Abstract
SARS-CoV-2 is the coronavirus that originated in Wuhan in December 2019 and has spread globally. Studies have shown that smokers are less likely to be diagnosed with or be hospitalized for COVID-19 but, once hospitalized, have higher odds for an adverse outcome. We have previously presented the potential interaction between SARS-CoV-2 Spike glycoprotein and nicotinic acetylcholine receptors (nAChRs), due to a “toxin-like” epitope on the Spike glycoprotein, with homology to a sequence of a snake venom toxin. This epitope coincides with the well-described cryptic epitope for the human anti-SARS-CoV antibody CR3022. In this study, we present the molecular complexes of both SARS-CoV and SARS-CoV-2 Spike glycoproteins, at their open or closed conformations, with the model of the human α7 nAChR. We found that all studied protein complexes' interface involves a large part of the “toxin-like” sequences of SARS-CoV and SARS-CoV-2 Spike glycoproteins and toxin binding site of human α7 nAChR. Our findings provide further support to the hypothesis about the protective role of nicotine and other cholinergic agonists. The potential therapeutic role of CR3022 and other similar monoclonal antibodies with increased affinity for SARS-CoV-2 Spike glycoprotein against the clinical effects originating from the dysregulated cholinergic pathway should be further explored.
Keywords: COVID-19, Cryptic epitope, CR3022, Molecular modeling, nAChRs, SARS-CoV-2, Spike glycoprotein
Abbreviations: aa, Amino Acids; ARDS, Acute Respiratory Distress Syndrome; ACE2, Angiotensin-Converting Enzyme 2; COVID-19, Corona Virus Disease 2019; ECD, Extracellular Domain; LBD, Ligand-binding domain; nAChR, Nicotinic Acetylcholine Receptor; NCS, Nicotinic Cholinergic System; RBD, Receptor Binding Domain; SARS-CoV, Severe Acute Respiratory Syndrome Coronavirus; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2
1. Introduction
Severe acute respiratory syndrome (SARS) is a viral respiratory infection caused by the coronavirus (SARS-CoV) associated with SARS. In February 2003, SARS was first reported in Asia. Before the global SARS outbreak of 2003 was contained, the disease spread to North America, South America, Europe, and Asia. No known SARS cases have been reported anywhere in the world after 2004 (Center for Disease Control and Prevention; Umesh and Anderson, 2004). In April 2003, rumors spread that smoking protected patients from developing SARS, which the authorities rejected (CNN International.com, 2003). Several studies suggested that smokers were under-represented among hospitalized SARS patients, but no firm conclusions about the association between smoking and SARS-CoV infection were drawn (Rainer et al., 2004; Tsui et al., 2003; Tsang et al., 2003; Ong et al., 2004).
While the global pandemic of Corona Virus Disease (2019) (COVID-19), a disease caused by the acute respiratory coronavirus 2 syndrome (SARS-CoV-2), progresses, it is crucial to reveal the pathophysiology and the risk and possibly protective factors associated with disease progression and severity in order to provide effective therapy (Tsatsakis et al., 2020). Published meta-analyses reporting an unusually low pooled prevalence of smoking among hospitalized COVID-19 patients compared with population smoking rates (Farsalinos et al., 2020; Farsalinos et al., 2020a; Farsalinos et al., 2020b) bolstered the debate about the connection between smoking and COVID-19. Other studies report that smokers are less likely to be diagnosed with or be hospitalized for COVID-19 (Farsalinos et al., 2020a; Farsalinos et al., 2020b; Hippisley-Cox et al., 2020; Kowall et al., 2020; Giannouchos et al., 1101; Rossato et al., 2020; Meini et al., 2020; Simons et al., 2020), but, once hospitalized, they have higher odds for an adverse outcome (Farsalinos et al., 2020b; Alqahtani et al., 2020; Vardavas and Nikitara, 2020). Cox et al. recently published a cohort study of 8.28 million participants, including 19,486 confirmed COVID-19 cases, and found that smoking was associated with lower odds for COVID-19 diagnosis and ICU admission (Hippisley-Cox et al., 2020). This unusual correlation of smoking with a potential protective effect for COVID-19 was mainly substantial for heavy and moderate smokers. In contradiction, other studies have shown that smokers, once hospitalized, are at higher risk for adverse outcomes (Farsalinos et al., 2020b).
For the first time in April 2020, we hypothesized that the nicotinic cholinergic system (NCS) might be involved in the pathophysiology of severe COVID-19 and recently built on this hypothesis (Farsalinos et al., 2020b, 2020c). In light of the above considerations, these reports raise the possibility that nicotine -through its action on the NCS- may protect patients against the development of severe COVID-19 that would require hospitalization. Such results should, however, be viewed cautiously before being experimentally verified.
Nicotinic acetylcholine receptors (nAChRs) are membrane proteins, consisting of five spanning-membrane subunits arranged around a central pore. They are divided into the muscle and the neuronal type. Muscle AChRs are located in the skeletal muscles, where they facilitate neuromuscular communication. Neuronal AChRs are found primarily in both the peripheral nervous system (PNS) and the central nervous system (CNS), but also in non-neuronal tissues. Several different nAChRs, consisting of a specific combination of subunits, facilitate discrete cellular physiological functions. Generally, expression of the neuronal form of nAChR differs in several cells of the nervous system. However, neuronal nAChRs are commonly expressed in peripheral ganglia and some brain regions, and in non-excitable cells, such as keratinocytes, epithelial cells, and immune cells (i.e., B cells, T cells, and macrophages). Among the various nAChR subtypes, the α7 receptor, which is widely distributed and overexpressed in the hippocampus, is the most important mediator of the anti-inflammatory properties of the cholinergic system due to its association with humoral and intrinsic immunity (Fujii et al., 2017; Kalamida et al., 2007).
The NCS is an essential pathway that regulates inflammatory response. Its effects on macrophages and other immune cells are mainly controlled by the vagus nerve and by α7 nicotinic acetylcholine receptors (nAChRs) (Tracey, 2002). This so-called “cholinergic anti-inflammatory pathway” has been found to be beneficial in animal models in the prevention of inflammatory conditions such as sepsis and Acute Respiratory Distress Syndrome (ARDS) (Wang et al., 2003). Thus, NCS dysregulation may be a potential cause of the uncontrolled inflammatory response in COVID-19. More clinical manifestations of COVID-19, such as anosmia and thromboembolic complications, could also be explained through its action (Fujii et al., 2017).
The plasma of recovered patients containing neutralizing Abs was a relatively safe and effective treatment choice during the first SARS-CoV and MERS-CoV outbreaks to decrease viral load and minimize mortality in extreme cases (Mair-Jenkins et al., 2015; Ko et al., 2018). Similarly, a limited number of COVID-19 patients treated with a plasma of SARS-CoV-2 convalescent patients have recently shown clinical progress and decreased viral load (Shen et al., 2020); but larger studies have failed to show clinical improvement (Agarwal et al., 2020; Simonovich et al., 2020). Up to now, several human Abs are found to recognize the SARS-CoV-2 S protein (Zost et al., 2020a; Robbiani et al., 2020; Rogers et al., 2020; Brouwer et al., 2020; Cao et al., 2020; Yuan et al., 2020; Li et al., 2020; Andreano et al., 2020; Wec et al., 2020; Pinto et al., 2020; Ju et al., 2020; Wu et al., 2020; Chi et al., 2020; Seydoux et al., 2020; Shi et al., 2020; Barnes et al., 2020), however, cross-neutralizing Abs are rarely found in patients with COVID-19 (Rogers et al., 2020; Brouwer et al., 2020; Ju et al., 2020; Lv et al., 2020). The scientific research emphasis has therefore changed from cross-neutralizing SARS-CoV Abs to isolating new SARS-CoV-2 neutralizing Abs from COVID-19 convalescent patients (Robbiani et al., 2020; Rogers et al., 2020; Ju et al., 2020; Chen et al., 2020a; Zost et al., 2020b).
In the study of Chen et al. (2020a), convalescent patients were clinically fully recovered and tested negatively for SARS-CoV-2 virus RNA with qPCR before blood sampling. Zost and colleagues (Zost et al., 2020b) isolated human Abs from blood samples of a married couple of Wuhan. PBMCs were obtained by leukapheresis 50 days after symptom onset. In Robbiani and colleagues' study (Robbiani et al., 2020), a total of 149 COVID-19-convalescent individuals were examined. All patients participating in the study were characterized asymptomatic for a 14-days period before blood sampling. Patients in this cohort exhibited typical (mild) symptoms that continued for approximately 12 days; however, 11 of the participants, while recovering, needed hospitalization.
