Abstract
Background: A number of genes are associated with the incidence of non-syndromic cleft lip/palate (NSCL/P). Studies have shown a significant association between polymorphisms of ATP-binding cassette, sub-family A, member 4 (ABCA4) with the risk of NSCL/P. The present meta-analysis assessed the association between ABCA4 polymorphisms (rs560426 and rs481931) and the NSCL/P risk by reviewing case-control studies. Methods: Four databases (Scopus; Cochrane Library; Web of Science; and PubMed) were searched for articles published up to June 2020. The Review Manager 5.3 software was used to calculate the crude odds ratio (OR) and 95% confidence interval (CI). Both subgroup analyses for ethnicity and source of controls and a meta-regression related to publication year were conducted. Results: Of 94 retrieved studies, 12 were analyzed in this meta-analysis (2859 NSCL/P patients and 3792 controls for ABCA4 rs560426 polymorphism and 1333 NSCL/P patients and 1884 controls for ABCA4 rs481931 polymorphism). Overall, there was no significant association between both polymorphisms and the risk of NSCL/P. However, subgroup analysis demonstrated that there was a higher risk of NSCL/P for specific models: the allelic model (OR = 1.13; p = 0.03), the homozygote model (OR = 1.53; p = 0.04), and the recessive model (OR = 1.30; p = 0.03) in the Asian ethnicity for the rs560426 polymorphism. Conclusion: The findings confirmed that the NSCL/P risk was significantly associated with the G allele and GG genotype of rs560426 polymorphism but not for rs481931 polymorphism. There were no associations between both polymorphisms (rs560426 and rs481931) and the NSCL/P risk in those of European descent and the mixed ethnicities.
Keywords: ABCA4, polymorphism, variation, non-syndromic cleft lip/palate, meta-analysis
1. Introduction
More than 70% of all cleft lips/palates (CL/Ps) present without further anomalies and are thus nonsyndromic [1]. Its prevalence ranges from 1/700 to 1/1000, depending on the geographical area and ethnicity [2]. Non-syndromic cleft lip/palate (NSCL/P) has a multifactorial etiology and therefore both environmental and genetic risk factors can affect its occurrence. However, the link between different environmental factors and the disease is contradictory [1,3], except for maternal smoking. Maternal smoking, alcohol consumption, folic acid and vitamin deficiencies especially during the first trimester of pregnancy have been reported to increase the incidence of NSCL/P [4]. In addition, differences in craniofacial characteristics can depend on demographic factors such as age [5,6] and gender [5]. Considering the complex etiology of NSCL/P, studies have shown that gene–gene and gene–environment interactions may be related to NSCL/P susceptibility [3,7,8]. Subsequent genome-wide association studies (GWAS) showed that a number of gene loci are closely associated with the incidence of NSCL/P, which was increased to 40 risk loci by identifying 14 novel loci in the Chinese population added to the 26 previously known risk loci [9,10,11,12]. In addition, an overlap between the genetics of nsCL/P and biologically relevant facial phenotypes was reported, suggesting that a decreased philtrum width can be caused by genetic risk SNPs of nsCL/P [13].
ATP-binding cassette, sub-family A, member 4 (ABCA4) belongs to the transmembrane protein superfamily [14], which is located on chromosome 1p22.1 [15,16]. The ABCA4 is an ATP-binding cassette transporter that is particularly expressed in the rod and cone photoreceptor cells of the vertebrate retina [14,15,16]. It is also expressed in the murine brain [17]. A systematic review reported the significant presence of ocular abnormalities in patients with NSCL/P [18]. Further, GWAS studies [19,20,21] confirmed an association between polymorphisms of ABCA4 and the NSCL/P risk, where one study [19] showed genome-wide significances of 8.14 × 10−8 and 5.01 × 10−12 for the rs560426 polymorphism in combined European and Asian cases and another study [20] reported 1.06 × 10−12 for the rs481931 polymorphism in Asian families. A third study [21], a genome-wide meta-analysis, showed a genome-wide significance of 3.14 × 10−12 for rs560426 in an Asian population. In addition, some studies showed a significant association between rs560426 and rs481931 polymorphisms of ABCA4 with the risk of NSCL/P [22]; in contrast, other studies found no significant association [8,23]. Given these contradictory results, it appears that the genetic basis of oral clefts is still unclear, and identification of additional risk factors for NSCL/P might greatly help in genetic counseling and may prevent the occurrence or further development of this condition [23]. There are no previous meta-analyses reporting the association of ABCA4 (rs560426 and rs481931) and the risk of NSCL/P. Therefore, the present meta-analysis aimed to evaluate the association between the most common polymorphisms of ABCA4 (rs560426 and rs481931) and the risk of NSCL/P in case-control studies.
2. Materials and Methods
This report conforms to the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) guidelines [24].
2.1. Literature Search Strategy
Four electronic databases (Web of Science; Cochrane Library; Scopus; and PubMed) were searched for articles published until 21 June 2020, without restrictions of language or publication date. The searched terms were: (“ABCA4” or “ATP-binding cassette subfamily A member 4” or “rs560426” or “rs481931”) and (“cleft” or “cleft lip” or “cleft palate” or “orofacial cleft” or “oral cleft”). In addition, we checked the references of retrieved articles, review articles, and GWAS studies to find potential articles.
2.2. Eligibility Criteria
One reviewer (M.S.) retrieved the studies from the databases, excluded the duplicate and irrelevant studies after reviewing the titles and abstracts. This was followed by full-text review of the eligible articles. The studies were included if they met the following inclusion criteria: (I) case-control design; (II) NSCL/P was the outcome of interest; (III) reporting ABCA4 rs560426 (A > G) and/or rs481931 (C > A) polymorphisms, and (IV) having the required data to calculate the odds ratios (ORs) with 95% confidence intervals (CIs). The studies were excluded if they did not have the required data regarding genotype distributions or were animal studies, review articles, letters to the editor reporting previous studies, and family-based studies. The second reviewer (M.M.I.) checked the relevant articles based on the eligibility criteria. The differences between the two reviewers were resolved by the third reviewer (S.K.T.).
2.3. Data Extraction
Two reviewers (M.M.I. and M.S.) independently extracted the data from each study. The information retrieved from the studies included: the first author’s name, the publication year, the ethnic group, the source of controls, the mean age, the number of males in the two groups, the number of patients and controls with each genotype, the genotyping method, and the p-value of the Hardy–Weinberg equilibrium in controls. If there was a disagreement between the reviewers, the problem was solved by a third reviewer (S.B.).
2.4. Quality of Assessment
One reviewer (M.M.I.) rated the quality of each included article using the Newcastle–Ottawa Quality Assessment Scale (NOS). The NOS involves assessment of each article for quality in three components: selection, comparability, and exposure. A case-control study could have a minimum and maximum total score of 0 and 9, respectively; a higher score demonstrates better quality [25].
2.5. Statistical Analyses
An analysis was performed by the Review Manager 5.3 using crude OR and 95% CI to show the association between ABCA4 polymorphisms and the risk of NSCL/P in the five genetic models [26]. The Z test was applied to evaluate the pooled OR significance. Heterogeneity across the studies was checked by the Cochrane Q test and I2 statistic. Heterogeneity was considered to be statistically significant if p < 0.1 or I2 > 50%. If there was no significant heterogeneity, the fixed-effects model (Mantel–Haenszel method) was used to estimate the values. Otherwise, we used the random-effects model (DerSimonian and Laird method). The Chi-square test was used to calculate the Hardy–Weinberg equilibrium in the control group of each study. Subgroup analysis was done according to the ethnicity and the source of controls. Meta-regression is a quantitative method used in meta-analysis to estimate the impact of moderator variables on the study effect size. The Comprehensive Meta-Analysis 2.0 was used to derive a funnel plot using the Egger’s and Begg’s tests and p < 0.05 indicated significant existence of publication bias. To evaluate the stability of the results, the following sensitivity analyses were applied: “cumulative analysis” and “one study removed”. The analysis was checked by three reviewers in a discussion (S.B., D.S.B., and M.S.).
3. Results
3.1. Study Selection
Among the 94 retrieved studies, after removing duplicate (61 studies) and excluding the book chapters, conference papers, and studies without reporting the association between the ABCA4 polymorphisms and NSCL/P risk or irrelevant studies (11 studies), a total of 23 studies were evaluated (Figure 1). Of 23 studies, eleven were excluded due to the following reasons: four had no control groups, four did not have the required data, two were letters to the editor reporting previous studies, and one evaluated the mothers of children. Finally, 12 studies were included in this meta-analysis.
