alpha and beta diversity. The Shannon diversity index and Pielou’s evenness score were calculated as metrics of alpha diversity. The Shannon diversity index provides a metric of community diversity based on the number of taxa present and the abundance of each species. Differences in genotype did not significantly affect Shannon diversity index (Figure 1A). In general, infection with H. felis resulted in a decreased diversity index (statistically significant for WT at 6 months (p < 0.05); Myd88−/− at 1 month (p < 0.05) and 3 months (p < 0.01); TrifLps2 at 3 months (p < 0.05); and DKO at 3 months (p < 0.01). For the three knockout genotypes, diversity was decreased in infected samples at 1 and 3 months but increased back to uninfected levels at 6 months. The opposite was observed in the WT genotype. Using Pielou’s evenness, which assesses how evenly distributed taxa are within a community, the evenness scores followed the same trends as the Shannon diversity index, as expected (Figure 1B).
Figure 1.
Changes in microbial diversity in the stomach across four different genotypes, with infection and time. Shannon diversity index (A) and Pielou’s evenness (B) values for each gastric community, divided by genotype and month. Top and bottom plots represent the same data, alpha diversity values, with lines on the bottom plots denoting average values for infected and uninfected communities over time. Statistical significance on bottom plots refers to differences between infected and uninfected, months 1–6 combined. (C) Principal component analysis of robust Aitchison distance values between communities, months 1–6 combined. Biplot arrows indicate operational taxonomic units (OTUs) driving separation between samples, with arrows labeled with the genus of the OTU. Arrows and genus labels are matched by color. All diversity metrics were calculated using QIIME2. Statistical significance determined by Student’s t-test for alpha diversity and permutational multivariate analysis of variance (PERMANOVA) with Benjamini-Hochberg FDR correction for beta diversity (* p < 0.05, ** p < 0.01, *** p < 0.005).
To assess the dissimilarity between the gastric microbial communities, we next analyzed the robust Aitchison distance between communities as a metric of beta diversity (Figure 1C). Aitchison distance is a compositional metric of the Euclidean distance between samples after centered log-ratio transformation [22]. Robust Aitchison distance analysis incorporates matrix completion to account for the large number of zeros in microbiome data sets due to the absence of individual taxa in samples [19]. PERMANOVA analysis of the robust Aitchison distance between samples showed significant separation between communities by all genotype pairs (p < 0.001) except WT and Myd88−/− (p = 0.20). Pairwise comparison (with Benjamini–Hochberg FDR correction) showed that infection groups within each genotype were significantly different from other groups (p < 0.05), with the exception of the following interesting pairs: WT uninfected/infected (p = 0.07); Myd88−/− uninfected/infected (p = 0.07); WT infected/Myd88−/− infected (p = 0.15); TrifLps2 infected/DKO uninfected (p = 0.42); and TrifLps2 infected/DKO infected (p = 0.05). Visualization of principal coordinate analysis (PCA) displayed that WT/Myd88−/− genotypes and Trif Lps2/DKO genotypes had more similar gastric communities, despite statistically significant differences (Figure 1C). A biplot of the robust Aitchison PCA allowed us to visualize the OTUs most heavily influencing community dissimilarity as vectors in the PCA axes (Figure 1C, arrows). These vectors revealed that the operational taxonomic units most heavily influencing the distance between samples were from the Helicobacter and Lactobacillus genera.
3.2. Variation in Abundance of Microbial Taxa in Different Genotypes over Time and after H. Felis Infections
Given the differences in community diversity between genotypes and infection status, we conducted taxonomic analysis of the 16S rRNA gene sequences in each sample. The predominant phyla across all genotypes, irrespective of infection status and time included, Bacteroidetes, Firmicutes and Proteobacteria (Figure 2). Bacteroidetes and Firmicutes were present both in uninfected and infected samples with insignificant variations between genotypes. Proteobacteria were observed in high abundance in infected samples as compared to uninfected samples in all genotypes as expected (Figure S1). However, the levels of Proteobacteria dropped significantly at 3 months and 6 months in infected Myd88−/− mice and at 6 months for DKO mice in infected samples. In contrast, in TrifLps2 and WT no such drop was observed. Moreover, the levels of Proteobacteria in TrifLps2 were similar at 1 month and 6 months, while peaking at 3 months (Figure S1).
