Abstract
Glycerol-3-phosphate acyltransferases (GPATs) play an important role in glycerolipid biosynthesis, and are mainly involved in oil production, flower development, and stress response. However, their roles in regulating plant height remain unreported. Here, we report that Arabidopsis GPAT1 is involved in the regulation of plant height. GUS assay and qRT-PCR analysis in Arabidopsis showed that GPAT1 is highly expressed in flowers, siliques, and seeds. A loss of function mutation in GPAT1 was shown to decrease seed yield but increase plant height through enhanced cell length. Transcriptomic and qRT-PCR data revealed that the expression levels of genes related to gibberellin (GA) biosynthesis and signaling, as well as those of cell wall organization and biogenesis, were significantly upregulated. These led to cell length elongation, and thus, an increase in plant height. Together, our data suggest that knockout of GPAT1 impairs glycerolipid metabolism in Arabidopsis, leading to reduced seed yield, but promotes the biosynthesis of GA, which ultimately enhances plant height. This study provides new evidence on the interplay between lipid and hormone metabolism in the regulation of plant height.
Keywords: glycerol-3-phosphate acyltransferase, oil biosynthesis, plant height, gibberellin metabolism, cell wall
1. Introduction
Plant height is closely correlated with yield trait owing to its crucial role in plant architecture, pod bearing number, and lodging resistance [1,2,3]. Plant height is a complex trait regulated by many endogenous and environmental factors, and gibberellin (GA), known as the “green revolution phytohormone”, plays a foremost role among these factors [2,4,5,6].
Bioactive GAs are plant diterpene hormones that are essential for multiple aspects of plant growth and development, including seed germination, leaf expansion, trichome development, stem elongation, flowering, male fertility, and fruit set [7,8,9]. GA biosynthesis in plants involves many enzymes and cellular compartments, as well as the isoprenoid biosynthetic pathway [9,10,11]. The plastid-specific methylerythritol phosphate (MEP) pathway is particularly responsible for GA generation, as it provides the precursor geranylgeranyl diphosphate (GGPP) required for GA biosynthesis [11]. GGPP is converted to bioactive GAs sequentially by the copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS), ent-kaurene oxidase (KO), ent-kaurenoic acid oxidase (KAO), and 2-oxoglutarate-dependent dioxygenases (2-OGDs) [9,10]. As an important part of its metabolism, deactivation of GA functions to regulate the concentration of bioactive GAs in plants can be achieved through GA2ox [12], GA methyl transferase [13], and cytochrome P450 monooxygenase [14]. Furthermore, components of GA signaling such as the GA receptor gibberellin insensitive dwarf 1 (GID1), GID2, and DELLA proteins, can regulate the levels of bioactive GAs to maintain plant growth and development [9,15]. GAs induce the expression of cell wall-associated genes, such as xyloglucan endotransglucosylase/hydrolases (XTHs), pectin methylesterase (PME), pectin methylesterase inhibitor (PMEI), and expansins (EXPs), to regulate cell expansion, which contributes to stem elongation [16,17,18,19].
While the implications of lipid metabolism in the regulation of plant height remain unclarified, a few genes involved in lipid transport [20] and hydrolysis [21,22] have been shown to influence this phenotype. Glycerol-3-phosphate acyltransferase (GPAT) is an important enzyme in lipid biosynthesis that transfers an acyl group to the sn-1 or sn-2 position of sn-glycerol-3-phosphate (Gro3P) to produce lysophosphatidic acid (lysoPtdOH). This is necessary for the production of intracellular lipids such as membrane lipids and storage triacylglycerol (TAG), as well as extracellular lipids such as suberin-associated waxes, cutin, and suberin [23,24]. The multifaceted functions of GPAT were revealed through a series of biochemical and mutational analyses. Ten GPATs have been reported in Arabidopsis thaliana, which are categorized into sn-1-GPAT (ATS1 and GPAT9) and sn-2-GPAT (GPAT1-8) [23,25]. The plastidial ATS1 and endoplasmic reticulum (ER)-localized GPAT9 contribute to the biosynthesis of glycerolipids but not extracellular lipids [26,27,28,29,30]. For the mitochondria-localized GPATs, GPAT1 is involved in glycerolipid biosynthesis, which is essential for tapetum differentiation and male fertility, while the functions of GPAT2 and GPAT3 are still unknown [25,31]. ER-bound GPAT4, 6, and 8, which have both sn-2 acyltransferase and phosphatase activities, are required for cutin biosynthesis [28,32,33]. GPAT4 and GPAT8 are functionally redundant and contribute to proper lateral root outgrowth [34], as well as prevent water loss and pathogen infection [35]. GPAT6 plays multiple roles in stamen development and fertility [36] and promotes salt tolerance [37]. ER-bound suberin-associated GPAT5 and GPAT7 only possess sn-2 acyltransferase activity [32,33,38,39]. GPAT5 plays a role in the production of suberin and suberin-associated root waxes, which contribute to the tolerance of plants to salt stress [38,39]. Lastly, GPAT7 may only contribute to suberin biosynthesis under stress, such as wounding [32]. Studies on plant GPATs have revealed that they are required for the synthesis of intracellular and extracellular lipids and have important physiological functions, yet their roles in hormone metabolism and hormone-mediated plant development are still unknown.
In a previous report, Arabidopsis GPAT1 was shown to act as an essential factor for tapetum and pollen development [31]. In this work, we studied the role of GPAT1 in regulating Arabidopsis plant height. Our results revealed that knockout of GPAT1 increased plant height by activating GA metabolism and signaling, which then stimulated cell wall organization and biogenesis. This study provides evidence on how GPAT-mediated glycerolipid metabolism influences plant height through GA.
2. Results
2.1. Expression Pattern of the Arabidopsis GPAT1 Gene
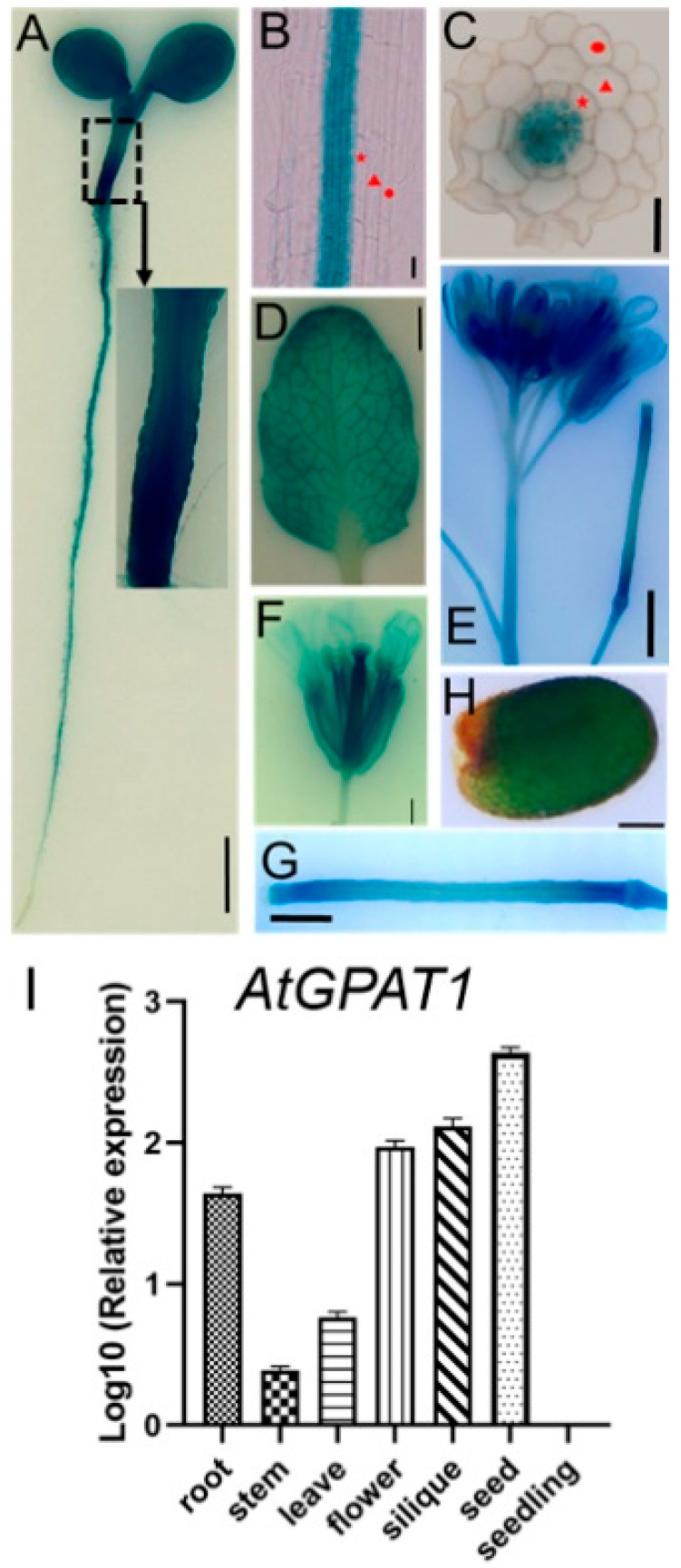
In previous studies using RNA gel blot analysis, GPAT1 was shown to be mainly expressed in developing siliques and flower buds [31]. To further determine the expression pattern of GPAT1, we made a construct where the GUS reporter was driven by the GPAT1 promoter, and transfected it into WT Arabidopsis plants. One transgenic ProGPAT1-GUS line was selected to analyze the expression of GPAT1 gene. GUS activity was detected in young seedlings, roots, leaves, inflorescence stems, flowers, and mature seeds by histochemical staining (Figure 1A–I). GUS staining in the hypocotyls was strongest in whole, seven-day-old seedlings, including their cotyledons and roots (Figure 1A). Expression of GPAT1 in roots was only detected in the stele, while there was no GUS activity in the epidermis, cortex, or endodermis of the roots (Figure 1B,C). GUS expression was also observed in rosette leaves, and leaf venation showed stronger GUS activity than other parts of the leaf (Figure 1D). GUS expression was also detectable in inflorescences and upper inflorescence stems of WT plants; strong staining was observed in the former and weak in the latter (Figure 1E). Though GUS activity was detected in upper stems, no GUS activity was observed in the middle and lower stems (Figure S1). In flowers, GUS activity was also detected in sepals, petals, and carpels (Figure 1F), while expression in stamens was demonstrated by in situ hybridization [36]. Figure 1G,H shows that GPAT1 was also expressed in young (about four days after pollination) siliques, as well as in mature seeds. To determine the relative expression levels of GPAT1 in different tissues of WT Arabidopsis plants, we performed qRT-PCR experiments. qRT-PCR indicated that the highest expression level of GPAT1 was in the seed, followed by a descending order of siliques, flowers, stems and leaves, roots, and seedlings (Figure 1I). The GPAT1 expression pattern indicated by qRT-PCR was similar to that presented by GUS staining. These results suggest that GPAT1 mRNA is significantly expressed in seeds, siliques, flowers, and steles, the only region in roots that expresses GPAT1.