Figure 1.
PRISMA flow-chart of the study selection.
3.2. Study Characteristics
Table 1 shows some features of the studies included in this meta-analysis. The studies were published between 2011 and 2018. Five studies [8,27,28,29,30] reported the risk of NSCL/P related to ABCA4 polymorphisms in the Asian ethnicity, four [7,9,23,31] in the mixed ethnicity, and three [32,33,34] in the European descent ethnicity. In six studies [23,27,29,30,32,33], the source of controls were hospitals, while for six other studies [7,8,9,29,31,34], the source of controls was the general population. All included studies reported the ABCA4 rs560426 polymorphism, but only six [7,8,30,32,33,34] reported the ABCA4 rs481931 polymorphism. There were different genotyping methods among the studies presented in Table 1. The Hardy–Weinberg equilibrium was not seen in controls of two studies [8,27] reporting the ABCA4 rs560426 polymorphism and two studies [8,33] reporting the ABCA4 rs481931 polymorphism. We included these studies in the analyses because by excluding them, the number of studies would have been reduced, while this could have been one important reason for an overall biased pattern of results. There were 2859 NSCL/P cases and 3792 controls in the meta-analysis of the ABCA4 rs560426 polymorphism and 1333 NSCL/P cases and 1884 controls in the meta-analysis of the ABCA4 rs481931 polymorphism.
Table 1.
Characteristics of the studies included in this meta-analysis (n = 12).
| First Author, (Year) | Ethnic Group | Source of Controls | Mean Age, Year (NSCL/P Patients to Controls) | No. of Males, (NSCL/P Patients to Controls) | ABCA4 rs560426 | ABCA4 rs481931 | Genotyping Method | p-Value for HWE in Controls | ||
|---|---|---|---|---|---|---|---|---|---|---|
| AA/AG/GG | CC/CA/AA | |||||||||
| Case | Control | Case | Control | |||||||
| Pan et al. (2011) [27] | Asian | HB | 5.54 to 5.49 | 246 to 242 | 145/175/51 | 167/160/57 | NA | NA | TaqMan | 0.071 |
| Fontoura et al. (2012) [32] | European descent | HB | 17.3 to 24.8 | 252 to 165 | 116/118/86 | 74/203/123 | 184/155/44 | 154/192/57 | TaqMan | 0.542/0.818 |
| Huang et al. (2012) [28] | Asian | HB | NA | 169 to 203 | 135/126/39 | 157/171/26 | NA | NA | MALDI-TOF MS (Sequenom) | 0.024 |
| Mostowska et al. (2012) [33] | European descent | HB | NA | NA | 62/105/39 | 120/230/96 | 79/98/29 | 156/196/94 | PCR-HRM | 0.467/0.028 |
| Bagordakis et al. (2013) [31] | Mixed | PB | NA | NA | 74/140/85 | 127/172/85 | NA | NA | Multiplex PCR | 0.067 |
| Zhong-wei et al. (2013) [29] | Asian | PB | NA | NA | 54/91/36 | 36/50/18 | NA | NA | TaqMan | 0.928 |
| Ludwig et al. (2014) [9] | Mixed | PB | NA | 102 to 111 | 37/73/33 | 100/163/66 | NA | NA | MALDI-TOF MS (Sequenom) | 0.977 |
| do Rego Borges et al. (2015) [23] | Mixed | HB | NA | NA | 76/152/65 | 74/187/91 | NA | NA | TaqMan | 0.223 |
| Babu Gurramkonda et al. (2015) [34] | European descent | PB | NA | NA | 46/72/26 | 61/80/35 | 41/68/35 | 57/87/32 | Kompetitive allele specific PCR (KASP) | 0.348/0.905 |
| Mi et al. (2015) [30] | Asian | HB | 4.98 to 5.20 | NA | 88/104/30 | 158/137/29 | 79/107/36 | 113/157/54 | Mini-sequencing (SNAPSHOT) | 0.928/0.965 |
| Velázquez-Aragón et al. (2016) [7] | Mixed | PB | 5.5 to 1.33 | 99 to 132 | 44/56/32 | 54/137/68 | 32/71/27 | 71/131/53 | Kompetitive allele specific PCR (KASP) | 0.326/0.602 |
| Wu et al. (2018) [8] | Asian | PB | NA | NA | 103/116/29 | 111/145/24 | 92/126/30 | 91/154/35 | PCR-RFLP | 0.014/0.015 |
Abbreviations: MALDI-TOF MS, Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; PCR, polymerase chain reaction; HRM, high resolution melting; RFLP, restriction fragment length polymorphism; NSCL/P, non-syndromic cleft lip/palate; HB, hospital-based; PB, population-based; HWE, Hardy–Weinberg equilibrium, NA, not available.
3.3. Quality Assessment
All studies were scored for quality (Table 2), and all had a score of ≥7.
Table 2.
Quality assessment scores for the studies included in this meta-analysis (n = 12).
| First Author, (Year) | Selection (Four Points) | Comparability (Two Points) | Exposure (Three Points) | Total Points |
|---|---|---|---|---|
| Pan et al. (2011) [27] | *** | ** | *** | 8 |
| Fontoura et al. (2012) [32] | *** | - | *** | 7 |
| Huang et al. (2012) [28] | *** | * | *** | 7 |
| Mostowska et al. (2012) [33] | *** | ** | *** | 8 |
| Bagordakis et al. (2013) [31] | **** | - | *** | 7 |
| Zhong-wei et al. (2013) [29] | **** | - | *** | 7 |
| Ludwig et al. (2014) [9] | **** | - | *** | 7 |
| do Rego Borges et al. (2015) [23] | *** | - | *** | 7 |
| Babu Gurramkonda et al. (2015) [34] | **** | ** | *** | 9 |
| Mi et al. (2015) [30] | *** | ** | *** | 8 |
| Velázquez-Aragón et al. (2016) [7] | **** | * | *** | 8 |
| Wu et al. (2018) [8] | **** | - | *** | 7 |
Each asterisk indicates one point. Selection: Is the case definition adequate? (one point), Representativeness of the cases (one point), Selection of Controls (one point), and Definition of Controls (one point). Comparability: Comparability of cases and controls on the basis of the design or analysis (two points). Exposure: Ascertainment of exposure (one point), Same method of ascertainment for cases and controls (one point), and Non-Response rate (one point).
3.4. Pooled Analysis
Table 3 shows the pooled results (combining data from each individual study) for the risk of NSCL/P in those with the ABCA4 rs560426 polymorphism from the twelve studies. The heterogeneity in allelic, homozygote, heterozygote, and dominant models was high (I2 > 50%), and therefore the random-effects model was used. In the recessive model, the fixed-effects model was used because of low heterogeneity (I2 < 50%). The pooled OR was 1.01 (95% CI: 0.88, 1.15; p = 0.92; I2 = 72% (Ph or Pheterogeneity < 0.0001)) in the allelic model (G vs. A), 1.08 (95% CI: 0.79, 1.47; p = 0.64; I2 = 77% (Ph < 0.00001)) in the homozygote model (GG vs. AA), 0.93 (95% CI: 0.73, 1.17; p = 0.53; I2 = 76% (Ph < 0.00001)) in the heterozygote model (AG vs. AA), 0.89 (95% CI: 0.70, 1.14; p = 0.37; I2 = 81% (Ph < 0.00001)) in the dominant model (AG + GG vs. AA), and 1.08 (95% CI: 0.91, 1.26; p = 0.38; I2 = 36% (Ph = 0.10)) in the recessive model (GG vs. AA + AG). Overall, individuals with the ABCA4 rs560426 polymorphism were not at higher risk for NSCL/P than those without the ABCA4 rs560426 polymorphism.
Table 3.