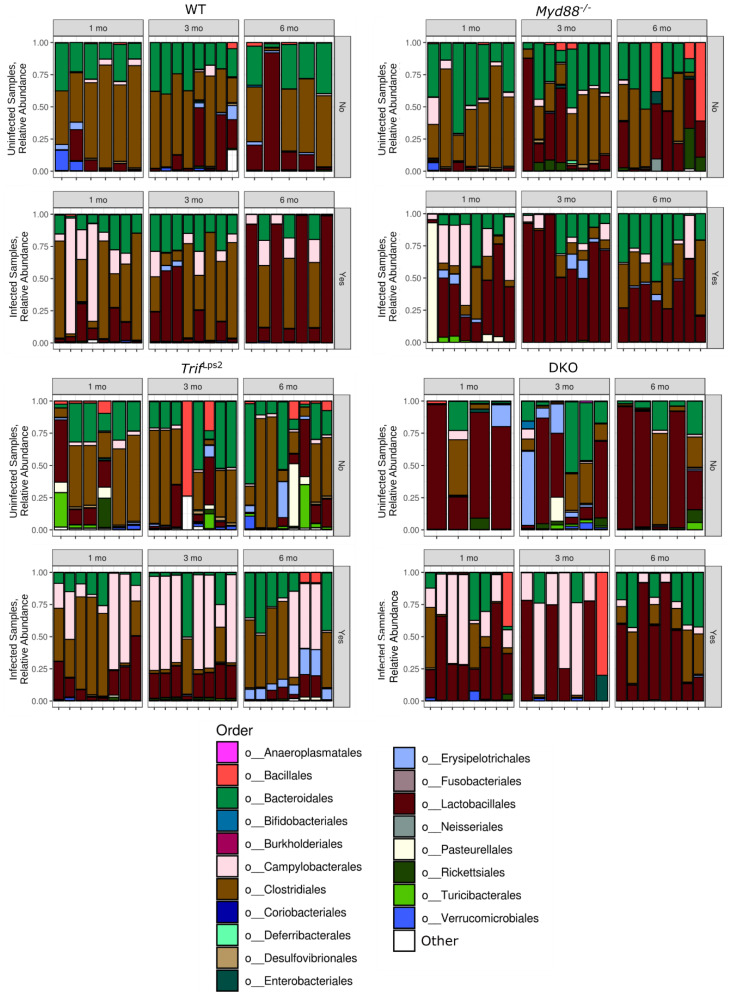
Figure 2.
Relative abundance of different phyla across four genotypes. Relative abundance of individual phyla from WT, Myd88−/−, TrifLps2, and double knockout (DKO) genotypes. The top eight phyla are shown in the legend. Samples are grouped into Helicobacter-infected and non-Helicobacter-infected and divided by time point. Sequencing data were processed in QIIME2, then plotted in PhyloSeq.
We observed predominance of five orders across all genotypes—Bacteroidales, Campylobacterales, Clostridiales, Lactobacillales, Erysipelotrichales (Figure 3). The order Bacteroidales were observed across all genotypes with no overall significant difference. Clostridiales were present in all genotypes but less abundant in DKO mice. However, it was observed that levels of Clostridiales dropped significantly in Myd88−/− post infection as compared to uninfected samples especially at 3 months. Campylobacterales were observed more predominantly in infected samples in all four genotypes, as expected since Helicobacter is a member of the Campylobacterales order. However, the levels declined significantly at 3 and 6 months in Myd88−/− mice and at 6 months in DKO mice although Campylobacterales can be seen throughout all time points in TrifLps2 infected mice. Lactobacillales were seen at highest relative abundance in Myd88−/− infected mice especially at 3 months followed by DKO infected mice, WT infected mice, and to a lesser extent TrifLps2 mice (Figure 3). Erysipelotrichales were predominantly seen at 6 months in TrifLps2 infected mice as compared to other genotypes. Apart from these orders, Rickettsiales were observed mainly in uninfected Myd88−/− and DKO mice as compared to TrifLps2 and WT mice and were almost lost upon infection with H. felis. In addition, the order Bacillales were observed in uninfected samples of TrifLps2, Myd88−/− and WT but not in DKO uninfected mice. However, they were observed in DKO post infection in one sample both at 1 month and 3 months and in two samples in TrifLps2 at 6 months post infection with H. felis.