Figure 1.
Analysis of GPAT1 expression in Arabidopsis wild-type plants. (A–H) Expression analysis of GPAT1 by ProGPAT1-GUS. (A) GUS staining in a five-day-old seedling grown on agar. Bar = 10 mm. (B,C) GUS staining in roots of seven-day-old seedlings grown on agar. The red solid circle, triangle and pentagram indicated the epidermis, cortex and endodermis, respectively. Bar = 20 μm. (D) GUS staining in rosette leaves. Bar = 10 mm. (E) GUS staining in inflorescences and inflorescence stems. Bar = 2 mm. (F) GUS staining in flowers. Bar = 1 mm. (G) GUS staining in 4-DAP siliques. DAP, day after pollination. Bar = 1 mm. (H) GUS staining in mature seeds. Bar = 100 μm. (I) Expression profiles of GPAT1 in different tissues of WT plants. Total RNA was extracted from different tissues of soil-grown plants for qRT-PCR. Values were standardized using the Arabidopsis Actin 2 gene. Values are the means of three independent experiments ± standard error (SE) (n = 3).
2.2. Knockout of GPAT1 Decreases TAG Content and Alters FA Composition in Seeds
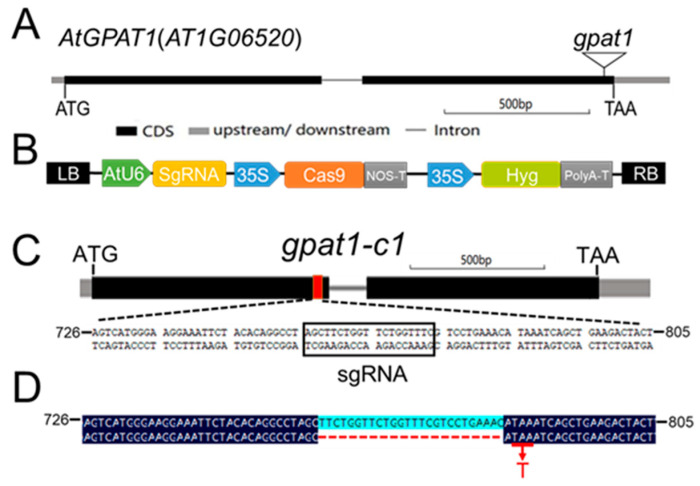
In a previous work, Zheng et al. showed that loss of GPAT1 caused no significant change in seed oil content, but reduced TAG content in flower buds [31]. The underlying mechanism controlling this phenomenon was not clear. On account of the high expression of GPAT1 in siliques and seeds, we inferred that GPAT1 might play an important role in seed oil biosynthesis. To test this, we ordered a proven loss-of-function mutant (SALK_052352) of GPAT1 (gpat1) [36], which had a T-DNA insertion in the second exon (Figure 2A), and created a new loss-of-function mutant by CRISPR/Cas9-mediated gene editing (gpat1-c1) to further target the first exon (Figure 2B,C). Through sequencing, we found that a 26-bp sequence near the sgRNA target was deleted in the gpat1-c1 mutant, which led to the early appearance of a stop codon TAA (Figure 2D).
Figure 2.
Mutant creation and identification of GPAT1. (A) Structure of GPAT1 gene and genomic organization of the gpat1 loci. The T-DNA insertion point is indicated as a triangle. (B) Structure of the pAtU6-26:sgRNA-23p35S:Cas9 pBlunt vector. (C) CRISPR/Cas9 sgRNA targets the first exon of GPAT1. The box indicates the target sequences. (D) Sequencing of the GPAT1 site targeted by sgRNA. Red hyphens indicate the deletions which led to the early appearance of the stop codon. Red letter T indicate the stop codon.
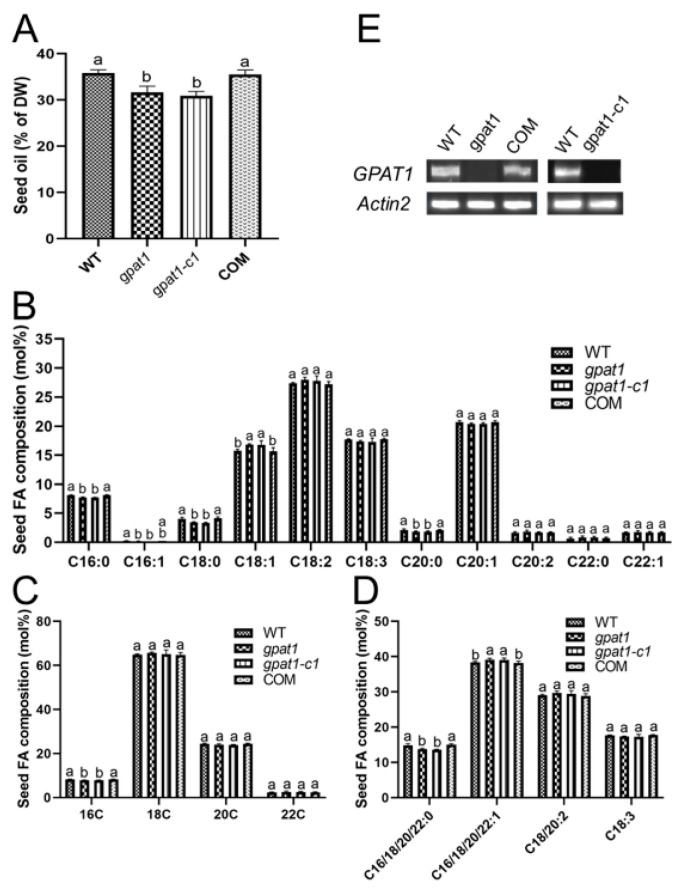
We then implemented a different protocol to determine the seed oil and FA contents in WT and mutants of GPAT1. Seed oil contents of gpat1 and gpat1-c1 mutants were reduced by 11.7% and 13.7% compared with that of WT, respectively (Figure 3A). We further analyzed the FA composition in seed TAG, which was significantly altered in mutant plants (Figure 3B). For example, palmitic acid (16:0), palmitoleic acid (16:1), stearic acid (18:0), and arachidic (20:0) contents in gpat1 seeds were 5.0%, 17.9%, 14.9%, and 13.9% lower than in WT seeds, respectively. In contrast, oleic acid (18:1) content was 7.1% higher in gpat1 seeds than in WT seeds. Similar results were found in the gpat1-c1 line. We also analyzed the contents of 16-, 18-, 20-, and 22-carbon FAs. The levels of 16-carbon FAs were markedly lower in seeds of mutant plants, while levels of 18-, 20-, and 22-carbon FAs were similar to those of WT seeds (Figure 3C). The total contents of saturated (16:0, 18:0, 20:0, and 22:0) FAs were significantly reduced, whereas the total contents of monounsaturated FAs (MUFAs) with one double bond (16:1, 18:1, 20:1, and 22:1) increased (Figure 3D). To further confirm the function of GPAT1 in seed oil biosynthesis, we developed a complementation transgenic line to the gpat1 mutant (COM), and measured the mRNA levels of GPAT1 in WT, gpat1, and COM by RT-PCR. The expression of GPAT1 in COM and WT was comparable while that in the gpat1 and gpat1-c1 was undetectable (Figure 3E), suggesting that COM was available. The oil content and levels of different FAs in seeds of the COM line were also comparable to those of WT plants (Figure 3A–D). We also studied the role of GPAT1 gene in cuticle formation via toluidine blue dye method [35], and analysis of the leaf cuticle of gpat1 mutant did not reveal any obvious cuticle defect (Figure S2) which suggested that disruption of GPAT1 gene might have no significant effect on cuticle formation. Taken together, these results suggest that knockout of GPAT1 leads to remarkable reduction in seed oil content and alteration of FA compositions, which is consistent with the expression profile of GPAT1.