The results of Forest plot analysis of NSCL/P risk related to the ABCA4 rs560426 polymorphism using the five genetic models.
| Genetic Model | First Author, Publication Year | NSCL/P | Control | Weight | Odds Ratio | ||
|---|---|---|---|---|---|---|---|
| Events | Total | Events | Total | M-H, Random, 95%CI | |||
| G vs. A | Pan, 2011 | 277 | 742 | 274 | 768 | 9.2% | 1.07 [0.87, 1.32] |
| Mostowska, 2012 | 183 | 412 | 422 | 892 | 8.7% | 0.89 [0.70, 1.13] | |
| Huang, 2012 | 204 | 600 | 223 | 708 | 8.8% | 1.12 [0.89, 1.41] | |
| Fontoura, 2012 | 290 | 640 | 449 | 800 | 9.2% | 0.65 [0.53, 0.80] | |
| Zhong-wei, 2013 | 163 | 362 | 86 | 208 | 6.7% | 1.16 [0.82, 1.64] | |
| Bagordakis, 2013 | 310 | 598 | 342 | 767 | 9.1% | 1.34 [1.08, 1.66] | |
| Ludwig, 2014 | 139 | 286 | 295 | 658 | 7.9% | 1.16 [0.88, 1.54] | |
| Mi, 2015 | 164 | 444 | 195 | 648 | 8.3% | 1.36 [1.05, 1.76] | |
| do Rego Borges, 2015 | 282 | 586 | 369 | 704 | 9.0% | 0.84 [0.68, 1.05] | |
| Babu Gurramkonda, 2015 | 124 | 288 | 150 | 352 | 7.2% | 1.02 [0.74, 1.39] | |
| Velázquez-Aragón, 2016 | 120 | 264 | 273 | 518 | 7.5% | 0.75 [0.56, 1.01] | |
| Wu, 2018 | 174 | 496 | 193 | 560 | 8.3% | 1.03 [0.80, 1.32] | |
| Subtotal (95%CI) | 5718 | 7583 | 100.0% | 1.01 [0.88, 1.15] | |||
| Total Events | 2430 | 3271 | |||||
| Heterogeneity: Tau2 = 0.04; Chi2 = 39.48, df = 11 (p < 0.0001); I2 = 72%; Test for overall effect: Z = 0.10 (p = 0.92) | |||||||
| GG vs. AA | Pan, 2011 | 51 | 226 | 57 | 224 | 9.1% | 0.85 [0.55, 1.32] |
| Mostowska, 2012 | 39 | 101 | 96 | 216 | 8.8% | 0.79 [0.49, 1.27] | |
| Huang, 2012 | 39 | 174 | 26 | 183 | 8.3% | 1.74 [1.01, 3.01] | |
| Fontoura, 2012 | 86 | 202 | 123 | 197 | 9.4% | 0.45 [0.30, 0.67] | |
| Zhong-wei, 2013 | 36 | 60 | 18 | 54 | 6.6% | 3.00 [1.39, 6.45] | |
| Bagordakis, 2013 | 85 | 159 | 85 | 212 | 9.3% | 1.72 [1.13, 2.60] | |
| Ludwig, 2014 | 33 | 70 | 66 | 166 | 8.1% | 1.35 [0.77, 2.37] | |
| Mi, 2015 | 30 | 118 | 29 | 187 | 8.1% | 1.86 [1.05, 3.29] | |
| do Rego Borges, 2015 | 65 | 141 | 91 | 165 | 9.0% | 0.70 [0.44, 1.09] | |
| Babu Gurramkonda, 2015 | 26 | 72 | 35 | 96 | 7.6% | 0.99 [0.52, 1.86] | |
| Velázquez-Aragón, 2016 | 32 | 76 | 68 | 122 | 8.0% | 0.58 [0.32, 1.03] | |
| Wu, 2018 | 29 | 132 | 24 | 135 | 7.8% | 1.30 [0.71, 2.38] | |
| Subtotal (95%CI) | 1531 | 1957 | 100.0% | 1.08 [0.79, 1.47] | |||
| Total EVENTS | 551 | 718 | |||||
| Heterogeneity: Tau2 = 0.23; Chi2 = 47.66, df = 11 (p < 0.00001); I2 = 77%; Test for overall effect: Z = 0.47 (p = 0.64) | |||||||
| AG vs. AA | Pan, 2011 | 175 | 320 | 160 | 327 | 9.3% | 1.26 [0.92, 1.72] |
| Mostowska, 2012 | 105 | 167 | 230 | 350 | 8.6% | 0.88 [0.60, 1.30] | |
| Huang, 2012 | 126 | 261 | 171 | 328 | 9.2% | 0.86 [0.62, 1.19] | |
| Fontoura, 2012 | 118 | 234 | 203 | 277 | 8.7% | 0.37 [0.26, 0.54] | |
| Zhong-wei, 2013 | 91 | 145 | 50 | 86 | 6.9% | 1.21 [0.70, 2.09] | |
| Bagordakis, 2013 | 140 | 214 | 172 | 299 | 8.8% | 1.40 [0.97, 2.01] | |
| Ludwig, 2014 | 73 | 110 | 163 | 263 | 7.7% | 1.21 [0.76, 1.93] | |
| Mi, 2015 | 104 | 192 | 137 | 295 | 8.8% | 1.36 [0.95, 1.96] | |
| Babu Gurramkonda, 2015 | 72 | 118 | 80 | 141 | 7.4% | 1.19 [0.73, 1.96] | |
| do Rego Borges, 2015 | 152 | 228 | 187 | 261 | 8.6% | 0.79 [0.54, 1.16] | |
| Velázquez-Aragón, 2016 | 56 | 100 | 137 | 191 | 7.3% | 0.50 [0.30, 0.83] | |
| Wu, 2018 | 116 | 219 | 145 | 256 | 8.8% | 0.86 [0.60, 1.24] | |
| Subtotal (95% CI) | 2308 | 3074 | 100.0% | 0.93 [0.73, 1.17] | |||
| Total Events | 1328 | 1835 | |||||
| Heterogeneity: Tau2 = 0.13; Chi2 = 46.53, df = 11 (p < 0.00001); I2 = 76%; Test for overall effect: Z = 0.63 (p = 0.53) | |||||||
| AG + GG vs. AA | Pan, 2011 | 226 | 371 | 217 | 384 | 9.1% | 1.20 [0.90, 1.60] |
| Fontoura, 2012 | 204 | 320 | 326 | 400 | 8.7% | 0.40 [0.28, 0.56] | |
| Huang, 2012 | 165 | 300 | 197 | 354 | 8.9% | 0.97 [0.71, 1.33] | |
| Mostowska, 2012 | 144 | 206 | 326 | 446 | 8.5% | 0.85 [0.59, 1.23] | |
| Zhong-wei, 2013 | 127 | 181 | 68 | 104 | 7.2% | 1.25 [0.74, 2.08] | |
| Bagordakis, 2013 | 225 | 299 | 257 | 384 | 8.7% | 1.50 [1.07, 2.11] | |
| Ludwig, 2014 | 106 | 143 | 229 | 329 | 7.8% | 1.25 [0.80, 1.95] | |
| Mi, 2015 | 134 | 222 | 166 | 324 | 8.6% | 1.45 [1.03, 2.05] | |
| do Rego Borges, 2015 | 217 | 293 | 278 | 352 | 8.5% | 0.76 [0.53, 1.10] | |
| Babu Gurramkonda, 2015 | 68 | 144 | 115 | 176 | 7.7% | 0.47 [0.30, 0.75] | |
| Velázquez-Aragón, 2016 | 88 | 132 | 205 | 259 | 7.6% | 0.53 [0.33, 0.84] | |
| Wu, 2018 | 145 | 248 | 169 | 280 | 8.6% | 0.92 [0.65, 1.31] | |
| Subtotal (95%CI) | 2859 | 3792 | 100.0% | 0.89 [0.70, 1.14] | |||
| Total Events | 1849 | 2553 | |||||
| Heterogeneity: Tau2 = 0.15; Chi2 = 59.28, df = 11 (p < 0.00001); I2 = 81%; Test for overall effect: Z = 0.90 (p = 0.37) | |||||||
| GG vs. AA + AG | Pan, 2011 | 51 | 371 | 57 | 384 | 9.6% | 0.91 [0.61, 1.37] |
| Fontoura, 2012 | 86 | 320 | 123 | 400 | 12.2% | 0.83 [0.60, 1.15] | |
| Huang, 2012 | 39 | 300 | 26 | 354 | 6.9% | 1.89 [1.12, 3.18] | |
| Mostowska, 2012 | 39 | 206 | 96 | 446 | 9.4% | 0.85 [0.56, 1.29] | |
| Bagordakis, 2013 | 85 | 299 | 85 | 384 | 11.4% | 1.40 [0.99, 1.98] | |
| Zhong-wei, 2013 | 36 | 181 | 18 | 104 | 5.3% | 1.19 [0.63, 2.22] | |
| Ludwig, 2014 | 33 | 143 | 66 | 329 | 7.9% | 1.20 [0.74, 1.92] | |
| do Rego Borges, 2015 | 65 | 293 | 91 | 352 | 10.9% | 0.82 [0.57, 1.18] | |
