Abstract
Introduction
The COVID-19 pandemic caused by SARS-CoV-2 places immense worldwide demand on healthcare services. Earlier identification of patients at risk of severe disease may allow intervention with experimental targeted treatments, mitigating the course of their disease and reducing critical care service demand.
Methods and analysis
This prospective observational study of patients tested or treated for SARS-CoV-2, who are under the care of the tertiary University Hospital Southampton NHS Foundation Trust (UHSFT), captured data from admission to discharge; data collection commenced on 7 March 2020. Core demographic and clinical information, as well as results of disease-defining characteristics, was captured and recorded electronically from hospital clinical record systems at the point of testing. Manual data were collected and recorded by the clinical research team for assessments which are not part of the structured electronic healthcare record, for example, symptom onset date. Thereafter, participant records were continuously updated during hospital stay and their follow-up period. Participants aged >16 years were given the opportunity to provide consent for excess clinical sample storage with optional further biological sampling. These anonymised samples were linked to the clinical data in the Real-time Analytics for Clinical Trials platform and were stored within a biorepository at UHSFT.
Ethics and dissemination
Ethical approval was obtained from the HRA Specific Review Board (REC 20/HRA/2986) for waiver of informed consent for the database-only cohort; the procedures conform with the Declaration of Helsinki. The study design, protocol and patient-facing documentation for the biobanking arm of the study have been approved by North West Research Ethics Committee (REC 17/NW/0632) as an amendment to the National Institute for Health Research Southampton Clinical Research Facility-managed Southampton Research Biorepository. This study will be published as peer-reviewed articles and presented at conferences, presentations and workshops.
Keywords: COVID-19, health informatics, protocols & guidelines, chronic airways disease, respiratory infections, respiratory medicine (see thoracic medicine)
Strengths and limitations of this study.
Close alignment of research and clinical practice in a near real-time manner.
Longitudinal data collection and sampling opportunities.
A single-centre study with data collection reflective of clinical need rather than a strict protocolised time frame.
Use of novel artificial intelligence techniques for data analysis.
Introduction
The outbreak of COVID-19 caused by the novel SARS-CoV-2 was declared a pandemic by the WHO on 11 March 2020.1 In the UK, 1 629 657 people have tested positive for SARS-CoV-2, of which 66 713 have died (https://www.gov.uk/guidance/coronavirus-covid-19-information-for-the-public; correct as of 17:00, 30 November 2020).2 So far, emerging effective treatments such as remdesivir and dexamethasone have only modest effects on clinical outcomes in a subcohort of patients,3 4 and any vaccines have yet to demonstrate their efficacy. While many patients recover from SARS-CoV-2 infection without need for hospitalisation, a small proportion go on to develop severe disease. The demand on healthcare services, especially critical care, is immense. Earlier identification of those patients at risk of severe disease may allow intervention with experimental targeted treatments mitigating the course of their disease and reducing demand on critical care services.
The need for rapid translation of treatments from development to clinical application is being addressed by urgent public health studies such as ACCORD-25 and RECOVERY trials.6 Despite the urgency to enrol patients in these studies, there are challenges in identifying suitable patients in a fluid, ‘real-time’ clinical environment within a novel disease. The longitudinal Research Evaluation Alongside Clinical Treatment in COVID-19 (REACT COVID-19) database and biosampling study captures the natural history of SARS-CoV-2 infection and its clinical response to current interventions, as well as characterises specific phenotypes and endotypes, with the potential to rapidly identify patients for novel treatment trials.
The Research Evaluation Alongside Clinical Treatment observational and biobanking study of COVID-19 (REACT COVID-19 study) evolved around a collaboration between the University of Southampton (UoS), University Hospital Southampton NHS Foundation Trust (UHSFT) and the digital Experimental Cancer Medicine Team (ECMT) and their work to create the REACT platform. The REACT platform is used for the determination of early clinical benefit of new cancer medicines in early-phase drug trials and has been adapted by the digital ECMT in collaboration with UHSFT to allow rapid upload and interpretation of clinical data in the context of the COVID-19 pandemic. This study is supported by the National Institute for Health Research (NIHR) Southampton Clinical Research Facility (CRF) and NIHR Southampton Biomedical Research Centre (BRC), which takes advantage of the expertise and resource in hosting large prospective cohort studies.
Objectives
Development of a real-time ‘parent’ database of well-characterised patients with COVID-19 to facilitate a better understanding of the natural history of SARS-CoV-2 infection from prehospital through to level 3 care and subsequent course of COVID-19-related complications from discharge through to clinic follow-up.