Figure 3.
Effect on seed oil content and FA composition by GPAT1 mutation. (A) TAG contents of dry seeds of WT, gpat1, gpat1-c1 and complementation line COM plants grown in soil. Data are means of the five replicates with SE. DW, dry weight. (B) Fatty-acid composition in TAGs of WT, gpat1, gpat1-c1 and COM lines. (C) Contents of 16-, 18-, 20-, and 22-carbon FAs in TAGs of WT, gpat1, gpat1-c1 and COM lines. (D) Contents of saturated FAs and unsaturated FAs with one, two, or three double bonds in TAGs of WT, gpat1, gpat1-c1 and COM lines. (E) Level of GPAT1 transcript in developing siliques at ∼20 days after flowering (DAF) in WT, gpat1, gpat1-c1 and COM plants. Values are means and SE based on one-way ANOVA (Duncan and Tukey test). Different letters indicate significant differences at p < 0.05. WT, wild type.
2.3. Knockout of GPAT1 Enhances Plant Height but Decreases Seed Yield
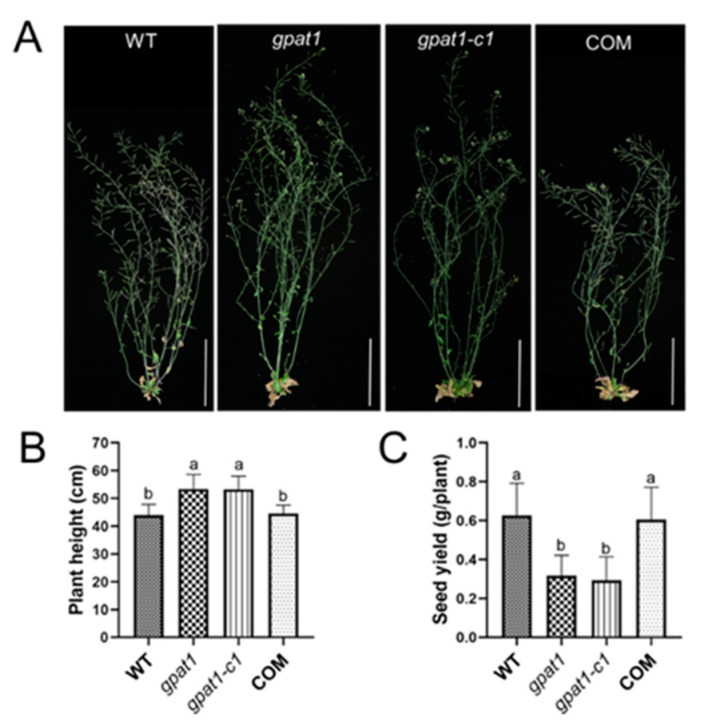
Mutation in GPAT1 resulted in reduced yield, as indicated by the reduced silique size and seed yield per silique [31]. In some situations, plant height is negatively correlated with seed yield [2]. To investigate whether Arabidopsis GPAT1 is associated with plant height, we compared the height among the mutants of GPAT1 and WT plants (Figure 4A). Compared with WT, plant heights of gpat1 and gpat1-c1 mutants were promoted by 21.5% and 21.3%, respectively, while that of the complementation transgenic COM plants was not altered (Figure 4B). We further scored the total seed yield per plant, which was severely reduced in gpat1 and gpat1-c1 mutants, by 49.7% and 53.2% compared to WT, respectively, while the total seed yield of the COM plants was comparable to that of WT (Figure 4C). Due to the high expression of GPAT1 in roots and hypocotyls (Figure 1A), we also analyzed the primary root length and hypocotyl length of WT and gpat1 mutant plants. There was no evident difference in primary root length or hypocotyl length found between WT and gpat1 mutant plants (Figure S3A–C). These results suggest that knockout of Arabidopsis GPAT1 increased plant height but decreased total seed yield.
Figure 4.
Effect on plant morphology and yield by knockout of GPAT1. (A) Ten-week-old plants of WT, gpat1, gpat1-c1 and the complementation transgenic line COM. Bar = 10 cm. (B) The gpat1 and gpat1-c1 mutant plants showed increased plant height compared with WT and COM plants. Plant height was determined from ten independent plants from five plots for each genotype (n = 10). (C) Total seed yield of individual plant from two plots (n = 12). Values are means ± SE based on one-way ANOVA (Duncan and Tukey test). Different letters indicate significant difference at p < 0.05. WT, wild type.
2.4. Knockout of GPAT1 Alters Cell Morphology of the Stem
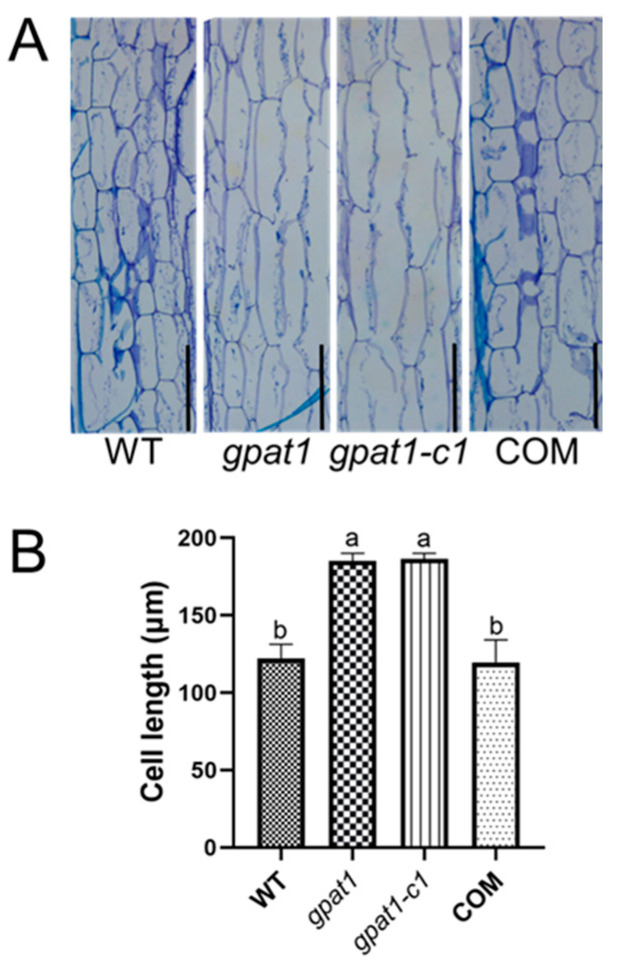
To investigate whether the cell morphology of the stem was affected by mutations in GPAT1, the basal nodes of stems from ten-week-old WT, gpat1, gpat1-c1, and the complementation transgenic COM plants were selected and embedded in paraffin sections, followed by an analysis of their anatomical details (Figure 5A). By measuring the cell length of longitudinal paraffin sections obtained from stems, we found that mutations in GPAT1 increased cell length significantly compared to WT plants (Figure 5B). The alteration of cell morphology brought by a loss-of-function in GPAT1 was recovered by introducing the whole genomic sequence of the GPAT1 gene into the gpat1 mutant, which confirmed that the mutation introduced to GPAT1 significantly contributed to the alteration of the cell morphology of the stems (Figure 5A,B). These results show that a loss of GPAT1 promotes cell elongation during stem development.
Figure 5.
Knockout of GPAT1 alters the stem cell morphology. (A) Longitudinal section of stems obtained from ten-week-old WT, gpat1, gpat1-c1 and COM plants. The basal nodes of stems were selected for embedding in paraffin sections. Bar = 200 μm. (B) The length of stem cells of ten-week-old WT, gpat1, gpat1-c1 and COM plants. Values are mean ± SD (n = 150 cells from five individual longitudinal sections of stems). Different letters indicate significant differences at p < 0.05, as determined by one-way ANOVA with Tukey’s post-test. WT, wild type.