Figure 3.
Relative abundance of different orders across four genotypes. Relative abundance of the top 15 orders from WT, Myd88−/−, TrifLps2, and DKO genotypes. Remaining phyla are grouped into “Other”. Samples are grouped into Helicobacter-infected and non-Helicobacter-infected and divided by time point. Sequencing data were processed in QIIME2, then plotted in PhyloSeq.
3.3. Association of Lactobacillales with Infection and Mouse Genotype
Given the increase in Lactobacillales seen in infected mice by 16S rRNA amplicon sequencing (Figure 3) and the identification of Lactobacillus as a genus driving community dissimilarity (Figure 1C), we further investigated the relationship between changes in Lactobacillales and disease progression. We used the Songbird [20] and Qurro [21] tools to analyze changes in Lactobacillales between conditions. Songbird calculates log ratios of relative abundance ratios between two taxonomic units for each sample, a compositional analysis that accounts for absolute microbial abundance differences between samples [20]. Qurro can then be used to visualize and compare these differentials between samples [21]. This analysis revealed a highly significant increase in the log ratio of Lactobacillales/Rickettsiales (L/R) in infected communities compared to uninfected communities (Figure 4A). Analyzing this change within individual genotypes showed the log (L/R) ratio was significantly increased in infected Myd88−/− and DKO mice compared to uninfected (Figure 4B). The ratio was not significantly different between infected and uninfected communities in TrifLps2 and WT mice. To confirm this finding, we repeated the analysis comparing Lactobacillales to other organisms present in all communities. Similar results were observed for log (Lactobacillales/Bacteroidales) (Figure S2A,B). No significant differences were seen in log ratios of the Clostridiales/Rickettsiales ratio (Figure 4C,D), and Clostridiales/Bacteroidales (Figure S2C,D) indicating the specificity of this finding to Lactobacillales. We therefore concluded that the order Lactobacillales is increased in infected communities in Myd88−/− and DKO, but not in WT and TrifLps2 genotypes.
Figure 4.
Log ratios between relevant orders across four genotypes. Log ratios of the Lactobacillales/Rickettsiales (A,B) and Clostridiales/Rickettsiales (C,D) relative abundance ratios between samples, months 1–6 combined. (A,C) show ratios by infection status, all genotypes combined. (B,D) show ratios by genotype and infection status. Log ratios were calculated and processed using Songbird and Qurro. Statistical significance was determined by analysis of variance (ANOVA) ** p < 0.01, *** p < 0.005).
3.4. Lactobacillales Are Associated with Gastric Cancer Progression
We next examined if there was an association between Lactobacillales levels and gastric disease progression in our fast-progressing model. Histological analysis revealed that Myd88−/− genotype displays the worst gastric disease following infection, followed by the DKO genotype. H&E stained stomach sections (Figure S3) from each mouse of each genotype (WT and Myd88−/−, TrifLps2and DKO) were evaluated on basis of pathology and scored on scale of 0–4 (Figure 5). Since these genotypes also possessed the highest levels of Lactobacillales, we hypothesized higher Lactobacillales levels would be associated with higher mucous histology scores. Analysis of the log(L/R) ratio and the mucous histology score of each sample with ordinal logistic regression demonstrated that mice with a higher gastric Lactobacillales level had a significantly higher likelihood of a higher histology metaplasia score (Figure 6A). For the regression analysis, histology scores were grouped into categories of 0, 1–2, and 3–4 to increase statistical power. No association between Clostridiales levels and histology scores was observed (Figure 6B).
Figure 5.
