Abstract
A putative novel clade within the genus Streptomyces was discovered following antifungal screening against Pseudogymnoascus destructans, the causative agent of white-nose syndrome, and described using multi-locus sequencing analysis. Swabs from both the cave myotis bat (Myotis velifer) and the Brazilian free-tailed bat (Tadarida brasiliensis) in southern New Mexico bore isolates AC536, AC541T and AC563, which were characterised using phylogenetic, morphological, and phenotypic analyses. Multi-locus sequence analysis positions AC541T with neighbors Streptomyces rubidus (NRRL B-24619T), Streptomyces guanduensis (NRRL B-24617T), and Streptomyces yeochonensis (NRRL B-24245T). A complete genome of the type strain was assembled to determine its taxonomy and secondary metabolite potential. ANI comparisons between all closely related types strains are shown to be well below the 95–96% species delineation. DNA-DNA relatedness between AC541T and its nearest neighbors ranged between 23.7 and 24.1% confirming novelty. Approximately 1.49 Mb or 17.76% of the whole genome is devoted to natural product biosynthesis. The DNA G + C content of the genomic DNA of the type strain is 73.13 mol %. Micromorphology depicts ovoid spores with smooth surfaces in flexuous chains. Strains presented an ivory to yellow hue on most ISP media except inorganic salts-starch agar (ISP4) and can grow on d-glucose, mannitol, and d-fructose, but exhibited little to no growth on l-arabinose, sucrose, d-xylose, inositol, l-rhamnose, d-raffinose, and cellulose. This clade possesses the capability to grow from 10 to 45 °C and 12.5% (w/v) NaCl. There was strain growth variation in pH, but all isolates thrive at alkaline levels. Based on our polyphasic study of AC541T, the strain warrants the assignment to a novel species, for which the name Streptomyces buecherae sp. nov. is proposed. The type strain is AC541T (= JCM 34263T, = ATCC TSD201T).
Keywords: Antifungal, White-nose syndrome, Bat, Streptomyces, Biosynthetic gene clusters
Introduction
Antifungals are of particular importance as there has been a dramatic increase of fungal infections in plants and animals in the last two decades (Fisher et al. 2020). Since 2006, white-nose syndrome (WNS), caused by the fungus Pseudogymnoascus destructans, has devastated bat populations, causing the death of over six million bats (Blehert 2012). Like many fungal pathogens, P. destructans exhibits generalist host capabilities (Zukal et al., 2014), high virulence (Turner et al. 2011) and resilience in the environment during host absence (Hoyt et al. 2015). High mortality rates of invasive fungal infections and spread of pathogens that threaten wildlife require a more aggressive search for novel broad-spectrum antifungals. Here, we present a novel clade with potential antifungal properties that were isolated from WNS-free bat external surfaces in a study by Hamm et al. (2017). Natural products produced by organisms composing the bat microbiome may serve as an alternative preventative measure or treatment for already infected WNS bats.
The current study uses a polyphasic taxonomic approach to describe this new species, for which the name Streptomyces buecherae sp. nov. is proposed. A combination of antiSMASH and BLAST bioinformatics applied to the genomic sequence illuminates the potential of S. buecherae sp. nov. to express novel biosynthetic gene clusters that may play a role in its antifungal capability.
