Abstract
The ongoing pandemic of severe acute respiratory syndrome (SARS), caused by the SARS-CoV-2 human coronavirus (HCoV), has brought the international scientific community before a state of emergency that needs to be addressed with intensive research for the discovery of pharmacological agents with antiviral activity. Potential antiviral natural products (NPs) have been discovered from plants of the global biodiversity, including extracts, compounds and categories of compounds with activity against several viruses of the respiratory tract such as HCoVs. However, the scarcity of natural products (NPs) and small-molecules (SMs) used as antiviral agents, especially for HCoVs, is notable. This is a review of 203 publications, which were selected using PubMed/MEDLINE, Web of Science, Scopus, and Google Scholar, evaluates the available literature since the discovery of the first human coronavirus in the 1960s; it summarizes important aspects of structure, function, and therapeutic targeting of HCoVs as well as NPs (19 total plant extracts and 204 isolated or semi-synthesized pure compounds) with anti-HCoV activity targeting viral and non-viral proteins, while focusing on the advances on the discovery of NPs with anti-SARS-CoV-2 activity, and providing a critical perspective.
Keywords: natural products, phytochemicals, antiviral protease inhibitors, ACE2, coronavirus, SARS-CoV, MERS-CoV, antiviral agents, cytopathic effect, virus-host interactome
1. Introduction
Coronaviruses (CoVs), are enveloped, positive strand RNA viruses, with a genome of 27–33 kb, the largest in all RNA viruses. Their virion is spherical (approx. 125 nm diameter), with club-shaped spike-proteins (S protein) that stick out from the surface and result in a crown-like appearance of the enveloped virion [1]. There are seven known human CoVs (HCoVs), (Figure 1) two of which (HCoV-229E, HCoV-NL63) belong to the alpha genera of the subfamily Orthocoronavirinae [2] and the remaining five, HCoV-OC43, HCoV-HKU1, MERS-CoV, SARS-CoV and SARS-CoV-2, belong to the beta genera. Most of the circulating HCoVs cause symptoms of common cold, although they occasionally can also cause severe or fatal disease. Three beta-CoVs, namely MERS-CoV, SARS-CoV and SARS-CoV-2, emerged in the last 20 years causing several epidemics of acute respiratory illness associated with high mortality: 10% CFR for SARS CoV-1 and 34% for MERS-CoV [3,4]. The SARS-CoV-2-induced COVID-19 pandemic has caused more than one million deaths since the onset of the disease on 12 December 2019 [5,6]. The genomic sequences of SARS-CoV and SARS-CoV-2 are 79.6% identical and their half-lives in aerosols and in plastic, metal and cardboard surfaces are reportedly similar [5,7]. The comparatively far higher contagiousness and pandemic potential of SARS-CoV-2 are thought to reflect in part the substantial prevalence of undocumented contagious infections compared to the documented ones [7]. The contagiousness of the virus renders its containment difficult and the demand for prophylactic and therapeutic agents an utmost necessity that drives the scientific community in a massive screening effort. In this scenario, bioactive molecules from the vegetable kingdom are a source worthful to mine. The modern tools of NPs chemistry (fast identification, dereplication, fast chemical profiling, in silico screening) and biological evaluation (high throughput in vitro screening assays, live infection assays, high throughput genomics and proteomics of host’s response to infection) provide ample means to explore plant biodiversity for discovery and/or development of NPs/SMs that can help cope with COVID-19 and here we summarize the efforts accomplished up to date.
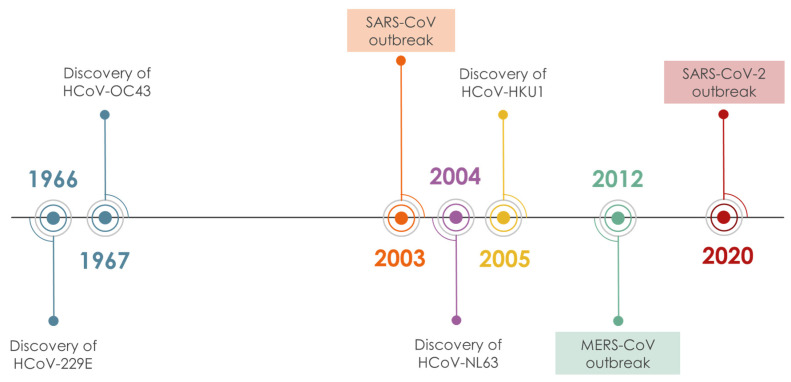
Figure 1.
Timeline of HCoV discovery.
The aim of this review is to summarize the anti-HCoV activity of natural products and derivatives thereof and their potential for prevention and/or treatment of coronavirus infections, COVID-19 in particular. We have reviewed the bibliography related to human coronaviruses and natural products since the discovery of the first HCoV in the 1960s, up to December 2020. Scopus, PubMed/MEDLINE, Web of Science, and Google Scholar, were employed for the literature search. A total of 135 references related to CoVs and NPs were assessed, while results corresponding to non-human coronaviruses were excluded. Finally, 52 original publications presenting results on anti-HCoV activity were incorporated in the review, corresponding to 19 total plant extracts and 204 isolated or semisynthesized pure compounds.
2. SARS-CoV-2 and SARS-CoV: Structural Aspects and Therapeutic Targeting
SARS-CoV is by far the most studied HCoV among the seven strains. It has a genome size of almost 30 kb [4]. Electron microscopy has shown that the viral particles have an average diameter of 80–140 nm and bear characteristic proteinaceous spikes (S) on the envelope. The surface S protein, encoded by the most variable structural gene of the genome [8], is involved in attachment and entry into the host cell, by interacting with key host cell receptor, the angiotensin-converting enzyme 2 (ACE2) [9], and thus it is the main target for antiviral peptides and antibodies. The ACE2 is a metalloprotease expressed in the lung, intestine, liver, heart, vascular endothelium, testis and kidney cells [4]. Entry into a host cell is an essential step of transmission of SARS-CoV. S protein binds to ACE2 through its S1 subunit but requires at least two protease cleavages to drive fusion through its S2 subunit. Proteolysis at the S1/S2 boundary and a second site within S2 is known to release a fusion peptide, which anchors on the host cell membrane to trigger a change of S2 conformation that promotes virus insertion into the target cell [10]. Several proteases, including extracellular proteases (e.g., elastases in the respiratory tract) and host cell surface proteases (e.g., transmembrane protease serine 2, TMPRSS2) could cleave S protein to render it fusion-competent. TMPRSS2 is reportedly requisite for S protein priming and S2-driven fusion of viral and host membranes [11,12]. However, SARS-CoV can also enter host cells through endocytosis and processing for fusion by endosomal cysteine proteases (e.g., cathepsin L), whose activity is however not essential in presence of TMPRSS2 [13,14]. A combination of a TMPRSS2 inhibitor and a cathepsin L inhibitor can effectively block SARS-CoV entry to host cells [15]. There are two major domains in S1, the N-terminal domain (S1-NTD), which is known to bind sugars, and the C-terminal domain (S1-CTD), which is responsible for recognizing the host receptor. ACE2 interacts specifically with a single region of S1-CTD, known as receptor binding domain (RBD). Host susceptibility to SARS-CoV infection is primarily determined by the affinity of RBD for ACE2 [16]. The RBD binds to the outer surface of the peptidase domain of ACE2, without involving or affecting the peptidase activity, which is not requisite for virus entry [10]. Determination of the crystal structure of SARS-CoV RBD complexed with ACE2 (PDB code: 2AJF) and functional studies have resolved several mechanistic aspects of ACE2 recognition by SARS-CoV and can help to develop effective vaccines.
SARS-CoV-2 is very closely related to SARS-CoV. The S1 and S2 subunits of SARS-CoV and SARS-CoV-2 are largely conserved, with the S2 subunits sharing the higher sequence identity (88%). Nevertheless, the SARS-CoV-2 S protein contains a furin-cleavage site that is involved in the biogenesis of the virus and differentiates it from all other SARS-like coronaviruses [17,18]. Like SARS-CoV, SARS-CoV-2 binds to ACE2 with high affinity [11,19,20]. The crystal structures of SARS-CoV and SARS-CoV-2 RBD bound to the S-binding domain of ACE2 are nearly identical [21]. The cryo-EM structure of SARS-CoV-2 S protein trimer is also reported [18,22]. The RBD is encoded by the most variable gene of SARS-CoV-2 genome. Although most of the amino acid residues that are essential for binding of SARS-CoV RBD to ACE2 are conserved in SARS-CoV-2 RBD, the latter is not recognized by several monoclonal antibodies directed to SARS-CoV RBD [22]. However, sera from mice immunized with SARS-CoV could inhibit SARS-CoV-2 host cell entry by nearly 90% [18]. Both SARS-CoV and SARS-CoV-2, use TMPRSS2 for S protein priming, and camostat mesylate (an approved TMPRSS2 inhibitor) was reported to partially inhibit SARS-CoV-2 cell entry. However, in cell lines expressing both TMPRSS2 and cathepsins B and L (CathB/L), full inhibition was observed with a combination of camostat mesylate and E-64d, an inhibitor of CathB/L [11].
The genome of both SARS-CoV and SARS-CoV-2 encodes also several non-structural proteins (Nsps), including RNA-dependent RNA polymerase (RdRp, nsp12), [23,24] helicase/NTPase (simply helicase) [25], and two cysteine proteases involved in viral nascent polyprotein processing in different sites, namely 3CLpro (chymotrypsin-like protease, also known as the main protease, Mpro) [26,27] and PLpro (papain-like protease) [28]. RdRp is the catalytic center of the replication/transcription complex formed by its interaction with multiple Nsps (nsp7, nsp8), [23] while it is the target of remdesivir, a nucleotide analogue that has recently been approved by FDA, for COVID-19 treatment [29,30]. The two cysteine proteases of the two SARS-CoVs, share a great similarity and are an attractive target for antiviral discovery. 3CLpro [31] and PLpro [32] participate in the proteolytic cleavage cascade of the viral polyproteins (1a and 1ab), which is essential for the maturation and replication of the virus. Specifically, 3CLpro cleaves pp1a and pp1b in more than 11 positions, and also releases the key-factors for replication RdRb and helicase [33]. PLpro cleaves the polyprotein in 3 sites and also has a deubiquitinating (DUB) and deISGylating (deISG) activity. This latter property poses implications for the host immune response in viral infection. It has been shown that ISGylation is important for viral clearance and protection, while ubiquitination is implicated in innate immune signaling pathways [34].