Histopathological scoring for mucous metaplasia. Following infection with H. felis for 1 month (A), 3 months (B) and 6 months (C), H&E-stained stomach sections from each mouse (WT and Myd88−/−, TrifLps2and DKO) were evaluated for indications of pathology. Mucous metaplasia was scored by a blinded comparative pathologist according to the criteria described in Materials and Methods. A p value of 0.05 was considered statistically significant. (A) 1month post infection, n = 14 for WT, n = 15 for Myd88−/−, n = 14 for TrifLps2, and n = 12 for DKO mice; (B) 3 months post infection, n = 16 for WT, n = 16 for Myd88−/−, n = 16 for TrifLps2, n = 13 for DKO mice; (C) 6 months post infection, n = 12 for WT, n = 16 for Myd88−/−, n = 16 for TrifLps2, n = 12 for DKO mice. Statistical significance was determined by Mann-Whitney test, *** p < 0.005).
Figure 6.
Predictive relationship between Lactobacillales and mucous metaplasia from Helicobacter infection. (A,B) Ordinal logistic regression analysis of log ratios of Lactobacillales/Rickettsiales (A), and Clostridiales/Rickettsiales (B) Relative abundance and gastric mucous histology score. The black circle marks the average of each category. Ordinal logistic regression was calculated using the polr command in R (*** p < 0.001). (C–E) ROC curve for log ratios of Campylobacterales, Lactobacillales, and Clostridiales to Rickettsiales. The blue line represents the performance of each ratio log fold-differential in predicting Helicobacter infection. The red line represents the result expected for a metric with a 50% chance of predicting infection. The area under the curve (AUC) value refers to the area under the blue line. Receiver operating characteristic (ROC) plots were constructed in Prism7 using log fold-differentials from Songbird and Qurro.
Given the association of Lactobacillales levels and mucous metaplasia, we examined the ability of Lactobacillales log ratios to predict if a mouse had been infected with H. felis. We constructed receiver operator characteristic (ROC) curves for log ratios of the orders Campylobacterales, Lactobacillales, and Clostridiales compared to Rickettsiales. Log (Lactobacillales/Rickettsiales) ratios predicted mouse infection status at a rate similar to log (Campylobacterales/Rickettsiales), the order containing Helicobacter and the logical “gold standard” for infection prediction (Figure 6C,D). In comparison, log (Clostridiales/Rickettsiales) ratios did not predict infection status at a rate better than random (Figure 6E). Together, these data indicate Lactobacillales correlates strongly with Helicobacter infection status and mucous histology score.
4. Discussion
Advancement in research on the gut microbiome has uncovered the existence of a diverse microbiome in the human gut in a delicate balance. These microbiota are vital for maintenance of human health and play an important role in energy metabolism, nutrient absorption and defense against pathogens [3,23,24,25]. However, if this balance is altered dysbiosis can lead to susceptibility to gastrointestinal pathogenesis and cancer [3]. The acidic environment of the stomach supports a smaller number of bacteria as compared to other parts of the gut, but dysbiosis due to various factors, such as genetic, environmental, or pathogen invasion can lead to gastric cancer [3]. The association of Helicobacter with gastric cancer has been well established, as it is characterized as a Type I carcinogen by the World Health Organization (WHO) [2]. Previous studies in insulin-gastrin (INS-GAS) mice have shown that H. pylori infections lead to an overall decrease in microbial density [26]. Our findings indicate there is also an overall decrease in microbiome diversity upon H. felis infections across all genotypes.
In this study we sampled the mouse stomach mucosal tissue instead of fecal samples to understand the changes specifically in the gastric microbiome in different genotypes, with respect to time as well as infection status. Analyzing stomach mucosal tissue provides a more accurate image of gastric microbial communities during gastric cancer progression than fecal samples. Moreover, as previously described Myd88−/− mice serve as a fast-progressing gastric cancer model, where gastric adenocarcinoma is reached within 6 months of infection with Helicobacter [8]. Therefore, analyzing gastric mucosa for the periods of 1 month, 3 months, and 6 months provides a clear picture of microbial diversity and composition fluctuations as disease progresses to gastric cancer.