Materials and methods
Isolation and cultivation
Strains AC536, AC541T and AC563 were isolated in June 2014 from bats’ skin and fur near Rattlesnake Springs in the Carlsbad Caverns National Park (32° 10′ 31″ N 104° 26′ 38″ W) in southeastern New Mexico as reported by Hamm et al. (2017). Both AC541T and AC536 were isolated from a female cave myotis bat (Myotis velifer). Isolate AC563 was isolated from a female Brazilian free-tailed bat (Tadarida brasiliensis). This putative novel Streptomyces species documented in Hamm et al. (2017) appears to have the ability to colonize multiple bat species that occur in Carlsbad Caverns National Park. Bats were captured using mist nests following methods described by Kunz et al. (2009). Nets were monitored throughout the night and bats were removed upon capture. Bats at Carlsbad Cavern National Park were sampled following approved protocols under collection permits: 2014 New Mexico Game and Fish Department Scientific Collecting Permit (SP670210, SCI#3423, SCI#3340), National Park Service Scientific Collecting Permit (CAVE-2014-SCI-0013), and an Institutional Animal Care and Use Committee (IACUC) Permit from the University of New Mexico (Protocol#12-100835-MCC) and from the National Park Service (Protocol#IMR_ELMA.PARA_Northup_Bat_2013.A2).
Phylogenetic analysis
Gene extraction for multi-locus sequence analysis (MLSA) and alignments for phylogenetic trees were made using BIGSdb software (Jolley and Maiden 2010) using data and methods previously described for the Streptomycetaceae family (Labeda et al. 2017). MLSA data for the representative strains AC536, AC541T and AC563 were previously reported (Hamm et al. 2017). The partial sequences of five house-keeping genes, atpD (ATP synthase F1, beta subunit), gyrB (DNA gyrase B subunit), rpoB (RNA polymerase beta subunit), recA (recombinase A) and trpB (tryptophan synthetase, beta subunit), were utilised in a MLSA to construct a phylogenetic tree using MEGA X software (Kumar et al. 2018). The near neighbors were selected on the basis of gyrB homology using the MLSA database (Labeda et al. 2017). Phylogenetic reconstructions were made using maximum-likelihood based on the General Time Reversible model (Nei and Kumar 2000); this model was chosen on the basis of the likelihood tests implemented in MEGA X. Measures of bootstrap support for internal branches were obtained from 1000 pseudoreplicates with a Neighbor-Joining (BioNJ) starting tree.
Genome sequencing and analysis
Genomic DNA was isolated from all strains using UltraClean® Microbial DNA isolation kits (MoBio Labs, Carlsbad, CA) following the instructions of the manufacturer. PacBio sequencing and library preparation were performed by CD-Genomics using standard PacBio sequel library preparation and sequencing. The PacBio reads were assembled using CLC Genomics Workbench (v20.0) with genome finishing plugin module (v1.9). The assembly was polished with Illumina reads using the same system. The Illumina reads were prepared using Nextera XT library preparation kit following the manufacturer’s suggested protocols. The prepared libraries were sequenced using a MiSeq DNA sequencer with the MiSeq V3 2×300 sequencing kit. The resulting reads were quality trimmed to the 95% confidence level. The final sequence yielded one chromosome that was deposited in NCBI GenBank under accession number CP060404. The representative strains AC536 and AC563 were only sequenced using the MiSeq V3 2×300 kit and accessioned into NCBI GenBank under the numbers JACOFO01 and JACOFP01, respectively. The average nucleotide identity (ANI) was determined using OrthoANI using default parameters on the website (Yoon et al. 2017). Additionally, an in silico digital DNA-DNA hybridisation (DDH) was used to confirm novelty from the closest neighboring type strains. Because isolates for this novel clade were discovered by antifungal screening, antiSMASH 5.1.1 and standard genome mining techniques (Blin et al. 2019) were used for biosynthetic gene cluster (BGC) prediction.
Morphological and phenotypic analysis
Scanning electron microscopy (SEM) was conducted on the type strain to deduce the micromorphology and sporulation. Spore morphology was assessed after 14 days at 28 °C on ISP 3 media. Spores were fixed with vapor over 4.0% osmium tetroxide solution for 24 h, then dehydrated using an acetone step gradient. The specimens were then dried in a carbon dioxide critical point dryer (Tousimis, Rockville, MD) and immediately placed in a sputter coater for a 2-min coating of gold (Structure Probe, Inc., West Chester, PA). The SEM images were obtained on a JEOL JSM-6010LA (JEOL Ltd. Tokyo, Japan). Typical operating conditions were an accelerating voltage of 10 kV and a spot size of 30.