In another study, Ju et al. (2020) isolated 206 Receptor Binding Domain (RBD) mAbs from 8 SARS-CoV-2 infected individuals. Serum samples were obtained approximately 20 days after disease initiation. Except for one who succumbed to COVID-19, all other patients fully recovered. Rogers et al. (2020) used a high-throughput pipeline to separate and classify mAbs from convalescent patients. Seventeen donors exhibiting typical COVID-19 symptoms and tested positive by a nasopharyngeal swab for SARS-CoV-2 by PCR were studied. In this cohort, disease severity ranged from mild to severe, with all patients eventually recovered healthy.
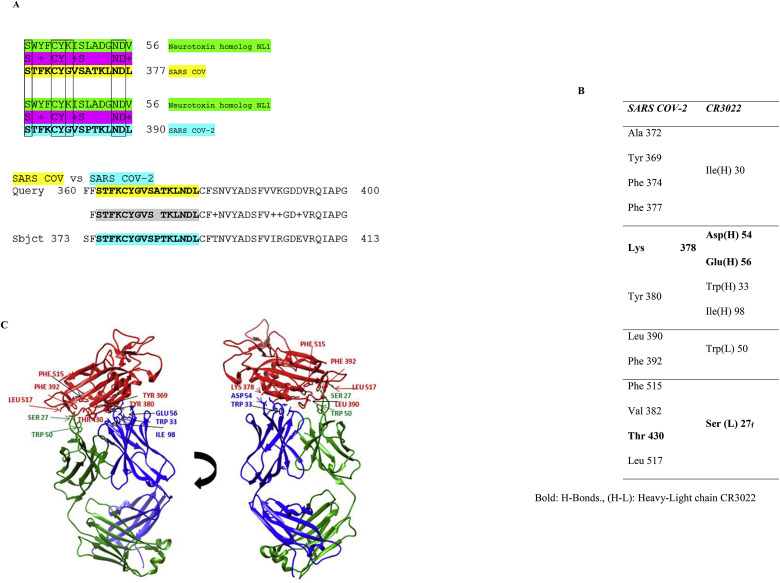
Yuan et al. recently reported the crystal structure of CR3022, a human antibody previously isolated from a convalescent SARS patient, in complex with the RBD of the SARS-CoV-2 Spike (S) glycoprotein (Yuan et al., 2020). The RBD epitope [aa ~333–527 in the SARS-CoV-2 Spike glycoprotein] facilitates the virus's receptor-mediated entry into the host cells. Huo et al. also reported that CR3022 binds to the RBD and presented the Fab/RBD complex's crystal structure at 2.4 Å resolution (Huo et al., 2020). The antibody's neutralizing potential is established for SARS-CoV but not for SARS-CoV-2, although it may exhibit some in vivo protection against the latter. Both groups recognized that the highly conserved and cryptic epitope for CR3022 (Fig. 1 ) is inaccessible in the prefusion Spike. Neither group could explain this epitope's exact function, but both proposed that this cryptic epitope could be therapeutically useful, possibly in synergy with an antibody that blocks angiotensin-converting enzyme-2 (ACE2) attachment. In SARS-CoV and SARS-CoV-2, the amino acids that form this epitope are highly conserved. Of the 28 residues of the epitope buried by CR3022, 24 are conserved between SARS-CoV and SARS-CoV-2. This explains the CR3022's cross-reactivity to SARS-CoV and SARS-CoV-2 RBD's. Despite its high epitope residue conservation, CR3022 Fab binds to SARS-CoV RBD with much greater affinity than SARS-CoV-2 RBD.
Fig. 1.

SARS-COV-2 RBD epitope sequence for the CR3022 mAb. Grey-shadowed amino acids show the CR3022 interacting residue epitopes in the SARS-CoV-2 RBD. (A, Ala; D, Asp; E, Glu; F, Phe; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr).
Structural alignment of the CR3022-SARS-CoV-2 RBD complex with the ACE2-SARS-CoV-2 RBD implies that the binding CR3022 does not interfere with ACE2 (Brouwer et al., 2020) and indicates that the mechanism by which CR3022 neutralizes SARS-CoV is not dependent on direct receptor binding blocking; an interpretation consistent with the finding that CR3022 does not compete with ACE2 for RBD binding (Tian et al., 2019).
Liu et al. recently presented a rare SARS-CoV-2 specific Ab exhibiting cross-neutralization activity against SARS-CoV, called COVA1-16, isolated from a convalescent COVID-19 patient, which binds to the same epitope (i.e., as the CR3022) (Liu et al., 2020). The affinity for SARS-CoV-2 Spike glycoprotein is higher than the affinity for SARS-CoV Spike glycoprotein, as expected. Brouwer and colleagues (Brouwer et al., 2020) initially isolated mAb COVA1-16 by analyzing a total of 403 mAbs from 3 -outpatient and clinically-convalescent COVID-19 patients, aged between 18 and 75 years. All patients were tested positive for SARS-CoV-2. Sample collection was carried out approximately four weeks after the onset of COVID-19 symptoms. Upper respiratory tract infection and mild pneumonia were found in two infected people. At the same time, the third patient participating in the study developed serious pneumonia, and became respiratory inadequate 1.5 weeks after symptom onset, requiring admission to the mechanical ventilation intensive care unit.
Through computational modeling and docking experiments, we have previously identified a key interaction between the SARS-CoV-2 Spike glycoprotein, mainly aa 381–386, and the nAChR alpha subunit (mainly aa 189–192) extracellular domain (ECD). This region forms the core of the nAChRs “toxin-binding site”. Similar to the interaction between the nAChR alpha subunit and α-bungarotoxin, this interaction reinforced the possibility of SARS-CoV-2 interacting with nAChRs (Farsalinos et al., 2020d). Remarkably, other research groups have identified such interaction through different epitopes of SARS-CoV-2 Spike glycoprotein (Oliveira et al., 2020).
This study examined the potential interaction between α7 nAChRs and SARS-CoV and SARS-CoV-2 Spike glycoproteins RBDs. For SARS-CoV-2, our previous study identified such an interaction located at aa 375–390, a homologous region to a sequence of the NL1 homologous neurotoxin, a well-established NCS inhibitor (Tsetlin and Hucho, 2004). Interestingly this epitope is part of the cryptic epitope mentioned above for the human antibody CR3022 and COVA1-16 (Pinto et al., 2020; Huo et al., 2020; Liu et al., 2020). Having presented the 3D structural location of this “toxin-like” sequence on the Spike glycoprotein and the superposition of the modeled structure of the Neurotoxin homolog NL1 and the SARS-CoV-2 Spike glycoprotein previously (Farsalinos et al., 2020d), we are extending our molecular modeling and docking experiments by presenting the complexes of both SARS-CoV and SARS-CoV-2 S glycoproteins with the ECD of the model of human α7 nAChR pentamer, in their “open” and “closed” conformations, ideally adopted by Spike glycoproteins.
2. Methods
2.1. Sequence retrieval and alignment
We compared amino acid sequences between SARS-CoV and SARS-CoV-2 Spike glycoproteins and snake venom neurotoxins. Sequence retrieval of the protein sequences of both virus-related Spike proteins and “three-finger” neurotoxins from various species was performed using the National Center for Biotechnology Information (NCBI, Bethesda, MD, USA) databases. BLAST searches were performed using Mega BLAST (Basic Local Alignment Sea) with the UniProt protein database (UniProt. UniProt- P0) with default parameters. Multiple sequence alignments were performed using the ClustalOmega program (Clustal-O) (Sievers et al., 2011).
2.2. Structure retrieval
The following 3D structures were downloaded from the Protein Data Bank (PDB): 1) PDB id: 6LZG: SARS-CoV-2 Spike glycoprotein (S1) 3D structure in complex with the human angiotensin-converting enzyme 2 (hACE2), 2) PDB id: 6M18: hACE2 (1R41, 1R42) cryo-EM determined complex of spike protein S-ACE2-B0AT1 neutral amino acid transporter, 3) PDB id: 6NB7: structure of a neutralizing to SARS-CoV mAb cross-reacting with complexed ACE2-S protein of SARS-CoV-2, 4) PDB id: 4UY2: ECD of the nAChR α9 subunit complexed with α-bungarotoxin., 5) PDB id:6W41: SARS-CoV-2 RBD complexed with CR3022, and 6) PDB id: 3SQ9: ligand-binding domain (LBD) of a chimera pentameric α7 nAChR.