Cross-sectional phenotypical characterisation of a cohort of patients with COVID-19 to describe the disease heterogeneity seen in clinical practice.
Identify potential risk factors for progression to severe disease measured by admission to level 2 or 3 care, increase in respiratory support or death (primary endpoint).
Use of samples stored in the biorepository (eg, blood, urine and sputum) to develop an endotype-level understanding of disease clusters.
Assess the efficacy of current best practice management strategies.
Identify subgroups of patients for novel treatment strategies in the form of independent formal randomised control trials.
Methods and analysis
Study design
The REACT COVID-19 study is a prospective observational study of patients tested or treated for SARS-CoV-2 infection who are under the care of UHSFT. The study is planned as a long-term research project with no limit to enrolment nor any time-defined end, with data collection commencing on 7 March 2020 with admission of the first SARS-CoV-2 patient to University Hospital Southampton (UHS). Patients are recruited into the study at the point of testing for SARS-CoV-2 at UHSFT. Core demographic and clinical information, as well as results of disease-defining characteristics, are captured and recorded electronically from hospital clinical record systems at the point of testing. Manual data are collected and recorded by the clinical research team for assessments which are not part of the electronic healthcare record, for example, symptom onset date. Thereafter, participants’ records are continuously updated during their hospital stay and for 12 months following discharge from acute admission, in their follow-up period under UHSFT. This pragmatic, opportunistic approach to data collection takes full advantage of the electronic healthcare records and seamlessly links with the REACT platform to reduce data collection burden on the patient, clinician or researcher. In addition, participants over the age of 16 years old are given the opportunity to provide consent for storage of excess sample taken as part of their routine clinical management with optional further biological sampling. These anonymised samples are linked to the anonymised clinical data in the REACT platform and stored within the Southampton Research Biorepository (SRB), managed by the NIHR CRF at UHSFT.
Patient and public involvement
Patient representatives were involved in and were part of the governance structure of the SRB, and patient representatives were involved in the design and ongoing management of the WATCH study, on which much of this observational biobank study was based. The SRB Oversight Committee has a lay representative to work alongside other members and may jointly be consulted in the different matters related to the construction and running of the biorepository.
Setting
UHSFT is an 1100-bed tertiary centre and, as such, has assessed and admitted a large number of SARS-CoV-2-positive patients.
Recruitment
All patients under the care of UHSFT who are tested or treated for SARS-CoV-2 are included in the REACT COVID-19 study. A subcohort of patients aged ≥16 years old are given the opportunity to provide consent for storage of excess sample taken as part of their routine clinical management with optional further biological sampling. They are offered the opportunity to provide this at any point during their care under UHSFT. So far, UHSFT has admitted 629 patients with confirmed COVID-19, correct as of 17 June 2020. Study enrolment will be continual into the future, aligned to the ongoing function of the clinical service with no discrete recruitment target or endpoint.
Data collection
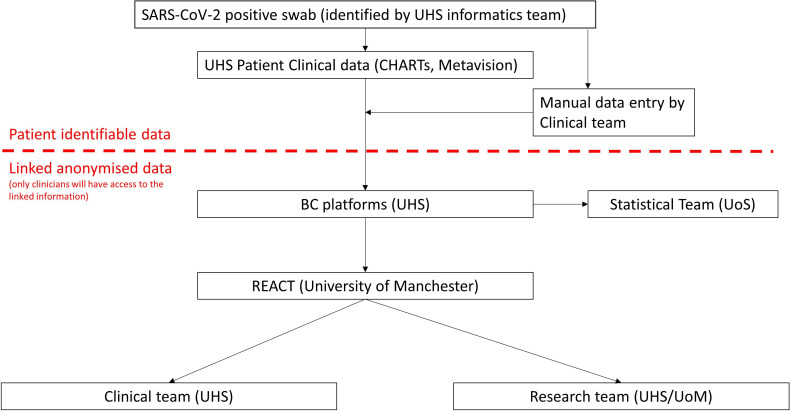
This study is divided into two parts and is conducted in conjunction with standard clinical care. The database component involves the assimilation of data collected as part of routine clinical care for patients of any age tested for or admitted to UHSFT for suspected SARS-CoV-2 infection. Core demographic and clinical data collected as part of standard clinical care will be extracted from the electronic hospital records. Where required, additional data relevant to COVID-19 clinical course is manually extracted from the clinical notes and uploaded to the database by clinical researchers where automatic electronic export is not possible. Data collected from study participants is collated in the highly secure, contemporary and encrypted data platform, BC|INSIGHT (Microsoft Data Centre, UK). Data are then uploaded to the REACT platform to allow rapid capture of the natural history of the disease, with clinicians able to visualise trends in patient trajectories and to identify patients who may be suitable for approach for intervention studies (summarised in figure 1.).