AC536, AC541T and AC563 were chosen as representatives of this novel clade for standard phenotypic observations on ISP media (Shirling and Gottlieb 1966). Carbohydrate utilisation was determined using a 1% concentration of each carbon source added to basal agar 5338 (Shirling and Gottlieb 1966) and measured for utilisation 14 days after inoculation and incubation at 25 °C. Temperature parameters were assessed using ISP 1 media at a range of 5–45 °C (Shirling and Gottlieb 1966). Tolerance to various pH levels (4–12) was tested in tubes with ISP2 medium (Shirling and Gottlieb 1966). Sodium chloride growth was measured using basal medium 5339 (casein peptone 10 g/L, yeast extract 5 g/L) supplemented with sodium chloride 0–15% (w/v), in 2.5% intervals. Phenotypic characterisation of S. rubidus (NRRL B-24619T) and S. guanduensis (NRRL B-24617T) were re-assessed and compared with the original descriptions (Xu et al. 2006).
Results and discussion
In addition to the three strains described in Hamm et al. (2017), a search of genomes deposited in GenBank identified a strain, Streptomyces sp. NA00687 (CP054929), as a likely conspecific member of this clade. Streptomyces sp. NA00687 was isolated from marine sediment in Hainan, China. The phylogeny of the subject strains and selected type strains was determined using a previously reported five gene MLSA in Hamm et al. (2017) (Fig. 1). The results show the three strains isolated in this study and Streptomyces sp. NA00687 all cluster together in a monophyletic clade. The MLSA distance computed from the partial sequences of these house-keeping loci is > 0.007 