2.3. Molecular modeling and docking experiments
The protein structure prediction of the ECD of human α7 nAChR was performed using ROSETTA software (Bender et al., 2016), applying an automated multi-step and multi-template homology modeling approach. The complexes between SARS-CoV and SARS-CoV-2 Spike 1 (S1) glycoprotein and ECD of the human pentameric α7 nAChR were modeled using the HADDOCK server (van Zundert and Bonvin, 2014). The predicted interaction surfaces and the ambiguous interaction restraints (AIRs) used to drive the HADDOCK process were automatically generated using the WHISCY software. The input data for the WHISCY prediction of the interaction surface between ECD of human pentameric α7 nAChR and SARS-CoV and SARS-CoV-2 S1 glycoproteins were the conserved residues that were experimentally identified to be involved in the interaction between α7 nAChR chimera and α-bungarotoxin. The binding affinity of biomolecular complexes was predicted using PRODIGY software (Xue et al., 2016). All the protein structures are visualized using UCSF chimera software (Goddard et al., 2005).
3. Results
3.1. Sequence alignment
We scrutinized the amino acid conservation between SARS-CoV-2 and other SARS-related coronavirus sequences in the Spike protein using the simple local alignment search tool BLASTP. The multiple amino acid sequence alignment of the Spike glycoprotein RBD of all SARS-related coronaviruses is shown in Fig. 2 . Phylogenetically, SARS-CoV-2 is closely linked to SARS-CoV, which triggered the human outbreak of 2003. As we can see in Fig. 2A, the RBD epitope is highly conserved between all SARS-related coronaviruses, including the SARS-CoV-2, suggesting a typical evolutionary pattern. Remarkably, the aa 375–395 RBD fragments containing the neurotoxin-like residues of the nAChRs-interacting snakes are among the most conserved regions of almost all CoVs related to SARS. Exceptions are human SARS-CoV OC43, SARS-CoV HKU1, and MERS-CoV, which have a somewhat different composition of RBD amino acids that indicate a distinction from the previously described strains of the viruses, particularly in the aa 375–395 region (aa similarity <50%) (Fig. 2B).
Fig. 2.
(A): Multiple amino acid sequence alignment of spike glycoprotein RBDs of SARS-CoV-2 and other SARS-related coronaviruses. Non-conserved residues among the RBDs of SARS-COV-2 and other SARS-related coronaviruses are shown in a red color font. Yellow highlighted panel indicates the highly conserved region of SARS-CoVs RBD (aa 375–395), bearing the snakes' neurotoxin-like residues, potentially resulting in binding to nAChRs and adversely affecting their function. (B): Sequence alignment of Spike glycoprotein RBDs of SARS-CoV-2 and the other human SARS-related CoVs (i.e., SARS-CoV 0C43, SARS-CoV HKU1, and MERS-CoV). Non-conserved residues are shown in red color. The red frame indicates the aa 375–395 region highly conserved in all other SARS-related CoVs but not to SARS-CoV 0C43, SARS-CoV HKU1, and MERS-CoV. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3A presents the sequence alignment of SARS-CoV and SARS-CoV-2 S1 glycoproteins (A7J8L4, P0DTC2) with Neurotoxin homolog NL1 (Q9DEQ3). We found a double “recombination” within the same S-protein sequence (aa 375–390), which is homologous in the Neurotoxin homolog NL1 sequence, part of the “three-finger” interacting motif of the toxins. This peptide fragment (aa 375–390) is part of the SARS CoV-2 Spikes RBD (aa 333–527) and is located adjacent to the ACE2 Receptor Binding Motif (RBM), through which the Spike glycoprotein recognizes the ACE2 receptor on the host's cell surface. Quite notably, this peptide is the main part of the epitope for the CR3022 antibody, as described before (Yuan et al., 2020; Huo et al., 2020). The main interacting amino acids between the RBD of SARS-CoV and mAb CR3022, as described in the crystal structure of CR3022 and SARS-CoV-2 Spike glycoprotein (Yuan et al., 2020), are shown in Fig. 3B. Molecular models of mAb CR3022 interacting with SARS-CoV are presented in Fig. 3C.
Fig. 3.
(A): Sequence alignment between the SARS-CoV and SARS-CoV-2 Spike glycoproteins and Neurotoxin homolog NL1 depicting amino acids within this sequence identical or functionally equivalent to Neurotoxin homolog NL1 toxin. (B): Amino acid interactions between SARS-CoV-2 RBD and SARS-CoV neutralizing mAb CR3022. (C): Spatial amino acid interactions between SARS-CoV-2 RBD (red) and CR3022 heavy (blue) and light chain (green) through different angles. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Molecular modeling of a toxin-like fragment of SARS-CoV and SARS-CoV-2 RBD
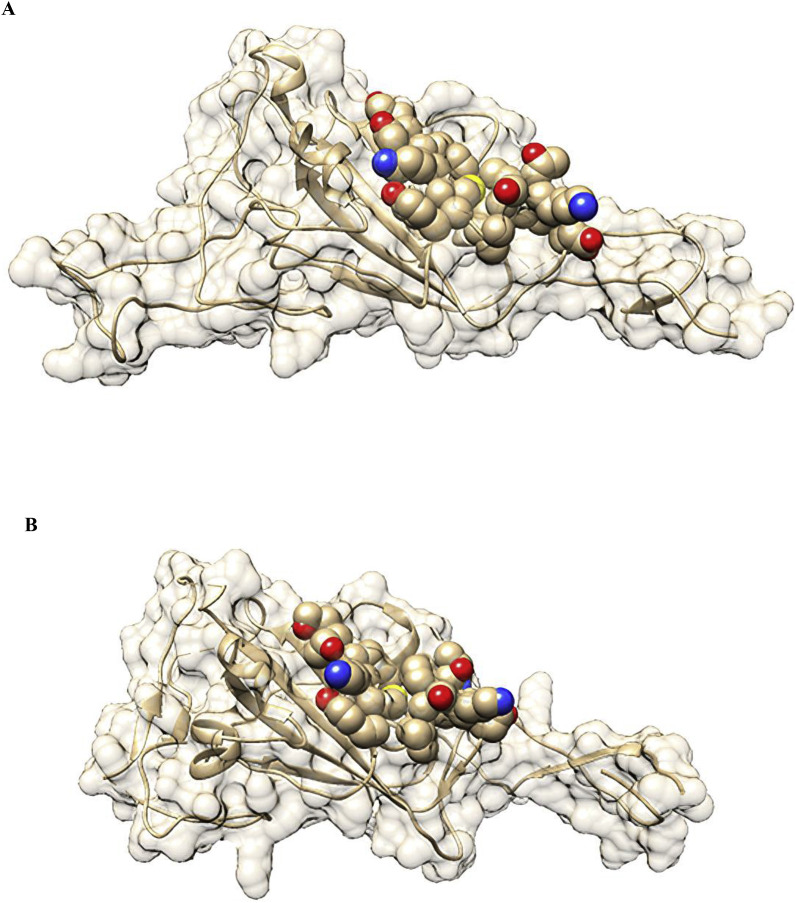
This “toxin-like” fragment on SARS-CoV (aa 362–377) and SARS-CoV-2 (aa 375–390) RBD, containing an amphipathic sequence of alternating polar and hydrophobic amino acid residues with selectively charged amino acids in a conserved order, lies on the spike protein surface and is not buried in the domain core. In ball and stick representation, the toxin-like sequence and its location in the protein surface are illustrated in Fig. 4 . Neighboring the ACE2 binding motif, this entity may interact with the human α7 nAChRs like neurotoxins.
Fig. 4.
Structural location of the toxin-like sequence within the SARS-CoV (A) and SARS-CoV-2 (B) Spike glycoprotein. The toxin-like sequence is illustrated in ball and stick format.
3.3. Interaction of SARS-CoV and SARS-CoV-2 S1 with the ECD of human α7 nAChR
We have previously identified the interaction between the SARS-CoV-2 S1 glycoprotein (aa 381–386) and the α9 subunit of nAChR ECD (aa 189–192), a region that forms the core of the nAChR “toxin-binding site”. The interaction between the two proteins is caused by the complementarity of the hydrogen bonds and shape (Farsalinos et al., 2020d). The interaction model is very similar to the interaction between α9 nAChR and both α-bungarotoxin and the homologous neurotoxin NL1 (snake venom toxins inhibiting nAChRs). Similar interacting surfaces were observed between the SARS-CoV and SARS-CoV-2 S1 and the LBD of the pentameric α7 nAChR chimera.
Herein, the HADDOCK models show that all studied protein complexes' interface involves most of the toxin-like sequences within SARS-CoV S proteins and toxin binding sites of human α7 nAChR. The binding affinity (ΔG, expressed in kcal mol−1), the dissociation constant (K d at 25 °C, expressed in Molar), electrostatic energy (expressed in kcal mol−1), and the buried surface area (expressed in Å2) for all the modeled protein complexes are presented in Table 1 . The dissociation constant of all SARS-α7 nAChR complexes is found to be in the nM range, comparable with experimental supported Kds of well-known enzymatic interacting partners that produce stable protein complexes [i.e., E2-E3 pairs in ubiquitination pathway (Chasapis et al., 2012)].