| Mi, 2015 | 30 | 222 | 29 | 324 | 6.6% | 1.59 [0.92, 2.73] | |
| Babu Gurramkonda, 2015 | 26 | 144 | 35 | 176 | 6.2% | 0.89 [0.51, 1.56] | |
| Velázquez-Aragón, 2016 | 32 | 132 | 68 | 259 | 7.7% | 0.90 [0.55, 1.46] | |
| Wu, 2018 | 29 | 248 | 24 | 280 | 6.1% | 1.41 [0.80, 2.50] | |
| Subtotal (95%CI) | 2859 | 3792 | 100.0% | 1.08 [0.91, 1.26] | |||
| Total Events | 551 | 718 | |||||
| Heterogeneity: Tau2 = 0.03; Chi2 = 17.14, df = 11 (p = 0.10); I2 = 36%; Test for overall effect: Z = 0.87 (p = 0.38) | |||||||
Abbreviations: NSCL/P, non-syndromic cleft lip with or without a cleft palate; CI, confidence interval. All models were analyzed based on random-effects model except for “GG vs. AA + AG” that was based on fixed-effects model
The pooled results for the risk of NSCL/P related to the ABCA4 rs481931 polymorphism based on the findings from the six studies are shown in Table 4. The heterogeneity in all analyses was low, and therefore, fixed-effect models were used (I2 < 50%). The pooled OR was 0.92 (95% CI: 0.83, 1.02; p = 0.12; I2 = 35% (Ph = 0.17)) in the allelic model (A vs. C), 0.85 (95%CI: 0.68, 1.05; p = 0.13; I2 = 33% (Ph = 0.19)) in the homozygote model (AA vs. CC), 0.88 (95%CI: 0.75, 1.03; p = 0.12; I2 = 16% (Ph = 0.31)) in the heterozygote model (CA vs. CC), 0.87 (95%CI: 0.75, 1.00; p = 0.06; I2 = 29% (Ph = 0.22)) in the dominant model (CA + AA vs. CC), and 0.89 (95%CI: 0.74, 1.09; p = 0.26; I2 = 22% (Ph = 0.27)) in the recessive model (AA vs. CC + CA). Based on the genetic models, there was no significant risk of NSCL/P in those with the ABCA4 rs481931 polymorphism.
Table 4.
The results of Forest plot analysis of NSCL/P risk related to the ABCA4 rs481931 polymorphism using the five genetic models.
| Genetic Model | First Author, Publication Year | NSCL/P | Control | Weight | Odds Ratio | ||
|---|---|---|---|---|---|---|---|
| Events | Total | Events | Total | M-H, Fixed, 95%CI | |||
| A vs. C | Fontoura, 2012 | 243 | 766 | 306 | 806 | 26.6% | 0.76 [0.62, 0.94] |
| Mostowska, 2012 | 183 | 412 | 422 | 892 | 19.4% | 0.89 [0.70, 1.13] | |
| Mi, 2015 | 179 | 444 | 265 | 648 | 16.8% | 0.98 [0.76, 1.25] | |
| Babu Gurramkonda, 2015 | 138 | 288 | 151 | 352 | 9.2% | 1.22 [0.90, 1.67] | |
| Velázquez-Aragón, 2016 | 125 | 260 | 237 | 510 | 10.9% | 1.07 [0.79, 1.44] | |
| Wu, 2018 | 186 | 496 | 224 | 560 | 17.2% | 0.90 [0.70, 1.15] | |
| Subtotal (95%CI) | 2666 | 3768 | 100.0% | 0.92 [0.83, 1.02] | |||
| Total Events | 1054 | 1605 | |||||
| Heterogeneity: Chi2 = 7.74, df = 5 (p = 0.17); I2 = 35%; Test for overall effect: Z = 1.57 (p = 0.12) | |||||||
| AA vs. CC | Fontoura, 2012 | 44 | 228 | 57 | 211 | 26.6% | 0.65 [0.41, 1.01] |
| Mostowska, 2012 | 29 | 108 | 94 | 250 | 23.1% | 0.61 [0.37, 1.00] | |
| Babu Gurramkonda, 2015 | 35 | 76 | 32 | 89 | 8.8% | 1.52 [0.81, 2.84] | |
| Mi, 2015 | 36 | 115 | 54 | 167 | 16.8% | 0.95 [0.57, 1.59] | |
| Velázquez-Aragón, 2016 | 27 | 59 | 53 | 124 | 10.3% | 1.13 [0.61, 2.11] | |
| Wu, 2018 | 30 | 122 | 35 | 126 | 14.4% | 0.85 [0.48, 1.50] | |
| Subtotal (95%CI) | 708 | 967 | 100.0% | 0.85 [0.68, 1.05] | |||
| Total Events | 201 | 325 | |||||
| Heterogeneity: Chi2 = 7.49, df = 5 (p = 0.19); I2 = 33%; Test for overall effect: Z = 1.52 (p = 0.13) | |||||||
| CA vs. CC | Fontoura, 2012 | 155 | 339 | 192 | 346 | 31.0% | 0.68 [0.50, 0.91] |
| Mostowska, 2012 | 98 | 177 | 196 | 352 | 17.6% | 0.99 [0.69, 1.42] | |
| Mi, 2015 | 107 | 186 | 157 | 270 | 16.3% | 0.97 [0.67, 1.42] | |
| Babu Gurramkonda, 2015 | 68 | 109 | 87 | 144 | 8.5% | 1.09 [0.65, 1.81] | |
| Velázquez-Aragón, 2016 | 71 | 103 | 131 | 202 | 8.3% | 1.20 [0.72, 2.00] | |
| Wu, 2018 | 126 | 218 | 154 | 245 | 18.4% | 0.81 [0.56, 1.18] | |
| Subtotal (95% CI) | 1132 | 1559 | 100.0% | 0.88 [0.75, 1.03] | |||
| Total Events | 625 | 917 | |||||
| Heterogeneity: Chi2 = 5.93, df = 5 (p = 0.31); I2 = 16%; Test for overall effect: Z = 1.57 (p = 0.12) | |||||||
| CA + AA vs. CC | Fontoura, 2012 | 199 | 383 | 249 | 403 | 31.1% | 0.67 [0.50, 0.89] |
| Mostowska, 2012 | 127 | 206 | 290 | 446 | 18.7% | 0.86 [0.61, 1.22] | |
| Babu Gurramkonda, 2015 | 103 | 144 | 119 | 176 | 8.1% | 1.20 [0.74, 1.95] | |
| Mi, 2015 | 143 | 222 | 211 | 324 | 16.3% | 0.97 [0.68, 1.39] | |
| Velázquez-Aragón, 2016 | 98 | 130 | 184 | 255 | 8.2% | 1.18 [0.73, 1.92] | |
| Wu, 2018 | 156 | 248 | 189 | 280 | 17.6% | 0.82 [0.57, 1.17] | |
| Subtotal (95%CI) | 1333 | 1884 | 100.0% | 0.87 [0.75, 1.00] | |||
| Total Events | 826 | 1242 | |||||
| Heterogeneity: Chi2 = 7.05, df = 5 (p = 0.22); I2 = 29%; Test for overall effect: Z = 1.91 (p = 0.06) | |||||||
| AA vs. CC + CA | Mostowska, 2012 | 29 | 206 | 94 | 446 | 23.6% | 0.61 [0.39, 0.97] |
| Fontoura, 2012 | 44 | 383 | 57 | 403 | 22.8% | 0.79 [0.52, 1.20] | |
| Babu Gurramkonda, 2015 | 35 | 144 | 32 | 176 | 10.1% | 1.44 [0.84, 2.48] | |
| Mi, 2015 | 36 | 222 | 54 | 324 | 17.0% | 0.97 [0.61, 1.53] | |
| Velázquez-Aragón, 2016 | 27 | 130 | 53 | 255 | 13.1% | 1.00 [0.59, 1.68] | |
| Wu, 2018 | 30 | 248 | 35 | 280 | 13.4% | 0.96 [0.57, 1.62] | |
| Subtotal (95%CI) | 1333 | 1884 | 100.0% | 0.89 [0.74, 1.09] | |||
| Total Events | 201 | 325 | |||||
| Heterogeneity: Chi2 = 6.39, df = 5 (p = 0.27); I2 = 22%; Test for overall effect: Z = 1.12 (p = 0.26) | |||||||
Abbreviations: NSCL/P, non-syndromic cleft lip with or without a cleft palate; CI, confidence interval.