Figure 1.
Data flow between systems and institutions within the REACT COVID-19 database. CRUK MI, Cancer Research UK, Manchester Institute; ECMT, Experimental Cancer Medicine Team; REACT, Real-time Analytics for Clinical Trials; UHS, University Hospital Southampton; UoS, University of Southamptom.
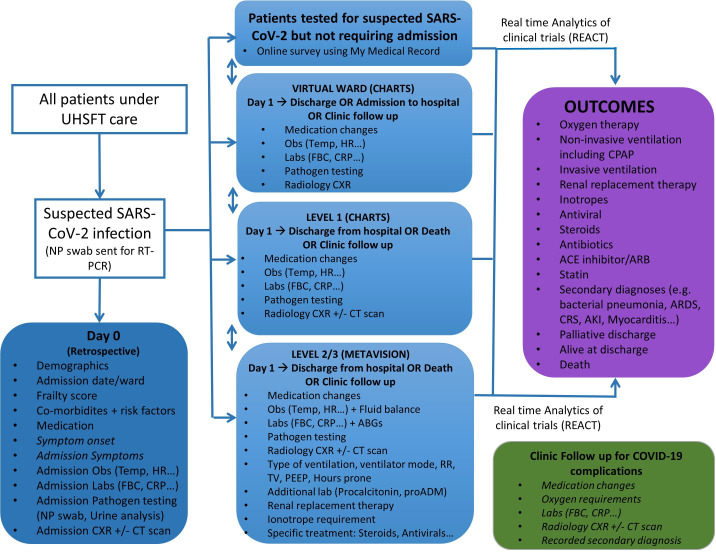
Data captured at point of testing for SARS-CoV-2 includes core demographic and clinical data relevant to COVID-19. Subsequent data capture follows clinical disease course and includes information on medication, vital signs, and pathological and radiological measurements. These data are captured in real time during the follow-up period under UHSFT care, including any period of hospitalisation, and for 12 months after admission to capture virtual follow-up or outpatient clinic follow-up data (summarised in figure 2). Any treatment interventions as part of routine clinical care are also captured (where possible electronically), and these form part of the visualisations in the REACT platform. Data for research purposes will be stored in an anonymised format.
Figure 2.
Data capture for all patients to be included in the database. Data variables will be recorded as part of routine clinical care and will be captured as part of the observational study. Data variables listed in italics will be manually captured and added to the database. CHARTS is an integrated user interface for electronic healthcare records. MetaVision is a clinical information system for critical care. ABG, arterial blood gas; AKI, acute kidney injury; ARB, angiotensin receptor blocker; ARDS, acute respiratory distress syndrome; CPAP, continuous positive airways pressure; CRP, C reactive protein; CRS, cytokine release syndrome; CXR, chest X-ray; HR, heart rate; FBC, full blood count; Obs, clinical observations; NP, nasopharyngeal; PEEP, positive end expiratory pressure; proADM, proadrenomedullin; RT-PCR, reverse transcriptase PCR; RR, respiratory rate; Temp, temperature; TV, tidal volume; UHSFT, University Hospital Southampton NHS Foundation Trust.
The majority of data collected is consistent with the data already being collected as part of routine clinical care of patients with COVID-19, therefore seamlessly linking with clinical practice. Clinical data to be captured are listed in table 1. The REACT database will host a clinically identifiable ‘front end’ for real-time patient management during the acute admission and for up to 12 months postdischarge, in which the link anonymised data will be made identifiable and visible only to the clinical team as determined by a statement of reference. Data for research purposes will be stored in an anonymised format.
Table 1.