3. NPs with Anti-HCoV Potential
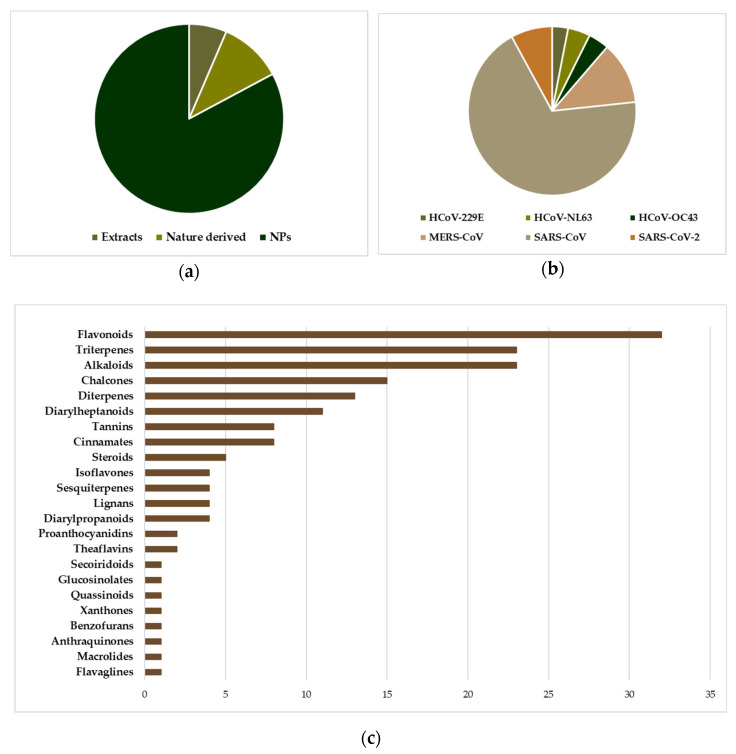
Before the emergence of SARS-CoV and MERS-CoV, the investigation of NPs as anti-HCoV agents was limited to only a few studies. The two epidemics have spurred however the interest in the discovery of anti-HCoV agents, including the investigation of agents active against the “common cold” HCoVs, 229E, NL63, OC43 and HKU1. Screening chemical libraries against target proteins and/or viral replication in cell-based assays have been extensively employed, resulting in the discovery of several antiviral natural products/small molecules (NPs/SMs) [35,36,37,38]. It was found that NPs/SMs are a rich source of drug potential leads against coronaviruses due to their pronounced structural diversity and complexity. Several compilations of pre-2020 findings on NPs/SMs with activity against coronaviruses were recently reported, some of which could serve as leads for the development of new drugs [39,40,41,42,43,44,45,46,47,48,49,50]. Among the identified molecules the majority is represented by NPs (Figure 2a) and most of them have been found as inhibitors of SARS-CoV (Figure 2b). Among them, the most numerous classes are flavonoids, triterpenes and alkaloids (Figure 2c), which is not surprising since in these classes reside the greatest number of NPs described as endowed with inhibitory activity against replication of multiple different viruses. In addition, flavonoids are a class of NPs present in almost every plant species and are strongly represented in various NP-based chemical libraries available for screening.
Figure 2.
The position of natural products implicated in anti-HCoV research. (a) Distribution of entities (extracts, NPs, Nature derived molecules) subjected to anti-HCoV assays (cell based or viral enzyme inhibition), in a total of 326 experiments corresponding to 197 entities. “Nature derived” refers to synthetic/semisynthetic NPs with a basic core inspired by NPs. (b) The prevalence of SARS-CoV testing among several NP entities. (c) The distribution of the 168 pure NPs tested in various anti-HCoV assays, with respect to the chemical category. Data derived from Tables S1–S4, Supporting information.
In the majority of studies, the initial screening involves the addition of the compound or extract to be tested in the normal cell culture, and a subsequent inoculation with the desired viral strain. The cytopathic morphology of the cells is evaluated under a microscope, while the cytotoxicity of the compound/extract is evaluated in normal cells and compared. Compounds with low cytotoxicity are then typically evaluated in multiple concentrations for their ability to inhibit viral replication (EC50) and host cell growth (CC50), and the selectivity index (SI, CC50/EC50 ratio) is used to identify the lead compounds, where higher SI refers to compounds more active than toxic. To a first step in investigating the molecular mechanisms underlying the antiviral activity, interactions of the lead compounds with identified targets, such as viral proteases, host proteases, or viral S proteins, are evaluated. Due to the additive complexity of the host/virus system, it is rather difficult to correlate cytopathic effects (CPE) to specific target interaction/inhibition, and this is the case for most natural products tested for their anti-HCoV potential, where extensive mechanistic studies are limited. Available in the Supporting Information are Table S1 (Plant extracts tested for anti HCoV activity in various strains), Table S2 (Pure natural products and nature-derived compounds tested for anti HCoV activity in various strains), Table S3 (Inhibitory activity of extracts against the main protease of SARS-CoV, 3CLpro) and S4 (Inhibition of HCoV enzymes from natural and nature-derived products), summarizing the results of anti-HCoV evaluation (viral enzyme inhibition, cell-based assays against viral propagation) of NPs against all strains.
3.1. “Common Cold” HCoVs
HCoV-229E (alpha-HCoV) and HCoV-OC43 (beta-HCoV) were identified in the mid-60s, as two of the strains causing the common cold [51,52,53]. The presence of non-typical respiratory viral strains was suspected since 1962, yet difficulties in cultivating viruses impeded scientists from discovering those strains earlier [52]. Van der Hoek, reviews the discovery and clinical manifestations of those “old HCoVs” [54]. Replication of both strains in the human body produces similar symptoms, such as excess nasal excretions, light cough, malaise and rarely, fever. Nevertheless, those strains were proven serologically unrelated, as antibodies isolated from OC43-infected persons, were unable to neutralize 229E. Lower respiratory tract infection was found more common in children and the elderly, manifested with bronchitis, bronchiolitis, croup, and pneumonia, while healthy adults presented mainly common cold. The prevalence of old CoVs infection in the general population has been studied in several cases, although difficult because of the varying clinical manifestations. Reverse-transcription PCR (RT-PCR) has been used for the identification of HCoV viral strains, in one of the largest studies among the UK population, revealing the autumn-winter seasonality of OC43, the lack of seasonality for 229E and its high detection in immunocompromised patients [55]. There are several indications that viral infection from 229E reprograms the host cell transcriptome with a subsequent fine-tuning of NF-κΒ, in order to ensure optimal replication [56].
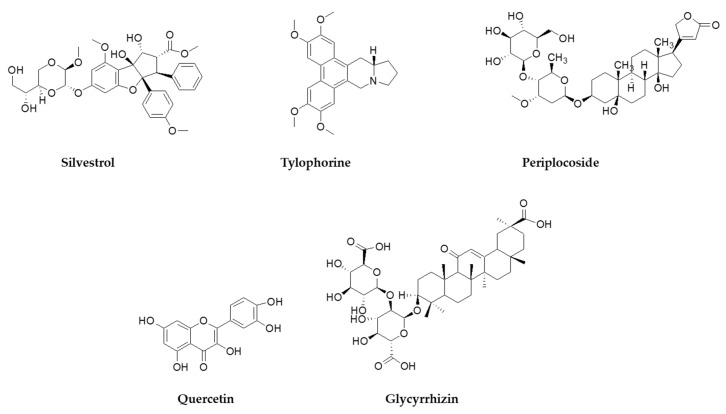
HCoV-229E uses human aminopeptidase N/CD13 glycoprotein (APN) to infect host cells. APN is a metalloprotease expressed in lung, intestinal, and kidney epithelium. The receptor was discovered in 2004, nearly 40 years after the discovery of the virus [57], while the X-ray crystal structures of the RBD of the S protein in complex with hAPN, as well as the electron cryomicroscopy structure of the 229E S-protein, was reported in 2019 [58]. Characterization and mapping of the fusion core of the S protein revealed that the domain HR1 folds into an unusually long helix, in post-fusion conformation, while there are very strong interactions between domains HR1 and HR2 that should be taken into account when designing antiviral agents mimicking HR2 binding to HR1 [59,60,61]. As in many CoVs, the S protein of 229E is cleaved by TMPRSS2, human airway trypsin-like protease (HAT) [62], and cathepsin L (CPL) [63]. Recent data show that cleavage into S1/S2 subunits by TMPRSS2 is not a prerequisite for infection, as the virus can use alternatively the endocytic pathway [64]. HCoV-NL63 is closely related to 229E and was discovered in 2004 in infants and immunocompromised patients [65,66]. NL63 uses the same receptor as SARS-CoV for viral entry (ACE2) [67], while the crystal structure of the S protein RBD in complex with ACE2 was reported in 2009, revealing that although NL63 and SARS recognize the same receptor regions, their RBD cores have no structural homology [68]. Currently, there are very few attempts to investigate the antiviral activity of extracts against 229E and NL63. A standardized extract of Pelargonium sidoides (Geraniaceae) displayed weak activity against 229E (EC50 = 44.50 μg/mL) and is approved in Germany as a phytotherapeutic against respiratory infections, was tested in a panel of respiratory viruses and showed a weak activity against 229E with an EC50 of 44.50 μg/mL [69]. In an early study, Stevia sp. extracts rich in steviosides used as a natural sweetener, showed virostatic and virucidal activity against 229E, yet there was no follow-up of the results reported [70]. Indole alkaloids indigodole B and tryptanthrine, isolated from Strobilanthes cusia (Acanthaceae) have been shown to reduce NL63 titers in early and late stages of LLC-MJ2 cells infection, while inhibiting enzymes vital for viral replication (PLP2, RNA polymerase) [71]. Isolated NPs/SMs that have been tested for their effect in 229E in vitro replication include saikosaponins isolated from Bupleurum and Heteromorpha species (Apiaceae) [72], pentacyclic triterpenes isolated from Euphorbia neriifolia (Euphorbiaceae) [73], xanthones isolated from Calophyllum blancoi (Guttiferae), [74] and the flavagline silvestrol that is isolated from plants of the genus Aglaia (Meliaceae) [75]. Silvestrol (Figure 3) in particular, had an EC50 in the nanomolar range (3.0 nM) in infected MRC-5 cells and an excellent Selectivity Index (>3300), showing no significant cytotoxic effect in the primary cells used [75]. Tacrolimus, an immunosuppressant drug isolated from the soil bacterium Streptomyces tsukubaiensis [76], was found to inhibit replication in NL63 and 229E with an EC50 of 5.1 and 5.4 μM, respectively [77].
Figure 3.
NPs active against HCoVs replication.
HCoV-OC43 is more similar to HCoV-HKU1 (beta-HCoV) that was recovered in 2005 from adult patients with pneumonia [78]. Their fundamental difference from all other HCoVs is that their virions have two surface projections participating in infection: the common in all HCoVs S protein, and protrusions comprised of hemagglutinin esterase (HE) [79]. In the case of both OC43 and HKU1 the S protein binds to 9-O-acetyl-sialic acids, attached to glycoproteins and lipids of the host cell membrane, in order to infect while the HCoV HE protrusions don’t seem to play a vital role [80,81].
The sialoglycan binding site of the S protein is conserved among all CoVs that bind to 9-O-acetyl-sialic acids, including the two HCoVs OC43 and HKU1. Sialic acids are important receptors in many human pathogens including Influenza viruses. Specifically, Influenza A/B hemagglutinin esterase (HE) and Influenza C/D hemagglutinin esterase fusion protein (HEF), all use modified sialic acids for infection. Also, it has been proposed that the HE protrusions of OC43 have a phylogenetic relationship with Influenza C HEF, while at the same time act in synergy with the 9-O-acetyl-sialic acids-binding domain of the OC43 S protein [79]. It is observed that ligand specificity of HKU1 and OC43 S protein and Influenza C/D HEF is similar, as all recognize 9-O-acetylsialic acids through hydrogen bonding with the 9-O-acetylcarbonyl moiety and formation of a hydrophobic pocket accommodating the 9-O-acetylmethyl group [82].
To our knowledge, no study on the antiviral potential of extracts against OC43 and HKU1 exists up to now, while no NPs were ever tested for HKU1 antiviral activity. Nevertheless, several studies exist concerning purified NPs/SMs, with emphasis on isoquinoline alkaloids. Cepharanthine, fangchinoline and tetrandrine isolated from Stephania tetrandra (Menispermaceae) have been very recently shown to inhibit OC-43 induced cell death of lung cells, in early stage of infection, and suppress viral replication [83,84]. Additionally, in a recent screening for broad spectrum anti-HCoV agents against NL63, OC43 and MERS, alkaloids lycorine (Table S1) and emetine inhibited viral replication of all strains with EC50 below 5 μM [84]. In the same study, the in vivo antiviral activity was established against a lethal intraperitoneal injection of OC43 in female BALB/c mice, where after administration of lycorine for 14 days post-inoculation (15 mg/kg) more than 80% of the mice were still alive [84].