or more than 70% DNA–DNA homology, a guideline empirically determined by Rong and Huang (2012). ANI was used to confirm the taxonomic assignments (Fig. 2). The results show the ANI comparisons between all closely related types strains to be well below the 95–96% threshold commonly used to delineate prokaryotic species (Richter and Rosselló-Móra 2009). This is supported with DDH (Supplementary Table 1).
Fig. 1.

Phylogenetic relationships among strains based on five genes (atpD, gyrB, rpoB, recA and trpB). Phylogenetic reconstructions were made using maximum-likelihood based on the General Time Reversible model (Nei and Kumar 2000); this model was chosen on the basis of the likelihood test implemented in MEGA X. Measures of bootstrap support for internal branches were obtained from 1000 pseudoreplicates with a Neighbor-Joining (BioNJ) starting tree. The bar is equal to an MLSA distance of 0.020. Bootstrap support values < 69 have been removed
Fig. 2.
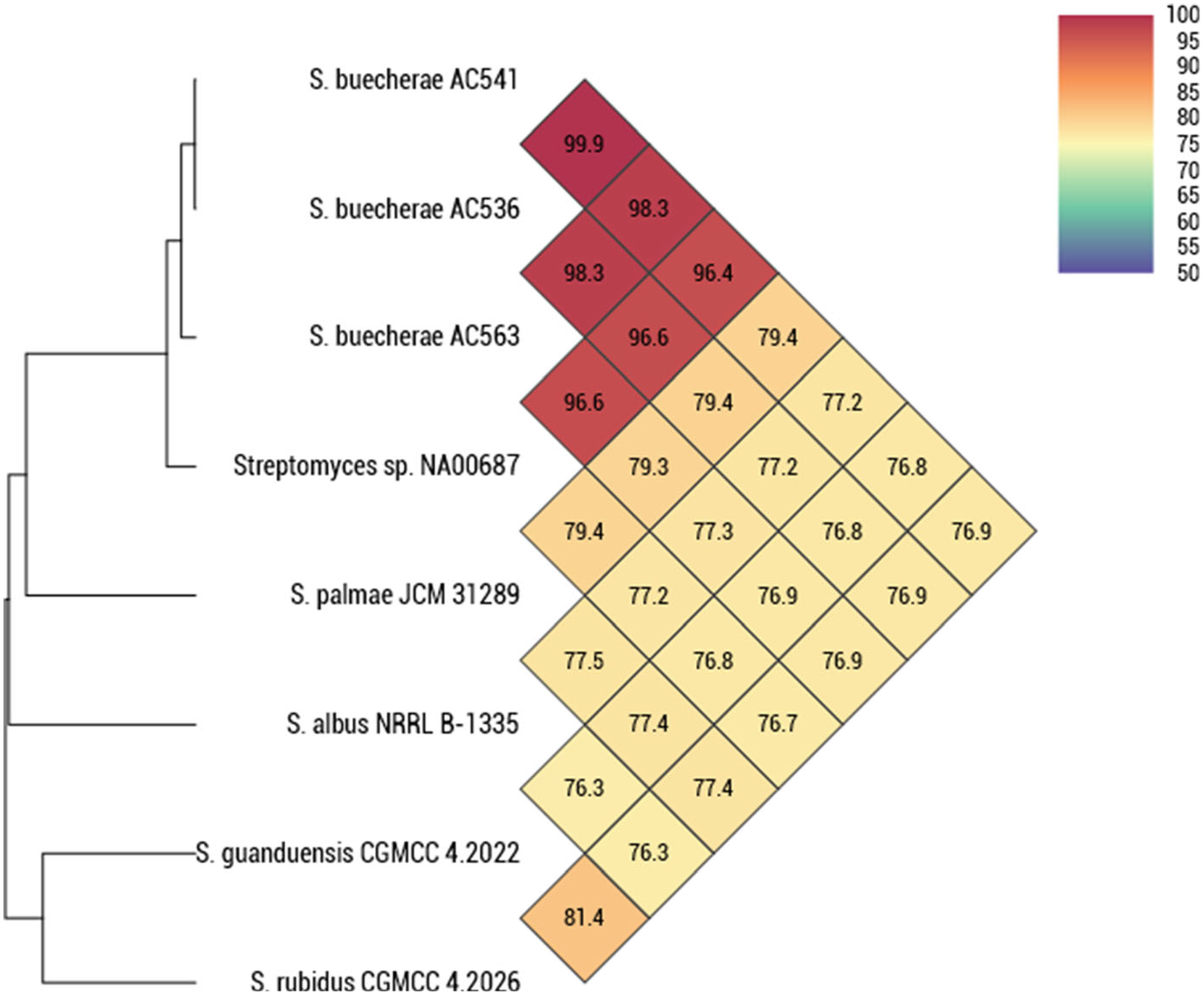
Average nucleotide identity (ANI) determined using OrthoANI (Yoon et al. 2017) of AC536, AC541T, AC563, and NA00678. The results show the ANI comparisons between all closely related types strains to be well below the 95–96% species delineation
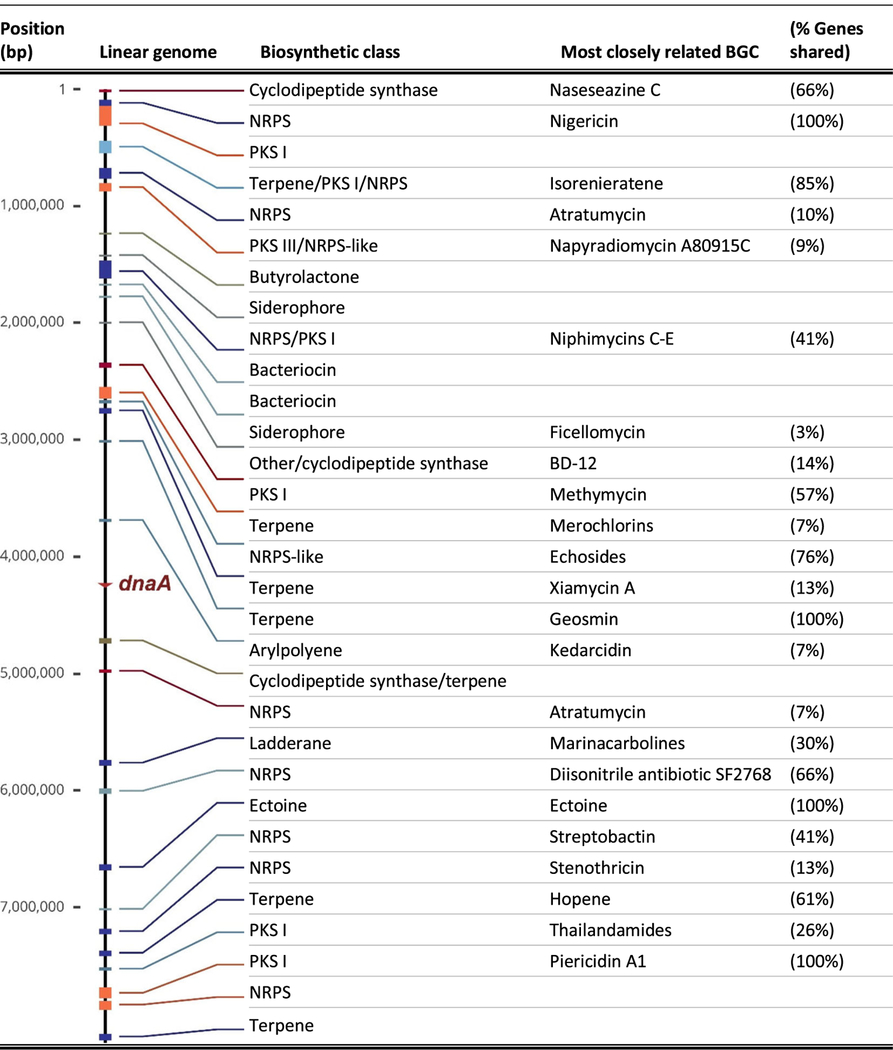
The chromosome of Streptomyces buecherae was assembled into a single, 8.4 Mb long, contiguous linear sequence with a GC content of 73.13%. The assembly process found no evidence for linear or circular plasmids. Linearity of the single genomic chromosome was evidenced by the presence of characteristic terminal inverted repeats (TIRs). The dnaA gene that initiates replication by unwinding DNA at the origin of replication, oriC, was found at 4.24 Mb, near the genome center (Table 1).