2.5. Transcriptome Differences between WT and gpat1 Mutant Plants
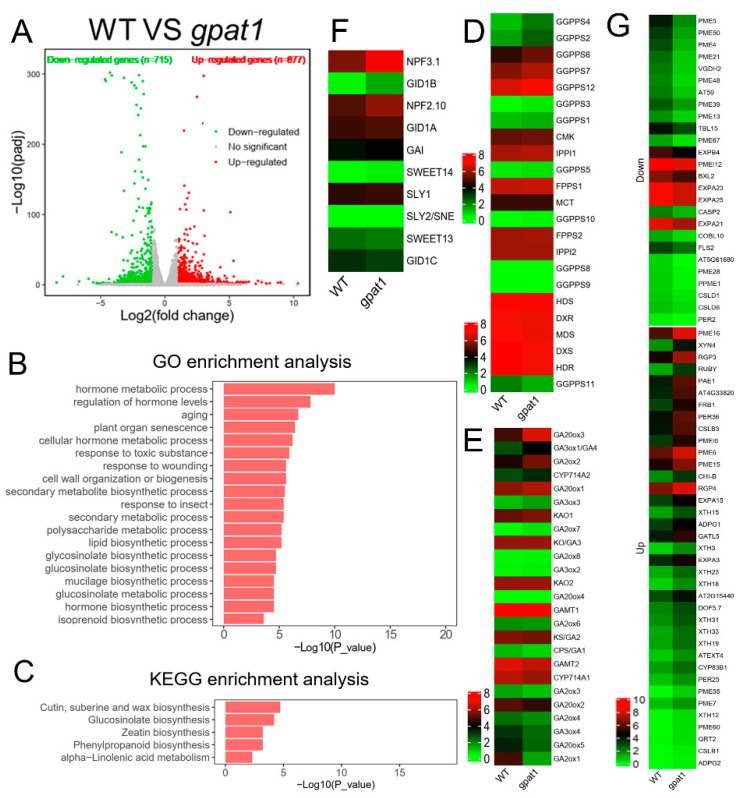
To uncover the mechanisms of stem lengthening and accompanying cell phenotypes in knockout mutants of GPAT1, RNA-Seq was conducted to identify DEGs and their associated GO (gene ontology) terms and KEGG (Kyoto encyclopedia of genes and genomes) pathways. A total of 1585 DEGs were identified between WT and gpat1 plants, with 872 genes upregulated and 713 genes downregulated (Figure 6A). GO and KEGG enrichment analyses were employed to gain insights into the biological functions of the upregulated DEGs, with the significantly enriched pathways shown in Figure 6B,C. For example, the hormone metabolic process and regulation of hormone levels pathways were the top two highly enriched GO terms, while GA biosynthesis-related isoprenoid biosynthetic pathway was also enriched. Cell wall-related metabolite processes were also enriched, as were the polysaccharide metabolic process, mucilage biosynthetic process, and cell wall organization or biogenesis process. There were five enriched KEGG pathways including cutin, suberine and wax biosynthesis, linolenic acid metabolism, phenylpropanoid biosynthesis, glucosinolate biosynthesis, and zeatin biosynthesis. The results from GO and KEGG analyses indicated that a loss of GPAT1 function may accelerate the hormone metabolic processes and cell wall-related secondary metabolite biosynthetic processes.
Figure 6.
RNA-Seq profiling of WT and gpat1 lines. (A) Volcano plot showing the DEGs between two libraries of WT and gpat1 lines. p-adjust < 0.05 & |log2FC| ≥ 1 were used as the threshold to determine the significance of DEGs. Green dots show down-regulated genes, red dots represent up-regulated genes, and grey dots indicate transcripts that did not change significantly in the gpat1 library compared with the WT. (B,C) GO and KEGG enrichment analysis of up-regulated DEGs between WT and gpat1 lines. (D–G) Heat map showed the genes expression in the MEP pathway, GA metabolism pathway, GA transport and signaling and cell wall organization or biogenesis.
Considering that GA plays an important role in regulating plant height, as well as the results from our GO and KEGG analyses, we set out to quantify the expression of genes in GA metabolism and signaling. The MEP pathway predominantly provides the precursor GGPP for GA biosynthesis in Arabidopsis [11]. Among the 23 genes in the MEP pathway in Arabidopsis, there were only two DEGs (GGPPS2 and GGPPS4) that were significantly upregulated (Figure 6D and Table S1). GGPP generated by the MEP pathway subsequently gets fluxed into the GA biosynthesis pathway. There were three GA biosynthesis-related DEGs including GA3ox1, GA20ox3, and GA20ox5, and three GA deactivation-related DEGs including GA2ox1, GA2ox2, and GA2ox4 found in gpat1 mutants versus WT lines (Figure 6E and Table S1). GA3ox1 and GA20ox3 were evidently upregulated while GA20ox5 was markedly downregulated. GA2oxs are mainly responsible for the deactivation of bioactive GAs, and we found that GA2ox1 and GA2ox4 were significantly downregulated while GA2ox2 was observably upregulated (Figure 6E and Table S1). DEGs related to GA transport and signaling were also found. The nitrate transporter 1/peptide transporter family (NPF) members NPF3.1 [40,41] and NPF2.10 [42,43,44], as well as the SWEET (sugar will eventually be exported transporter) family members SWEET13 and SWEET14 [45] were reported to transport GAs in Arabidopsis, and we found NPF3.1 was significantly upregulated (Figure 6F and Table S1). The DELLA protein GAI and GA receptor protein GID are extremely important for GA signaling, as well as the F-box proteins SLY1 and SLY2 [9,46], among which GID1B was identified as the only DEG that was significantly upregulated (Figure 6F and Table S1).
GAs are important plant hormones that function in the promotion of cell elongation. Plant cell walls determine cell wall sizes and shapes, and are thus essential for plant growth and development [47,48,49]. GAs can regulate cell wall-related genes to control cell elongation [50]. In line with this, the GO term “cell wall organization or biogenesis” was significantly enriched in our upregulated clustering analysis (Figure 6B). Upon further analysis, there were more upregulated DEGs from this GO term than those that were downregulated (Figure 6G). Out of the 37 total upregulated DEGs, the xyloglucan endotransglucosylase/hydrolase (XTH) gene family was most enriched, followed by genes involved in pectin modification such as PME (pectin methylesterase), PMEI (pectin methylesterase inhibitor), and PAE (pectin acetylesterase), and other polysaccharide-related genes such as polygalacturonase, galacturonosyltransferase, glycosyltransferase, cellulose synthase, endo-1,4-β-xylanase, O-fucosyltransferase, and endochitinase. In addition, other cell wall-associated genes such as expansin, extensin, and oxidase (i.e., peroxidase, cytochrome P450, galactose oxidase) were all included in the upregulated DEGs. Out of the 26 downregulated DEGs, there were 16 pectin-associated genes, 4 expansin genes, and 2 cellulose synthase genes. Furthermore, cell wall-related genes such as BXL2 (beta-D-xylosidase 2), TBL15 (trichome birefringence-like 15), CASP2 (casparian strip membrane domain protein 2), COBL10 (COBRA-like 10), PER2 (peroxidase 2), and FLS2 (flagellin-sensitive 2) also belonged to the downregulated DEGs.
Overall, transcriptomic analysis demonstrated that the upregulated genes in various pathways, including isoprenoid biosynthesis, GA biosynthesis and signaling, and cell wall organization or biogenesis, might be related to enhanced cell elongation, and thus, plant height in gpat1 mutant plants.
2.6. Validation of DEGs Using qRT-PCR
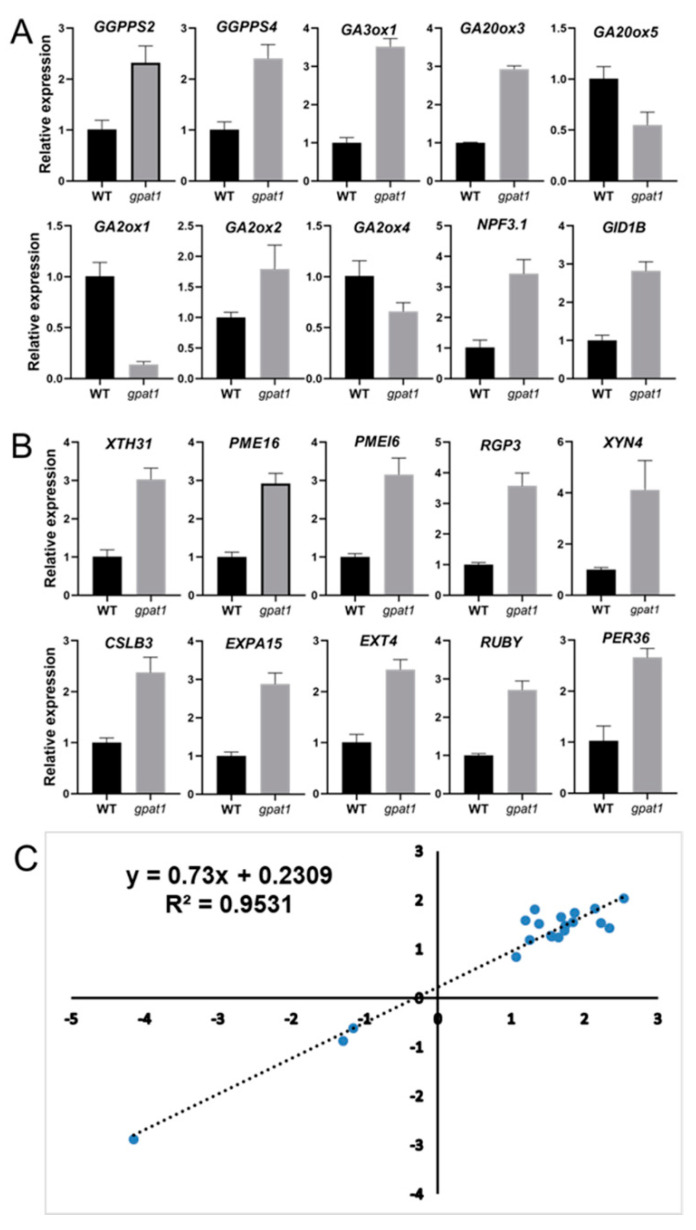
We selected and verified ten DEGs in pathways of isoprenoid biosynthesis and GA metabolism and signaling through qRT-PCR analysis. For GGPPS genes, the transcript levels of GGPPS2 and GGPPS4 were increased by about 1.3-fold and 1.4-fold in the gpat1 mutant lines, respectively (Figure 7A). For GA biosynthesis, the expression levels of GA3ox1 and GA20ox3 were respectively enhanced by almost 2.5- and 2-fold, while that of GA20ox5 was decreased by about 0.5-fold in the gpat1 mutant lines (Figure 7A). For GA inactivation, the mRNA levels of GA2ox1 and GA2ox4 were respectively reduced by 86.4% and 34.6%, while that of GA2ox2 was increased by 78.9% in the gpat1 mutant plants (Figure 7A). The transcript level of NPF3.1 was promoted by almost 2.4-fold in the gpat1 mutant plants (Figure 7A), while that of GID1B was also upregulated by approximately 2-fold (Figure 7A).