In our study, Campylobacterales were observed in infected mice across all genotypes, which is expected as Helicobacter belongs to the order Campylobacterales. However, in Myd88−/−, our fast-progressing gastric cancer model, we observed a reduction in Campylobacterales abundance at 3 months and 6 months. This could be attributed to the fact that the advancement of gastric cancer lesions results in an increase in mucosal atrophy and decrease in acid secretion, potentially hindering Helicobacter colonization and facilitating an increase in abundance of other bacteria. In agreement with our study, patients with advanced atrophic gastritis have been found to have hypo-chlorohydric stomach mucosa and low abundance of H. pylori, with the gastric microbiome dominated by non-Helicobacter species [27,28]. A separate study by Basir et al. [29] revealed that increase in H. pylori colonization showed high correlation with severe chronic gastritis in human subjects. Similar correlations were observed by other groups [30,31] in their respective studies in gastric cancer patients. However, a study carried out on 273 human gastric biopsies revealed no relationship between H pylori density and chronic gastritis [32]. Thus, conflicting results have been observed when correlating Helicobacter density to severity of disease.
However, the low abundance of H. pylori appears to facilitate the dominance of other organisms of the microbiome. We observed an increase in Lactobacillales in infected Myd88−/− mice and DKO mice at 3 and 6 months. Previous studies on gastric cancer patients also showed increased abundance of Lactobacillales, supporting their possible role in gastric cancer progression [27,33] and in our case in a fast-progressing form of gastric cancer. Even though Lactobacillus species are utilized in probiotics and are commonly thought to be beneficial for the host, high levels of lactic acid can be detrimental in case of gastric cancer. Lactate can serve as a source of energy for tumor cells, which can lead to increased ATP production and promotion of inflammation [34,35,36,37,38]. Previous studies in INS-GAS mice have shown that mice harboring a complex microbiome develop gastric cancer in 7 months post infection with H. pylori as compared to H. pylori infections in germ free mice where development of gastric cancer is prolonged. In addition, supplementation of INS-GAS germ-free mice with a simplified microbiome of Lactobacillus, Clostridium and Bacteroides species was sufficient to promote development of gastric cancer [26]. This suggests the role of certain species in the gut microbiota in promoting gastric cancer progression.
Comparison of the gastric microbiome from Myd88−/− mice to WT, TrifLps2 and DKO mice, we were able to intensively analyze how changes in Lactobacillales could be connected to gastric cancer development and progression. Myd88−/− and DKO mice had significantly higher levels of Lactobacillales upon infection with H. felis, while WT and TrifLp2 mice did not (Figure 4). Myd88−/− and DKO mice also had significantly worse disease development than WT and TrifLp2, as demonstrated by mucous metaplasia scores (Figure 5). These findings indicate a correlation between Lactobacillales levels and gastric cancer development that holds across genotypes. Ordinal logistic regression analysis and ROC curves (Figure 6) further demonstrate that the log-fold differential of Lactobacillales/Rickettsiales relative abundance ratio allows prediction of the infection status of a sample, irrespective of genotype. These data strongly indicate that Lactobacillales and gastric cancer progression are linked in these mouse models. A study carried out on gastric cancer patients from high-risk groups in Singapore and Malaysia revealed a high relative abundance of lactic acid-producing bacteria such as Lactococcus and Lactobacillus, and as well as oral cavity bacteria including Fusobacterium, Veillonella, Leptotrichia, Haemophilus, and Campylobacter [34]. This is in agreement with our findings that show a potential connection between gastric cancer and Lactobacillus, and further indicates that the Myd88−/− model of fast-progressing gastric cancer recapitulates a gastric microbiome change noted in human populations.
Recent studies in Taiwan, have reported that gastric cancer patients show increased colonization of Clostridium and Fusobacterium [39]. Other studies have shown that INS-GAS germ free mice develop gastric cancer when supplemented with Lactobacillus sp., Clostridium sp., and Bacteroides sp. [25]. In contrast to this, in our study the levels of Clostridiales significantly decreased in our fast progressing gastric cancer model as compared to other genotypes, suggesting that even though Clostridiales may have previously been shown to play a role in gastric cancer progression in other studies, it does not play a significant role in our fast-progressing gastric cancer model.