Data points to be captured and frequency of capture
| Data variable | Data source | Frequency of capture |
| Demographics | ||
| Sex at birth (male/female) | Electronic | Once |
| Date of birth and age | Electronic | Once |
| Pregnancy status (yes/no) | Electronic | Once |
| Admission weight (kg) | Electronic | Once |
| Admission height (m) | Electronic | Once |
| Ethnicity | Electronic | Once |
| Profession/employment status | Electronic/manual | Once |
| Disease onset and admission to UHSFT care | ||
| Symptom onset date | Notes | Once |
| Admission date | Electronic | Once |
| Admission ward and level of care | Electronic | Once |
| Frailty score | Electronic | Once |
| Comorbidities and risk factors | ||
| Chronic obstructive pulmonary disease | Electronic | Once |
| Asthma | Electronic | Once |
| Interstitial lung disease | Electronic | Once |
| Bronchiectasis | Electronic | Once |
| Hypertension | Electronic | Once |
| Chronic cardiac disease (not hypertension) | Electronic | Once |
| Chronic kidney disease | Electronic | Once |
| Chronic Liver disease | Electronic | Once |
| Chronic neurological disorder | Electronic | Once |
| Metastatic solid tumour | Electronic | Once |
| Malignant neoplasm (including leukaemia and lymphoma) | Electronic | Once |
| Diabetes | Electronic | Once |
| Obesity (body mass index score>30) | Electronic | Once |
| AIDS/HIV infection | Electronic | Once |
| Rheumatological disorder | Electronic | Once |
| Dementia | Electronic | Once |
| Other diagnoses, including immunosuppression | Notes/electronic | Once |
| Vital signs at admission to UHSFT care | ||
| Temperature (°C) | Electronic | Continuous |
| Heart rate (beats/min) | Electronic | Continuous |
| Respiratory rate (breaths/min) | Electronic | Continuous |
| Systolic blood pressure (mm Hg) | Electronic | Continuous |
| Diastolic blood pressure (mm Hg) | Electronic | Continuous |
| Oxygen saturations (%) | Electronic | Continuous |
| Fraction of inspired oxygen (%) | Electronic | Continuous |
| Symptoms at admission to UHSFT care | ||
| Anosmia | Notes | Once |
| History of fever | Notes | Once |
| Cough | Notes | Once |
| Shortness of breath | Notes | Once |
| Myalgia | Notes | Once |
| Arthralgia | Notes | Once |
| Fatigue/malaise | Notes | Once |
| Headache | Notes | Once |
| Altered consciousness/confusion | Notes | Once |
| Cold symptoms (sore throat, rhinitis and ear pain) | Notes | Once |
| Wheezing | Notes | Once |
| Chest pain | Notes | Once |
| Seizures | Notes | Once |
| Abdominal pain | Notes | Once |
| Diarrhoea | Notes | Once |
| Vomiting | Notes | Once |
| Conjunctivitis | Notes | Once |
| Skin rash | Notes | Once |
| Ulcers | Notes | Once |
| Lymphadenopathy | Notes | Once |
| Laboratory results | ||
| Haemoglobin (g/L) | Electronic | Daily |
| White blood cell count (109/L) | Electronic | Daily |
| Neutrophil count (109/L) | Electronic | Daily |
| Lymphocyte count (109/L) | Electronic | Daily |
| Eosinophil count (109/L) | Electronic | Daily |
| Platelet count (109/L) | Electronic | Daily |
| International normalised ratio | Electronic | Daily |
| Alanine aminotransferase (units/L) | Electronic | Daily |
| Aspartate transaminase (units/L) | Electronic | Daily |
| Bilirubin (µmol/L) | Electronic | Daily |
| Alkaline phosphatase (units/L) | Electronic | Daily |
| Sodium (mmol/L) | Electronic | Daily |
| Potassium (mmol/L) | Electronic | Daily |
| Urea (mmol/L) | Electronic | Daily |
| Creatinine (mmol/L) | Electronic | Daily |
| Glucose (mmol/L) | Electronic | Daily |
| C reactive protein (mg/L) | Electronic | Daily |
| Lactate dehydrogenase (units/L) | Electronic | Daily |
| Ferritin (ng/L) | Electronic | Daily |
| Triglycerides | Electronic | Daily |
| D-dimer | Electronic | Daily |
| Troponin (ng/L) | Electronic | Daily |
| Vitamin D | Electronic | Daily |
| Arterial blood gas measurements | Electronic | Daily |
| Fraction of inspired oxygen (%) | ||
| Partial pressure of oxygen (kPa) | ||
| Partial pressure of carbon dioxide (kPa) | ||
| pH | ||
| Bicarbonate | ||
| Base excess | ||
| Lactate | ||
| Chest X-ray on admission to UHSFT care | ||
| Are infiltrates present? | Manual | Daily |
| Check all quadrants where infiltrates are present | Manual | Daily |