Figure 7.
qRT-PCR profiling of DEGs between WT and gpat1 lines. (A) The expression levels of genes in the MEP pathway, GA metabolism and signaling pathway. (B) The transcript levels of genes involved in cell wall biogenesis or organization. (C) Correlation analysis of gene expression pattern by RNA-seq and qRT-PCR. Expression values are relative to that of the EV control. Values are means ± SE (n = 3 technical replicates).
We also selected and verified ten representatively upregulated DEGs involved in cell wall organization through qRT-PCR analysis. The mRNA levels of XTH31, PME16, PMEI6, and EXPA15 were upregulated by almost 2-fold in the gpat1 mutant(Figure 7B), while those of RGP3, XYN4, and CSLB3, which are involved in cell wall polysaccharide biosynthesis or modification, were also upregulated by almost 1.5- to 3-fold, respectively, in the gpat1 mutant (Figure 7B). The transcript levels of EXT4 and the galactose oxidase RUBY were also enhanced by 1.5-fold, while that of PER36 increased by 1.6-fold in the gpat1 mutants (Figure 7B). After verifying the accuracy of our transcriptomics data through qRT-PCR, we determined that our RNA-Seq and qRT-PCR data were highly correlated, with an R2 of 0.9531 (Figure 7C).
Together, these gene expression data suggest that loss of GPAT1 upregulates genes involved in GA metabolism and signaling, as well as in cell wall organization and biogenesis, which may ultimately lead to cell elongation and increased plant height.
3. Discussion
Previous studies have suggested that GPAT1 is involved in the glycerolipid metabolism and plays a prominent role in tapetum differentiation and male fertility [31]. We report here that knockout of GPAT1 in Arabidopsis impaired glycerolipid metabolism while accelerating GA biosynthesis and cell wall organization, thus promoting stem cell length and plant height.
GPAT1 is highly expressed in flowers, siliques, and seeds (Figure 1), indicating that GPAT1 may play a significant role in the development of these organs. It was reported that knockout of GPAT1 in Arabidopsis impaired mitochondrial membrane biogenesis, which led to mitochondrial dysfunction and a delay in tapetal degeneration [31]. Considering the high expression of GPAT1 in siliques and seeds, seed oil content was quantified following a method which is different from that taken by Zheng et al. [31]. The procedure for seed oil extraction was based on methods described by Hara and Radin [51]. For transmethylation, we used the triacylglycerol internal standard (TAG-17:0) instead of free fatty acid or its methyl esters, which can minimize errors brought about by variation in the efficiency of methylation. Oil content was then calculated using the formula provided by Li et al. [52], which showed significant reduction in mutants of GPAT1 compared with WT and COM plants (Figure 3A). Furthermore, the FA composition of seed TAG was also significantly altered in mutant plants (Figure 3B–D). These results suggest that GPAT1 is essential for oil biosynthesis not just in flowers but also in different tissues of the plant, which is consistent with the high expression pattern of GPAT1 in siliques and seeds.
Loss of GPAT1 led to a decrease in seed yield, which may have disorganized the normal energy distribution in plants. We speculated that impairment of reproductive development may be conducive to vegetative development. Thus, we assessed the morphology of gpat1, gpat1-c1, and WT plants. By comparison, we found that knockout of GPAT1 increased plant height markedly (Figure 4). The complementation experiment reversed this phenotype, strongly implying that GPAT1 played a role in the regulation of plant height. Plant height is often correlated with cell elongation. To investigate whether loss of GPAT1 affected the cell morphology of stems to result in altered plant height, we measured the cell length of longitudinal paraffin sections obtained from stems. Cell length of stems from GPAT1 mutant plants was dramatically higher than those of WT and COM plants (Figure 5A,B). These results indicated that knockout of GPAT1 contributed to cell elongation in stems of Arabidopsis.
GAs act as a key factor in controlling plant height by regulating cell elongation [53], and are generated from GGPP, which is produced by GGPPS [9,54,55]. Loss of GPAT1 increased the expression of GGPPS2 and GGPPS4 significantly (Figure 6D and Figure 7A and Table S2), which may provide more GGPP for GA biosynthesis. GA20ox and GA3ox are responsible for the final formation of bioactive GAs, which are dominant in the regulation of GA biosynthesis [56,57,58,59,60]. Among the four GA3ox genes identified in Arabidopsis, GA3ox1 and GA3ox2 function redundantly in the production of bioactive GAs required for stem elongation, while GA3ox1 plays a predominant role [57]. The increased expression of GA3ox1 (Figure 6E and Figure 7A and Table S2) indicated that it may contribute to the stem elongation of gpat1 mutant plants. Among the five GA20oxs in Arabidopsis, GA20ox1 and GA20ox3 were the top two abundantly expressed genes in stems, and while GA20ox1 has been demonstrated to function in stem elongation, GA20ox3 maybe also have unreported functions related to this due to its expression pattern [59]. Our results show that the transcript level of GA20ox1 was enhanced in gpat1 mutants, although the increase was not marked according to RNA-seq data, the expression of GA20ox3 was significantly promoted (Figure 6E and Figure 7A and Table S2), indicating that GA20ox3 may play a key role in stem elongation in the case of gpat1 mutants. Bioactive GA concentrations are regulated through its biosynthesis and inactivation, and GA inactivation can be achieved through the GA methyl transferase (GAMT), gibberellin 2 oxidase (GA2ox), and CYP714A catabolic enzymes [10]. In gpat1 mutants, the mRNA levels of GA2ox1 and GA2ox4 were evidently reduced while that of GA2ox2 increased significantly (Figure 6E and Figure 7A, and Table S2). The antagonism between these three genes may contribute to reduced production of inactive GAs. However, we were not able to determine why these GA2oxs exhibited different expression levels in the gpat1 mutant compared with WT.
The movement and distribution of bioactive GAs are important for plant growth and development [10]. The mRNA level of NPF1.3, a GA influx transporter gene, was markedly higher in the gpat1 mutant than in WT (Figure 6F and Figure 7A, and Table S2), suggesting that NPF1.3 may play a role in the increased distribution of bioactive GAs in stems of gpat1 mutants. GA signaling is essential for the physiological functions of GAs, in which the hub repressor DELLA, GA receptor GID1, and the F-box proteins SLY1 and SNEEZY (SLY2/SNE) in Arabidopsis and GID2 in rice are regarded as key components [46]. In gpat1 mutants, the expression of DELLA protein GAI, SLY1, and SLY2 were not altered, while one of the three GID1 genes, GID1B, was more highly expressed than in WT. Among the three GID1 genes, GID1A contributed most to GA responses [61], and its transcript level was slightly higher in gpat1 mutants compared with WT (Figure 6F and Figure 7A and Table S2), indicating that both GID1A and GID1B may contribute to the stem elongation of gpat1 mutants.
In plants, cell elongation in apical and intercalary meristems brings about stem elongation, and the biogenesis and modification of cell wall has a significant effect on cell elongation. XTHs involved in hemicellulose synthesis contribute to cell wall extension by cutting and/or rejoining xyloglucan chains [62,63,64]. XTH31 was reported to be involved in cell wall modification and cell elongation [65], and was significantly expressed in the gpat1 mutant, further confirming the role of XTHs in increasing plant height (Figure 6G and Figure 7B, and Table S2). The pectin esterification level in cell walls is associated with cell length, and the interplay between PME and PMEI plays a pivotal role in regulating pectin esterification levels [19]. The enhanced expression levels of PME16 and PMEI6 in stems [66,67] suggest that both genes may contribute to the elongation of stems in gpat1 mutants (Figure 6G and Figure 7B, and Table S2). Reversibly glycosylated proteins (RGPs) act as UDP-L-Ara mutases that catalyze the formation of UDP-Araf from UDP-Arap, and may be involved in cell wall elongation and thickening [68,69]. Endo-1,4-b-xylanases (XYNs) belong to the glycoside hydrolase family and appear to be involved in xylan modification in cell walls [70,71]. Cellulose synthase like (CSL) proteins are involved in the synthesis of carbohydrate-based polymers such as cellulose, pectins, and hemicelluloses, and therefore plant cell wall formation and cell elongation [72,73]. In the gpat1 mutant plants, the mRNA levels of RGP3, XYN4, and CSLB3 were significantly enhanced (Figure 6G and Figure 7B and Table S2), suggesting that these cell wall polysaccharide-related genes may be beneficial for stem elongation of these plants. Furthermore, loss of GPAT1 led to increased expression of the expansin gene EXPA15, extensin EXT4, galactose oxidase gene RUBY, and peroxidase gene PER36, which are all associated with cell elongation [74,75,76,77] (Figure 6G and Figure 7B and Table S2), indicating that these genes may function to enhance the height of gpat1 mutant plants.