5. Conclusions
Our study suggests that differences in genotypes help define the stomach microbiome diversity as different genotypes have significantly dissimilar communities. However, the correlation between Lactobacillales levels and Helicobacter-induced gastric cancer progression holds across multiple genotypes. This sustained connection indicates Lactobacillales shares a close relationship with gastric cancer. The presence of Lactobacillales in the gastric microbiome needs to be further investigated to understand their probable role in the fast-progressing form of gastric cancer, as well as the stomach microbiome as a whole. Moreover, future studies need to be carried to observe any further alterations in gastric microbiome when cancer has fully developed helping us in understanding the role of the microbiome on disease outcome.
Acknowledgments
This work is supported by the National Cancer Institute of the National Institute of Health under award R21CA210227. We thank M. Corr (UCSD) for contributing mice, which were originally generated by S. Akira (Osaka University, Osaka, Japan) and backcrossed 10 times onto a C57BL/6 background, bred, and maintained at UCSD. JC received support from the NIH grants 5T32GM127235-02 and 1F30HL152666-01.
Abbreviations
| H. pylori | Helicobacter pylori |
| WHO | World Health Organization |
| Myd88 −/− | Myeloid differentiation primary response 88- deficient |
| H. felis | Helicobacter felis |
| TRIF | Toll/IL-1R (TIR) domain-containing adaptor-inducing interferon-β |
| Trif Lps2 | TRIF deficient |
| WT | Wild Type |
| DKO | double knockout |
| BHI | Brain Heart Infusion |
| QIIME2 | Quantitative Insights into Microbial Ecology |
| TIR | Toll/interleukin-1 receptor |
| SEPP | SATé-enabled phylogenetic placement |
| OTU | Operational taxonomic unit |
| ROC | Receiver operating characteristic |
| FDR | False-discovery rate |
| INS-GAS | insulin-gastrin |
| IFN | interferon |
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/1/189/s1, Figure S1: Relative abundance of individual phyla, Figure S2: Log-fold differentials of Lactobacillales and Clostridiales with Bacteroidales, Figure S3: Histopathology of gastric disease induced by H. felis infection in different genotypes.
Author Contributions
Conceptualization, P.B., J.C., I.L.-P., K.Z., M.O.; methodology, P.B., J.C., K.Z., M.O.; validation, J.C., P.B.; formal analysis, J.C., P.B.; investigation, P.B., J.C., I.L.-P.; resources, K.Z., M.O.; data curation, J.C.; writing—original draft preparation, P.B., J.C.; writing—review and editing, P.B., J.C., K.Z., M.O.; visualization, P.B., J.C., I.L.-P., K.Z., M.O.; supervision, K.Z., M.O.; project administration, K.Z., M.O.; funding acquisition, J.C., M.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institutes of Health (NIH), grant numbers 5T32GM127235-02 and 1F30HL152666-01, and the National Cancer Institute of the NIH, grant number R21CA210227.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Animal Care and Use Committee at the University of California, San Diego (Protocol # S202243, approved on 05/05/2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw sequencing data presented in this study are openly available in the NCBI Sequence Read Archive (SRA) at accession number PRJNA685500.
Conflicts of Interest
The authors state they have no conflict of interest to declare.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization Cancer. WHO. [(accessed on 12 September 2018)]; Available online: https://www.who.int/news-room/fact-sheets/detail/cancer.