3.2. MERS-CoV
A limited number of NPs have been tested for anti-MERS-CoV activity in cell-based infection assays, while their activity has not been associated with a specific target. The majority of the NPs/Sms tested exhibited good activity (EC50 < 20 μΜ), nevertheless, SI was found relatively low for most compounds. Alkaloids like emetine (ipecac alkaloids), lycorine (Amaryllidaceae alkaloids), harmine (β-carboline alkaloids), and conessine (steroidal alkaloids) exhibit increased potency with EC50 < 5 μΜ. Emetine in particular, was found to inhibit viral S-mediated entry [84] Notably, silvestrol, a flavagline isolated from the tree bark of Aglaia sp. (Meliaceae) [85] is the only NP with an EC50 in the nanomolar range (1.3 nM), by inhibiting the expression of MERS N-protein and nsp8 [75]. Only one original publication investigates a large panel of secondary metabolites isolated from Broussonetia papyrifera (Moraceae), for their ability to inhibit MERS two major proteases PLpro and 3CLpro. Limited potency was found for all tested compounds, against both proteases (IC50 > 27 μΜ) [86]. Recently, a library containing 502 NPs/SMs of various origin was screened for anti-MERS activity, in a MERS pseudovirus pre- infection assay. After confirmation of the actives in pre- and post- infection assays with MERS, dihydrotanshinone was identified to both block the viral entry by binding to the S protein of MERS, and possibly inhibiting viral replication [87].
3.3. SARS-CoV
Limited studies exist examining the anti-SARS-CoV activity of plant extracts, while some promising results of antiviral activity have no follow-up to determine the mechanism of action and specific targets [88,89,90,91,92,93,94]. A study on more than 200 plant extracts against two SARS-CoV viral strains (BJ-001 and BJ-006) revealed that the ethanol cortex extract of Lycoris radiata (Amaryllidaceae) inhibited the virus-induced CPE in both strains (EC50 2.4 and 2.1 μΜ, respectively) and had good SI (>350). After fractionation of the active extract, the antiviral activity was attributed to the alkaloid fraction that contains lycorine, although the specific mechanism of action has not been identified [89].
The anti-herpes activity of aloe emodin, a known anthraquinone isolated from Rheum palmatum (Polygonaceae), has prompted studies for anti-HCoV activity. Aloe emodin has been studied in multiple aspects of viral infection, although no studies of cell-based direct anti-SARS-CoV activity have been performed with the use of a real SARS strain. Aloe emodin is shown to bind the S protein of SARS-CoV thus possibly inhibiting host cell entry [95], inhibit the ion channel formed by the 3a protein of SARS and OC43 strains possibly inhibiting virus release, [96] and very weakly inhibit the activity of SARS 3CLpro [97]. Other examples of NPs/SMs with rather well-defined modes of anti-HCoV action tetra-O-galloyl-β-D-glucose, which binds to the S protein of SARS, inhibiting host cell entry of the virus [98], and cyclosporine A and derivatives thereof, which are known to suppress SARS-CoV and MERS-CoV replication by inhibiting cyclophilin A [99].
Notable anti-SARS-CoV activity is attributed to the phenanthraindolizidine alkaloids tylophorine (Figure 3) and tylophorine N-oxide isolated from Tylophora indica (Apocynaceae), with EC50 0.018 and 0.34 μΜ, respectively [100]. As shown from published data, natural and semisynthetic pentacyclic triterpenes have shown very good anti-SARS-CoV activity, with betulonic acid being the most active with an EC50 of 0.63 μΜ [37,101,102,103]. Interestingly, the 3β-OH analogue of betulonic acid (betulinic acid), had a weak antiviral activity (EC50 > 10 μΜ), although it strongly inhibited SARS-CoV 3CLpro (IC50 = 10 μΜ), contrary to betulonic acid (IC50 > 100 μΜ) [102]. Diterpenoids with various structures have also a significant potency against SARS-CoV, with the abietane diterpene ferruginol isolated from Chamaecyparis obtusa (Cupressaceae) showing an EC50 of 1.39 μΜ, while the labdane diterpene pinusolidic acid inhibited SARS-CoV infection with an EC50 of 4.71 μΜ and an excellent SI (159) [102].
Some other structurally diverse NPs /SMs have shown anti-SARS-CoV activity with EC50 close or below 10 μΜ (reserpine, β-yohimbine [37], gallic acid [98], honokiol, forskolin, magnolol [102], luteolin [98]), yet most of the compounds tested exhibited limited activity (EC50 > 20 μΜ) [37,91,102,104,105].
Significant efforts towards the discovery of viral cysteine protease inhibitors from NPs/SMs have been made. 3CLpro is inhibited by several NPs with diverse structures such as biflavonoids (amentoflavone, ginkgetin, sciadopitysin, bilobetin) [106], flavonoids (apigenin, quercetin, hyperoside, quercetine L-fucose derivatives, herbacetin, luteolin, pectolinarin, rhoifolin, hesperetin, kazinol A, kazinol B, broussoflavan A, papyriflavonol A, kaempferol, 4-hydroxyisolonchocarpin [86,97,106,107], isolflavones (daidzein) [97], pentacyclic triterpenes (betulinic acid, celastrol, iguesterin, pristimerin, tingenone) [102,108], phytosterols (β-sitosterol) [109], lignans (savinin) [102], indole alkaloids (indican, indigo, indirubin, semisynthetic isatin analogues) [97,110,111], glucosinolates (sinigrin) [97], anthraquinones (emodin, aloe emodin) [109], theaflavins (3-Isotheaflavin-3-gallate, theaflavin-3,3′-digallate) [112], phenanthrenes (crytotanshinone, tanshinone IIa, dihydrotanshinone I, tanshinone I, methyl tanshinonate, tanshinone IIB, rosmariquinone) [113], phlorotannins (dieckol, 7-phloroeckol, 2-phloroeckol, dioxinodehydroeckol, phlorofucofuroeckol A, fucodiphloroethol G) [114], chalcones (broussochalcone B, 3′-(3-methylbut-2-enyl)-3′,4,7-trihydroxyflavane, isoliquiritigenin, broussochalcone A) [86,115], (diarylheptanoids (hirsutenone, hirsutanonol, rubranoside B, rubranoside A, rubranol, oregonin, platyphyllenone, platyphyllonol 5-O-β-D-xylopyranoside, platyphyllone, curcumin) [102,116], and diarylpropanoids (kazinol F) [86]. Among the NPs tested, the pentacyclic triterpene iguestrin, isolated from the root methanol extract of Tripterygium regeli (Celastraceae) was the most active with an IC50 of 2.6 μΜ, followed by pristimerin (5.5 μΜ) [108], amentoflavone (8.3 μΜ) [106], and 3-theaflavin-3-gallate (9.8 μΜ) [112]. Notably, semisynthetic diversely substituted analogues of isatin, a small molecule of natural origin derived from the oxidation of the purple dye indigo, were found very active towards 3CLpro, with IC50 ranging from the lower micromolar range (0.95 μΜ), to 23.5 μΜ [110,111].
Similarly, several NPs/SMs inhibit SARS-CoV PLpro, like chalcones (bavachalcones, broussochalcones) [86,115,117], cinnamates, [118] coumarins, [117] diarylheptanoids (hirsutenone, curcumin, hirsutanonol, rubranosides A and B, rubranol, oregonin, isoliquiritigenin, papyriflavonol A) [86,116], diarylpropanoids (kazinols) [86], flavonoids (tomentins, quercetin, diplacone, mimulone, kaempferol, hyperoside, etc.) [86,119], isoflavones (corylifol A, neobavaisoflavone, bavachinin) [117], and phenanthrenes (tanshinones, rosmariquinone) [113]. Notably, tanshinone derivatives isolated from Salvia miltiorrhiza (Lamiaceae) have been found to exhibit very good inhibitory activity against PLpro, with cryptotanshinone having an IC50 of 0.8 μΜ [113], while an unusual a chalcone derivative with a hyperoxide group isolated from the ethanol leaves extract of Angelica keiskei (Apiaceae) showed an IC50 of 1.2 μΜ [115].
Finally, only one study investigates the inhibition of SARS-CoV helicase from NPs. The known flavonoids scutellarein and myricetin were found to inhibit nsp13 with IC50 0.86 and 2.71 μΜ, respectively [120].
Very few NPs or NPs-inspired SMs/SMs, have been investigated for their in vivo anti-HCoV effect. In a 2006 study, several in vitro viral inhibitors were also tested for in vivo efficacy. Chloroquine and phosphate salts, amodiaquine and pentoxifylline were first evaluated in vitro against SARS-CoV replication, with EC50 ranging from 1 to 10 μΜ [121]. Chloroquine and amodiaquine are quinoline analogues of quinine, the latter isolated from the bark of Cinchona sp. (Rubiaceae), and pentoxifylline is a synthetic analogue of theophylline present in tea and cocoa tee. The in vitro promising results were not verified in the BALB/c SARS-CoV infection model, where all the aforementioned compounds did not reduce the virus titer in the lungs post-infection, in a statistically significant manner [122]. More recently, a study of the effect of compassionate use of hydroxychloroquine or chloroquine in-hospital outcomes for COVID-19 was unable to confirm the benefit of such treatment [123]. The discovery of most of these NPs/SMs was based on high throughput 3CLpro inhibitory assays. For instance, herbacetin, pectolinarin and rhoifolin were found to efficiently block the enzymatic activity of SARS-CoV 3CLpro following a screening of a large flavonoid library against purified 3CLpro using a tryptophan-based fluorescence method to monitor flavonoid-dependent inhibition of proteolysis of a custom-synthesised fluorogenic substrate [124].
Glycyrrhizin (Figure 3) was found to inhibit both SARS-CoV penetration in host cells and virus replication (SI 67). A 2003 study investigating the effect of glycyrrhizin during several stages of infection of Vero cells i.e., during virus absorption, after absorption, and both during and after absorption reported that glycyrrhizin was most effective both during and after adsorption (EC50 300 mg/L) [125]. This study prompted for the development of several glycyrrhizin analogues that showed very potent SARS replication inhibitory activity and, in some cases, SI > 41. For carbamido- analogues of glycyrrhizin, the EC50 was significantly low, and that lead to the speculation that those analogues bind to the S protein of SARS through the glucosamine moiety, thus blocking viral entry to the cell [101]. When tested against HCoV-NL63, glycyrrhizin showed no antiviral effect [126]. Glycyrrhizin moderately inhibits infection of Vero cells with porcine epidemic diarrhea virus (PEDV), a coronavirus prevalent in the swine industry. It inhibits viral entry and replication, while virus assembly and viral release remain unaffected. Interestingly, it was found that glycyrrhizin significantly decreases the mRNA of proinflammatory cytokines, namely IL-6, IL-8 and TNF-a, suggesting that it reduces the proinflammatory response of the host cells during viral infection [127]. This reduction in the proinflammatory effects by glycyrrhizin, has been confirmed also with infection of lung epithelial cells with the highly pathogenic H5N1 influenza strain [128]. For SARS, the antiviral mechanism of action of glycyrrhizin is unclear and may include both the interaction with a specific molecular target, and the attenuation of proinflammatory host cell response, as suggested by the previous studies.