In our work, loss of function of GPAT1 was shown to impair glycerolipid metabolism in Arabidopsis, leading to reduced total seed yield, but promote stem cell elongation and plant height. RNA-seq and qRT-PCR data suggest that loss of GPAT1 resulted in increased expression of genes in the MEP pathway, GA biosynthesis and signaling, and pathways involved in cell wall organization and biogenesis, which may explain the elongated cell length and enhanced plant height observed. GPAT1 catalyzes the first step of glycerolipid biosynthesis by acylating glycerol-3-phosphate at the sn-1 or sn-2 hydroxyl with an acyl donor, while the fatty acyls are offered by FAs. In FA biosynthesis, pyruvate is an important precursor for producing FAs, and is also the precursor for the MEP pathway that contributes GA biosynthesis. Thus, we speculate that the GPAT1 mutation-mediated glycerolipid metabolism impairment may reduce the utilization of pyruvate for the MEP pathway. This may therefore activate GA biosynthesis and cell wall organization to accelerate cell elongation and promote plant height. However, this inference needs further experiments to be authenticated.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
WT (ecotype Columbia-0), gpat1 (SALK_052352) and CRISPR-generated gpat1-c1 mutants, and COM (transgenic complementation lines of gpat1) A. thaliana seeds were used in this study. Seeds were sown on Murashige and Skoog (MS) medium (1% sucrose, 1% agar), imbibed at 4 °C for 2–3 days in the dark, and transferred to a growth chamber with a light intensity of 200 μmol m−2 s−1 (16/8 h of light/dark at 22 °C). Ten-day-old seedlings grown on plates were transferred to potting soil under controlled growth conditions. The mutant SALK_052352 was bought from ABRC.
4.2. β-Glucuronidase (GUS) Staining Assay and Light Microscopy
For the GUS staining assay, a 2045-bp sequence upstream of the first ATG in the GPAT1 gene was cloned into the binary vector pCAMBIA1301 carrying the GUS gene downstream of the inserted promoter [78]. The construct ProGPAT1::GUS was transformed into WT Arabidopsis (Col-0) plants via the floral dip method [79]. Transgenic plants were screened by hygromycin (25 μg/mL) selection, and the homozygous transgenic lines were used for GUS analysis. GUS histochemical staining was performed according to procedures described previously [80]. Images of whole mount tissues were taken with a Leica DVM6a stereoscope. The primers used for amplification of genomic DNA fragments are listed in Supplementary Table S1.
4.3. qRT-PCR and Reverse Transcription PCR
Total RNA was extracted from various tissues of WT Arabidopsis plants using RNAiso plus reagent (TaKaRa, Dalian, China) following the manufacturer’s instructions. cDNA obtained using the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa) was used for quantitative real-time PCR (qRT-PCR) and reverse transcription PCR (RT-PCR). qRT-PCR was performed using SYBR® Premix Ex Taq™ (Tli RNaseH Plus) and the CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA), as specified by the manufacturer. The primers used for RT-PCR and qRT-PCR are listed in Supplementary Table S1.
4.4. Mutant Creation, Identification and Complementation
To create a mutant of the GPAT1 gene with the CRISPR/Cas9 gene editing system, we designed an sgRNA target sequence using the online tool CRISPR-P2.0 (http://crispr.hzau.edu.cn/CRISPR2/) [81]. The selected target sequence was cloned into the final pAtU6-26:sgRNA-23p35S:Cas9 pBlunt vector [82]. The construct was confirmed by sequencing and then introduced into WT Arabidopsis plants via the floral dip method [79]. For genotyping, the target sequences of each GPAT1 gene were amplified from WT and transgenic lines, after which PCR products were directly sequenced for analysis. The mutated sequences of each GPAT1 gene in the transgenic lines were revealed by aligning the sequences between WT and the transgenic lines. T-DNA insertional line SALK_052352 for GPAT1 was verified using the procedures provided by the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/tdnaprimers.2.html). For complementation, the full-length GPAT1 fragment was amplified from WT Arabidopsis, cloned into the pCambia1300 vector, and then introduced into gpat1 homozygote mutants via the floral dip method. The primers used above are outlined in Supplementary Table S1.
4.5. Cytological Observation
For histological analysis, the first basal node of stems above the rosette leaves were collected from WT, gpat1, gpat1-c1, and complementation transgenic lines and fixed in FAA (75% ethanol:acetic acid:formaldehyde, 90:5:5, v/v/v) for 24 h, dehydrated with a graded series of ethanol (75%, 85%, 90%, 95%, and 100%—50 min each step), followed by an immersion in 100% xylene and embedding in paraffin. Samples were cut into 8-µm thick transverse serial sections with a Leica RM2016 microtome and mounted on glass slides. The sections were stained with toluidine blue for 5 min, washed three times with water, and sealed with resin when dried. Samples were photographed with a Leica DM1000. The images obtained were used for cell length measurement with Image J software.
4.6. Seed TAG and Fatty Acid (FA) Analysis
TAG extraction from Arabidopsis seeds was performed as described previously [51,83]. Approximately 10 mg of dried seeds was used for TAG extraction, and the lipid extracts were evaporated under nitrogen for direct methylation. For transmethylation, methanol containing 5% (w/v) sulfuric acid, 0.01% (w/v) BHT (2,6-Di-tert-butyl-4-methylpheno, Sigma, St. Louis, MO, USA), 20% (v/v) toluene, and TAG-17:0 internal standard (NU-CHEK, Elysian, MN, USA) were added to the combined lipid extracts and incubated for 2 h at 90 °C, followed by addition of 1.5 mL NaCl (0.9% w/v) to quench the transmethylation. The seed TAG was extracted with 1.5 mL of hexane and subjected to gas chromatography analysis. The total TAG content in seeds was calculated as described [51]. FA composition was expressed as a molar percentage.
4.7. RNA-Sequencing and Data Analysis
Total RNA of tender inflorescence stems from WT and gpat1 plants was extracted for RNA-sequencing. cDNA library construction and Illumina sequencing were performed by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). The RNA-seq data were aligned to TAIR10 reference genome using hisat2 [84] with default parameters. The alignments were passed through StringTie [85] for transcript assembly, while RSEM [86] was used to quantify the expression levels of all transcripts, which were normalized with Transcripts Per Million (TPM). We identified significantly differentially expressed genes (DEGs) using the R package DEseq2 [87] with a cut-off of adjusted p-value (FDR) < 0.05 and |log2FC| ≥ 1 between WT and gpat1 mutants. An enrichment analysis was performed to predict the biological processes and KEGG pathways of the DEGs via the online tool Metascape [88].
Abbreviations
| GPAT | glycerol-3-phosphate acyltransferase |
| GA | gibberellin |
| MEP | methylerythritol phosphate |
| GGPP | geranylgeranyl diphosphate |
| CPS | copalyl diphosphate synthase |
| KS | ent-kaurene synthase |
| KO | ent-kaurene oxidase |
| KAO | ent-kaurenoic acid oxidase |
| 2-OGD | 2-oxoglutarate-dependent dioxygenase |
| GID1 | gibberellin insensitive dwarf 1 |
| XTH | xyloglucan endotransglucosylase/hydrolase |
| PME | pectin methylesterase |
| PMEI | pectin methylesterase inhibitor |
| EXP | expansin |
| TAG | triacylglycerol |
| FA | fatty acid |
| GO | gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| DEGs | differentially expressed genes |
| TPM | Transcripts Per Million |
| NPF | nitrate transporter 1/peptide transporter family |
| SWEET | sugar will eventually be exported transporter |
| RGP | reversibly glycosylated protein |
| XYN | endo-1,4-b-xylanase |
| CSL | cellulose synthase like proteins |
| EXT | extensin |
| GUS | β-glucuronidase |
| BHT | 2,6-Di-tert-butyl-4-methylpheno |
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/22/2/785/s1, Figure S1: Analysis of GPAT1 expression in the middle and lower stems of Arabidopsis wild-type plants by GUS staining, Figure S2: Permeability of cuticles of WT and gpat1 mutant to toluidine blue, Figure S3: The effects of disruption of GPAT1 gene on root development, Table S1. Details of the primers used in this article. Table S2. Expression patterns of genes in selected pathways.