| Chest X-ray report | ||
| CT scan of chest under UHSFT care | ||
| Ground-glass opacification (bilateral? Location, eg, peripheral or basal) | Manual | Daily |
| Consolidation (bilateral? Location, eg, peripheral or basal) | Manual | Daily |
| Bronchovascular thickening within lesions? | Manual | Daily |
| CT report | Manual | Daily |
| Pathogen testing | ||
| Nasopharyngeal swab (SARS-CoV-2) positive/negative and viral titre | Electronic | Daily |
| Nasopharyngeal swab respiratory virus panel positive/negative and viral titre | Electronic | Daily |
| COVID-19 serology | Electronic | Daily |
| Sputum microscopy culture and sensitivity | Electronic | Daily |
| Blood culture | Electronic | Daily |
| Urine culture | Electronic | Daily |
| Medication | ||
| Each individual medication: name, dose, frequency, start date, stop date | Electronic | Daily |
| Data to be collected from patients not requiring admission to UHSFT | ||
| Online survey via my medical record | Electronic online | Daily until discharge or symptoms resolve (maximum of 28 days) |
| Level 2/3 (data from MetaVision, a clinical information system for critical care) | ||
| Invasive/non-invasive ventilation | Electronic | Hourly |
| Ventilator mode | Electronic | Hourly |
| Respiratory rate | Electronic | Hourly |
| Tidal volume | Electronic | Hourly |
| Positive end expiratory pressure | Electronic | Hourly |
| Hours prone | Electronic | Hourly |
| Fraction of inspired oxygen (from ABG) | Electronic | 3 Hourly |
| Partial pressure of oxygen (from ABG) | Electronic | 3 Hourly |
| Renal replacement therapy | Electronic | Daily |
| Procalcitonin | Electronic | Every 2 days |
| proADM | Electronic | Every 2 days |
| Number of inotropes | Electronic | Continuous |
| Steroids (yes/no) | Electronic | Daily |
| Antivirals (yes/no) | Electronic | Daily |
| Heart rate (beats/min) | Electronic | Hourly |
| Blood pressure (mean) | Electronic | Hourly |
| Fluid balance/24 hours | Electronic | Daily |
| Temp | Electronic | Hourly |
| Glasgow Coma Scale score | Electronic | Hourly |
| Richmond Agitation–Sedation Scale score | Electronic | Hourly |
| Confusion assessment method for the ICU score | Electronic | Hourly |
| Outcomes | ||
| Secondary diagnosis | ||
| Bacterial pneumonia | Manual | Once |
| Acute respiratory distress syndrome | Manual | Once |
| Cytokine release syndrome | Manual | Once |
| Acute kidney injury | Manual | Once |
| Acute liver injury | Manual | Once |
| Myocarditis | Manual | Once |
| Cardiomyopathy | Manual | Once |
| Pulmonary embolus | Manual | Once |
| Stroke | Manual | Once |
| Treatment during admission | ||
| Antiviral agent (yes/no) | Electronic | Once |
| Antibiotic (yes/no) Route of administration |
Electronic | Once |
| Oral corticosteroid (yes/no) | Electronic | Once |
| Intravenous corticosteroid (yes/no) | Electronic | Once |
| ACE inhibitor/ARB (yes/no) | Electronic | Once |
| Statin (yes/no) | Electronic | Once |
| Level 2/3 admission (yes/no) | Electronic | Once |
| Treatment escalation plan ceiling of care | Manual | Once |
Oxygen therapy
|
Electronic | Once |
Non-invasive ventilation
|
Electronic | Once |
Non-invasive ventilation
|
Electronic | Once |
Invasive ventilation
|
Electronic | Once |
Renal replacement therapy
|
Electronic | Once |
Inotropes
|
Electronic | Once |
| Plasma exchange (yes/no) | Electronic | Once |
| Intravenous immunoglobulin (yes/no) | Electronic | Once |
| Destination of discharge from UHSFT care | ||
| Palliative discharge | Electronic | Once |
| Alive at discharge | Electronic | Once |
| Death | Electronic | Once |
| 28-day mortality | Electronic | Once |
| Readmission within 28 days | Electronic | Once |
| Post COVID-19 complications | Manual | Once |
| Clinic follow-up for COVID-19 complications | ||
| Medication changes | Manual | Once |
| Long-term oxygen requirements | Manual | Once |
| Pathology results | Manual | Once |
| Chest X-ray and CT chest scan results | Manual | Once |
| Recorded secondary diagnosis | Manual | Once |
| Exercise tolerance | Manual | Once |
ABG, arterial blood gas; ARB, angiotensin receptor blocker; ICU, intensive care unit; proADM, proadrenomedullin; UHSFT, University Hospital Southampton NHS Foundation Trust.