SARS infection inflicts a dysregulation in the immune response of the host and is followed by upregulation of proinflammatory cytokines and activation NF-κB. NPs inhibiting NF-κB, and thus having the potential to diminish the severity of SARS infection, have been studied in mouse models. Caffeic acid phenethyl ester (CAPE) and parthenolide, although having no impact on SARS replication, were found to significantly decrease the levels of proinflammatory cytokines and lung pathology of SARS infected BALB/c mice [129]. Most importantly, treatment with these drugs that inhibited NF-κB activation led to reduction in inflammation and lung pathology in both SARS-CoV-infected cultured cells and mice and significantly increased mouse survival after SARS-CoV infection. These data indicated that activation of the NF-κB signaling pathway represents a major contribution to the inflammation-induced after SARS-CoV infection and that NF-κB inhibitors are promising antivirals in infections caused by SARS-CoV and potentially other pathogenic human coronaviruses.
3.4. SARS-CoV-2
3.4.1. Virtual Screening Approaches
Crystallography of HCoV enzymes and reports on their potential ligands, have assisted computational methods (docking-scoring calculations of protein-ligand interactions using similarity to known actives) to discover SARS-CoV-2 inhibitors [27]. Virtual screening is extensively employed for the moment, to identify potential ligands from SMs and in-house NPs libraries [130,131,132].
Regarding 3CLpro, recent efforts, have identified potential inhibitors among: alkaloids [133,134,135,136,137,138], and especially indole alkaloids [139,140,141,142,143,144]; terpenoids [138], and especially diterpenes [133,145]; sesquiterpenes [134,146,147], sesquiterpene lactones [148], and triterpenes [133,136,140,149]; anthocyanins [150,151] and proanthocyanidins [152,153]; ellagitannins [153,154]; flavonoids [133,137,140,144,147,148,152,155,156,157,158,159,160,161], biflavonoids [162], and macrocyclic flavonoids [148]; isoflavones [144,148]; chalcones [163,164]; lignans [133,134]; coumarins [159]; caffeic acid esters and cinnamates [144,153,165,166]; stilbenes [148]; diarylheptanoids [165,167]; polyketides [138]; quinones [168]; oligosaccharides [157]; depsipeptides [169]; and xanthones [164].
In a more limited number of studies, potential affinity to PLpro has been suggested for flavonoids and biflavonoids [133]; chalcones [163]; caffeic acid esters [133]; terpenoids and sesquiterpene lactones [133,138]; polyketides [138]; and alkaloids [133,138].
The active site of RdRp has been also investigated as a possible target of inhibition finding multiple possible ligands, such as flavonoids, xanthones [133], polyketides [138], terpenes and sesquiterpene lactones [133,138], and alkaloids [138,143]. Finally, alkaloids, polyketides and terpenes have shown affinity for non-structural protein 15 (nsp15) of SARS-CoV-2 [138], while several NPs have been investigated as potential ligands to virulence factors Nsp1 (suppresses type-I IFN expression), Nsp3c (promotes replication of the viral genome and transcription of viral mRNA) and ORF7a (inhibits virus restriction) [133].
Homology modeling is a tool that was also used for the identification of NPs that interfere with the S protein interface with the human receptor ACE2. The only NP that was identified through virtual screening to potentially interfere with ACE2-mediated host cell entry, was hesperidin [170], although selected flavonoids and epigallocatechins have shown an affinity for the S protein. Recent research indicated that the S protein of SARS-CoV-2 contains a furin-like cleavage site that is absent from SARS-CoV, pointing to the need of developing specific furin inhibitors [17]. This is a new finding that is currently under intensive investigation [171].
It has been demonstrated that the serine protease TMPRSS2 is needed for S protein priming and host cell entry [11]. Virtual screening of natural SM libraries against a TMPRSS2 structure built by homology modeling detected several potential protease inhibitors, including norsesquiterpenes, diterpenes and xanthones, [133,164,172] alkaloids, [173] chalcones, [164] coumarins, [173] and flavonoids. [174] Interestingly, SARS-CoV infectivity is reportedly associated TMPRSS2 expression levels, which are induced by androgens and suppressed by estrogens, implying that natural estrogen receptor agonists (e.g., genistein) and androgen receptor antagonists (e.g., atraric acid, indole-3-carbinol, niphatenone B) could impact SARS-CoV-2 infection [175,176,177,178].
3.4.2. Actual Screening of NPs for Anti-SARS-CoV-2 Activity
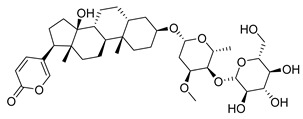
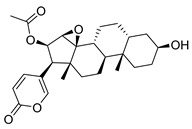
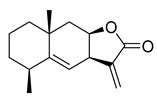
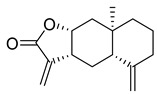
The first results from the physical screening of NPs for anti-SARS-CoV-2 using host cells and/or viral enzymes have been published very recently. A high throughput cell-based screening assay was established by Zhang et al. [179] according to which the effect of potential inhibitors in the CPE in Vero-E6 cells is assessed in the entire viral cell cycle. Known inhibitors were used as positive controls (remdesivir, chloroquine, neutralizing human antibody CB62 and IFN-α), while multiple concentrations of DMSO in the culturing media was used in order to determine potential solvent effects on CPE. A library of 1058 compounds was screened, in a two-level assay: the first screening revealed 30 hits (>50% protection from CPE), 17 of which have never been associated with SARS-CoV-2 through different assays. (Table 1) Further evaluation showed that viral propagation was inhibited in a dose-dependent manner, while EC50 values ranged between 0.011 and 11.03 μΜ. In an attempt to identify possible mechanisms of action of active compounds bufalin and digoxin, it was postulated that those compounds target the ion transport function of Na+/K+-ATPase, and intracellular ion homeostasis [179], with the most potent inhibitor represented by the quassinoid derivative bruceine A (Table 1) that displayed an EC50 value of 0.011 μΜ and a SI of 2854.
Table 1.
Antiviral activity of selected NPs against HCoV-NL63, HCoV-OC43, MERS-CoV, SARS-CoV, and SARS-CoV-2.
| Structure | Name | Source | Target | Assay | Ref. | |
|---|---|---|---|---|---|---|
|
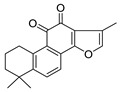
Cryptotanshinone |
Salvia miltiorrhiza Lamiaceae |
SARS-CoV | Enzyme inhibition (IC50) | PLpro 0.8 μΜ, 3CLpro 226.7 μΜ | [113] |
| SARS-CoV-2 | CPE inhibition (EC50) | 5.024 μΜ | [179] | |||
|
Tanshinone IIA |
Salvia miltiorrhiza Lamiaceae |
SARS-CoV | Enzyme inhibition (IC50) | PLpro 1.6 μΜ, 3CLpro 89.1 μΜ | [113] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
|
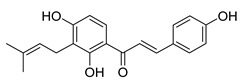
Isobavachalcone |
Psoralea sp. Fabaceae |
SARS-CoV | Enzyme inhibition (IC50) | PLpro 7.3 μΜ | [117] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
|
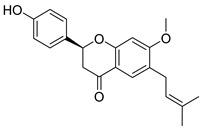
Bavachin |
Psoralea sp. Fabaceae |
SARS-CoV | Enzyme inhibition (IC50) | PLpro 38.4 μΜ | [117] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
|
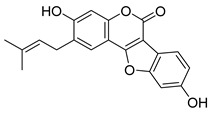
Psoralidin |
Psoralea sp. Fabaceae |
SARS-CoV | Enzyme inhibition (IC50) | PLpro 4.2 μΜ | [117] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
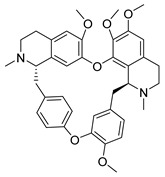
|
Tetrandrine |
Stephania tetrandra Menispermaceae |
HCoV-NL63 | CPE inhibition (EC50) | 2.05 μΜ | [84] |
| HCoV-OC43 | 0.29 μΜ/0.33 μΜ | [83,84] | ||||
| MERS-CoV | 12.68 μΜ | [84] | ||||
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||

|
Cepharanthine |
Stephania tetrandra Menispermaceae |
HCoV-OC43 | CPE inhibition (EC50) | 0.83 μΜ | [83] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
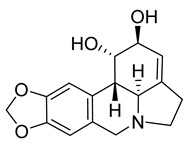
|
Lycorine |
Lycoris sp. Amaryllidaceae |
HCoV-NL63 | CPE inhibition (EC50) | 0.47 μΜ | [84] |
| HCoV-OC43 | 0.15 μΜ | |||||
| MERS-CoV | 1.63 μΜ | |||||
| SARS-CoV | 169.8 μΜ | [89] | ||||
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
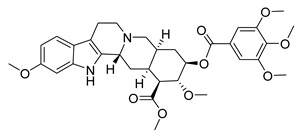
|
Reserpine |
Rauvolfia serpentina Apocynaceae |
SARS-CoV | CPE inhibition (EC50) | 3.4 μΜ | [37] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
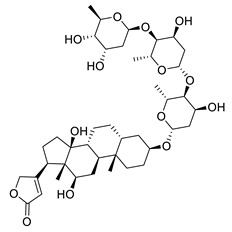
|
Digoxin |
Digitalis sp. Plantaginaceae |
SARS-CoV-2 | CPE inhibition (EC50) | 0.1541 μΜ | [179] |
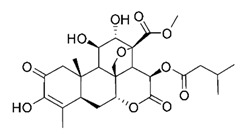
|
Bruceine A |
Brucea javanica Simaroubaceae |
0.011 μΜ | |||
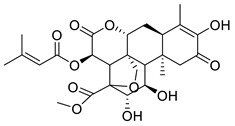
|
Brusatol |
Brucea javanica Simaroubaceae |
0.0492 μΜ | |||
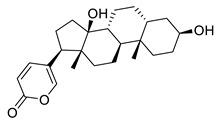
|
Bufalin | Toad venom Bufonidae |
0.018 μΜ | |||
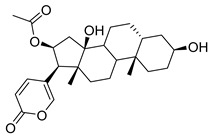
|
Bufotalin | Toad venom Bufonidae |
0.0259 μΜ | |||
|
Toad venom Bufonidae |
0.0657 μΜ | ||||
|
Cinobufagin | Toad venom Bufonidae |
0.018 μΜ | |||
|
Alantolactone |
Inula helenium Asteraceae |
1.724 μΜ | |||
|
Isoalantolactone |
Inula helenium Asteraceae |
1.483 μΜ | |||
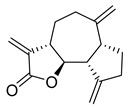
|
Dehydrocostus lactone |
Saussurea costus Asteraceae |
2.322 μΜ | |||
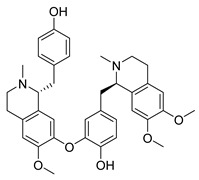
|
Liensinine |
Nelumbo nucifera Nelumbonaceae |
2.537 μΜ | |||
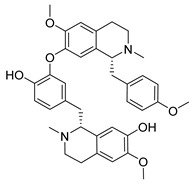
|
Isoliensinine |
Nelumbo nucifera Nelumbonaceae |
1.615 μΜ | |||
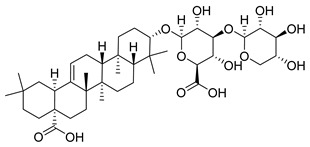
|
Momordinic |
Bassia scoparia Amaranthaceae |
3.529 μΜ | |||
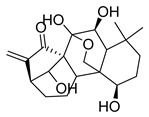
|
Oridonin |
Isodon sp. Lamiaceae |
1.462 μΜ | |||
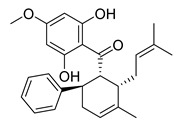
|
Panduratin A |
Boesenbergia rotunda Zingiberaceae |
SARS-CoV-2 | CPE inhibition (EC50) | 0.81 μΜ | [180] |
The only existing study for associating anti-SARS-CoV-2 activity of an extract with isolated components, concerns the investigation of plant biodiversity of Thailand. Three extracts from respective plants are reported to have viral infection inhibition, namely Andrographis paniculata (Acanthaceae), Zingiber officinale (Zingiberaceae), and Boesenbergia rotunda (Zingiberaceae), are reported to display viral infection inhibition. The extract of B. rotunda and the isolate panduratin A (Table 1) suppressed SARS-CoV-2 infectivity in Vero E6 cells with EC50 of 3.62 μg/ mL and 0.81 μM, respectively [180].