- 2.Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. [(accessed on 15 January 2021)];IARC Monogr. Eval. Carcinog. Risks Hum. 1994 61:1–241. Available online: https://www.ncbi.nlm.nih.gov/books/NBK487782/ [PMC free article] [PubMed] [Google Scholar]
- 3.Noto J.M., Peek R.M., Jr. The gastric microbiome, its interaction with Helicobacter pylori, and its potential role in the progression to stomach cancer. PLoS Pathog. 2017;13:e1006573. doi: 10.1371/journal.ppat.1006573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aziz F., Chakarobaty A., Liu K., Yoshitomi H., Li X., Monts J., Xu G., Li Y., Bai R., Bode A.M., et al. Gastric tumorigenesis induced either by Helicobacter pylori infection or chronic alcohol consumption through IL-10 inhibition. Res. Sq. 2020 doi: 10.21203/rs.2.23065/v1. [DOI] [Google Scholar]
- 5.Majmudar K., Golemi I., Tafur A.J., Toro J.D., Visonà A., Falgá C., Sahuquillo J.C., Lorente M.A., Tufano A., Weinberg I., et al. RIETE Investigators. Outcomes after venous thromboembolism in patients with gastric cancer: Analysis of the RIETE Registry. Vasc. Med. 2020;25:210–217. doi: 10.1177/1358863X19893432. [DOI] [PubMed] [Google Scholar]
- 6.Petra C.V., Rus A., Dumitraşcu D.L. Gastric microbiota: Tracing the culprit. Clujul Med. 2017;90:369–376. doi: 10.15386/cjmed-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delgado S., Cabrera-Rubio R., Mira A., Suárez A., Mayo B. Microbiological Survey of the Human Gastric Ecosystem Using Culturing and Pyrosequencing. Methods Microb. Ecol. 2013;65:763–772. doi: 10.1007/s00248-013-0192-5. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee A., Thamphiwatana S., Carmona E.M., Rickman B., Doran K.S., Obonyo M. Deficiency of the myeloid differentiation primary response molecule MyD88 leads to an early and rapid development of Helicobacter-induced gastric malignancy. Infect. Immun. 2014;82:356–363. doi: 10.1128/IAI.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang S., Moss S.F. Rodent models of Helicobacter infection, inflammation, and disease. Methods Mol. Biol. 2012;921:89–98. doi: 10.1007/978-1-62703-005-2_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai X., Carlson J., Stoicov C., Li H., Wang T.C., Houghton J. Helicobacter felis eradication restores normal architecture and inhibits gastric cancer progression in C57BL/6 mice. Gastroenterology. 2005;128:1937–1952. doi: 10.1053/j.gastro.2005.02.066. [DOI] [PubMed] [Google Scholar]
- 11.Correa P., Haenszel W., Cuello C., Tannenbaum S., Archer M. A model for gastric cancer epidemiology. Lancet. 1975;2:58–60. doi: 10.1016/S0140-6736(75)90498-5. [DOI] [PubMed] [Google Scholar]
- 12.Obonyo M., Rickman B., Guiney D.G. Effects of myeloid differentiation primary response gene 88 (MyD88) activation on Helicobacter infection in vivo and induction of a Th17 response. Helicobacter. 2011;16:398–404. doi: 10.1111/j.1523-5378.2011.00861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mejias-Luque R., Lozano-Pope I., Wanisch A., Heikenwalder M., Gerhard M., Obonyo M. Increased LIGHT expression and activation of non-canonical NF-kappaB are observed in gastric lesions of MyD88-deficient mice upon Helicobacter felis infection. Sci. Rep. 2019;9:1–9. doi: 10.1038/s41598-019-43417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers A.B., Taylor N.S., Whary M.T., Stefanich E.D., Wang T.C., Fox J.G. Helicobacter pylori but not high salt induces gastric intraepithelial neoplasia in B6129 mice. Cancer Res. 2005;65:10709–10715. doi: 10.1158/0008-5472.CAN-05-1846. [DOI] [PubMed] [Google Scholar]
- 15.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z., Kightley E.P., Thompson L.R., Hyde E.R., Gonzalez A., et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems. 2017;2:e00191-16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen S., McDonald D., Gonzalez A., Navas-Molina J.A., Jiang L., Zech Xu Z., Winker K., Kado M.D., Orwoll E., Manary M., et al. Phylogenetic Placement of Exact Amplicon Sequences Improves Associations with Clinical Information. mSystems. 2018;3:e00021-18. doi: 10.1128/mSystems.00021-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMurdie P.J., Holmes S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martino C., Morton J.T., Marotz C.A., Thompson L.R., Tripathi A., Knight R., Zengler K. A novel sparse compositional technique reveals microbial perturbations. mSystems. 