Table 1.
Haddock parameters of SARS-CoV S1 and SARS-CoV-2 S1 with ECD of human α7 nAChR pentamer.
| SARS-CoV S1 |
SARS-CoV-2 S1 |
|||
|---|---|---|---|---|
| Open | Closed | Open | Closed | |
| with human α7 nAChR | ||||
| ΔG (kcal mol−1) | −8,5 | −10,6 | −10 | −9,4 |
| Kd (Molar) at 25.0 °C | 5.6E-07 | 1.6E-08 | 4.6E-08 | 1.3E-07 |
| HADDOCK score | −117.6 (8.5) | −96.2 (4.4) | −38.9 (12.0) | −46.2 (7.9) |
| Electrostatic Energy (kcal mol−1) | −174.4 (31.4) | −114.6 (28.9) | −198.9 (19.6) | −41.6 (14.5) |
| Buried Surface Area (Å2) | 1777.7 (251.6) | 1845.9 (109.3) | 1561.8 (60.3) | 1677.1 (122.5) |
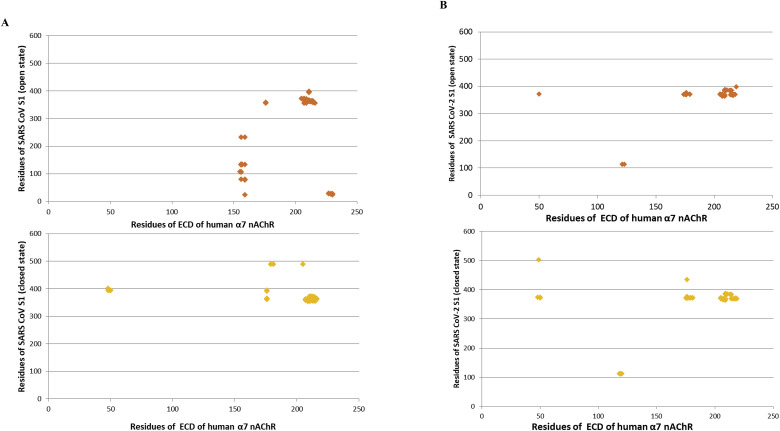
Fig. 5 shows the clusters of intermolecular contacts (ICs) at the interface (within the threshold distance of 5.5 Å) for the complexes between SARS-CoV S1 (A) and SARS-CoV-2 S1 (B) glycoproteins and the LBD of the human α7 nAChR. The main ICs cluster for each interaction involves the regions: 355Phe-372Cys, 365Cys-375Ser and 383Ser-388Cys, and 207Glu-217Tyr of SARS-CoV Spike, SARS-CoV-2 Spike protein, and the LBD of the human α7 nAChR, respectively.
Fig. 5.
The cluster of intermolecular contacts (ICs) at the interface (within the threshold distance of 5.5 Å) for the complexes between SARS-CoV S1 (A) and SARS-CoV-2 S1 (B) glycoproteins in open and closed conformation with the LBD of the human ECD of α7 nAChR.
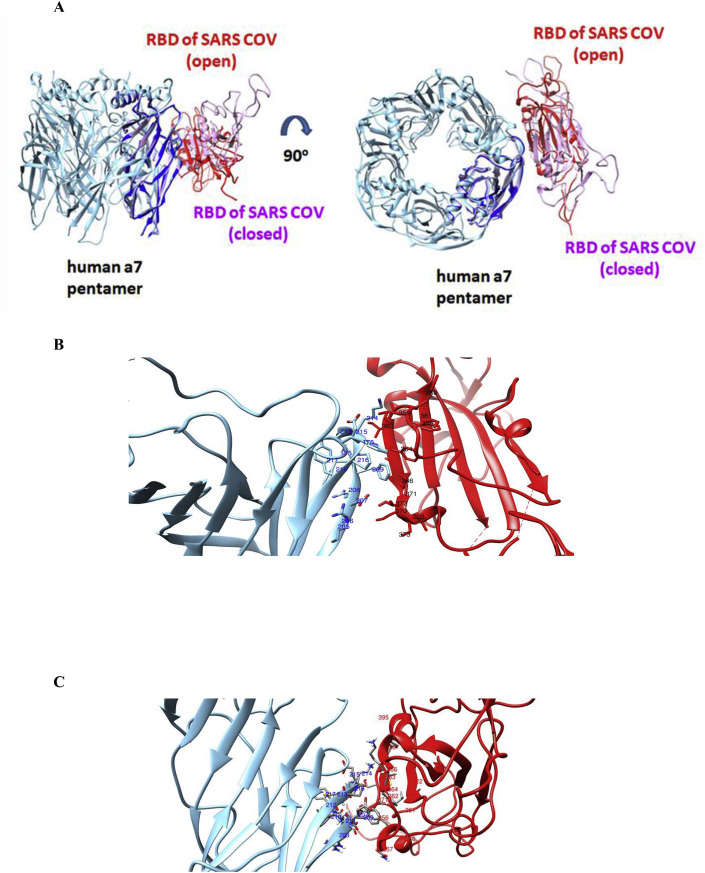
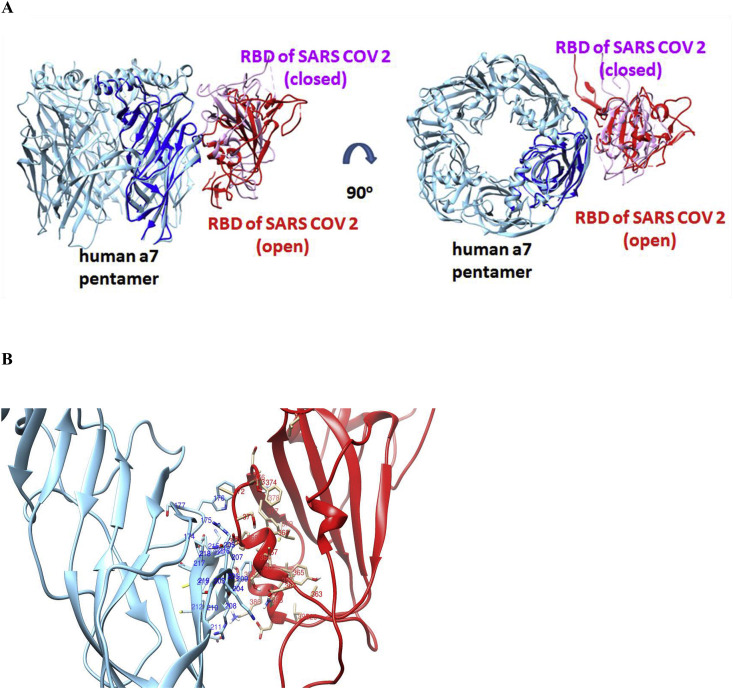
The relative orientation of RBD spike protein in SARS-CoV and LBD of α7 nAChR differs significantly between the open and closed conformation of their complexes (Fig. 6 ). Slightly different orientations of the protein interactions were observed between the open and closed complexes of SARS-CoV-2 Spike with the LBD of the α7 nicotinic receptor (Fig. 7 ). Both open and closed conformation, the SARS CoV-2 RBD orientation when interacting with α7 subunit, is more similar to the closed, rather than the open, conformation of the SARS CoV RBD.
Fig. 6.
(A): HADDOCK complexes of the RBD of SARS-CoV in its open and closed state with one subunit of the pentameric extracellular domain of human α7 nAChR. (B): The interaction interface between the RBD of SARS-CoV in an open state (red color) with the extracellular domain of human α7 nAChR (cyan color). (C): The interaction interface between the RBD of SARS-CoV in a closed state (red color) with the extracellular domain of human α7 nAChR (cyan color). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 7.