Concerning the inhibition of SARS-CoV-2 main protease 3CLpro, only three publications exist for the moment, reporting the flavonoids baicalin and baicalein, as potent inhibitors with an IC50 of 6.41 and 0.94 μΜ, respectively, as well as inhibitors of SARS-CoV-2 replication in Vero E6 cells [181]. Baicalin has also been investigated for the in vitro propagation of SARS-CoV in Vero-E6 cells, though no significant activity was found (EC50 > 100 μM) [105]. Additionally, the labdane diterpene andrographolide and a semisynthetic derivative displayed inhibitory activity against 3CLpro [182], while tannic acid was found active with an IC50 of 2.1 μΜ [183].
Finally, the well-known flavonoid quercetin (Figure 3) has been proposed as a SARS-CoV-2 3CLpro inhibitor (Ki = 7.4 μM), while molecular simulations showed that it binds to the active site of the enzyme [184]. Flavonoids have been highlighted in the past as HCoVs protease inhibitors. Quercetin itself weakly inhibits SARS-CoV 3CLpro (IC50 23.8 μΜ), and shows no inhibition in MERS-CoV 3CLpro, while it is reported to potently inhibit SARS-CoV PLpro (IC50 8.6 μΜ) [106]. Although these results may seem promising, quercetin has not been tested in cell viral infection assays, and additionally, like many dietary flavonoids and polypenols, has poor oral bioavailability [185].
NPs have not yet been investigated thoroughly for anti-HCoV activity. Nevertheless, NPs such as the Nobel Prize awarded antimalarial drug artemisinin, isolated from Artemisia annua (Asteraceae), have shown notable bioactivity against viruses of the Herpesviridae family (e.g., herpes simplex virus type 1 and Epstein-Barr virus), hepatitis B virus, hepatitis C virus, and bovine viral diarrhea virus [186].
3.4.3. Host Interactions and Future Prospects
Compounds targeting host cell components may help to treat SARS-CoV-2 infection. It has been reported that certain NPs (cholesterol, β-sitosterol, betulinic acid, hopane and glycyrrhizin), may reduce SARS-CoV-2 infectivity by inhibiting lipid-dependent attachment of the virus to host cells [187,188]. It is also reported that 25-hydroxy-cholesterol shows broad antiviral activity by blocking membrane fusion in the viral infection stage [189]. A recent study of SARS-CoV-2 human protein interactome reported that as many as 332 human proteins are intercepted by SARS-CoV-2 proteins, with several of these interceptions potentially targeted by NPs/SMs already approved by FDA or in preclinical or clinical development [190].
This study [190] revealed that the SARS-CoV-2 infection, replication and biogenesis program interfered with host components involved in DNA replication, regulation of gene expression, RNA processing and translation, protein expression and ubiquitination, ER/Golgi function and vesicle trafficking, nuclear transport, mitochondrial import receptors, extracellular matrix cytoskeletal function, IFN signaling, NF-kB-mediated inflammatory response and innate immune response and lipid modification. Some of these interactions could be targeted by NPs/SMs. For instance, the non-structural protein 6 (Nsp6) component of the viral replication complex may interfere with the function of vacuolar ATPase ATP6AP1 and the endoplasmic reticulum endomembrane compartments to favor coronavirus replication, an interference that might be inhibited by the macrolide antibiotic bafilomycin A1, an ATPase inhibitor in preclinical development. With regard to host proteases, the interactome study reported that 3CLpro may interfere with histone deacetylase 2-mediated inflammation and interferon response to SARS-CoV-2 infection in a manner inhibited by the fungal metabolite apicidin, a histone deacetylase inhibitor in preclinical development [191].
Virus-encoded helicase interacts with the centrosomal protein CEP250 and the interaction might be targeted by the natural polyketide WDB002. Virus-encoded 3′-5′ exonuclease interacts with the purine biosynthesis enzyme IMPDH2, a target of cyclophilin A, which in turn is involved in the packaging of the viral capsid; the IMPDH2-CypA interaction is modulated by the natural product sanglifehrin A [190].
At the beginning of the pandemic, the use of kinase inhibitors was proposed as a therapeutic strategy against COVID-19. Some kinase inhibitors, such as imatinib, dasatinib and trametinib, that have been shown to inhibit viral replication in vitro [192], may rely on an indirect inhibition of TMPRSS2 function. Indeed, although there is no demonstrated association between TMPRSS2 and kinase inhibition, it has been postulated that kinase inhibition could result in the inhibition of TMPRSS2 function, localization, or activity, and the observed blocking of the infection. Abl kinase and the ERK/MAPK, and PI3K/AKT/mTOR signaling pathways have been shown to play a role in the relevant MERS-CoV and SARS-CoV infection [192,193]. Imatinib, a BCR-Abl kinase inhibitor, is under clinical investigation for having beneficial results in hospitalized adults with COVID-19 [194], although it has been shown that it has no antiviral activity [195].
The role of kinases in the pathology of COVID-19, and especially in the inflammatory responses, is investigated intensively. In a recent study [196] it was found a causal link between life-threatening COVID-19 and high expression of TYK2, the gene encoding Tyrosine Kinase 2, a member of the JAK family kinases. TYK2 abnormalities have been established in several autoimmune diseases such as systemic lupus [197], rheumatoid arthritis [198], and multiple sclerosis [199]. Small molecule TYK2 inhibitors are under clinical trials for their safety and efficacy in psoriasis [200,201], and other autoimmune diseases associated with inflammation [202]. The natural product parthenolide is shown to inhibit all three Janus kinases (JAK1, JAK2 and TYK2), thus inhibiting STAT3 signaling [203]. Interestingly, parthenolide and caffeic acid phenyl ethyl ether (CAPE), were tested as NF-κΒ inhibitors in a mouse in vivo model of SARS-CoV, and found to significantly reduce lung inflammation and pathology [129].
4. Conclusions
Natural products have played an active and important role in drug discovery up until today. Nevertheless, their valorization as antiviral agents remains limited. It is indicative that antiviral NP research has peaked only around imminent threats, as happened with SARS-CoV. Our review of the bibliography covering the study of NPs against HCoVs, revealed that there are many possibilities in examining more thoroughly available NPs in order to discover new antiviral agents. NP databases and NP libraries with physical samples available for bioactivity screening may have shortened the time needed for a compound to reach bioactivity evaluation, but as shown from the literature survey, tested compounds tend to revolve around very common and not very diverse structures. In terms of anti HCoV activity and NPs, the available literature is limited in order to draw sound conclusions about structure-activity relationships. Nevertheless, the data show that there is a trend for alkaloids, triterpenoids/triterpene saponins, and polyhydroxylated flavonoids. HCoVs and NPs research has been fragmented up to this point, driven by states of emergency, such as the emergence of SARS-CoV in 2002, and the current threat of SARS-CoV-2. Although there are a number of published results, there is a lack of systematic investigation of NPs showing promising activity, and this needs to be addressed by research efforts defining the mechanism of action of these compounds. Under this scope, compounds based on NPs scaffolds with higher potency, more favorable physicochemical properties, and diminished toxicity, can be designed, thus providing a multidisciplinary approach that antiviral discovery needs. NPs offer great chemodiversity that needs to be further exploited, especially under the current pressure of this global pandemic.
Acknowledgments
On behalf of the University of Athens, the present work was conducted under the European Union (ERDF) and Greek national Operational Program “Competitiveness, Entrepreneurship and Innovation”, call “STRENGTHENING RESEARCH AND INNOVATION INFRASTRUCTURES” (project code: 5002803). AC and ET conducted this research under the project “EXaSCale smArt pLatform Against paThogEns for Corona Virus—Exscalate4CoV” founded by the EU’s H2020-SC1-PHE-CORONAVIRUS-2020 call, grant N. 101003551.
Supplementary Materials
Table S1: Natural products tested for anti-HCoV activity in cell based assays; Table S2: Natural products tested for HCoV viral enzyme inhibition. Table S3: Inhibitory activity of extracts against the main protease of SARS-CoV, 3CLpro; Inhibition of HCoV enzymes from natural and nature-derived products; Table S4: Inhibition of HCoV enzymes from natural and nature-derived products.
Author Contributions
K.V.; writing—original draft preparation, K.V., A.C.; writing—review and editing, K.V., A.-L.S.; review, editing and supervision, M.N.A.; writing, review and supervision, A.C., E.T.; visualization, supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Maier H.J., Bickerton E., Britton P. Coronaviruses: Methods and Protocols. Humana Press; Heidelberg, Germany: 2015. pp. 1–282. [Google Scholar]
- 2.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Memish Z.A., Perlman S., Van Kerkhove M.D., Zumla A. Middle East respiratory syndrome. Lancet. 2020;395:1063–1077. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng V.C.C., Lau S.K.P., Woo P.C.Y., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laha S., Chakraborty J., Das S., Manna S.K., Biswas S., Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect. Genet. Evol. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 Cell entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient Activation of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein by the Transmembrane Protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirato K., Kawase M., Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous Treatment of Human Bronchial Epithelial Cells with Serine and Cysteine Protease Inhibitors Prevents Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-Amino Acid Fragment of the SARS Coronavirus S Protein Efficiently Binds Angiotensin-converting Enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 22.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahn D.-G., Choi J.-K., Taylor D.R., Oh J.-W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012;157:2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanner J.A., Watt R.M., Chai Y.-B., Lu L.-Y., Lin M.C., Peiris J.S.M., Poon L.L.M., Kung H.-F., Huang J.-D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratia K., Saikatendu K.S., Santarsiero B.D., Barreto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like-protease: Structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. [(accessed on 15 January 2021)]; Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment.
- 30. [(accessed on 15 January 2021)]; Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/214787Orig1s000lbl.pdf.