3.5. Subgroup Analysis
The first subgroup analysis evaluated the effect of ethnicity and the source of controls in the association of NSCL/P with the ABCA4 rs560426 polymorphism (Table 5). A significant risk for NSCL/P was observed in the Asian ethnicity for the allelic model (OR = 1.13; 95% CI: 1.01, 1.27; p = 0.03), the homozygote model (OR = 1.53; 95% CI: 1.01, 2.31; p = 0.04), and the recessive model (OR = 1.30; 95% CI: 1.03, 1.63; p = 0.03). The results did not demonstrate a risk of NSCL/P related to the rs560426 polymorphism among people of European descent and mixed ethnicities (such as Brazilian and Mexican ethnicities). Furthermore, the source of controls did not seem to have any effect on the association of NSCL/P and the ABCA4 rs560426 polymorphism.
Table 5.
Analysis of non-syndromic cleft lip/palate risk related to the rs560426 polymorphism according to ethnicity and source of controls.
| Variable (N) | G vs. A | GG vs. AA | AG vs. AA | AG + GG vs. AA | GG vs. AA + AG |
|---|---|---|---|---|---|
| OR (95%CI), I2 (%), Ph | OR (95%CI), I2 (%), Ph | OR (95%CI), I2 (%), Ph | OR (95%CI), I2 (%), Ph | OR (95%CI), I2 (%), Ph | |
| Overall (12) | 1.01 (0.88, 1.15), 72, <0.0001 | 1.08 (0.79, 1.47), 77, <0.00001 | 0.93 (0.73, 1.16), 76, <0.00001 | 0.89 (0.70, 1.14), 81, <0.00001 | 1.08 (0.91, 1.26), 36, 0.10 |
| Ethnicity | |||||
| Asian (5) | 1.13 (1.01, 1.27), 0, 0.59 | 1.53 (1.01, 2.31), 62, 0.03 | 1.08 (0.92, 1.26), 35, 0.19 | 1.13 (0.97, 1.32), 10, 0.35 | 1.30 (1.03, 1.63), 27, 0.24 |
| European Descent (3) | 0.82 (0.63, 1.08), 71, 0.03 | 0.67 (0.42, 1.09), 64, 0.06 | 0.72 (0.36, 1.45), 88, 0.0002 | 0.55 (0.34, 0.88), 79, 0.009 | 0.85 (0.67, 1.07), 0, 0.98 |
| Mixed (4) | 1.00 (0.76, 1.31), 79, 0.003 | 0.99 (0.59, 1.68), 78, 0.004 | 0.92 (0.60, 1.42), 76, 0.006 | 0.94 (0.60, 1.49), 81, 0.001 | 1.07 (0.87, 1.30), 41, 0.17 |
| Source of Controls | |||||
| Hospital-Based (6) | 0.94 (0.86, 1.03), 80, 0.0001 | 0.83 (0.69, 1.01), 80, 0.0002 | 0.87 (0.76, 1.01), 84, <0.00001 | 0.89 (0.78, 1.02), 85, <0.00001 | 0.98 (0.83, 1.15), 57, 0.04 |
| Population-Based (6) | 1.07 (0.91, 1.26), 52, 0.06 | 1.29 (0.86, 1.94), 66, 0.01 | 1.02 (0.76, 1.36), 60, 0.03 | 0.91 (0.62, 1.33), 80, 0.0002 | 1.18 (0.97, 1.44), 0, 0.63 |
Bold numbers mean statistically significant (p < 0.05). Ph indicates Pheterogeneity. Abbreviations: OR, odds ratio; CI, confidence interval. “N” shows the number of studies.
The second subgroup analysis assessed the risk of NSCL/P related to the ABCA4 rs481931 polymorphism according to the ethnicity and the source of controls (Table 6). There was no association between the risk of NSCL/P and the rs481931 polymorphism based on ethnicity or source of controls.
Table 6.
Analysis of non-syndromic cleft lip/palate risk related to the rs481931 polymorphism according to ethnicity and source of controls.
| Variable (N) | A vs. C | AA vs. CC | CA vs. CC | CA + AA vs. CC | AA vs. CC + CA |
|---|---|---|---|---|---|
| OR (95%CI), I2 (%), Ph | OR (95%CI), I2 (%), Ph | OR (95%CI), I2 (%), Ph | OR (95%CI), I2 (%), Ph | OR (95%CI), I2 (%), Ph | |
| Overall (6) | 0.92 (0.83, 1.02), 35, 0.17 | 0.85 (0.68, 1.05), 33, 0.19 | 0.88 (0.75, 1.03), 16, 0.31 | 0.87 (0.75, 1.00), 0.29, 0.22 | 0.89 (0.74, 1.09), 22, 0.27 |
| Ethnicity | |||||
| Asian (2) | 0.94 (0.79, 1.12), 0, 0.65 | 0.90 (0.62, 1.32), 0, 0.76 | 0.89 (0.68, 1.16), 0, 0.49 | 0.89 (0.69, 1.15), 0, 0.51 | 0.97 (0.68, 1.36), 0, 0.99 |
| European Descent (3) | 0.92 (0.71, 1.18), 68, 0.04 | 0.81 (0.48, 1.36), 67, 0.05 | 0.83 (0.67, 1.03), 46, 0.15 | 0.85 (0.62, 1.16), 56, 0.11 | 0.87 (0.55, 1.38), 66, 0.05 |
| Mixed (1) | 1.07 (0.79, 1.44) | 1.13 (0.61, 2.11) | 1.20 (0.72, 2.00) | 1.18 (0.73, 1.92) | 1.00 (0.59, 1.68) |
| Source of Controls | |||||
| Hospital-Based (2) | 0.85 (0.67, 1.09), 57, 0.13 | 0.77 (0.55, 1.07), 21, 0.26 | 0.80 (0.56, 1.14), 55, 0.14 | 0.79 (0.55, 1.14), 61, 0.11 | 0.86 (0.63, 1.18), 0, 0.52 |
| Population-Based (4) | 0.98 (0.86, 1.12), 11, 0.34 | 0.91 (0.68, 1.20), 47, 0.13 | 0.97, (0.79, 1.20), 0, 0.62 | 0.95 (0.78, 1.16), 0, 0.44 | 0.91 (0.71, 1.17), 49, 0.12 |
p-Value > 0.05 in all analyses and Ph indicates Pheterogeneity. Abbreviations: OR, odds ratio; CI, confidence interval. “N” shows the number of studies.
3.6. Meta-Regression
The results of the meta-regression for finding the effect of the publication year and the number of participants on the results showed that publication year was not a significant confounding on the association between both rs560426 and rs481931 polymorphisms and the risk of NSCL/P (Table 7). In addition, the number of participants was not a significant confounding factor on the association between rs560426 polymorphism and the risk of NSCL/P, but the number of participants was a significant confounding factor for the rs481931 polymorphism on the pooled ORs in allele, homozygote, and dominant models.
Table 7.