Table 1.
Descriptions and positions of the biosynthetic gene clusters on the complete linear genome of Streptomyces buecherae AC541T
|
BGC Biosynthetic gene cluster, NRPS non-ribosomal peptide synthetase, PKS polyketide synthase
BGC analysis indicated the presence of between 31 and 35 biosynthetic gene clusters (Table 1; neighboring BGCs are sometimes misidentified as single hybrid BGCs). AntiSMASH analyses matched four BGCs in S. buecherae AC541T to experimentally validated BGCs found in the MiBIG database (Kautsar et al. 2020). These four clusters contain 100% of the genes (not 100% homologous) involved in the biosynthesis of nigericin, geosmin, ectoine, and piericidin A. Additional BGCs were identified that shared ≥ 65% of their gene content with known BGCs encoding naseseazine C, isorenieratene, echosides, diisonitrile antibiotic SF2768, and hopene. The 20 remaining BGCs likely encode for natural products of significant novelty, with 13 biosynthetic regions sharing limited similarity (< 61%) to known classes and seven biosynthetic regions without significant similarity to any currently known BGCs. The biosynthetic capacity of AC541T includes eight NRPS, four PKSs, two bacteriocins, five terpenes, two siderophores, one ladderane, one cyclodipeptide (2,5-diketopiperazines), one butyrolactone, one arylpolyene, one ectoine, and five hybrid clusters. Approximately 1.49 Mb or 17.76% of the whole genome is devoted to natural product biosynthesis.