- 31.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., et al. Structures of Two Coronavirus Main Proteases: Implications for Substrate Binding and Antiviral Drug Design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Báez-Santos Y.M., John S.E.S., Mesecar A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong T.R. Therapies for coronaviruses. Part 2: Inhibitors of intracellular life cycle. Expert Opin. Ther. Pat. 2009;19:415–431. doi: 10.1517/13543770802600698. [DOI] [PubMed] [Google Scholar]
- 34.Mielech A.M., Chen Y., Mesecar A.D., Baker S.C. Nidovirus papain-like proteases: Multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong B., Gui C.-S., Xu X.-Y., Luo C., Chen J., Luo H.-B., Chen L.-L., Li G.-W., Sun T., Yu C.-Y., et al. A 3D model of SARS_CoV 3CL proteinase and its inhibitors design by virtual screening. Acta Pharm. Sin. 2003;24:497–504+619. [PubMed] [Google Scholar]
- 36.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu C.-Y., Jan J.-T., Ma S.-H., Kuo C.-J., Juan H.-F., Cheng Y.-S.E., Hsu H.-H., Huang H.-C., Wu D., Brik A., et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao R.Y., Tsui W.H.W., Lee T.S.W., Tanner J.A., Watt R.M., Huang J.-D., Hu L., Chen G., Chen Z., Zhang L., et al. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Islam M.T., Sarkar C., El-Kersh D.M., Jamaddar S., Uddin S.J., Shilpi J.A., Mubarak M.S. Natural products and their derivatives against coronavirus: A review of the non-clinical and pre-clinical data. Phyther. Res. 2020;34:2471–2492. doi: 10.1002/ptr.6700. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y., Islam M.S., Wang J., Li Y., Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): A review and perspective. Int. J. Biol. Sci. 2020;16:1708–1717. doi: 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Hatamleh M.A.I., Hatmal M.M., Sattar K., Ahmad S., Mustafa M.Z., Bittencourt M.D.C., Mohamud R. Antiviral and Immunomodulatory Effects of Phytochemicals from Honey against COVID-19: Potential mechanisms of action and future Directions. Molecules. 2020;25:5017. doi: 10.3390/molecules25215017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boozari M., Hosseinzadeh H. Natural products for COVID-19 prevention and treatment regarding to previous coronavirus infections and novel studies. Phyther. Res. 2020 doi: 10.1002/ptr.6873. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y., Liang C., Xin L., Ren X., Tian L., Ju X., Li H., Wang Y., Zhao Q., Liu H., et al. The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020;206:112711. doi: 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiwari V., Beer J.C., Sankaranarayanan N.V., Swanson-Mungerson M., Desai U.R. Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov. Today. 2020;25:1535–1544. doi: 10.1016/j.drudis.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J., Huang J. Current Findings Regarding Natural Components with Potential Anti-2019-nCoV Activity. Front. Cell Dev. Biol. 2020;8:589. doi: 10.3389/fcell.2020.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.da Silva Antonio A., Wiedemann L.S.M., Veiga-Junior V.F. Natural products’ role against COVID-19. RSC Adv. 2020;10:23379–23393. doi: 10.1039/D0RA03774E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khalifa S.A.M., Yosri N., El-Mallah M.F., Ghonaim R., Guo Z., Musharraf S.G., Du M., Khatib A., Xiao J., Saeed A., et al. Screening for natural and derived bio-active compounds in preclinical and clinical studies: One of the frontlines of fighting the coronaviruses pandemic. Phytomedicine. 2020:153311. doi: 10.1016/j.phymed.2020.153311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verma S., Twilley D., Esmear T., Oosthuizen C.B., Reid A.M., Nel M., Lall N. Anti-SARS-CoV Natural Products with the Potential to Inhibit SARS-CoV-2 (COVID-19) Front. Pharmacol. 2020;11:561334. doi: 10.3389/fphar.2020.561334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adhikari B., Marasini B.P., Rayamajhee B., Bhattarai B.R., Lamichhane G., Khadayat K., Adhikari A., Khanal S., Parajuli N. Potential roles of medicinal plants for the treatment of viral diseases focusing on COVID-19: A review. Phyther. Res. 2020 doi: 10.1002/ptr.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z., Yang L. Turning the Tide: Natural Products and Natural-Product-Inspired Chemicals as Potential Counters to SARS-CoV-2 Infection. Front. Pharmacol. 2020;11:1013. doi: 10.3389/fphar.2020.01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamre D., Procknow J.J. A New Virus Isolated froiii the Human Respiratory Tract. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 52.McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. USA. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McIntosh K., Becker W.B., Chanock R.M. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. USA. 1967;58:2268–2273. doi: 10.1073/pnas.58.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Der Hoek L. Human coronaviruses: What do they cause? Antivir. Ther. 2007;12:651–658. [PubMed] [Google Scholar]
- 55.Gaunt E.R., Hardie A., Claas E.C.J., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poppe M., Wittig S., Jurida L., Bartkuhn M., Wilhelm J., Müller H., Beuerlein K., Karl N., Bhuju S., Ziebuhr J., et al. The NF-κB-dependent and -independent transcriptome and chromatin landscapes of human coronavirus 229E-infected cells. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z., Tomlinson A.C., Wong A.H., Zhou D., Desforges M., Talbot P.J., Benlekbir S., Rubinstein J.L., Rini J.M. The human coronavirus HCoV-229E S-protein structure and receptor binding. Elife. 2019;8 doi: 10.7554/eLife.51230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu C., Feng Y., Gao F., Zhang Q., Wang M. Characterization of HCoV-229E fusion core: Implications for structure basis of coronavirus membrane fusion. Biochem. Biophys. Res. Commun. 2006;345:1108–1115. doi: 10.1016/j.bbrc.2006.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang W., Zheng Q., Yan M., Chen X., Yang H., Zhou W., Rao Z. Structural characterization of the HCoV-229E fusion core. Biochem. Biophys. Res. Commun. 2018;497:705–712. doi: 10.1016/j.bbrc.2018.02.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan L., Meng B., Xiang J., Wilson I.A., Yang B. Crystal structure of the post-fusion core of the Human coronavirus 229E spike protein at 1.86 Å resolution. Acta Crystallogr. Sect. D Struct. Biol. 2018;74:841–851. doi: 10.1107/S2059798318008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H., et al. TMPRSS2 Activates the Human Coronavirus 229E for Cathepsin-Independent Host Cell Entry and Is Expressed in Viral Target Cells in the Respiratory Epithelium. J. Virol. 2013;87:6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawase M., Shirato K., Matsuyama S., Taguchi F. Protease-Mediated Entry via the Endosome of Human Coronavirus 229E. J. Virol. 2009;83:712–721. doi: 10.1128/JVI.01933-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bonnin A., Danneels A., Dubuisson J., Goffard A., Belouzard S. HCoV-229E spike protein fusion activation by trypsin-like serine proteases is mediated by proteolytic processing in the S2′ region. J. Gen. Virol. 2018;99:908–912. doi: 10.1099/jgv.0.001074. [DOI] [PubMed] [Google Scholar]
- 65.van der Hoek L., Pyrc K., Berkhout B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 2006;30:760–773. doi: 10.1111/j.1574-6976.2006.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J.M., Wolthers K.C., Wertheim-van Dillen P.M.E., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hofmann H., Pyrc K., Van Der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu K., Li W., Peng G., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. USA. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Michaelis M., Doerr H.W., Cinatl J. Investigation of the influence of EPs® 7630, a herbal drug preparation from Pelargonium sidoides, on replication of a broad panel of respiratory viruses. Phytomedicine. 2011;18:384–386. doi: 10.1016/j.phymed.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kedik S.A., Yartsev E.I., Stanishevskaya I.E. Antiviral activity of dried extract of Stevia. Pharm. Chem. J. 2009;43:198–199. doi: 10.1007/s11094-009-0270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai Y.-C., Lee C.-L., Yen H.-R., Chang Y.-S., Lin Y.-P., Huang S.-H., Lin C.-W. Antiviral action of tryptanthrin isolated from strobilanthes cusia leaf against human coronavirus nl63. Biomolecules. 2020;10:366. doi: 10.3390/biom10030366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng P.-W., Ng L.-T., Chiang L.-C., Lin C.-C. Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin. Exp. Pharmacol. Physiol. 2006;33:612–616. doi: 10.1111/j.1440-1681.2006.04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang F.-R., Yen C.-T., Ei-Shazly M., Lin W.-H., Yen M.-H., Lin K.-H., Wu Y.-C. Anti-human coronavirus (anti-HCoV) triterpenoids from the leaves of Euphorbia neriifolia. Nat. Prod. Commun. 2012;7:1415–1417. doi: 10.1177/1934578X1200701103. [DOI] [PubMed] [Google Scholar]
- 74.Shen Y.-C., Wang L.-T., Khalil A.T., Chiang L.C., Cheng P.-W. Bioactive pyranoxanthones from the roots of Calophyllum blancoi. Chem. Pharm. Bull. 2005;53:244–247. doi: 10.1248/cpb.53.244. [DOI] [PubMed] [Google Scholar]
- 75.Müller C., Schulte F.W., Lange-Grünweller K., Obermann W., Madhugiri R., Pleschka S., Ziebuhr J., Hartmann R.K., Grünweller A. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antivir. Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hatanaka H., Iwami M., Kino T., Goto T., Okuhara M. FR-900520 and FR-900523, novel immunosuppressants isolated from a Streptomyces. I. Taxonomy of the producing strain. J. Antibiot. (Tokyo) 1988;41:1586–1591. doi: 10.7164/antibiotics.41.1586. [DOI] [PubMed] [Google Scholar]
- 77.Carbajo-Lozoya J., Müller M.A., Kallies S., Thiel V., Drosten C., Von Brunn A. Replication of human coronaviruses SARS-CoV, HCoV-NL63 and HCoV-229E is inhibited by the drug FK506. Virus Res. 2012;165:112–117. doi: 10.1016/j.virusres.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woo P.C.Y., Lau S.K.P., Chu C.-M., Chan K.-H., Tsoi H.-W., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W.-k., et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeng Q., Langereis M.A., Van Vliet A.L.W., Huizinga E.G., De Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. USA. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q.K., Marasco W.A., Baric R.S., Sims A.C., Pyrc K., et al. Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hulswit R.J.G., Lang Y., Bakkers M.J.G., Li W., Li Z., Schouten A., Ophorst B., Van Kuppeveld F.J.M., Boons G.-J., Bosch B.-J., et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. USA. 2019;116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G.-J., Bosch B.-J., Rey F.A., de Groot R.J., et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim D.E., Min J.S., Jang M.S., Lee J.Y., Shin Y.S., Park C.M., Song J.H., Kim H.R., Kim S., Jin Y.-H., et al. Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus oc43 infection of mrc-5 human lung cells. Biomolecules. 2019;9:696. doi: 10.3390/biom9110696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N., Deng Y., Wang H., Ye F., Cen S., et al. High-Throughput Screening and Identification of Potent Broad-Spectrum Inhibitors of Coronaviruses. J. Virol. 2019;93 doi: 10.1128/JVI.00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hwang B.Y., Su B.-N., Chai H., Mi Q., Kardono L.B.S., Afriastini J.J., Riswan S., Santarsiero B.D., Mesecar A.D., Wild R., et al. Silvestrol and Episilvestrol, Potential Anticancer Rocaglate Derivatives from Aglaia silvestris. J. Org. Chem. 2004;69:3350–3358. doi: 10.1021/jo040120f. [DOI] [PubMed] [Google Scholar]