Meta-regression analysis based on publication year for association between rs560426 and rs481931 polymorphisms and the risk of non-syndromic cleft lip/palate.
| Variable | Polymorphism | Allele | Homozygote | Heterozygote | Recessive | Dominant | |
|---|---|---|---|---|---|---|---|
| Publication Year | rs560426 | R | 0.025 | 0.047 | 0.113 | 0.172 | 0.075 |
| Adjusted R2 | −0.099 | −0.098 | −0.086 | −0.068 | −0.094 | ||
| P | 0.937 | 0.884 | 0.726 | 0.593 | 0.816 | ||
| rs481931 | R | 0.406 | 0.456 | 0.243 | 0.388 | 0.495 | |
| Adjusted R2 | −0.044 | 0.010 | −0.176 | −0.062 | 0.057 | ||
| P | 0.424 | 0.364 | 0.643 | 0.447 | 0.318 | ||
| Number of Participants | rs560426 | R | 0.098 | 0.410 | 0.171 | 0.118 | 0.036 |
| Adjusted R2 | −0.089 | 0.085 | −0.068 | −0.085 | −0.099 | ||
| P | 0.761 | 0.185 | 0.594 | 0.714 | 0.912 | ||
| rs481931 | R | 0.953 | 0.913 | 0.810 | 0.650 | 0.814 | |
| Adjusted R2 | 0.886 | 0.793 | 0.570 | 0.279 | 0.579 | ||
| P | 0.003 | 0.011 | 0.051 | 0.162 | 0.049 |
R: Correlation coefficient. Bold numbers mean statistically significant (p < 0.05).
3.7. Sensitivity Analysis
The sensitivity analyses (“cumulative analysis” and “one study removed” analyses), identified that the pooled ORs under all genetic models were stable and trustworthy (the values of the pooled ORs did not change). Further, excluding two studies [8,28] with p-values < 0.05 for the Hardy–Weinberg equilibrium in controls did not change the overall result regarding the risk of NSCL/P in relation to the ABCA4 rs560426 polymorphism. Excluding two other studies [8,33] did not change the overall result regarding the risk of NSCL/P with the ABCA4 rs481931 polymorphism (not presented in tables). The quality score of all studies was high (≥7) and therefore we could not use a sensitivity analysis (excluding the studies with low quality) for this topic.
3.8. Publication Bias
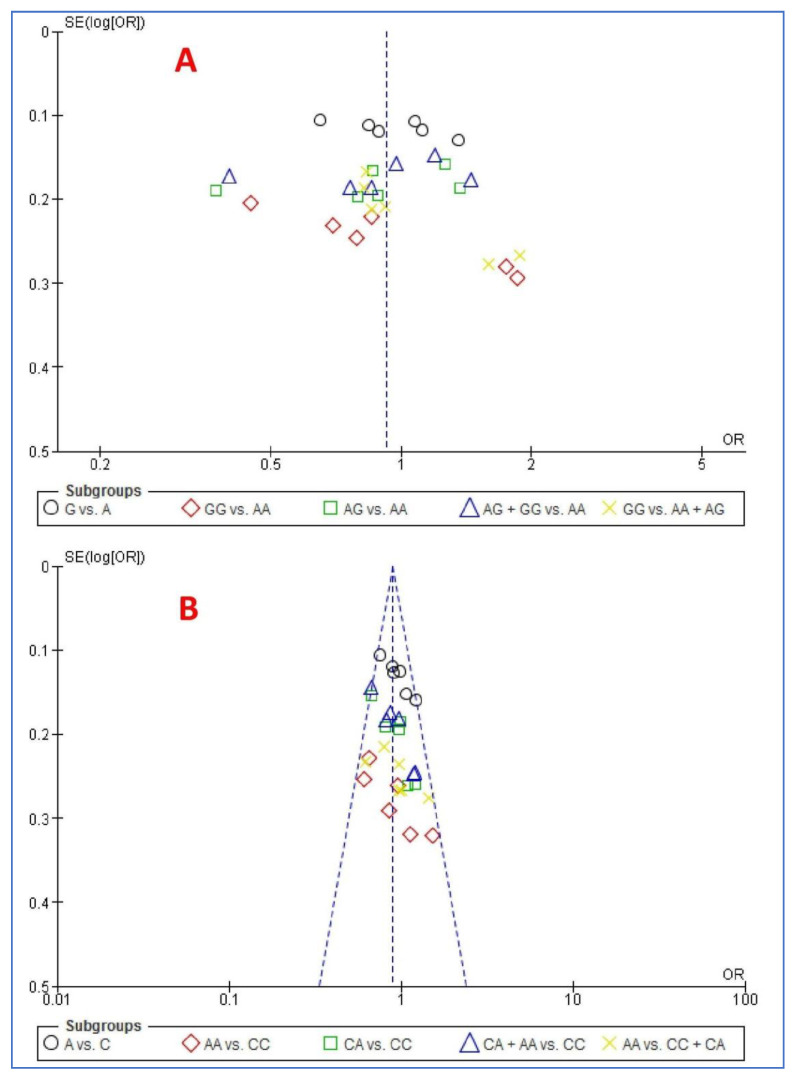
Figure 2 shows the funnel plot of the association between ABCA4 polymorphisms and the risk of NSCL/P using the five genetic models. The Egger’s and Begg’s tests did not reveal any publication bias (p > 0.05) except for four states. Both tests for allele and homozygote models revealed a publication bias and Egger’s test for heterozygote and recessive models revealed a publication bias (p < 0.05).
Figure 2.
Funnel plot of the association between ABCA4 polymorphisms and risk of NSCL/P using the five genetic models: (A) rs560426 and (B) rs481931.
4. Discussion
The present meta-analysis evaluated the associations between rs560426 and rs481931 polymorphisms of ABCA4 and the risk of NSCL/P based on five genetic models. The key findings were as follows: While there was no significant association between both polymorphisms and the risk of NSCL/P, in contrast, subgroup analyses demonstrated that there was a higher risk of NSCL/P for the allelic model, the homozygote model, and the recessive model in the Asian ethnicity for the rs560426 polymorphism. Further, there were no associations between both polymorphisms (rs560426 and rs481931) and the NSCL/P risk in the European descent and the mixed ethnicities. The present results showed association of the rs560426 polymorphism and the risk of NSCL/P among the Asian, but not among the European, descent. Further, the meta-regression showed that the number of participants was a confounding factor for the association between the rs481931 polymorphism and the risk of NSCL/P. Overall, the ethnicity and the number of participants may act as significant confounders in this association.
NSCL/P is a complex congenital anomaly that shows both clinical and genetic heterogeneity and the genetic basis of NSCL/P has remained unclear [23].
Further, there is little biological evidence to support the role of ABCA4 in facial cranial morphogenesis, especially since Abca4 null mice do not show cleft palate [35]. It turned out that the rs560426 G allele showed a 1.36- [30] and 1.34-fold [31] increase in the risk of NSCL/P, when compared to the A allele. Further, the rs560426 GG genotype showed 1.74- [28], 3.0- [29], 1.86- [30], and 1.72-fold [31] elevated risks of NSCL/P compared to the AA genotype. In contrast, the G allele (compared to the A allele), GG genotype [32] (compared to the AA genotype), AG genotype [7,32] (compared to the AA genotype), A allele [32] (compared to the C allele), AA genotype [32] (compared to the CC genotype), and the CA genotype [32] (compared to the CC genotype) had protective roles against NSCL/P. Further, when ethnicity was not considered, the present meta-analysis did not yield any significant risk of NSCL/P related to ABCA4 polymorphisms (rs560426 and rs481931). In contrast, when ethnicity was considered, among Asians, both the G allele (compared to the A allele) and GG genotype (compared to AA genotype) were associated with a significantly increased risk of NSCL/P. Next, there is a need to further investigate the possible role of the ABCA4 gene in the etiology of NSCL/P. This need stems from the following observations: First, some studies [30,31,32] indicated a link between the ABCA4 gene and the risk of a NSCL/P; second, two GWAS studies [19,36] underscored the associations between the ABCA4 gene with the risk of CL/P; third, the association between the ABCA4 gene and the risk of NSCL/P was observed among at least three different populations, that is to say, among Taiwanese, Honduran and Columbian populations [20,35,37]. Next, Fontoura et al. [32] found significant associations between ABCA4 rs481931 alleles and the status of bilateral and unilateral NSCL/P. Last, the replication of ABCA4 polymorphisms among independent families from various populations revealed that Asian families showed evidence of a higher risk of NSCL/P related to polymorphisms of the ABCA4 gene when compared to the European or American families [28]. Importantly, the present meta-analysis confirmed a stronger evidence of risk of NSCL/P related to the rs560426 polymorphism of the ABCA4 gene in Asian compared to other ethnicities.