Morphology of AC536, AC541T and AC563 were the same on ISP media. Isolates from this novel clade appear to have an ivory white to yellow color on the description media with good growth on most media except inorganic salts-starch agar (ISP4) (Table 2). Spore morphology was ovoid spores with smooth surfaces in flexuous chains (Fig. 3). The physiological properties of the strains are summarised in Table 3. This clade possesses the capability to grow at 10 °C and 45 °C, as well as 12.5% (w/v) NaCl, except AC563 which only grew at 10% (w/v). Conversely, its nearest phylogenetic neighbors, S. rubidus and S. guanduensis have a much more limited heat tolerance and do not possess the ability to withstand high salt environments. This clade exhibits slight growth variation in pH, but all thrive at alkaline levels. Streptomyces buecheraeT had fruitful growth on d-glucose, mannitol, and d-fructose, but it had minimal growth on l-arabinose, sucrose, d-xylose, inositol, l-rhamnose, d-raffinose, and cellulose. Another notable difference from its near neighbors, S. rubidus and S. guanduensis, is the ability of AC536, AC541T and AC563 to grow on l-arabinose, sucrose, d-xylose, and d-raffinose.
Table 2.
Growth and culture characteristics of strains AC541T on standard agar media after incubation for 14 days at 25 °C
| Media | Growth | Substrate mycelium | Aerial mycelium | Pigment |
|---|---|---|---|---|
| Yeast extract-malt extract agar (ISP2) | Good | Ivory | None | None |
| Oatmeal agar (ISP3) | Good | Signal white | Signal white | None |
| Inorganic salts-starch agar (ISP4) | None | N/A | N/A | N/A |
| Glycerol-asparagine agar (ISP5) | Sparse | Ivory | None | None |
| Peptone-yeast extract-iron agar (ISP6) | Good | Golden yellow | None | None |
| Tyrosine agar (ISP7) | Good | Ivory | None | None |
Fig. 3.

Scanning electron micrograph of strain AC541T grown on ISP 3 for 14 days at 28 °C. Spores are ovoid with smooth surfaces in flexuous chains. The bar is equal to 1 μm
Table 3.
Phenotypic and genomic characteristics of Streptomyces buecherae isolates AC536, AC541T and AC563 and type strains of S. guanduensis (NRRL B-24617T) and S. rubidus (NRRL B-24619T)
| Characteristic | AC541T | AC536 | AC563 | S. rubidus B-24619T | S. guanduensis B-24617T |
|---|---|---|---|---|---|
| Spore shape | Smooth | Smooth | Smooth | Smootha | Smootha |
| Melanin on ISP6 | − | − | − | − | − |
| Pigment on ISP7 | − | − | − | − | − |
| Genome | |||||
| Size (Mbp) | 8.4 | 7.93 | 8.16 | 8.08 | 8.28 |
| Proteins | 6347 | 6321 | 6656 | 6975 | 6954 |
| G + C % | 73.13% | 72.50% | 70.80% | ||
| NaCl tolerance (w/v) | |||||
| 2.5% | + | + | + | − | − |
| 5.0% | + | + | + | − | − |
| 7.5% | + | + | + | − | − |
| 10% | + | + | + | − | − |
| 12.5% | + | + | − | − | − |
| 15% | − | − | − | − | − |
| Heat tolerance | |||||
| 5 °C | − | − | + | − | − |
| 10 °C | + | + | + | + (−a) | − (−a) |
| 25 °C | + | + | + | + | + |
| 45 °C | + | + | + | − | − |
| pH tolerance | |||||
| Min pH | 6 | 5 | 6 | 5 (4.5a) | 5 (4.5a) |
| Max pH | 12 | 10 | 12 | ND | 9 |
| Carbohydrate 1% (w/v) | |||||
| d-Glucose | + | + | + | +a | +a |
| l-Arabinose | − | ⊖ | − | +a | +a |
| Sucrose | − | − | − | +a | +a |
| d-Xylose | − | − | − | +a | +a |
| Inositol | − | ⊖ | − | −a | ⊕a |
| Mannitol | + | + | + | ND | ND |
| d-Fructose | ⊕ | + | + | +a | +a |
| l-Rhamnose | − | − | − | −a | +a |
| d-Raffinose | − | − | ⊖ | +a | +a |
| Cellulose | − | ⊖ | ⊖ | ND | ND |
+ Good growth and positive utilization, ⊕ poor to fair growth, ⊖ faint growth and probably no utilization, − no growth, ND no data