2019;4:1–13. doi: 10.1128/mSystems.00016-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morton J.T., Marot C., Washburne A., Silverman J., Zaramela L.S., Edlund A., Zengler K., Knight R. Establishing microbial composition measurement standards with reference frames. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fedarko M.W., Martino C., Morton J.T., Gonzalez A., Rahman G., Marotz C.A., Minich J.J., Allen E.E., Knight R. Visualizing ’omic feature rankings and log-ratios using Qurro. NAR Genom. Bioinform. 2020;2:lqaa023. doi: 10.1093/nargab/lqaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gloor G.B., Macklaim J.M., Pawlowsky-Glahn V., Egozcue J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017;8:2224. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 25.Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lofgren J.L., Whary M.T., Ge Z., Muthupalani S., Taylor N.S., Mobley M., Potter A., Varro A., Eibach D., Suerbaum S., et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140:210–220. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dicksved J., Lindberg M., Rosenquist M., Enroth H., Jansson J.K., Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J. Med. Microbiol. 2009;58:509–516. doi: 10.1099/jmm.0.007302-0. [DOI] [PubMed] [Google Scholar]
- 28.Houben G.M., Stockbrügger R.W. Bacteria in the aetio-pathogenesis of gastric cancer: A review. Scand. J. Gastroenterol. Suppl. 1995;212:13–18. doi: 10.3109/00365529509090296. [DOI] [PubMed] [Google Scholar]
- 29.Basir H.R.G., Ghobakhlou M., Akbari P., Dehghan A., Rabiei M.A.S. Correlation between the intensity of Helicobacter pylori colonization and severity of gastritis. Gastroenterol. Res. Pract. 2017;10:1155–1159. doi: 10.1155/2017/8320496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayin S. The Relation between Helicobacter Pylori Density and Gastritis Severity. Int. Arch. Intern. Med. 2019;3:019. [Google Scholar]
- 31.Yakoob M.Y., Hussainy A.S. Chronic gastritis and Helicobacter pylori: A histopathological study of gastric mucosal biopsies. J. Coll. Physicians Surg. Pak. 2010;20:773–775. [PubMed] [Google Scholar]
- 32.Ardakani A., Mohammadizadeh F. The study of relationship between Helicobacter pylori density in gastric mucosa and the severity and activity of chronic gastritis. JRMS. 2006;11:282. [Google Scholar]
- 33.Eun C.S., Kim B.K., Han D.S., Kim S.Y., Kim K.M., Choi B.Y., Song K.S., Kim Y.S., Kim J.F. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19:407–416. doi: 10.1111/hel.12145. [DOI] [PubMed] [Google Scholar]
- 34.Castaño-Rodríguez N., Goh K.H., Fock M.K., Mitchell H.M., Kaakoush N.O. Dysbiosis of the microbiome in gastric carcinogenesis. Nat. Sci. Rep. 2017;7:1–9. doi: 10.1038/s41598-017-16289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doherty J.R., Cleveland J.L. Targeting lactate metabolism for cancer therapeutics. J. Clin. Investig. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennedy K.M., Scarbrough P.M., Ribeiro A., Richardson R., Yuan H., Sonveaux P., Landon C.D., Chi J.-T., Pizzo S., Schroeder T., et al. Catabolism of exogenous lactate reveals it as a legitimate metabolic substrate in breast cancer. PLoS ONE. 2013;8:e75154. doi: 10.1371/journal.pone.0075154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonveaux P., Copetti T., Saedeleer C.J.D., Végran F., Verrax J., Kennedy K.M., Moon E.J., Dhup S., Danhier P., Frérart F., et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS ONE. 2013;7:e33418. doi: 10.1371/journal.pone.0033418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonveaux P., Végran F., Schroeder T., Wergin M.C., Verrax J., Rabbani Z.N., Saedeleer C.J.D., Kennedy K.M., Diepart C., Jordan B.F., et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Investig. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsieh Y., Tung S., Pan H., Yen C.-W., Xu H.-W., Lin Y.-J., Deng Y.-F., Hsu W.-T., Wu C.-S., Li C. Increased Abundance of Clostridium and Fusobacterium in Gastric Microbiota of Patients with Gastric Cancer in Taiwan. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-017-18596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data presented in this study are openly available in the NCBI Sequence Read Archive (SRA) at accession number PRJNA685500.