(A): HADDOCK complexes of RBDs of SARS-CoV-2 in their open and closed state with one subunit of the pentameric extracellular domain of human α7 nAChR. (B): The interaction interface between the RBD spike of SARS-CoV-2 in the open state (red color) with the extracellular domain of human α7 nAChR (cyan color). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
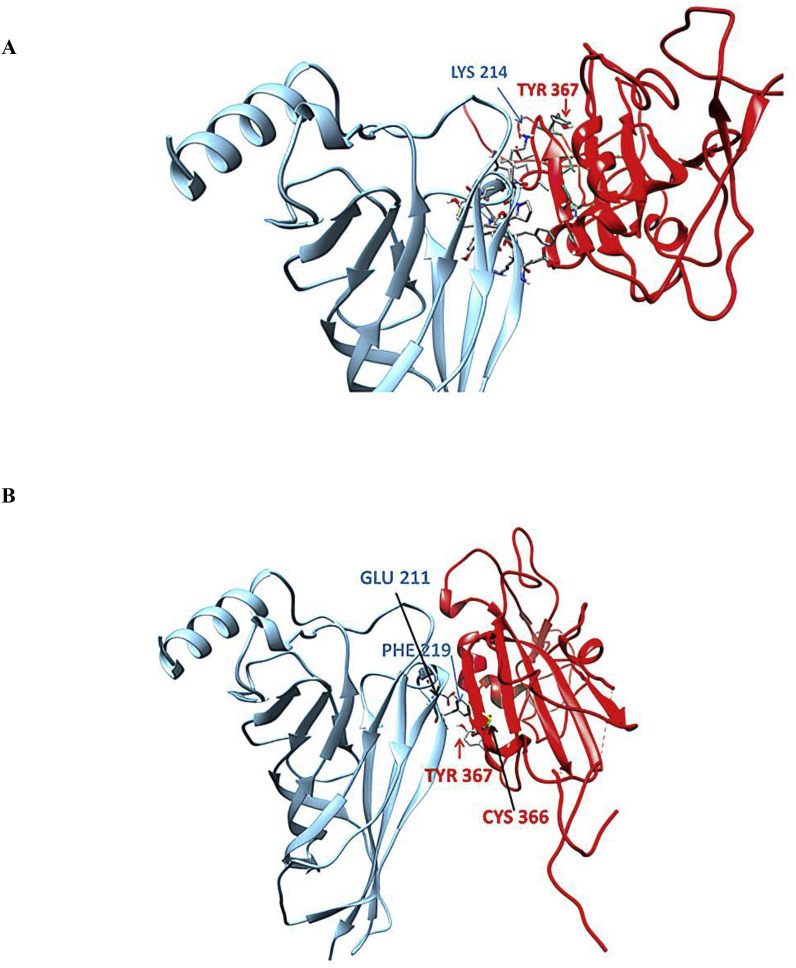
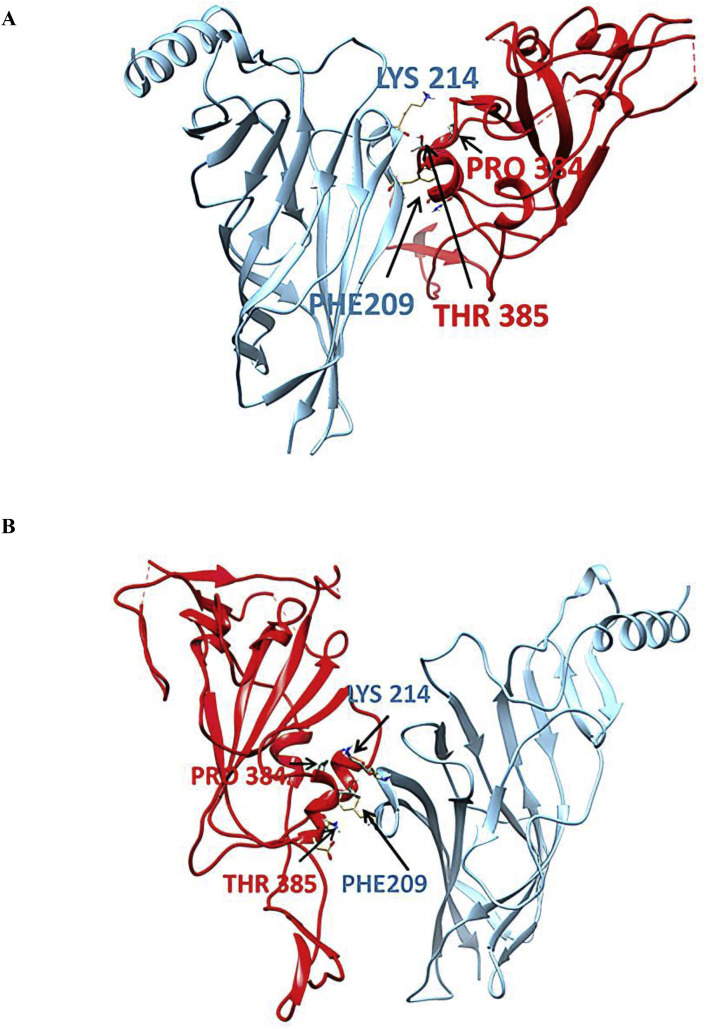
Detailed interacting surfaces of SARS-CoV and SARS-CoV-2 S1 RBDs and α7 nAChRs complexes are illustrated in Fig. 5, Fig. 6B. Two residues conserved in SARS cryptic epitopes (366Cys and 367Tyr) are critical for their interaction with the α7 subunit (Fig. 7). Specifically, SARS CoV-α7 contacts 366Cys - 214Lys and 367Tyr - 214Lys in the closed conformation, and 366Cys - 209Phe and 367Tyr - 211Glu in open conformation were identified (Fig. 8 ). Also, the 384Pro and 385Thr, which are conserved residues in the SARS CoV-2 cryptic epitope, are in close contact with 214Lys and 209 Phe of the α7 subunit (Fig. 9 ).
Fig. 8.
Contacts between 366Cys and 367Tyr of SARS-CoV cryptic epitope with residues of the α7 subunit in closed (A) and open (B) state of their complexes.
Fig. 9.
Contacts between 384Pro and 385Thr of SARS-CoV-2 cryptic epitope with the α7 subunit's residues in closed (A) and open (B) state of their complexes.
4. Discussion
Smoking is a risk factor for respiratory infections and increases pulmonary vulnerability and severity (Burton et al., 2012; Matos et al., 2020; Arcavi and Benowitz, 2004). Therefore, it was reasonably expected that smoking would be associated with COVID-19 susceptibility and severity. Smoking-related comorbidities such as COPD and cardiovascular disease also appear to be risk factors for severe COVID-19 (Leung et al., 2020). Recent studies explored the interaction between smoking (and nicotine) and ACE2 expression, suggesting that smoking up-regulates ACE2 expression, promoting viral cell entry (Leung et al., 2020; Blake et al., 2020a; Cai et al., 2020). Leung et al. studied the ACE2 gene expression levels and concluded that COPD and current smokers had substantially increased ACE2 expression (Leung et al., 2020). This and other studies focused on the concept that up-regulation of smoking-induced ACE2 would promote viral invasion and cell entry, leading to increased susceptibility and severity of COVID-19 (Blake et al., 2020b). However, some studies have strongly questioned that notion. Evidence from in vitro SARS-CoV studies indicates that viral replication causes ACE2 down-regulation, resulting in adverse effects due to unregulated angiotensin II accumulation and activity (Kuba et al., 2005). A similar mechanism may be implicated in SARS-CoV-2 infection (Vaduganathan et al., 2020).
Besides, many case-series of patients in the United States, Europe, and Asia reported a lack of association between the use of ACE-inhibitors (and angiotensin receptor blockers) and COVID-19 diagnosis and adverse outcome (Vaduganathan et al., 2020; Mehta et al., 2020). In one study, ACE-inhibitors were associated with a lower risk of in-hospital death (Reynolds et al., 2020). Also, ACE2 deficiency has been observed with age, diabetes mellitus, and heart disease, which tend to be risk factors for severe COVID-19 (Mehra et al., 2020; Xie et al., 2006; Pal and Bhansali, 2020). In contrast, children and young women have higher levels of ACE2 than older people, yet they are generally experiencing milder disease symptoms (Kassiri et al., 2009). So, it is possible that ACE2 up-regulation may protect against extreme COVID-19 in these cases (Ciaglia et al., 2020).
Until recently, none had examined the possibility of direct interaction between SARS-CoV-2 and the NCS. The nicotinic cholinergic system is an important pathway that regulates the inflammatory response, mainly through the vagus nerve and α7 nAChRs on several immune cells (Tracey, 2002). Experimental studies have shown beneficial effects of stimulating the cholinergic anti-inflammatory pathway in animal models of sepsis and ARDS (Fujii et al., 2017; Wang et al., 2003; Mabley et al., 2011). Alpha7 nAChRs, are also expressed in human bronchial epithelial and endothelial cells (Verdecchia et al., 2020), which are targeted by SARS-CoV-2. Thus, the uncontrolled inflammatory response and other clinical symptoms of COVID-19 (e.g., anosmia and thromboembolic complications) may be caused by NCS dysregulation (Farsalinos et al., 2020b).