Description of Streptomyces buecherae sp. nov
Streptomyces buecherae (bue’che.rae) is named in honor of Debbie Buecher, a notable bat biologist in the Southwest United States. Streptomyces buecherae is composed of four isolates (AC536, AC541T, AC563 and NA00687). On ISP media, colonies are ivory white to yellow. Spores are ovoid with smooth surfaces in flexuous chains. Plentiful growth was observed on d-glucose, mannitol, and d-fructose, but it had no to very little growth on l-arabinose, sucrose, d-xylose, inositol, l-rhamnose, d-raffinose, and cellulose. Isolates in this clade have the capability to thrive from pH 5.0–12.0, at the presence of 12.5% NaCl, and from 10 to 45 °C.
The type strain AC541T (= JCM 34263T, = ATCC TSD201T) was isolated from a female cave myotis bat (Myotis velifer) in Carlsbad Caverns National Park, located in southeastern New Mexico. The G + C mol content of the genomic DNA of the type strain is 73.13%. Approximately 1.49 Mb or 17.76% of the whole genome is devoted to natural product biosynthesis.
Supplementary Material
Acknowledgements
PSH and APA thank support from Western Illinois University and their Research Inspiring Student Excellence (RISE) and Women in Science (WIS) programs. CAD was supported by ARS National Programs 303, 304 and 306. DEN gratefully acknowledges the support of the staff at Carlsbad Caverns National Park with logistical support and assistance with the NPS Materials Transfer Agreement implementation. We would like to thank Dr. Ernest W. Valdez for providing expertise on bats. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the U.S. Department of Agriculture or the National Science Foundation. The mention of firm names or trade products does not imply that they are endorsed or recommended by the USDA over other firms or similar products not mentioned. USDA is an equal opportunity provider and employer.
Funding Initial funding was provided by the Eppley Foundation (DEN). Additional funding was provided by the IDNR (APA) and the Colorado Plateau Cooperative Ecosystem Studies Unit (CPCESU) (DEN). The lead author (PSH) would like to thank the Mycological Society of America for funding to present her research. This publication was also supported by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) under Award Number F32CA221327 (MWM) and the National Center for Complementary and Integrative Health (NCCIH) of the NIH under Award Number R01AT009143 (RJT, NLK). This material is based upon work supported by the National Science Foundation (APA).
Footnotes
Conflict of interest The authors declare no conflicts of interests in the manuscript.
The GenBank accession number for complete genome of Streptomyces buecherae AC541T is CP060404.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s10482-020-01493-4) contains supplementary material, which is available to authorized users.
Contributor Information
Paris S. Hamm, Department of Biological Sciences, Western Illinois University, Macomb, IL 61455, USA; Department of Biology, University of New Mexico, Albuquerque, NM 87131, USA.
Christopher A. Dunlap, U. S. Department of Agriculture, Peoria, IL 61604, USA
Michael W. Mullowney, Department of Chemistry, Northwestern University, Evanston, IL 60208, USA
Nicole A. Caimi, Department of Biology, University of New Mexico, Albuquerque, NM 87131, USA
Regan J. Thomson, Department of Chemistry, Northwestern University, Evanston, IL 60208, USA
Andrea Porras-Alfaro, Institute for Environmental Studies, Western Illinois University, Macomb, IL 61455, USA.