Biobanking
A subcohort of patients aged ≥16 years old are given the opportunity to provide consent for storage of excess sample taken as part of their routine clinical management with optional further biological sampling (ie, blood, urine, induced sputum, nasal samples, exhaled breath or bronchial wash samples; see table 2). They are offered the opportunity to provide this at any point during their care under UHSFT. Care is taken to offer this study to participants after or alongside recruitment to the national NIHR Urgent Public Health Priority observational studies (International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC), Diagnosis and Management of Febrile Illness using RNA Personalised Molecular Signature Diagnosis (DIAMONDS) and NIHR Bioresource). These samples are stored within the SRB (managed under the UoS Human Tissue Act Licence) and analysed accordingly to enable biomarker measures related to COVID-19, including genomics, transcriptomics, proteomics, lipidomics and biochemical studies. These measures and the clinical data are reciprocally iterative and inform knowledge as they feed into the clinical care in determining treatment pathways and the response to treatment, as well as the natural history of SARS-CoV-2 infection. Samples are stored as coded samples, without participant names, that can be linked to clinical data (linked anonymised).
Table 2.
Biological samples to be collected for storage within the Southampton Research Biorepository
| Biological sample | Proposed analysis |
| Blood | Whole blood sample for genotyping (10 mL EDTA tube), serum (5×10 mL serum tube), plasma (10 mL EDTA tube), mRNA (PAXgene) (total ~80 mL). In addition to these blood samples taken universally, subgroups of participants may be invited to donate a further sample of blood (up to 100 mL) for isolation of immune cells for ex vivo experimental study and for gene array. Serial sampling may be required (up to once a day); however, no more than 210 mL will be taken within a 2-week period. |
| Urine | Up to 50 mL of urine may be taken for characterisation of biomarkers of disease. |
| Saliva | Up to five samples within a 14-day period |
| Nasal samples | Nasal brushing (for microbial carriage analysis and culture of nasal epithelial cells) and nasosorption for mucosal sampling (for inflammatory indices) may be performed as part of the study. Serial sampling may be required, but no more than one nasal brushing, and six nasosorption samples will be taken in a 2-week period. |
| Exhaled breath samples | Exhaled breath condensate may be collected for assessment of volatile compounds which may influence disease progression. Serial sampling may be required, but no more than two samples will be taken within a 2-week period. |
| Sputum samples | Excess sputum samples collected as part of routine clinical practice may be processed and analysed for presence of microbes, as well as inflammatory cell infiltrate, gene expression and other immune/inflammatory indices. |
| Bronchoscopy samples | Non-directed bronchial wash via a suction catheter may be performed in intubated and ventilated patients only. Samples will be processed for microbial analysis, cell culture and molecular biology. Serial sampling may be required, but no more than one sample per day will be taken. |
Data management
Data are captured longitudinally, with change over time treated as explicit. After admission, data capture will occur longitudinally in parallel with participant ongoing clinical follow-up for up to 12 months of their discharge following their acute COVID-19 admission. Active participation in the database would end 12 months after the participant was discharged from UHSFT following their acute admission with COVID-19 19 or if there was no further involvement of UHSFT clinical teams in their care before this time. The Database will continue to aquire data until no further patients are seen at UHSFT with COVID-19 19 and all patients have been discharged from UHSFT care. Storage of data for research purposes will be in an anonymised format. Clinical and research data collected from the cohort in the study are kept in a highly secure contemporary encrypted data platform BC|INSIGHT (within the Clinical Informatics Research Unit, UoS) that was set up in a Microsoft Data Centre in South UK.
Missing data
The majority of data capture is electronic, and therefore we expect missing data to be minimal. For all data other than symptom onset, any missing data will be characterised as ‘missing’ and will be excluded from analysis. In keeping with the data capture running in parallel with clinical care rather than via independent research ‘visits’, the data collection may not include certain variables for all patients if the capture of that information or a sample subset was not clinically indicated.
For symptom onset date, which is used for the purposes of analysis as day 0, a valid date is necessary. Data will be extracted manually from the medical records and will be classified as the following:
Clear date.
Unclear: in this scenario, admission date will be used.
Nosocomial infection (acquired while in the hospital): date of first +swab will be used if symptom onset date is not otherwise available.
These three groups will be identified within the dataset and analysed for significant differences prior to assessment as a whole cohort for a specific outcome.
Analysis of the whole dataset will be both cross-sectional and longitudinal according to specific study questions. Specific datasets can be extracted from the database to investigate specific hypothesis.
This cohort can also be interrogated for inclusion criteria towards additional trials occurring in parallel to the REACT COVID-19 study. Participation in clinical trials will not exclude patients from the cohort but may omit patients from some analysis depending on the research question.
Statistical analysis
A variety of statistical methods will be employed to support a range of assessment needs. Associations to potential risk factors will also be tested via multiple logistic regression analyses. Statistical significance levels will be set at a p value of <0.05.