Spike protein is a crucial component of SARS-CoV-2, thus an important vaccine-development target protein (Wang et al., 2001). The virus determines host cell receptor binding through a conformational change of the Spike RBD domain (Chen et al., 2020a; Turab Naqvi et al., 2020). The Spike protein's overall structure of SARS-CoV-2 closely resembles the SARS-CoV, while the RBDs of almost all SARS-related coronaviruses display a highly conserved amino acid sequence (i.e., >70%). According to our hypothesis, it seems that SARS-CoV-2 Spike glycoprotein has a “toxin-like” sequence in its RBD that could bind to the nAChRs alpha subunit's toxin-binding domain. This binding may produce various adverse effects by dysregulating the NCS, which involves mostly α7 nAChRs. One of the consequences may be the disruption of the anti-inflammatory cholinergic pathway, leading to cytokine storm and inability to return to homeostasis for the immune response. Cholinergic dysfunction could explain several clinical manifestations of the disease (Kuba et al., 2005). While a positive association between smoking and adverse outcomes among hospitalized COVID-19 patients has been observed, this does not necessarily reject our hypothesis. Smokers experience abrupt nicotine cessation once hospitalized (except in the unlikely scenario that they receive nicotine replacement therapies). This will result in the rapid elimination of plasma nicotine levels within a few hours after hospital admission removing any hypothesized protective effect (Farsalinos et al., 2020a, 2020b). Moreover, some smokers suffer from comorbidities, which may blunt the hypothesized benefits of nicotine. Overall, these findings could have significant therapeutic implications if proven in vivo, as nicotine may partially reverse the virus's binding to host receptors, while other compounds may also compete for binding with SARS-CoV-2 Spike glycoprotein. The idea of the use of medicinal nicotine for the treatment of several pathologies has been previously documented (Chen et al., 2020b; Villafane et al., 2007).
The possible advantages of nicotine in battling negative SARS-CoV-2 symptoms were recently stressed by Kumar et al., through interactions between SARS-CoV-2 and ACE2 (Newhouse et al., 2012). Also, the interactions with the ACE2 receptor of two critical S protein-active sites (i.e., 6LZG, 6VW1), using two alkaloids, nicotine, and caffeine, suggest that the compounds may interact with the S and ACE2 proteins and interfere with their binding by blocking the active sites (Selvaa Kumar et al., 2020). To this end, a double randomized clinical trial on the efficacy of nicotine in COVID-19 prevention in caregivers was launched during the preparation of this manuscript (ClinicalTrials.gov). More in vitro and clinical studies are therefore required to investigate these interactions and nicotinic agonists' effects. Herein we extend our previous findings of interaction between SARS-CoV-2 Spike glycoprotein (aa 381–386) and the nAChR α9 subunit by presenting the complexes of both SARS-CoV and SARS-CoV-2 Spike glycoproteins with the ECD of the model of the human α7 nAChR pentamer, in their “open” and “closed” conformations.
Yan et al. characterized a conserved cryptic epitope on the spike glycoprotein, recognized by the human mAb CR3022, which does not appear to inhibit the binding to the ACE2 protein (Yuan et al., 2020). Similarly, Liu et al. identified and characterized the COVA1-16 mAb, which binds to SARS-CoV-2 Spike glycoprotein to a similar region (Liu et al., 2020). Through antigenic mutation, most RNA viruses can avoid the adaptive immunity of the host (Mohammadi et al., 2020). The error-prone of RNA polymerases enables the formation of sequence variation in each virus cycle replication, allowing the generation of mutant species (Alcami and szinowski, 2000). Many protein sites on the viral surface are nevertheless less prone to mutation (Duffy et al., 2008; Wang et al., 1995; Sahini et al., 2010). Typically, these unmutated sites relate to functions that are necessary for viral infection, such as receptor binding or membrane fusion (Ashkenazi et al., 2013). Quite surprisingly, the cryptic epitope on the SARS-CoV and SARS-CoV-2 Spike glycoproteins is highly conserved and coincides with the toxin-like sequence interacting with nAChR. According to this hypothesis, mutants of CR3022 with higher affinity to SARS-CoV-2 Spike glycoprotein might protect the epitope from interacting with the nAChRs.
5. Conclusions
These in silico findings support our recently published hypothesis that SARS-CoV-2 could interact with nAChRs causing dysregulation of the NCS and the cholinergic anti-inflammatory pathway. Such interactions may lead to uncontrolled immune response and cytokine storm, implicated in severe COVID-19 pathophysiology. Our efforts towards elucidating at atomic-level the potential molecular interactions between SARS-CoV-2 S glycoprotein and human nAChRs could provide a basis for an accelerated vaccine, immunotherapy, and diagnostic strategies against SARS-CoV-2 and other associated beta coronaviruses that could pose a significant global human threat in the future. It is important to emphasize that smoking has well-established detrimental health effects and cannot be recommended as a protective measure for any disease. Nevertheless, our findings suggest that nicotine's potential benefits using approved pharmaceutical products need to be examined.
Funding
No funding was provided for this study.
CRediT authorship contribution statement
George Lagoumintzis: Methodology, Investigation, Software, Reviewing results and data interpretation, Writing-review, and editing, contributed equally to this work. Christos T. Chasapis: Methodology, Investigation, Software, Reviewing results and data interpretation, Writing-review, and editing, contributed equally to this work. Nikolaos Alexandris: Methodology, Investigation, Software, Writing-review, and editing, contributed equally to this work. Dimitrios Kouretas: Methodology, review results and data interpretation, Writing-review, and editing. Socrates Tzartos: Methodology, Writing-review, and editing. Elias Eliopoulos: Methodology, Software, review results and data interpretation, Supervision. Konstantinos Farsalinos: Conceptualization, Writing-review, and editing. Konstantinos Poulas: Writing-review, and editing, Conceptualization, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Socrates J. Tzartos, Elias Eliopoulos, Konstantinos Poulas and Konstantinos Farsalinos are listed as inventors on pending patent application for cholinergic agonists and anti-SPIKE antibodies.
Acknowledgements
The authors are grateful to the “National Research Infrastructures on Integrated Structural Biology, Drug Screening Efforts and Drug Target Functional Characterization (INSPIRED)” for personnel's financial support.
Handling editor: Dr. Jose Luis Domingo
References
- Agarwal A., Mukherjee A., Kumar G., Chatterjee P., Bhatnagar T., Malhotra P., Placid Trial Collaborators Convalescent plasma in the management of moderate covid-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial) BMJ. 2020 Oct 22:371. doi: 10.1136/bmj.m3939. m3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami A., szinowski U.H. Viral mechanisms of immune evasion. Immunol. Today. 2000;21:447–455. doi: 10.1056/NEJMsr2005760. N EnglJ Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqahtani J.S., Oyelade T., Aldhahir A.M., Alghamdi S.M., Almehmadi M., Alqahtani A.S., Quaderi S., Mandal S., Hurst J.R. Prevalence, severity and mortality associated with COPD and smoking in patients with COVID-19: a rapid systematic review and meta-analysis. PloS One. 2020 May 11;15(5) doi: 10.1371/journal.pone.0233147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., et al. Identification of neutralizing human monoclonal antibodies from Italian Covid-19 convalescent patients. bioRxiv. 2020 doi: 10.1101/2020.05.05.078154. [DOI] [Google Scholar]
- Arcavi L., Benowitz N.L. Cigarette smoking and infection. Arch. Intern. Med. 2004 Nov 8;164(20):2206–2216. doi: 10.1001/archinte.164.20.2206. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A., Faingold O., Kaushansky N., Ben-Nun A., Shai Y. A highly conserved sequence associated with the HIV gp41loop region is an immunomodulator of antigen-specific T cells in mice. Blood. 2013;121:2244–2252. doi: 10.1182/blood-2012-11-468900. [DOI] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020 doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basic local alignment search tool. http://www.ncbi.nlm.nih.gov/BLAST/ Available online:
- Bender B.J., Cisneros A., 3rd, Duran A.M., Finn J.A., Fu D., Lokits A.D., Mueller B.K., Sangha A.K., Sauer M.F., Sevy A.M., Sliwoski G., Sheehan J.H., DiMaio F., Meiler J., Moretti R. Protocols for molecular modeling with Rosetta3 and RosettaScripts. Biochemistry. 2016;55(34):4748–4763. doi: 10.1021/acs.biochem.6b00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake S.J., Barnsley K., Lu W., McAlinden K.D., Eapen M.S., Sohal S.S. Smoking up-regulates angiotensin-converting enzyme-2 receptor: a potential adhesion site for novel coronavirus SARS-CoV-2 (Covid-19) J. Clin. Med. 2020;9(3):841. doi: 10.3390/jcm9030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake S.J., Barnsley K., Lu W., McAlinden K.D., Eapen M.S., Sohal S.S. Smoking upregulates angiotensin-converting enzyme-2 receptor: a potential adhesion site for novel coronavirus SARS-CoV-2 (Covid-19) J. Clin. Med. 2020;9(3):841. doi: 10.3390/jcm9030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020 doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Poignard P., Stanfield R.L., Wilson I.A. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Bossé Y., Xiao F., Kheradmand F., Amos C.I. Tobacco smoking increases the lung gene expression of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020 Apr 24 doi: 10.1164/rccm.202003-0693LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Disease Control and Prevention https://www.cdc.gov/sars/index.html
- Chasapis C.T., Kandias N.G., Episkopou V., Bentrop D., Spyroulias G.A. NMR-based insights into the conformational and interaction properties of Arkadia RING-H2E3 Ub ligase. Proteins. 2012;80(5):1484–1489. doi: 10.1002/prot.24048. [DOI] [PubMed] [Google Scholar]
- Chen X., Li R., Pan Z., et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaglia E., Vecchione C., Puca A.A. COVID-19 infection and the predictive ACE2 soluble levels: the favourable protection of children and women. Front. Pediatr. 2020 doi: 10.3389/fped.2020.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CNN International.com. Hong Kong squashes SARS smoking 'cure'. [cited 2003 Apr 20]. Available at: https://edition.cnn.com/2003/WORLD/asiapcf/east/04/18/china.sars.smoking/(accessed on August 14, 2020).
- Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Farsalinos K., Barbouni A., Niaura R. Systematic review of the prevalence of current smoking among hospitalized COVID-19 patients in China: could nicotine be a therapeutic option? Intern Emerg Med. 2020 May 9 doi: 10.1007/s11739-020-02355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsalinos K., Barbouni A., Niaura R. 2020. Smoking, Vaping and Hospitalization for COVID-19. Qeios ID: Z69O8A.11. [DOI] [Google Scholar]
- Farsalinos K., Barbouni A., Poulas K., Polosa R., Caponnetto P., Niaura R. Current smoking, former smoking, and adverse outcome among hospitalized COVID-19 patients: a systematic review and meta-analysis. Therapeutic Advances in Chronic Disease. 2020 doi: 10.1177/2040622320935765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsalinos K., Niaura R., Le Houezec J., Barbouni A., Tsatsakis A., Kouretas D., Vantarakis A., Poulas K. Editorial: nicotine and SARS-CoV-2: COVID-19 may be a disease of the nicotinic cholinergic system. Toxicol Rep. 2020 Apr 30 doi: 10.1016/j.toxrep.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsalinos K., Eliopoulos E., Leonidas D., Papadopoulos G., Tzartos S., Poulas K. Molecular nicotinic cholinergic system and COVID-19: in silico identification of an interaction between SARS-CoV-2 and nicotinic receptors with potential therapeutic targeting implications. Int. J. Mol. Sci. 2020;21(16):5807. doi: 10.3390/ijms21165807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Mashimo M., Moriwaki Y., Misawa H., Ono S., Horiguchi K., Kawashima K. Expression and function of the cholinergic system in immune cells. Front. Immunol. 2017 Sep 6;8:1085. doi: 10.3389/fimmu.2017.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannouchos TV, Sussman RA, Mier JM, Poulas K, Farsalinos K. Characteristics and risk factors for COVID-19 diagnosis and adverse outcomes in Mexico: an analysis of 89,756 laboratory-confirmed COVID-19 cases. Medrxiv. 10.1101/2020.06.04.20122481. [DOI] [PMC free article] [PubMed]
- Goddard T.D., Huang C.C., Ferrin T.E. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13(3):473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Hippisley-Cox J., Young D., Coupland C., et al. 31 July 2020. Risk of Severe COVID-19 Disease with ACE Inhibitors and Angiotensin Receptor Blockers: Cohort Study Including 8.3 Million People Heart. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Zhao Y., et al. Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020 doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kalamida D., Poulas K., Avramopoulou V., Fostieri E., Lagoumintzis G., Lazaridis K., Sideri A., Zouridakis M., Tzartos S.J. Muscle and neuronal nicotinic acetylcholine receptors. Structure, function and pathogenicity. FEBS J. 2007 Aug;274(15):3799–3845. doi: 10.1111/j.1742-4658.2007.05935.x. [DOI] [PubMed] [Google Scholar]
- Kassiri Z., Zhong J., Guo D., Basu R., Wang X., Liu P.P., Scholey J.W., Penninger J.M., Oudit G.Y. Loss of angiotensin-converting enzyme 2 accelerates maladaptive left ventricular remodeling in response to myocardial infarction. Circ Heart Fail. 2009;2:446–455. doi: 10.1161/CIRCHEARTFAILURE.108.840124. [DOI] [PubMed] [Google Scholar]
- Ko Jae-Hoon, Seok Hyeri, Sun Young, Cho Young, Eun Ha, Baek Jin Yang, Kim So Hyun, et al. Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: a single centre experience. Antivir. Ther. 2018;23:617–622. doi: 10.3851/IMP3243. [DOI] [PubMed] [Google Scholar]
- Kowall B., Nonnemacher M., Brune B., Brinkmann M., Dudda M., Böttcher J., Schmidt B., Standl F., Stolpe S., Dittmer U., Jöckel K.H., Stanga A. A model to identify individuals with a high probability of a SARS-CoV-2 infection. J. Infect. 2020 doi: 10.1016/j.jinf.2020.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J.M., Yang C.X., Tam A., Shaipanich T., Hackett T.L., Singhera G.K., Dorscheid D.R., Sin D.D. ACE2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur. Respir. J. 2020 Apr 8:2000688. doi: 10.1183/13993003.00688-2020. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Drelich A., Martinez D.R., Gralinski L., Chen C., Sun Z., et al. Potent neutralization of SARS-CoV-2 in vitro and in an animal model by a human monoclonal antibody. bioRxiv. 2020 doi: 10.1101/2020.05.13.093088. [DOI] [Google Scholar]
- Liu H., Wu N.C., Yuan M., Bangaru S., Torres J.L., Caniels T.G., van Schooten J., Zhu X., Lee C.D., Brouwer P.J.M., van Gils M.J., Sanders R.W., Ward A.B., Wilson I.A. Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity. 2020 Nov 21;S1074–7613(20):30464–30467. doi: 10.1016/j.immuni.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H., Wu N.C., Tsang O.T., Yuan M., Perera R., Leung W.S., So R., Chan J., Yip G.K., Chik T., Wang Y., Choi C., Lin Y., Ng W.W., Zhao J., Poon L., Peiris J., Wilson I.A., Mok C. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31(9):107725. doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabley J., Gordon S., Pacher P. Nicotine exerts an anti-inflammatory effect in a murine model of acute lung injury. Inflammation. 2011;34(4):231–237. doi: 10.1007/s10753-010-9228-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair-Jenkins John, Saavedra-Campos Maria, Kenneth Baillie J., Paul Cleary, Khaw Fu-Meng, Lim Wei Shen, et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 1 January 2015;211(Issue 1):80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos C.P., Boléo-Tomé J.P., Rosa P., Morais A. Pulmonology, Advance online publication; 2020. Tobacco and COVID-19: A position from Sociedade Portuguesa de Pneumologia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra M.R., Desai S.S., Kuy S., Henry T.D., Patel A.N. Cardiovascular disease, Drug therapy, and mortality in covid-19. N. Engl. J. Med. 2020 May 1 doi: 10.1056/NEJMoa2007621. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mehta N., Kalra A., Nowacki A.S., Anjewierden S., Han Z., Bhat P., Carmona-Rubio A.E., Jacob M., Procop G.W., Harrington S., Milinovich A., Svensson L.G., Jehi L., Young J.B., Chung M.K. Association of use of angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers with testing positive for coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020 May 5 doi: 10.1001/jamacardio.2020.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meini S., Fortini A., Andreini R., Sechi L.A., Tascini C. The paradox of the low prevalence of current smokers among covid-19 patients hospitalized in non-intensive care wards: results from an Italian multicenter case-control study. Nicotine Tob. Res. 2020 Sep 23:ntaa188. doi: 10.1093/ntr/ntaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi S., Heidarizadeh M., Entesari M., Esmailpour A., Esmailpour M., Moradi R., Sakhaee N., Doustkhah E. In silico investigation on the inhibiting role of nicotine/caffeine by blocking the S protein of SARS-CoV-2 versus ACE2 receptor. Microorganisms. 2020;8:160. doi: 10.3390/microorganisms8101600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhouse P., Kellar K., Aisen P., White H., Wesnes K., Coderre E., et al. Nicotine treatment of mild cognitive impairment: a 6-month double-blind pilot. clinical trial Neurology. 2012;78(2):91–101. doi: 10.1212/WNL.0b013e31823efcbb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A., Ibarra A.A., Bermudez I., Casalino L., Gaieb Z., Shoemark D.K., Gallagher T., Sessions R.B., Amaro R.E., Mulholland A.J. Simulations support the interaction of the SARS-CoV-2 spike protein with nicotinic acetylcholine receptors and suggest subtype specificity. bioRxiv. 2020:206680. doi: 10.1101/2020.07.16.206680. 2020.7.16. [DOI] [Google Scholar]
- Ong K.-C., Ng A.W.-K., Lee L.S.-U., Kaw G., Kwek S.-K., Leow M.K.S., Earnest A. Pulmonary function and exercise capacity in survivors of severe acute respiratory syndrome. Eur. Respir. J. Suppl. 2004;24(3):436–442. doi: 10.1183/09031936.04.00007104. [DOI] [PubMed] [Google Scholar]