Diana E. Northup, Department of Biology, University of New Mexico, Albuquerque, NM 87131, USA
References
- Blehert DS (2012) Fungal disease and the developing story of bat white-nose syndrome. PLoS Path 8:e1002779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Less SY, Medema MH, Webber T (2019) Antismash 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. 10.1093/nar/gkz310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Gurr SJ, Cuomo CA, Blehert DS, Jin H, Stukenbrock EH, Stajich JE, Kahmann R, Boone C, Denning DW, Gow NA (2020) Threats posed by the Fungal Kingdom to humans, wildlife, and agriculture. mBio 11:e00449 10.1128/mBio.00449-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm PS, Caimi NA, Northup DE, Valdez EW, Buecher DC, Dunlap CA, Labeda DP, Lueschow SR, Porras-Alfaro A (2017) Western bats as a reservoir of novel Streptomyces species with antifungal activity. Appl Environ Microbiol 83:e03057–16. 10.1128/AEM.03057-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt JR, Langwig KE, Okoniewski J, Frick WF, Stone WB, Kilpatrick AM (2015) Long-term persistence of Pseudogymnoascus destructans, the causative agent of white-nose syndrome, in the absence of bats. EcoHealth 12:330–333. 10.1007/s10393-014-0981-4 [DOI] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform 11:595 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, van de Hooft JJJ, van Santen JA, Tracanna V, Suarez-Duran HG, Pascal-Andreu V, Selem-Mojica N, Alanjary M, Robinson SL, Lund G, Epstein SC, Sisto AC, Charkoudian LK, Collemare J, Linington RG, Weber T, Medema MH (2020) MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res 48:D454–D458. 10.1093/nar/gkz882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz TH, Hodgkison R, Weise CD (2009) Methods of capturing and handling bats In: Kunz TH, Parsons S (eds) Ecological and behavioral methods for the study of bats, 2nd edn. The John Hopkins University Press, Baltimore, pp 3–35 [Google Scholar]
- Labeda DP, Dunlap CA, Rong X, Huang Y, Doroghazi JR, Ju K-S, Metcalf WW (2017) Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Van Leeuwenhoek 110:563–583. 10.1007/s10482-016-0824-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York [Google Scholar]
- Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. PNAS 106:19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong X, Huang Y (2012) Taxonomic evaluation of the Streptomyces hygroscopicus clade using multi-locus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for the systematics of the whole genus. Syst Appl Microbiol 35:7–18 [DOI] [PubMed] [Google Scholar]
- Shirling EB, Gottlieb DA (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340 [Google Scholar]
- Turner GG, Reeder DM, Coleman JTH (2011) A five-year assessment of mortality and geographic spread of white-nose syndrome in North American bats and a look to the future. Bat Res News 52:13–27 [Google Scholar]
- Xu C, Wang W, Cui Q, Huang Y, Liu Z, Zheng G, Goodfellow M (2006) Neutrotolerant acidiphilic Streptomyces species isolated from acidic-soils in China: Streptomyces guanduensis sp. nov., Streptomyces paucisporeus sp. nov., Streptomyces rubidus sp. nov. and Streptomyces yanglinensis sp. nov. Int J Syst Evol Microbiol 56:1109–1115 [DOI] [PubMed] [Google Scholar]
- Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. 10.1007/s10482-017-0844-4 [DOI] [PubMed] [Google Scholar]
- Zukal J, Bandouchova H, Bartonička T, Berkova H, Brack V, Brichta J, Dolinay M, Jaron KS, Kovacova V, Kovarik M, Martínková N, Ondracek K, Rehák Z, Turner GG, Pikula J (2014) White-nose syndrome fungus: a generalist pathogen of hibernating bats. PLoS ONE 9:e97224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