Cluster analysis methodology will also be used to determine the heterogeneous nature of COVID-19 through an unbiased approach. Tests of correlation will be used to ensure that excessive collinearity of cluster variables does not bias the clustering process. Associations of disease clusters/phenotypes with cluster variables across resulting clusters will use analysis of variance for continuous variables and χ2 tests for binary variables. Subsequent tests of association to identify patterns of disease morbidity for disease clusters will be conducted. Further tests of association for disease clusters with potential epidemiological and pathophysiological risk factors will also be undertaken.
To facilitate artificial intelligence and machine intelligence learning from this dataset, the following will be undertaken:
-
Mapping summary statistical data from existing intensive care unit studies (Observational Health Data Sciences and Informatics and similar), as well as demographics data, using these to contextualise and enrich the patterns observed for participants with COVID-19 at UHSFT.
The contextualisation component would use existing studies as a reference frame to contrast and compare similarities and differences among UHSFT and external patient cohorts.
The enrichment component would use existing studies and demographics as priors to support Bayesian inference over UHSFT data.
-
Codesign and evaluate a Bayesian inference model.
With a possible extension of the Bayesian framework into a causal inference setting.
Provide a critical analysis of the ethical and safety aspects related to the clinical application of Bayesian inference, that is, supporting the clinical decision-making process.
Other innovative artificial intelligence and machine learning methods will also be explored to identify and predict those patients who have worse outcomes.
Ethics and dissemination
The study design, protocol and patient-facing documentation have been approved by North West Research Ethics Committee (REC 17/NW/0632) as an amendment to the NIHR Southampton CRF-managed SRB. No patient identifiable data are included in this paper. Ethics approval for the study was obtained from HRA Specific Review Board (REC 20/HRA/2986) for waiver of informed consent for the database-only cohort. The database will be conducted in accordance with the principles of good clinical practice. Database documents (paper and electronic) will be collected and retained in accordance with the General Data Protection Regulations 2018 in a secure location during and after the trial has finished. All essential documents including source documents will be retained for a minimum period of 5 years following the end of the study. Clinical and research data collected in the database will be kept in a highly secure contemporary encrypted data platform BC|INSIGHT that was set up in a Microsoft Data Centre in South UK. Access to the study’s data is only given to study team members that have signed the delegation logs. Data access is via an encrypted web service with username/password-type authentication. The data held in the server are backed up weekly using the snapshot technology provided by Microsoft Azure, and the retention period of the snapshot storage is 2 years. Storage of data for research purposes will be in an anonymised format. Anonymised data may be released to individuals or organisations outside the UK, following approval by the REACT COVID-19 data access committee. A REACT COVID-19 Data Access Management Committee will be established to prioritise and ensure appropriate governance of requests to access linked anonymised clinical data. REACT Committee membership will include a chief investigator and principal investigators, the scientific lead for REACT and a representative from UHSFT Research and Development and/or Academic Health Sciences Centre prioritisation committee.
Written informed consent will be obtained from all study participants who agree to biobanking of excess clinical sample±additional sampling, as per approvals for amendment to the SRB reviewed by North West Research Ethics Committee (REC 17/NW/0632). For patients who have impaired cognition, informed consent will be sought from their legally acceptable representative, with retrospective consent sought for those who regain capacity to consent following recovery from acute illness. The findings from this study will be disseminated locally and internationally through manuscript publications in peer-reviewed journals.
Discussion
SARS-CoV-2 infection is presenting a heterogenous pattern of disease, with varying presentations from asymptomatic carriage, as demonstrated among healthcare workers7 to severe disease with mortality rates of 50% in patients requiring intensive care.8 Emerging understanding is identifying some comorbidities and risk factors for progression to severe disease, but there is still a wide breadth of severity among patients displaying the same risk profile.
The longitudinal, mainly automated data collection of the REACT COVID-19 study enables a granularity of data that has potential to identify early markers of disease severity and the relevance of their trajectory of change on disease progression. The REACT COVID-19 database can be used to rapidly and easily identify participants who may be appropriate for clinical trials and offers visual interpretation of large amounts of data for hypothesis generation.
The database provides a rich dataset to support the biobanking subcohort, within which there is the potential to recruit to smaller, focused basic science studies investigating the mechanisms driving disease processes resulting from infection with SARS-CoV-2.
The REACT COVID-19 study is mindful of the national and international efforts to bring together data from multiple centres in order to enhance the power of studies, be they clinical, observation, mechanistic or intervention studies. The study will therefore be aligning itself with the broader UHS and UoS research project of Enabling new treatment approaches for COVID-19 Treatment, supported by the Southampton Coronavirus Support Fund, NIHR Southampton BRC and NIHR Southampton CRF.