- Pal R., Bhansali A. COVID-19, diabetes mellitus and ACE2: the conundrum. Diabetes Res. Clin. Pract. 2020 doi: 10.1016/j.diabres.2020.108132:108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020 doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Rainer T., Smit D., Cameron P. Smoking and severe acute respiratory syndrome. Hong Kong J. Emerg. Med. 2004;11(3):143–145. doi: 10.1177/102490790401100303. [DOI] [Google Scholar]
- Reynolds H.R., Adhikari S., Pulgarin C., Troxel A.B., Iturrate E., Johnson S.B., Hausvater A., Newman J.D., Berger J.S., Bangalore S., Katz S.D., Fishman G.I., Kunichoff D., Chen Y., Ogedegbe G., Hochman J.S. Renin-angiotensin-aldosterone system inhibitors and risk of covid-19. N. Engl. J. Med. 2020 May 1 doi: 10.1056/NEJMoa2008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W-t, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020 Aug 21;369(6506):956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossato M., Russo L., Mazzocut S., Di Vincenzo A., Fioretto P., Vettor R. Current smoking is not associated with COVID-19. Eur. Respir. J. 2020 Jun;55(6):2010. doi: 10.1183/13993003.01290-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahini L., Tempczyk-Russell A., Agarwal R. Large-scale sequence analysis of hemagglutinin of influenza A virus identifies conserved regions suitable for targeting an anti-viral response. PloS One. 2010;5:e9268. doi: 10.1371/journal.pone.0009268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaa Kumar C., Senthil Arun Kumar, Haiyan Wei, Comparative docking studies to understand the binding affinity of nicotine with soluble ACE2 (sACE2)-SARS-CoV-2 complex over sACE2, Toxicology Reports, Volume 7, 2020, Pages 1366-1372. [DOI] [PMC free article] [PubMed]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity. 2020;53:98–105. doi: 10.1101/2020.05.12.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. J. Am. Med. Assoc. 2020 Apr 28;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovich V.A., Burgos Pratx L.D., Scibona P., Beruto M.V., Vallone M.G., Vázquez C., Savoy N., Giunta D.H., Pérez L.G., Sánchez M.D.L., Gamarnik A.V., Ojeda D.S., Santoro D.M., Camino P.J., Antelo S., Rainero K., Vidiella G.P., Miyazaki E.A., Cornistein W., Trabadelo O.A., Ross F.M., Spotti M., Funtowicz G., Scordo W.E., Losso M.H., Ferniot I., Pardo P.E., Rodriguez E., Rucci P., Pasquali J., Fuentes N.A., Esperatti M., Speroni G.A., Nannini E.C., Matteaccio A., Michelangelo H.G., Follmann D., Lane H.C., Belloso W.H., PlasmAr Study Group A randomized trial of convalescent plasma in covid-19 severe pneumonia. N. Engl. J. Med. 2020 Nov 24 doi: 10.1056/NEJMoa2031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons D., Shahab L., Brown J., Perski O. The association of smoking status with SARS-CoV-2 infection, hospitalization and mortality from COVID-19: a living rapid evidence review with Bayesian meta-analyses (version 7) Addiction. 2020 Oct 2 doi: 10.1111/add.15276. 10.1111/add.15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- X. Tian, C. Li, A. Huang, S. Xia, S. Lu, Z. Shi, L. Lu, S. Jiang, Z. Yang, Y. Wu, T. Ying, Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody Emerging Microb. Infect., 9:1, 382-385, DOI: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed]
- Tracey K.J. The inflammatory reflex. Nature. 2002;420:853–859. doi: 10.1038/nature01321. [DOI] [PubMed] [Google Scholar]
- Tsang K., Ho P., Ooi G., et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- Tsatsakis A., Calina D., Falzone L., Petrakis D., Mitrut R., Siokas V., Pennisi M., Lanza G., Libra M., Doukas S.G., Doukas P.G., Kavali L., Bukhari A., Gadiparthi C., Vageli D.P., Kofteridis D.P., Spandidos D.A., Paoliello M.M.B., Aschner M., Docea A.O. SARS-CoV-2 pathophysiology and its clinical implications: an integrative overview of the pharmacotherapeutic management of COVID-19. Food Chem. Toxicol. 2020 Dec;146:111769. doi: 10.1016/j.fct.2020.111769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetlin V.I., Hucho F. Snake and snail toxins acting on nicotinic acetylcholine receptors: fundamental aspects and medical applications. FEBS Lett. 2004 Jan 16;557(1–3):9–13. doi: 10.1016/s0014-5793(03)01454-6. [DOI] [PubMed] [Google Scholar]
- Tsui P.T., Kwok M.L., Yuen H., Lai S.T. Severe acute respiratory syndrome: clinical outcome and prognostic correlates. Emerg. Infect. Dis. 2003;9(9):1064–1069. doi: 10.3201/eid0909.030362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turab Naqvi Ahmad Abu, Fatima Kisa, Mohammad Taj, Fatima Urooj, Singh Indrakant K., Singh Archana, Muhammad Atif Shaikh, Hariprasad Gururao, Mustafa Hasan Gulam, Imtaiyaz Hassan Md. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866(10):165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesh D Parashar, Anderson Larry J. Severe acute respiratory syndrome: review and lessons of the 2003 outbreak. Int. J. Epidemiol. August 2004;33(4):628–634. doi: 10.1093/ije/dyh198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt. UniProtKB - P0DTC2 (SPIKE_SARS2).
- Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin-angiotensin-aldosterone system inhibitors in patients with covid-19. N. Engl. J. Med. 2020;382(17):1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zundert Gydo C.P., Bonvin Alexandre M.J. J. Modeling protein-protein complexes using the HADDOCK webserver "modeling protein complexes with HADDOCK. Methods Mol. Biol. 01/2014;1137:163–179. doi: 10.1007/978-1-4939-0366-5_12. [DOI] [PubMed] [Google Scholar]
- Vardavas C.I., Nikitara K. COVID-19 and smoking: a systematic review of the evidence. Tob. Induc. Dis. 2020 Mar 20;18:20. doi: 10.18332/tid/119324.Ko. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020 Apr 20;(20):30151–30155. doi: 10.1016/j.ejim.2020.04.037. pii: S0953-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villafane G., Cesaro P., Rialland A., Baloul S., Azimi S., Bourdet C., et al. Chronic high dose transdermal nicotine in Parkinson's disease: an open trial. Eur. J. Neurol. 2007;14(12):1313–1316. doi: 10.1111/j.1468-1331.2007.01949.x. [DOI] [PubMed] [Google Scholar]
- Wang L., Parr R.L., King D.J., Collisson E.W. A highly conserved epitope on the spike protein of infectious bronchitis virus. Arch. Virol. 1995;140:2201–2213. doi: 10.1007/BF01323240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Pereira E.F., Maus A.D., Ostlie N.S., Navaneetham D., Lei S., Albuquerque E.X., Conti-Fine B.M. Human bronchial epithelial and endothelial cells express alpha7 nicotinic acetylcholine receptors. Mol. Pharmacol. 2001 Dec;60(6):1201–1209. doi: 10.1124/mol.60.6.1201. [DOI] [PubMed] [Google Scholar]
- Wang H., Yu M., Ochani M., Amella C.A., Tanovic M., Susarla S., Li J.H., Wang H., Yang H., Ulloa L., Al-Abed Y., Czura C.J., Tracey K.J. Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature. 2003 Jan 23;421(6921):384–388. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D., Haslwanter D., Sakharkar M., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020 doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Chen J., Wang X., Zhang F., Liu Y. Age- and gender-related difference of ACE2 expression in rat lung. Life Sci. 2006;78:2166–2171. doi: 10.1016/j.lfs.2005.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Li C., Rodrigues João Pglm, Kastritis Panagiotis L. Alexandre Mjj Bonvin, Anna Vangone, PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics. 1 December 2016;32(Issue 23):3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., et al. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020 doi: 10.1101/2020.05.12.091462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]