One study [27] suggested that the strong correlation between the ABCA4 gene and the risk of NSCL/P may be due to the impact of nearby genes (such as Rho GTPase Activating Protein 29 (ARHGAP29), and not due to the impact of a single locus. ARHGAP29 is expressed in the palate and lips of mice, its expression depends on Interferon Regulatory Factor 6 (IRF6), and coding variants in ARHGAP29 are related to CL/P [12]. Therefore, evidence showed that ARHGAP29 is the risk gene at 1p22 [38]. It follows that it is possible that the associated variants in ABCA4 are in linkage disequilibrium with a causal variant included in a neighboring gene, and that associated variants in ABCA4 act as indirect surrogates for a real etiologic variant in individuals with NSCL/P [32,33,37].
Besides a purely genetic-based explanation of the etiology of NSCL/P, both gene–gene and gene–environment interactions showed that there was an interaction between the ABCA4 rs560426 polymorphism and folic acid consumption and the V-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B rs11696257 polymorphism [7]. Further, the different frequencies of the G allele of rs560426 among different populations may identify the complex genetic etiology of NSCL/P and this polymorphism alone has no effect on the pathogenesis of the NSCL/P [30]. Transmission analysis using case-parent core pedigrees showed that the C allele of rs481931 was significantly over-transmitted from parents to children, indicating that the C allele is associated with the NSCL/P risk. In addition, the C (rs481931)-G (rs560426) haplotype was significantly involved in the occurrence of NSCL/P [8].
Last, a significantly different rate of the G allele of the rs560426 polymorphism among Asians, but not among Europeans, showed that the genetic variations can be associated with the pathogenesis of NSCL/P, but the role of ethnicity on genetic variations should be taken into consideration.
To summarize, the main findings of the present meta-analysis are as follows: the discrepancy between the results of GWAS studies, this meta-analysis, and individual studies suggest that ABCA4 polymorphisms may be in an imbalance linked to polymorphism(s) located in other genes and ABCA4 can be an indirect substitute for NSCL/P etiology. However, the novelty should be balanced against the following limitations. First, the number of studies was rather small. Second, samples were not further investigated for age, sex, and genotyping methods. Third, not all studies reported the Hardy–Weinberg equilibrium. Likewise, fourth, some sources of controls and ethnicities evaluated in the studies were different. However, sensitivity analysis identified the stability of the results, and also the heterogeneity across the studies was minimal.
5. Conclusions
The findings of the present meta-analysis confirmed that the G allele and GG genotype of the rs560426 polymorphism were significantly associated with the NSCL/P risk in the Asian population, while no such association was observed for the rs481931 polymorphism. There was no association between both polymorphisms (rs560426 and rs481931) and the risk of NSCL/P in the European descent and the mixed ethnicities. There were no differences in the risk of NSCL/P with rs560426 and rs481931 polymorphisms in relation to the source of the controls. Therefore, the ethnicity and the number of participants may act as significant factors in this association. It follows that further studies are needed, focusing on the gene–gene interactions of ABCA4 and ARHGAP29 with larger sample sizes in different ethnicities to understand their contribution to the present pattern of results.
Acknowledgments
Santosh Kumar Tadakamadla is supported by National Health and Medical Research Council Early Career Fellowship, Australia.
Abbreviations
| ABCA4 | ATP-binding cassette, sub-family A, member 4 |
| CI | Confidence interval |
| GWAS | genome-wide association |
| NSCL/P | Non-syndromic cleft lip/palate |
| OR | Odds ratio |
| ARHGAP29 | Rho GTPase Activating Protein 29 |
| IRF6 | Interferon Regulatory Factor 6 |
Author Contributions
Conceptualization, M.M.I. and M.S.; methodology, M.S.; software, M.S.; validation, M.M.I. and S.B.; formal analysis, M.S.; investigation, D.S.B.; resources, S.B.; data curation, M.S.; writing—original draft preparation, S.B.; writing—review and editing, M.S., S.K.T., A.B., M.T., and S.B.; visualization, S.K.T.; supervision, M.M.I.; project administration, S.B.; funding acquisition, M.M.I. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Kermanshah University of Medical Sciences, Kermanshah, Iran (Grant number: 980760).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mossey P.A., Little J., Munger R.G., Dixon M.J., Shaw W.C. Cleft lip and palate. Lancet. 2009;374:1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- 2.Mossey P.A., Modell B. Epidemiology of oral clefts 2012: An international perspective. Front. Oral Biol. 2012;16:1–18. doi: 10.1159/000337464. [DOI] [PubMed] [Google Scholar]
- 3.Dixon M.J., Marazita M.L., Beaty T.H., Murray J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bezerra J.F., Oliveira G.H.M., Soares C.D., Cardoso M.L., Ururahy M.A.G., Neto F.P.F., Lima-Neto L.G., Luchessi A.D., Silbiger V.N., Fajardo C.M., et al. Genetic and non-genetic factors that increase the risk of non-syndromic cleft lip and/or palate development. Oral Dis. 2015;21:393–399. doi: 10.1111/odi.12292. [DOI] [PubMed] [Google Scholar]
- 5.Perillo L., Isola G., Esercizio D., Iovane M., Triolo G., Matarese G. Differences in craniofacial characteristics in Southern Italian children from Naples: A retrospective study by cephalometric analysis. Eur. J. Paediatr. Dent. 2013;14:195–198. [PubMed] [Google Scholar]
- 6.Leonardi R., Aboulazm K., Giudice A.L., Ronsivalle V., D’Antò V., Lagravère M., Isola G. Evaluation of mandibular changes after rapid maxillary expansion: A CBCT study in youngsters with unilateral posterior crossbite using a surface-to-surface matching technique. Clin. Oral Investig. 2020:1–11. doi: 10.1007/s00784-020-03480-5. [DOI] [PubMed] [Google Scholar]
- 7.Velázquez-Aragón J.A., Alcántara-Ortigoza M.A., Estandia-Ortega B., Reyna-Fabián M.E., Méndez-Adame C., Angel A.G.-D. Gene Interactions Provide Evidence for Signaling Pathways Involved in Cleft Lip/Palate in Humans. J. Dent. Res. 2016;95:1257–1264. doi: 10.1177/0022034516647034. [DOI] [PubMed] [Google Scholar]
- 8.Wu N., Lu Y., Liu K., Li Z., Liu Q., Lu L. Associations of ABCA4 and MAFB with Nonsyndromic Cleft Lip with or without Cleft Palate in a Northeastern Chinese Population. J. Hard Tissue Biol. 2018;27:181–184. doi: 10.2485/jhtb.27.181. [DOI] [Google Scholar]
- 9.Ludwig K.U., Wahle P., Reutter H., Paredes-Zenteno M., Munoz-Jimenez S.G., Ortiz-López R., Böhmer A.C., Tessmann P., Nowak S., Nöthen M.M., et al. Evaluating eight newly identified susceptibility loci for nonsyndromic cleft lip with or without cleft palate in a Mesoamerican population. Birth Defects Res. Part A Clin. Mol. Teratol. 2013;100:43–47. doi: 10.1002/bdra.23209. [DOI] [PubMed] [Google Scholar]
- 10.Böhmer A.C., Gölz L., Kreusch T., Kramer F.J., Pötzsch B., Nöthen M.M., Jäger A., Mangold E., Knapp M., Ludwig K.U. Investigation of dominant and recessive inheritance models in GWAS data ofnonsyndromic cleft lip with or without cleft palate. Birth Defects Res. 2018;110:336–341. doi: 10.1002/bdr2.1144. [DOI] [PubMed] [Google Scholar]