- 86.Park J.-Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H., Ryu Y.B., Lee W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzyme Inhib. Med. Chem. 2017;32:504–512. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim J.Y., Kim Y., II, Park S.J., Kim L.K., Choi Y.K., Kim S.-H. Safe, high-throughput screening of natural compounds of MERS-CoV entry inhibitors using a pseudovirus expressing MERS-CoV spike protein. Int. J. Antimicrob. Agents. 2018;52:730–732. doi: 10.1016/j.ijantimicag.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wen C.-C., Shyur L.-F., Jan J.-T., Liang P.-H., Kuo C.-J., Arulselvan P., Wu J.-B., Kuo S.C., Yang N.-S. Traditional chinese medicine herbal extracts of cibotium barometz, gentiana scabra, dioscorea batatas, cassia tora, and taxillus chinensis inhibit sars-cov replication. J. Tradit. Complement. Med. 2011;1:41–50. doi: 10.1016/S2225-4110(16)30055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li S.-Y., Chen C., Zhang H.-Q., Guo H.-Y., Wang H., Wang L., Zhang X., Hua S.-N., Yu J., Xiao P.-G. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005;67:18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen C.-J., Michaelis M., Hsu H.-K., Tsai C.-C., Yang K.D., Wu Y.-C., Cinatl J., Doerr H.W. Toona sinensis Roem tender leaf extract inhibits SARS coronavirus replication. J. Ethnopharmacol. 2008;120:108–111. doi: 10.1016/j.jep.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhuang M., Jiang H., Suzuki Y., Li X., Xiao P., Tanaka T., Ling H., Yang B., Saitoh H., Zhang L., et al. Procyanidins and butanol extract of Cinnamomi Cortex inhibit SARS-CoV infection. Antivir. Res. 2009;82:73–81. doi: 10.1016/j.antiviral.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Keyaerts E., Li S., Vijgen L., Rysman E., Verbeeck J., Van Ranst M., Maes P. Antiviral Activity of Chloroquine against Human Coronavirus OC43 Infection in Newborn Mice. Antimicrob. Agents Chemother. 2009;53:3416–3421. doi: 10.1128/AAC.01509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luo W., Su X., Gong S., Qin Y., Liu W., Li J., Yu H., Xu Q. Anti-SARS coronavirus 3C-like protease effects of Rheum palmatum L. extracts. Biosci. Trends. 2009;3:124–126. [PubMed] [Google Scholar]
- 94.Kumar V., Jung Y.-S., Liang P.-H. Anti-SARS coronavirus agents: A patent review (2008 – present) Expert Opin. Ther. Pat. 2013;23:1337–1348. doi: 10.1517/13543776.2013.823159. [DOI] [PubMed] [Google Scholar]
- 95.Ho T.-Y., Wu S.-L., Chen J.-C., Li C.-C., Hsiang C.-Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2007;74:92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schwarz S., Wang K., Yu W., Sun B., Schwarz W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antivir. Res. 2011;90:64–69. doi: 10.1016/j.antiviral.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin C.-W., Tsai F.-J., Tsai C.-H., Lai C.-C., Wan L., Ho T.-Y., Hsieh C.-C., Chao P.-D.L. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005;68:36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G., Zhang H., Luo H., Zhu L., Jiang P., et al. Small Molecules Blocking the Entry of Severe Acute Respiratory Syndrome Coronavirus into Host Cells. J. Virol. 2004;78:11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.De Wilde A., Raj V., Oudshoorn D., Bestebroer T., Van Nieuwkoop S., Limpens R.W.A.L., Posthuma C., Van Der Meer Y., Bárcena M., Haagmans B., et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang C.-W., Lee Y.-Z., Kang I.-J., Barnard D.L., Jan J.-T., Lin D., Huang C.-W., Yeh T.-K., Chao Y.-S., Lee S.-J. Identification of phenanthroindolizines and phenanthroquinolizidines as novel potent anti-coronaviral agents for porcine enteropathogenic coronavirus transmissible gastroenteritis virus and human severe acute respiratory syndrome coronavirus. Antivir. Res. 2010;88:160–168. doi: 10.1016/j.antiviral.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoever G., Baltina L., Michaelis M., Kondratenko R., Tolstikov G.A., Doerr H.W., Cinatl J. Antiviral Activity of Glycyrrhizic Acid Derivatives against SARS−Coronavirus. J. Med. Chem. 2005;48:1256–1259. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- 102.Wen C.-C., Kuo Y.-H., Jan J.-T., Liang P.-H., Wang S.-Y., Liu H.-G., Lee C.-K., Chang S.-T., Kuo C.-J., Lee S.-S., et al. Specific Plant Terpenoids and Lignoids Possess Potent Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus. J. Med. Chem. 2007;50:4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 103.Kim H., Eo E.-Y., Park H., Kim Y.-C., Park S., Shin H., Kim K. Medicinal herbal extracts of Sophorae radix, Acanthopanacis cortex, Sanguisorbae radix and Torilis fructus inhibit coronavirus replication in vitro. Antivir. Ther. 2010;15:697–709. doi: 10.3851/IMP1615. [DOI] [PubMed] [Google Scholar]
- 104.Yang Q.-Y., Tian X.-Y., Fang W.-S. Bioactive coumarins from Boenninghausenia sessilicarpa. J. Asian Nat. Prod. Res. 2007;9:59–65. doi: 10.1080/10286020500382397. [DOI] [PubMed] [Google Scholar]
- 105.Chen S., Chan K., Jiang Y., Kao R., Lu H., Fan K., Cheng V., Tsui W., Hung I., Lee T. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004;31:69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.-Y., Kim D., Naguyen T.T.H., Park S.-J., Chang J.S., Park K.H. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen L., Li J., Luo C., Liu H., Xu W., Chen G., Liew O.W., Zhu W., Puah C., Shen X., et al. Binding interaction of quercetin-3-β-galactoside and its synthetic derivatives with SARS-CoV 3CLpro: Structure–activity relationship studies reveal salient pharmacophore features. Bioorg. Med. Chem. 2006;14:8295–8306. doi: 10.1016/j.bmc.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ryu Y.B., Park S.-J., Kim Y.M., Lee J.-Y., Seo W.D., Chang J.S., Park K.H., Rho M.-C., Lee W.S. SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii. Bioorg. Med. Chem. Lett. 2010;20:1873–1876. doi: 10.1016/j.bmcl.2010.01.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen C., Zuckerman D.M., Brantley S., Sharpe M., Childress K., Hoiczyk E., Pendleton A.R. Sambucus nigra extracts inhibit infectious bronchitis virus at an early point during replication. BMC Veter Res. 2014;10:24. doi: 10.1186/1746-6148-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen L.-R., Wang Y.-C., Lin Y.W., Chou S.-Y., Chen S.-F., Liu L.T., Wu Y.-T., Kuo C.-J., Chen T.S.-S., Juang S.-H. Synthesis and evaluation of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorg. Med. Chem. Lett. 2005;15:3058–3062. doi: 10.1016/j.bmcl.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu W., Zhu H.-M., Niu G.-J., Shi E.-Z., Chen J., Sun B., Chen W., Zhou H., Yang C. Synthesis, modification and docking studies of 5-sulfonyl isatin derivatives as SARS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2014;22:292–302. doi: 10.1016/j.bmc.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schmidtke M., Meier C., Schacke M., Helbig B., Makarov V., Rabenau H.F., Cinatl J., Wutzler P. Antiviral activity of phenolic polymers and cycloSal-pronucleotides against a SARS-associated coronavirus. Chemother. J. 2005;14:16–21. [Google Scholar]
- 113.Park J.-Y., Kim J.H., Kim Y.M., Jeong H.J., Kim D.W., Park K.H., Kwon H.-J., Park S.-J., Lee W.S., Ryu Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012;20:5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Park J.-Y., Kim J.H., Kwon J.M., Kwon H.-J., Jeong H.J., Kim Y.M., Kim D., Lee W.S., Ryu Y.B. Dieckol, a SARS-CoV 3CLpro inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg. Med. Chem. 2013;21:3730–3737. doi: 10.1016/j.bmc.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Park J.-Y., Ko J.-A., Kim D.W., Kim Y.M., Kwon H.-J., Jeong H.J., Kim C.Y., Park K.H., Lee W.S., Ryu Y.B. Chalcones isolated fromAngelica keiskeiinhibit cysteine proteases of SARS-CoV. J. Enzym. Inhib. Med. Chem. 2016;31:23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- 116.Park J.-Y., Jeong H.J., Kim J.H., Kim Y.M., Park S.-J., Kim D., Park K.H., Lee W.S., Ryu Y.B. Diarylheptanoids from Alnus japonica Inhibit Papain-Like Protease of Severe Acute Respiratory Syndrome Coronavirus. Biol. Pharm. Bull. 2012;35:2036–2042. doi: 10.1248/bpb.b12-00623. [DOI] [PubMed] [Google Scholar]
- 117.Kim D.W., Seo K.H., Curtis-Long M.J., Oh K.Y., Oh J.-W., Cho J.K., Lee K.H., Park K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzym. Inhib. Med. Chem. 2013;29:59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- 118.Song Y.H., Kim D.W., Curtis-Long M.J., Yuk H.J., Wang Y., Zhuang N., Lee K.H., Jeon K.S., Park K.H. Papain-Like Protease (PLpro) Inhibitory Effects of Cinnamic Amides from Tribulus terrestris Fruits. Biol. Pharm. Bull. 2014;37:1021–1028. doi: 10.1248/bpb.b14-00026. [DOI] [PubMed] [Google Scholar]
- 119.Cho J.K., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21:3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu M.-S., Lee J., Lee J.M., Kim Y., Chin Y.-W., Jee J.-G., Keum Y.-S., Jeong Y.-J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Chan P.K., Sidwell R.W. Evaluation of Immunomodulators, Interferons and Knownin VitroSARS-CoV Inhibitors for Inhibition of SARS-Cov Replication in BALB/c Mice. Antivir. Chem. Chemother. 2006;17:275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- 123.Mehra M.R., Desai S.S., Ruschitzka F., Patel A.N. RETRACTED: Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: A multinational registry analysis. Lancet. 2020 doi: 10.1016/s0140-6736(20)31180-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 124.Jo S., Kim S., Shin D.H., Kim M.-S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pyrc K., Bosch B.J., Berkhout B., Jebbink M.F., Dijkman R., Rottier P., Van Der Hoek L. Inhibition of Human Coronavirus NL63 Infection at Early Stages of the Replication Cycle. Antimicrob. Agents Chemother. 2006;50:2000–2008. doi: 10.1128/AAC.01598-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huan C.-C., Wang H.-X., Sheng X.-X., Wang R., Wang X., Mao X. Glycyrrhizin inhibits porcine epidemic diarrhea virus infection and attenuates the proinflammatory responses by inhibition of high mobility group box-1 protein. Arch. Virol. 2017;162:1467–1476. doi: 10.1007/s00705-017-3259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Michaelis M., Geiler J., Naczk P., Sithisarn P., Leutz A., Doerr H.W., Cinatl J. Glycyrrhizin Exerts Antioxidative Effects in H5N1 Influenza A Virus-Infected Cells and Inhibits Virus Replication and Pro-Inflammatory Gene Expression. PLoS ONE. 2011;6:e19705. doi: 10.1371/journal.pone.0019705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.DeDiego M.L., Nieto-Torres J.L., Regla-Nava J.A., Jimenez-Guardeño J.M., Fernandez-Delgado R., Fett C., Castaño-Rodriguez C., Perlman S., Enjuanes L. Inhibition of NF-B-Mediated Inflammation in Severe Acute Respiratory Syndrome Coronavirus-Infected Mice Increases Survival. J. Virol. 2014;88:913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khare P., Sahu U., Pandey S.C., Samant M. Current approaches for target-specific drug discovery using natural com-pounds against SARS-CoV-2 infection. Virus Res. 2020;290:198169. doi: 10.1016/j.virusres.2020.198169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Naik B., Gupta N., Ojha R., Singh S., Prajapati V.K., Prusty D. High throughput virtual screening reveals SARS-CoV-2 multi-target binding natural compounds to lead instant therapy for COVID-19 treatment. Int. J. Biol. Macromol. 2020;160:1–17. doi: 10.1016/j.ijbiomac.2020.05.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chávez-Hernández A.L., Sánchez-Cruz N., Medina-Franco J.L. Fragment Library of Natural Products and Compound Databases for Drug Discovery. Biomolecules. 2020;10:1518. doi: 10.3390/biom10111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rakib A., Paul A., Chy N.U., Sami S.A., Baral S.K., Majumder M., Tareq A.M., Amin M.N., Shahriar A., Uddin Z., et al. Biochemical and Computational Approach of Selected Phytocompounds from Tinospora crispa in the Management of COVID-19. Molecules. 