Author Contributions
C.L. designed the experiment. Y.B., Y.S., Z.Z., Q.J., M.X., T.Z., H.F., X.Y., L.L., D.L. and X.Q. performed the experiments and statistical analysis. Y.B., Y.S. and Z.Z. wrote the manuscript. Z.C., S.W. and Q.Z. provided the guidance. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the National Science Foundation of China (No. 31701461) to Y.S. and Jiangsu Key Laboratory for the Research and Utilization of Plant Resources (JSPKLB202029) to Y.B.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Akhter M., Sneller C.H. Yield and yield components of early maturing soybean genotypes in the Mid-South. Crop Sci. 1996;36:877–882. doi: 10.2135/cropsci1996.0011183X0036000400010x. [DOI] [Google Scholar]
- 2.Liu F., Wang P., Zhang X., Li X., Yan X., Fu D., Wu G. The genetic and molecular basis of crop height based on a rice model. Planta. 2018;247:1–26. doi: 10.1007/s00425-017-2798-1. [DOI] [PubMed] [Google Scholar]
- 3.Cai G., Yang Q., Chen H., Yang Q., Zhang C., Fan C., Zhou Y. Genetic dissection of plant architecture and yield-related traits in Brassica napus. Sci. Rep. 2016;6:21625. doi: 10.1038/srep21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J., Song Q., Cregan P.B., Nelson R.L., Wang X., Wu J., Jiang G.L. Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genom. 2015;16:217. doi: 10.1186/s12864-015-1441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eshed Y., Lippman Z.B. Revolutions in agriculture chart a coursresee for targeted breeding of old and new crops. Science. 2019;366:eaax0025. doi: 10.1126/science.aax0025. [DOI] [PubMed] [Google Scholar]
- 6.Hedden P. The genes of the Green Revolution. Trends Genet. 2003;19:5–9. doi: 10.1016/S0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 7.Gantait S., Sinniah U.R., Ali M.N., Sahu N.C. Gibberellins-a multifaceted hormone in plant growth regulatory network. Curr. Protein Pept. Sci. 2015;16:406–412. doi: 10.2174/1389203716666150330125439. [DOI] [PubMed] [Google Scholar]
- 8.Hedden P., Sponsel V. A century of gibberellin research. J. Plant Growth Regul. 2015;34:740–760. doi: 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernández-García J., Briones-Moreno A., Blázquez M.A. Origin and evolution of gibberellin signaling and metabolism in plants. Semin. Cell Dev. Biol. 2020;12:S1084-9521. doi: 10.1016/j.semcdb.2020.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Rizza A., Jones A.M. The makings of a gradient: Spatiotemporal distribution of gibberellins in plant development. Curr. Opin. Plant Biol. 2019;47:9–15. doi: 10.1016/j.pbi.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasahara H., Hanada A., Kuzuyama T., Takagi M., Kamiya Y., Yamaguchi S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J. Biol. Chem. 2002;277:45188–45194. doi: 10.1074/jbc.M208659200. [DOI] [PubMed] [Google Scholar]
- 12.Rieu I., Eriksson S., Powers S.J., Gong F., Griffiths J., Woolley L., Benlloch R., Nilsson O., Thomas S.G., Hedden P., et al. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell. 2008;20:2420–2436. doi: 10.1105/tpc.108.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varbanova M., Yamaguchi S., Yang Y., McKelvey K., Hanada A., Borochov R., Yu F., Jikumaru Y., Ross J., Cortes D., et al. Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell. 2007;19:32–45. doi: 10.1105/tpc.106.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., Zhang B., Yan D., Dong W., Yang W., Li Q., Zeng L., Wang J., Wang L., Hicks L.M., et al. Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J. 2011;67:342–353. doi: 10.1111/j.1365-313X.2011.04596.x. [DOI] [PubMed] [Google Scholar]
- 15.Ferrero-Serrano Á., Cantos C., Assmann S.M. The role of dwarfing traits in historical and modern agriculture with a focus on rice. Cold Spring Harb. Perspect. Biol. 2019;11:a034645. doi: 10.1101/cshperspect.a034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Knaap E., Kim J.H., Kende H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000;122:695–704. doi: 10.1104/pp.122.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y., Kende H. Expression of alpha-expansin and expansin-like genes in deepwater rice. Plant Physiol. 2002;130:1396–1405. doi: 10.1104/pp.008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jan A., Yang G., Nakamura H., Ichikawa H., Kitano H., Matsuoka M., Matsumoto H., Komatsu S. Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol. 2004;136:3670–3681. doi: 10.1104/pp.104.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derbyshire P., McCann M.C., Roberts K. Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biol. 2007;7:31. doi: 10.1186/1471-2229-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng W., Li R., Xu Y., Mao R., Chen S., Chen L., Chen L., Liu Y.G., Chen Y. A lipid transfer protein variant with a mutant eight-cysteine motif causes photoperiod- and thermo-sensitive dwarfism in rice. J. Exp. Bot. 2020;71:1294–1305. doi: 10.1093/jxb/erz500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang J.H., Bae E.K., Choi Y.I., Lee O.R. Ginseng-derived patatin-related phospholipase PgpPLAIIIβ alters plant growth and lignification of xylem in hybrid poplars. Plant Sci. 2019;288:110224. doi: 10.1016/j.plantsci.2019.110224. [DOI] [PubMed] [Google Scholar]
- 22.Lu S., Fadlalla T., Tang S., Li L., Ali U., Li Q., Guo L. Genome-wide analysis of phospholipase D gene family and profiling of phospholipids under abiotic stresses in Brassica napus. Plant Cell Physiol. 2019;60:1556–1566. doi: 10.1093/pcp/pcz071. [DOI] [PubMed] [Google Scholar]
- 23.Jayawardhane K.N., Singer S.D., Weselake R.J., Chen G. Plant sn-glycerol-3-phosphate acyltransferases: Biocatalysts involved in the biosynthesis of intracellular and extracellular lipids. Lipids. 2018;53:469–480. doi: 10.1002/lipd.12049. [DOI] [PubMed] [Google Scholar]
- 24.Li-Beisson Y., Shorrosh B., Beisson F., Andersson M.X., Arondel V., Bates P.D., Baud S., Bird D., Debono A., Durrett T.P., et al. Arabidopsis Book. Volume 11. The American Society of Plant Biologists; Rockville, MA, USA: 2013. Acyl-lipid metabolism; p. e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X., Snyder C.L., Truksa M., Shah S., Weselake R.J. sn-Glycerol-3-phosphate acyltransferases in plants. Plant Signal. Behav. 2011;6:1695–1699. doi: 10.4161/psb.6.11.17777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunst L., Browse J., Somerville C. Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. USA. 1988;85:4143–4147. doi: 10.1073/pnas.85.12.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishida I., Tasaka Y., Shiraishi H., Murata N. The gene and the RNA for the precursor to the plastid-located glycerol-3-phosphate acyltransferase of Arabidopsis thaliana. Plant Mol. Biol. 1993;21:267–277. doi: 10.1007/BF00019943. [DOI] [PubMed] [Google Scholar]
- 28.Gidda S.K., Shockey J.M., Rothstein S.J., Dyer J.M., Mullen R.T. Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: Functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol. Biochem. 2009;47:867–879. doi: 10.1016/j.plaphy.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Shockey J., Regmi A., Cotton K., Adhikari N., Browse J., Bates P.D. Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. 2016;170:163–179. doi: 10.1104/pp.15.01563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer S.D., Chen G., Mietkiewska E., Tomasi P., Jayawardhane K., Dyer J.M., Weselake R.J. Arabidopsis GPAT9 contributes to synthesis of intracellular glycerolipids but not surface lipids. J. Exp. Bot. 2016;67:4627–4638. doi: 10.1093/jxb/erw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Z., Xia Q., Dauk M., Shen W., Selvaraj G., Zou J. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell. 2003;15:1872–1887. doi: 10.1105/tpc.012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W., Simpson J.P., Li-Beisson Y., Beisson F., Pollard M., Ohlrogge J.B. A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: Substrate specificity, sn-2 preference, and evolution. Plant Physiol. 2012;160:638–652. doi: 10.1104/pp.112.201996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W., Pollard M., Li-Beisson Y., Beisson F., Feig M., Ohlrogge J. A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proc. Natl. Acad. Sci. USA. 2010;107:12040–12045. doi: 10.1073/pnas.0914149107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berhin A., de Bellis D., Franke R.B., Buono R.A., Nowack M.K., Nawrath C. The root cap cuticle: A cell wall structure for seedling establishment and lateral root formation. Cell. 2019;176:1367–1378.e8. doi: 10.1016/j.cell.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Beisson F., Koo A.J., Molina I., Pollard M., Ohlrogge J. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc. Natl. Acad. Sci. USA. 2007;104:18339–18344. doi: 10.1073/pnas.0706984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X.C., Zhu J., Yang J., Zhang G.R., Xing W.F., Zhang S., Yang Z.N. Glycerol-3-phosphate acyltransferase 6 (GPAT6) is important for tapetum development in Arabidopsis and plays multiple roles in plant fertility. Mol. Plant. 2012;5:131–142. doi: 10.1093/mp/ssr057. [DOI] [PubMed] [Google Scholar]
- 37.Sui N., Tian S., Wang W., Wang M., Fan H. Overexpression of glycerol-3-phosphate acyltransferase from suaeda salsa improves salt tolerance in Arabidopsis. Front. Plant Sci. 2017;8:1337. doi: 10.3389/fpls.2017.01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beisson F., Li Y., Bonaventure G., Pollard M., Ohlrogge J.B. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell. 2007;19:351–368. doi: 10.1105/tpc.106.048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y., Beisson F., Ohlrogge J., Pollard M. Monoacylglycerols are components of root waxes and can be produced in the aerial cuticle by ectopic expression of a suberin-associated acyltransferase. Plant Physiol. 2007;144:1267–1277. doi: 10.1104/pp.107.099432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.David L.C., Berquin P., Kanno Y., Seo M., Daniel-Vedele F., Ferrario-Méry S. N availability modulates the role of NPF3.1, a gibberellin transporter, in GA-mediated phenotypes in Arabidopsis. Planta. 2016;244:1315–1328. doi: 10.1007/s00425-016-2588-1. [DOI] [PubMed] [Google Scholar]