- 11.Beaty T.H., Taub M.A., Scott A.F., Murray J.C., Marazita M.L., Schwender H., Parker M.M., Hetmanski J.B., Balakrishnan P., Mansilla M.A., et al. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum. Genet. 2013;132:771–781. doi: 10.1007/s00439-013-1283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leslie E.J., Mansilla M.A., Biggs L.C., Schuette K., Bullard S., Cooper M., Dunnwald M., Lidral A.C., Marazita M.L., Beaty T.H., et al. Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res. Part A Clin. Mol. Teratol. 2012;94:934–942. doi: 10.1002/bdra.23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howe L.J., Lee M.K., Sharp G.C., Smith G.D., Pourcain B.S., Shaffer J.R., Ludwig K.U., Mangold E., Marazita M.L., Feingold E., et al. Investigating the shared genetics of non-syndromic cleft lip/palate and facial morphology. PLoS Genet. 2018;14:e1007501. doi: 10.1371/journal.pgen.1007501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molday R.S., Zhong M., Quazi F. The role of the photoreceptor ABC transporter ABCA4 in lipid transport and Stargardt macular degeneration. Biochim. Biophys. Acta. 2009;1791:573–583. doi: 10.1016/j.bbalip.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun H., Molday R.S., Nathans J. Retinal stimulates ATP hydrolysis by purified and reconstituted ABCR, the photoreceptor-specific ATP-binding cassette transporter responsible for Stargardt disease. J. Biol. Chem. 1999;274:8269–8281. doi: 10.1074/jbc.274.12.8269. [DOI] [PubMed] [Google Scholar]
- 16.Cideciyan A.V., Swider M., Aleman T.S., Tsybovsky Y., Schwartz S.B., Windsor E.A., Roman A.J., Sumaroka A., Steinberg J.D., Jacobson S.G., et al. ABCA4 disease progression and a proposed strategy for gene therapy. Hum. Molec. Genet. 2009;18:931–941. doi: 10.1093/hmg/ddn421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhongsatiern J., Ohtsuki S., Tachikawa M., Hori S., Terasaki T. Retinal-specific ATP-binding cassette transporter (ABCR/ABCA4) is expressed at the choroid plexus in rat brain. J. Neurochem. 2005;92:1277–1280. doi: 10.1111/j.1471-4159.2004.02941.x. [DOI] [PubMed] [Google Scholar]
- 18.Násser L.S., Martelli D.R.B., Swerts M.S.O., Popoff D.A.V., De Barros L.M., Martelli H., Jr. Ophthalmic changes in cleft lip and palate. Rev. Bras. Talmol. 2016;75:94–98. doi: 10.5935/0034-7280.20160021. [DOI] [Google Scholar]
- 19.Beaty T.H., Murray J.C., Marazita M.L., Munger R.G., Ruczinski I., Hetmanski J.B., Liang K.Y., Wu T., Murray T., Fallin M.D., et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y., Zuo X., He M., Gao J., Fu Y., Qin C., Meng L., Wang W., Song Y., Cheng Y., et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat. Commun. 2017;8:14364. doi: 10.1038/ncomms14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig K.U., Mangold E., Herms S., Nowak S., Reutter H., Paul A., Becker J., Herberz R., AlChawa T., Nasser E., et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet. 2012;44:968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng H.-H., Chang N.-C., Chen K.-T., Lu J.-J., Chang P.-Y., Chang S.-C., Wu-Chou Y.-H., Chou Y.-T., Phang W., Cheng P.-J. Nonsynonymous variants in MYH9 and ABCA4 are the most frequent risk loci associated with nonsyndromic orofacial cleft in Taiwanese population. BMC Med. Genet. 2016;17:1–8. doi: 10.1186/s12881-016-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Do Rego Borges A., Sá J., Hoshi R., Viena C.S., Mariano L.C., De Castro Veiga P., Medrado A.P., Machado R.A., De Aquino S.N., Messetti A.C., et al. Genetic risk factors for nonsyndromic cleft lip with or without cleft palate in a Brazilian population with high African ancestry. Am. J. Med. Genet. Part A. 2015;167:2344–2349. doi: 10.1002/ajmg.a.37181. [DOI] [PubMed] [Google Scholar]
- 24.Moher D., Liberati A., Tetzlaff J., Altman D.G. The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells G.A., Shea B., O’Connell D., Robertson J., Peterson J., Welch V., Losos M., Tugwell P. The Newcastle-Ottawa Scale (Nos) for Assessing the Quality of Non-Randomised Studies in Meta-Analyses. Ottawa Hospital Research Institute; Ottawa, ON, Canada: 2011. [(accessed on 12 January 2016)]. Available online: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. [Google Scholar]
- 26.Lei S., Huang L., Liu Y., Xu L., Wang D., Yang L. Association between polymorphisms of heat-shock protein 70 genes and noise-induced hearing loss: A meta-analysis. PLoS ONE. 2017;12:e0188539. doi: 10.1371/journal.pone.0188539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Y., Zhang W., Du Y., Tong N., Han Y., Zhang H., Wang M., Ma J., Wan L., Wang L. Different roles of two novel susceptibility loci for nonsyndromic orofacial clefts in a Chinese Han population. Am. J. Med. Genet. Part A. 2011;155:2180–2185. doi: 10.1002/ajmg.a.34170. [DOI] [PubMed] [Google Scholar]
- 28.Huang E., Cheng H., Xu M., Shu S., Tang S. Association between single-nucleotide polymorphisms on chromosome 1p22 and 20q12 and nonsyndromic cleft lip with or without cleft palate: New data in Han Chinese and meta-analysis. Birth Defects Res. Part A Clin. Mol. Teratol. 2012;94:469–476. doi: 10.1002/bdra.23013. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Z.-W., Yang X., Wan Y.-B., Xin Y.-H., Zhai K., Ma J., Huang Y.-Q., Jiang M., Wang Y.-R. Associations of chromosomes 17q22, 10q25.3 and ABCA4 gene polymorphisms with non-syndromic cleft lip/palate in Ningxia Hui and Han population. J. Shandong Univ. 2013;51:103–108. [Google Scholar]
- 30.Mi N., Hao Y., Jiao X., Zheng X., Shi J., Chen Y. A polymorphic marker associated with non-syndromic cleft lip with or without cleft palate in a population in Heilongjiang Province, northern China. Arch. Oral Biol. 2015;60:357–361. doi: 10.1016/j.archoralbio.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Bagordakis E., Paranaíba L.M.R., Brito L.A., De Aquino S.N., Messetti A.C., Martelli-Júnior H., Swerts M.S.O., Graner E., Passos-Bueno M.R., Coletta R.D. Polymorphisms at regions 1p22.1 (rs560426) and 8q24 (rs1530300) are risk markers for nonsyndromic cleft lip and/or palate in the Brazilian population. Am. J. Med. Genet. Part A. 2013;161:1177–1180. doi: 10.1002/ajmg.a.35830. [DOI] [PubMed] [Google Scholar]
- 32.Fontoura C., Silva R.M., Granjeiro J.M., Letra A. Further evidence of association of the ABCA4 gene with cleft lip/palate. Eur. J. Oral Sci. 2012;120:553–557. doi: 10.1111/eos.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mostowska A., Hozyasz K.K., Wójcicka K., Biedziak B., Jagodziński P.P. Polymorphic variants at 10q25.3 and 17q22 loci and the risk of non-syndromic cleft lip and palate in the Polish population. Birth Defects Res. Part A Clin. Mol. Teratol. 2011;94:42–46. doi: 10.1002/bdra.22862. [DOI] [PubMed] [Google Scholar]
- 34.Gurramkonda V.B., Syed A.H., Murthy J., Chaubey G., Bhaskar Lakkakula V.K. Polymorphic variants near 1p22 and 20q11.2 loci and the risk of non-syndromic cleft lip and palate in South Indian population. Int. J. Pediatr. Otorhinolaryngol. 2015;79:2389–2393. doi: 10.1016/j.ijporl.2015.10.055. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Q., Blanton S.H., Hecht J.T. Association of ABCA4 and MAFB with non-syndromic cleft lip with or without cleft palate. Am. J. Med. Genet. Part A. 2011;155:1469–1471. doi: 10.1002/ajmg.a.33940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weng J., Mata N.L., Azarian S.M., Tzekov R.T., Birch D.G., Travis G.H. Insights into the function of Rim protein in photoreceptors and etiology of Stargardt’s disease from the phenotype in Abcr knockout mice. Cell. 1999;98:13–23. doi: 10.1016/S0092-8674(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 37.Lennon C.J., Birkeland A.C., Nuñez J.A., Su G.H., Lanzano P., Guzman E., Celis K., Eisig S., Hoffman D., Rendon M.R.G., et al. Association of candidate genes with nonsyndromic clefts in Honduran and Colombian populations. Laryngoscope. 2012;122:2082–2087. doi: 10.1002/lary.23394. [DOI] [PubMed] [Google Scholar]
- 38.Liu H., Leslie E.J., Carlson J.C., Beaty T.H., Marazita M.L., Lidral A.C., Cornell R.A. Identification of common non-coding variants at 1p22 that are functional for non-syndromic orofacial clefting. Nat. Commun. 2017;8:14759. doi: 10.1038/ncomms14759. [DOI] [PMC free article] [PubMed] [Google Scholar]