The ability of the REACT COVID-19 study to capture the granularity of the longitudinal path of clinical care is both a major strength and a potential limitation. There is no strict study ‘visit’ protocol to follow, with data capture occurring in a more fluid manner. Therefore, there is the potential for a greater number of incomplete datasets, with some patients having a greater amount of available data. This is likely to be reflective of the level of care required, such as in those patients who required intensive care support. Any analysis will allow this, and populations may be subcohorted to answer specific questions if the data are only available for a specific group.
The simplicity of data visualisation within the REACT COVID-19 study belies the complexity of the concepts behind this project, facilitated through the unique collaboration between UHSFT, the UoS and the digital ECMT. This collaboration possesses the requisite expertise in their respective specialisms to deliver all aspects of this study, alongside the infrastructure provided by the NIHR Southampton CRF and NIHR Southampton BRC. The REACT COVID-19 study enables visualisation of detailed clinical data and the application of novel artificial intelligence methods, which will allow greater understanding of the natural history of this novel disease through both longitudinal and cross-sectional analyses.
Supplementary Material
Acknowledgments
The authors acknowledge the National Institute for Health Research (NIHR) Southampton Clinical Research Facility (CRF) and University Hospital Southampton NHS Foundation Trust Research Nursing teams for their support in the set-up of this study, Dr Dave Stockley (National Institute for Health Research CRF Southampton Research Biorepository Manager) and Dr Alastair Watson for their help in proofreading and preparation of the manuscript for submission.
Footnotes
Correction notice: This article has been corrected since it first published. The provenance and peer review statement has been included.
Contributors: HB and AF designed the protocol and drafted the manuscript; AD and MC were involved in protocol design; JB, HP and FB were involved in the realisation of data extraction, integration, transformation and upload processes; CK, GT and SNF were involved in protocol design for the biobanking subcohort; NS and SW are involved in manual data collection processes; DL, PF, HP and JB were involved in the design and adaptation of the Real-time Analytics for Clinical Trials platform; and TW was involved in study conception and protocol design. All authors reviewed the final manuscript.
Funding: The REACT platform has been supported by the digital Experimental Cancer Medicine Team free of charge. The biobanking subcohort is supported the NIHR Southampton CRF and NIHR Southampton Biomedical Research Centre at University Hospital Southampton NHS Foundation Trust and as part of a broader effort (Enabling New Treatment Approaches for COVID-19 Treatment) by the University of Southampton (UoS) charity (Office of Development and Alumni Relations). In addition the Clinical Informatics Research Unit, UoS has supported infrastructure costs. The support described above was not provided from a specific award or grant.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were involved in the design, conduct, reporting or dissemination plans of this research. Refer to the Methods and analysis section for further details.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
No data are available.
Ethics statements
Patient consent for publication
Not required.
References
- 1.World Health Organisation . Who coronavirus disease (COVID-19) Dashboard. 2020. Available: https://covid19.who.int/?gclid=EAIaIQobChMIz5Xu84OJ6gIVCLrtCh3WlQ8KEAAYASAAEgIohvD_BwE [Accessed 16 June 2020].
- 2.UK Governement guidance, 2020. Available: https://www.gov.uk/guidance/coronavirus-covid-19-information-for-the-public [Accessed 18 June 2020].
- 3.Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the Treatment of Covid-19 - Preliminary Report. N Engl J Med 2020. [DOI] [PubMed] [Google Scholar]
- 4.Oxford University News Release . Low-Cost dexamethasone reduces death by up to one third in hospitalised patients with severe respiratory complications of COVID-19, 2020. Available: https://www.recoverytrial.net/files/recovery_dexamethasone_statement_160620_v2final.pdf [Accessed 16 June 2020].
- 5.ACCORD . Accelerating COVID-19 research and development. 2020. Available: https://www.nihr.ac.uk/news/national-collaborative-platform-launched-to-fast-track-potential-covid-19-treatments/24678 [Accessed 18 June 2020].
- 6.RECOVERY , 2020. Available: https://www.recoverytrial.net [Accessed 18 June 2020].
- 7.Rivett L, Sridhar S, Sparkes D, et al. Screening of healthcare workers for SARS-CoV-2 highlights the role of asymptomatic carriage in COVID-19 transmission. eLife 2020;9. 10.7554/eLife.58728. [Epub ahead of print: 11 05 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Docherty AB, Harrison EM, Green CA, et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: prospective observational cohort study. BMJ 2020;369:m1985. 10.1136/bmj.m1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data are available.