- 41.Tal I., Zhang Y., Jørgensen M.E., Pisanty O., Barbosa I.C., Zourelidou M., Regnault T., Crocoll C., Olsen C.E., Weinstain R., et al. The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 2016;7:11486. doi: 10.1038/ncomms11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishimaru Y., Washiyama K., Oikawa T., Hamamoto S., Uozumi N., Ueda M. Dimerization of GTR1 regulates their plasma membrane localization. Plant Signal. Behav. 2017;12:e1334749. doi: 10.1080/15592324.2017.1334749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jørgensen M.E., Nour-Eldin H.H., Halkier B.A. Transport of defense compounds from source to sink: Lessons learned from glucosinolates. Trends Plant Sci. 2015;20:508–514. doi: 10.1016/j.tplants.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 44.Saito H., Oikawa T., Hamamoto S., Ishimaru Y., Kanamori-Sato M., Sasaki-Sekimoto Y., Utsumi T., Chen J., Kanno Y., Masuda S., et al. The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat. Commun. 2015;6:6095. doi: 10.1038/ncomms7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanno Y., Oikawa T., Chiba Y., Ishimaru Y., Shimizu T., Sano N., Koshiba T., Kamiya Y., Ueda M., Seo M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016;7:13245. doi: 10.1038/ncomms13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davière J.M., Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 47.Szymanski D.B., Cosgrove D.J. Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 2009;19:R800-11. doi: 10.1016/j.cub.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 48.Wolf S., Hématy K., Höfte H. Growth control and cell wall signaling in plants. Annu. Rev. Plant Biol. 2012;63:381–407. doi: 10.1146/annurev-arplant-042811-105449. [DOI] [PubMed] [Google Scholar]
- 49.Bashline L., Lei L., Li S., Gu Y. Cell wall, cytoskeleton, and cell expansion in higher plants. Mol. Plant. 2014;7:586–600. doi: 10.1093/mp/ssu018. [DOI] [PubMed] [Google Scholar]
- 50.Sánchez-Rodríguez C., Rubio-Somoza I., Sibout R., Persson S. Phytohormones and the cell wall in Arabidopsis during seedling growth. Trends Plant Sci. 2010;15:291–301. doi: 10.1016/j.tplants.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Hara A., Radin N.S. Lipid extraction of tissues with a low-toxicity solvent. Anal. Biochem. 1978;90:420–426. doi: 10.1016/0003-2697(78)90046-5. [DOI] [PubMed] [Google Scholar]
- 52.Li Y., Beisson F., Pollard M., Ohlrogge J. Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry. 2006;67:904–915. doi: 10.1016/j.phytochem.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y., Zhao J., Lu W., Deng D. Gibberellin in plant height control: Old player, new story. Plant Cell Rep. 2017;36:391–398. doi: 10.1007/s00299-017-2104-5. [DOI] [PubMed] [Google Scholar]
- 54.Vandermoten S., Haubruge E., Cusson M. New insights into short-chain prenyltransferases: Structural features, evolutionary history and potential for selective inhibition. Cell. Mol. Life Sci. 2009;66:3685–3695. doi: 10.1007/s00018-009-0100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beck G., Coman D., Herren E., Ruiz-Sola M.A., Rodríguez-Concepción M., Gruissem W., Vranová E. Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant Mol. Biol. 2013;82:393–416. doi: 10.1007/s11103-013-0070-z. [DOI] [PubMed] [Google Scholar]
- 56.Fleet C.M., Yamaguchi S., Hanada A., Kawaide H., David C.J., Kamiya Y., Sun T.P. Overexpression of AtCPS and AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol. 2003;132:830–839. doi: 10.1104/pp.103.021725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchum M.G., Yamaguchi S., Hanada A., Kuwahara A., Yoshioka Y., Kato T., Tabata S., Kamiya Y., Sun T.P. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;45:804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- 58.Hu J., Mitchum M.G., Barnaby N., Ayele B.T., Ogawa M., Nam E., Lai W.C., Hanada A., Alonso J.M., Ecker J.R., et al. Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell. 2008;20:320–336. doi: 10.1105/tpc.107.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rieu I., Ruiz-Rivero O., Fernandez-Garcia N., Griffiths J., Powers S.J., Gong F., Linhartova T., Eriksson S., Nilsson O., Thomas S.G., et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008;53:488–504. doi: 10.1111/j.1365-313X.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- 60.Plackett A.R., Powers S.J., Fernandez-Garcia N., Urbanova T., Takebayashi Y., Seo M., Jikumaru Y., Benlloch R., Nilsson O., Ruiz-Rivero O., et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell. 2012;24:941–960. doi: 10.1105/tpc.111.095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.P., et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miedes E., Suslov D., Vandenbussche F., Kenobi K., Ivakov A., Van Der Straeten D., Lorences E.P., Mellerowicz E.J., Verbelen J.P., Vissenberg K. Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. J. Exp. Bot. 2013;64:2481–2497. doi: 10.1093/jxb/ert107. [DOI] [PubMed] [Google Scholar]
- 63.Hara Y., Yokoyama R., Osakabe K., Toki S., Nishitani K. Function of xyloglucan endotransglucosylase/hydrolases in rice. Ann. Bot. 2014;114:1309–1318. doi: 10.1093/aob/mct292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Sandt V.S., Suslov D., Verbelen J.P., Vissenberg K. Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. 2007;100:1467–1473. doi: 10.1093/aob/mcm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu X.F., Shi Y.Z., Lei G.J., Fry S.C., Zhang B.C., Zhou Y.H., Braam J., Jiang T., Xu X.Y., Mao C.Z., et al. XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell. 2012;24:4731–4747. doi: 10.1105/tpc.112.106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macquet A., Ralet M.C., Loudet O., Kronenberger J., Mouille G., Marion-Poll A., North H.M. A naturally occurring mutation in an Arabidopsis accession affects a beta-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell. 2007;19:3990–4006. doi: 10.1105/tpc.107.050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levesque-Tremblay G., Müller K., Mansfield S.D., Haughn G.W. HIGHLY METHYL ESTERIFIED SEEDS is a pectin methyl esterase involved in embryo development. Plant Physiol. 2015;167:725–737. doi: 10.1104/pp.114.255604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rautengarten C., Ebert B., Herter T., Petzold C.J., Ishii T., Mukhopadhyay A., Usadel B., Scheller H.V. The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell. 2011;23:1373–1390. doi: 10.1105/tpc.111.083931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao G.R., Liu J.Y. Isolation of a cotton RGP gene: A homolog of reversibly glycosylated polypeptide highly expressed during fiber development. Biochim. Biophys. Acta. 2002;1574:370–374. doi: 10.1016/S0167-4781(01)00311-6. [DOI] [PubMed] [Google Scholar]
- 70.Simpson D.J., Fincher G.B., Huang A.H.C., Cameron-Mills V. Structure and function of cereal and related higher plant (1→4)-β-xylan endohydrolases. J. Cereal Sci. 2003;37:111–127. doi: 10.1006/jcrs.2002.0488. [DOI] [Google Scholar]
- 71.Derba-Maceluch M., Awano T., Takahashi J., Lucenius J., Ratke C., Kontro I., Busse-Wicher M., Kosik O., Tanaka R., Winzéll A. Suppression of xylan endotransglycosylase PtxtXyn10A affects cellulose microfibril angle in secondary wall in aspen wood. New Phytol. 2015;205:666–681. doi: 10.1111/nph.13099. [DOI] [PubMed] [Google Scholar]
- 72.Speicher T.L., Li P.Z., Wallace I.S. Phosphoregulation of the plant cellulose synthase complex and cellulose synthase-like proteins. Plants. 2018;7:52. doi: 10.3390/plants7030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu H., Zhang R., Tang Y., Peng C., Wu L., Feng S., Chen P., Wang Y., Du X., Peng L. Cotton CSLD3 restores cell elongation and cell wall integrity mainly by enhancing primary cellulose production in the Arabidopsis cesa6 mutant. Plant Mol. Biol. 2019;101:389–401. doi: 10.1007/s11103-019-00910-1. [DOI] [PubMed] [Google Scholar]
- 74.Marowa P., Ding A., Kong Y. Expansins: Roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 2016;35:949–965. doi: 10.1007/s00299-016-1948-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Herger A., Dünser K., Kleine-Vehn J., Ringli C. Leucine-rich repeat extensin proteins and their role in cell wall sensing. Curr. Biol. 2019;29:R851–R858. doi: 10.1016/j.cub.2019.07.039. [DOI] [PubMed] [Google Scholar]
- 76.Šola K., Gilchrist E.J., Ropartz D., Wang L., Feussner I., Mansfield S.D., Ralet M.-C., Haughn G.W. RUBY, a Putative Galactose Oxidase, Influences pectin properties and promotes cell-to-cell adhesion in the seed coat epidermis of Arabidopsis. Plant Cell. 2019;31:809–831. doi: 10.1105/tpc.18.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Irshad M., Canut H., Borderies G., Pont-Lezica R., Jamet E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 2008;8:94. doi: 10.1186/1471-2229-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen Y., Shen L., Shen Z., Jing W., Ge H., Zhao J., Zhang W. The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ. 2015;38:2766–2779. doi: 10.1111/pce.12586. [DOI] [PubMed] [Google Scholar]
- 79.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 80.Jefferson R.A. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Report. 1987;5:387–405. doi: 10.1007/BF02667740. [DOI] [Google Scholar]
- 81.Lei Y., Lu L., Liu H.Y., Li S., Xing F., Chen L.L. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant. 2014;7:1494–1496. doi: 10.1093/mp/ssu044. [DOI] [PubMed] [Google Scholar]
- 82.Li B., Li Y., Liu F., Tan X., Rui Q., Tong Y., Qiao L., Gao R., Li G., Shi R., et al. Overexpressed tomosyn binds syntaxins and blocks secretion during pollen development. Plant Physiol. 2019;181:1114–1126. doi: 10.1104/pp.19.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bai Y., Zhu X., Guo X., Zhang W., Zhang G., Chen H., Zhang Q. Molecular cloning and functional characterization of GmAAPTs from soybean (Glycine max) Plant Signal. Behav. 2020;16:1845048. doi: 10.1080/15592324.2020.1845048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.