2020;25:3936. doi: 10.3390/molecules25173936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ibrahim M.A.A., Abdeljawaad K.A.A., Abdelrahman A.H.M., Hegazy M.E.F. Natural-like products as potential SARS-CoV-2 Mpro inhibitors: In-silico drug discovery. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1790037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gyebi G.A., Ogunro O.B., Adegunloye A.P., Ogunyemi O.M., Afolabi S.O. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): An in silico screening of alkaloids and terpenoids from African medicinal plants. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Das S., Sarmah S., Lyndem S., Roy A.S. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Quimque M.T.J., Notarte K.I.R., Fernandez R.A.T., Mendoza M.A.O., Liman R.A.D., Lim J.A.K., Pilapil L.A.E., Ong J.K.H., Pastrana A.M., Khan A., et al. Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. J. Biomol. Struct. Dyn. 2020:1–18. doi: 10.1080/07391102.2020.1776639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Badavath V.N., Kumar A., Samanta P.K., Maji S., Das A., Blum G., Jha A., Sen A. Determination of potential inhibitors based on isatin derivatives against SARS-CoV-2 main protease (mpro): A molecular docking, molecular dynamics and structure-activity relationship studies. J. Biomol. Struct. Dyn. 2020:1–19. doi: 10.1080/07391102.2020.1845800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Narkhede R.R., Pise A.V., Cheke R.S., Shinde S.D. Recognition of Natural Products as Potential Inhibitors of COVID-19 Main Protease (Mpro): In-Silico Evidences. Nat. Prod. Bioprospecting. 2020;10:297–306. doi: 10.1007/s13659-020-00253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Abdelrheem D.A., Ahmed S.A., El-Mageed H.R.A., Mohamed H.S., Rahman A.A., Elsayed K.N.M., Ahmed S.A. The inhibitory effect of some natural bioactive compounds against SARS-CoV-2 main protease: Insights from molecular docking analysis and molecular dynamic simulation. J. Environ. Sci. Heal. Part A. 2020;55:1373–1386. doi: 10.1080/10934529.2020.1826192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tripathi N., Goel B., Bhardwaj N., Sahu B., Kumar H., Jain S.K. Virtual screening and molecular simulation study of natural products database for lead identification of novel coronavirus main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1848630. [DOI] [PubMed] [Google Scholar]
- 143.Borquaye L.S., Gasu E.N., Ampomah G.B., Kyei L.K., Amarh M.A., Mensah C.N., Nartey D., Commodore M., Adomako A.K., Acheampong P., et al. Alkaloids from Cryptolepis sanguinolenta as Potential Inhibitors of SARS-CoV-2 Viral Proteins: An In Silico Study. BioMed Res. Int. 2020;2020:1–14. doi: 10.1155/2020/5324560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Qamar M.T.U., Alqahtani S.M., Alamri M.A., Chen L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020;10:313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rodrigues G.C.S., Maia M.D.S., De Menezes R.P.B., Cavalcanti A.B.S., De Sousa N.F., De Moura É.P., Monteiro A.F.M., Scotti L., Scotti M.T. Ligand and Structure-based Virtual Screening of Lamiaceae Diterpenes with Potential Activity against a Novel Coronavirus (2019-nCoV) Curr. Top. Med. Chem. 2020;20:2126–2145. doi: 10.2174/1568026620666200716114546. [DOI] [PubMed] [Google Scholar]
- 146.Muhammad I., Muangchoo K., Muhammad A., Ajingi Y.S., Muhammad I., Umar I.D., Muhammad A.B. A Computational Study to Identify Potential Inhibitors of SARS-CoV-2 Main Protease (Mpro) from Eucalyptus Active Compounds. Computation. 2020;8:79. doi: 10.3390/computation8030079. [DOI] [Google Scholar]
- 147.Ebada S.S., Al-Jawabri N.A., Youssef F.S., El-Kashef D.H., Knedel T.-O., Albohy A., Korinek M., Hwang T.-L., Chen B.-H., Lin G.-H., et al. Anti-inflammatory, antiallergic and COVID-19 protease inhibitory activities of phytochemicals from the Jordanian hawksbeard: Identification, structure–activity relationships, molecular modeling and impact on its folk medicinal uses. RSC Adv. 2020;10:38128–38141. doi: 10.1039/D0RA04876C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., Chugh A., Giri A.P., Kulkarni M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lakshmi S.A., Rajamohamed B.S., Priya A., Pandian S.K. Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: Using structure-based drug discovery approach. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1745282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fakhar Z., Faramarzi B., Pacifico S., Faramarzi S. Anthocyanin derivatives as potent inhibitors of SARS-CoV-2 main protease: An in-silico perspective of therapeutic targets against COVID-19 pandemic. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1801510. [DOI] [PubMed] [Google Scholar]
- 151.Khalifa I., Nawaz A., Sobhy R., Althwab S.A., Barakat H. Polyacylated anthocyanins constructively network with catalytic dyad residues of 3CLpro of 2019-nCoV than monomeric anthocyanins: A structural-relationship activity study with 10 anthocyanins using in-silico approaches. J. Mol. Graph. Model. 2020;100:107690. doi: 10.1016/j.jmgm.2020.107690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Iheagwam F.N., Rotimi S.O. Computer-Aided Analysis of Multiple SARS-CoV-2 Therapeutic Targets: Identification of Potent Molecules from African Medicinal Plants. Scientifica. 2020;2020:1–25. doi: 10.1155/2020/1878410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gahlawat A., Kumar N., Kumar R., Sandhu H., Singh I.P., Singh S., Sjöstedt A., Garg P. Structure-Based Virtual Screening to Discover Potential Lead Molecules for the SARS-CoV-2 Main Protease. J. Chem. Inf. Model. 2020;60:5781–5793. doi: 10.1021/acs.jcim.0c00546. [DOI] [PubMed] [Google Scholar]
- 154.Santos-Filho O. Identification of Potential Inhibitors of Severe Acute Respiratory Syndrome-Related Coronavirus 2 (SARS-CoV-2) Main Protease from Non-Natural and Natural Sources: A Molecular Docking Study. J. Braz. Chem. Soc. 2020;31:2638–2643. doi: 10.21577/0103-5053.20200139. [DOI] [Google Scholar]
- 155.Sampangi-Ramaiah M.H., Vishwakarma R., Shaanker R.U. Molecular docking analysis of selected natural products from plants for inhibition of SARS-CoV-2 main protease. Curr. Sci. 2020;118:1087–1092. doi: 10.18520/cs/v118/i7/1087-1092. [DOI] [Google Scholar]
- 156.Owis A.I., El-Hawary M.S., El Amir D., Aly O.M., Abdelmohsen U.R., Kamel M.S. Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv. 2020;10:19570–19575. doi: 10.1039/D0RA03582C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Karankumar B., Faheem, Sekhar K.V.G.C., Ojha R., Prajapati V.K., Pai A., Murugesan S. Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach for identification of prospective SARS-CoV-2 inhibitor from natural product databases. J. Biomol. Struct. Dyn. 2020:1–24. doi: 10.1080/07391102.2020.1824814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Majumder R., Mandal M. Screening of plant-based natural compounds as a potential COVID-19 main protease inhibitor: An in silico docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1817787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Khan S.A., Zia K., Ashraf S., Uddin R., Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1751298. [DOI] [PubMed] [Google Scholar]
- 160.Joshi T., Sharma P., Mathpal S., Pundir H., Bhatt V., Chandra S. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4529–4536. doi: 10.26355/eurrev_202004_21036. [DOI] [PubMed] [Google Scholar]
- 161.El-Mordy F.M.A., El-Hamouly M.M., Ibrahim M.T., El-Rheem G.A., Aly O.M., El-Kader A.M.A., Youssif K.A., Abdelmohsen U.R. Inhibition of SARS-CoV-2 main protease by phenolic compounds from Manilkara hexandra (Roxb.) Dubard assisted by metabolite profiling and in silico virtual screening. RSC Adv. 2020;10:32148–32155. doi: 10.1039/D0RA05679K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mazzini S., Musso L., Dallavalle S., Artali R. Putative SARS-CoV-2 Mpro Inhibitors from an In-House Library of Natural and Nature-Inspired Products: A Virtual Screening and Molecular Docking Study. Molecules. 2020;25:3745. doi: 10.3390/molecules25163745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mitra K., Ghanta P., Acharya S., Chakrapani G., Ramaiah B., Doble M. Dual inhibitors of SARS-CoV-2 proteases: Pharmacophore and molecular dynamics based drug repositioning and phytochemical leads. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1796802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Singh R., Gautam A., Chandel S., Ghosh A., Dey D., Roy S., Ravichandiran V., Ghosh D. Protease Inhibitory Effect of Natural Polyphenolic Compounds on SARS-CoV-2: An In Silico Study. Molecules. 2020;25:4604. doi: 10.3390/molecules25204604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ibrahim M.A.A., Abdelrahman A.H., Hussien T.A., Badr E.A., Mohamed T.A., El-Seedi H.R., Pare P.W., Efferth T., Hegazy M.-E.F. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Comput. Biol. Med. 2020;126:104046. doi: 10.1016/j.compbiomed.2020.104046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Costa A.N., De Sá É.R.A., Bezerra R.D.S., Souza J.L., Lima F.D.C.A. Constituents of buriti oil (Mauritia flexuosa L.) like inhibitors of the SARS-Coronavirus main peptidase: An investigation by docking and molecular dynamics. J. Biomol. Struct. Dyn. 2020:1–8. doi: 10.1080/07391102.2020.1778538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang J., Chiang J.-H., Tsai S., Hsu Y.-M., Bau D.-T., Lee K.-H., Tsai F.-J. In Silico De Novo Curcuminoid Derivatives from the Compound Library of Natural Products Research Laboratories Inhibit COVID-19 3CLpro Activity. Nat. Prod. Commun. 2020;15 doi: 10.1177/1934578x20953262. [DOI] [Google Scholar]
- 168.Caruso F., Rossi M., Pedersen J.Z., Incerpi S. Computational studies reveal mechanism by which quinone derivatives can inhibit SARS-CoV-2. Study of embelin and two therapeutic compounds of interest, methyl prednisolone and dexamethasone. J. Infect. Public Heal. 2020;13:1868–1877. doi: 10.1016/j.jiph.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Didemnins Inhibit COVID-19 Main Protease (Mpro) Biointerface Res. Appl. Chem. 2020;11:8204–8209. doi: 10.33263/BRIAC111.82048209. [DOI] [Google Scholar]
- 170.Basu A., Sarkar A., Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci. Rep. 2020;10:1–15. doi: 10.1038/s41598-020-74715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hasan A., Paray B.A., Hussain A., Qadir F.A., Attar F., Aziz F.M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rahman N., Basharat Z., Yousuf M., Castaldo G., Rastrelli L., Khan H. Virtual Screening of Natural Products against Type II Transmembrane Serine Protease (TMPRSS2), the Priming Agent of Coronavirus 2 (SARS-CoV-2) Molecules. 2020;25:2271. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vivek-Ananth R., Rana A., Rajan N., Biswal H.S., Samal A. In Silico Identification of Potential Natural Product Inhibitors of Human Proteases Key to SARS-CoV-2 Infection. Molecules. 2020;25:3822. doi: 10.3390/molecules25173822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chikhale R.V., Gupta V.K., E Eldesoky G., Wabaidur S.M., A Patil S., Islam A. Identification of potential anti-TMPRSS2 natural products through homology modelling, virtual screening and molecular dynamics simulation studies. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1798813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Papaioannou M., Schleich S., Prade I., Degen S., Roell D., Schubert U., Tanner T., Claessens F., Matusch R., Baniahmad A. The natural compound atraric acid is an antagonist of the human androgen receptor inhibiting cellular invasiveness and prostate cancer cell growth. J. Cell. Mol. Med. 2008;13:2210–2223. doi: 10.1111/j.1582-4934.2008.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Meimetis L.G., Williams D.E., Mawji N.R., Banuelos C.A., Lal A.A., Park J.J., Tien A.H., Fernandez J.G., De Voogd N.J., Sadar M.D., et al. Niphatenones, Glycerol Ethers from the Sponge Niphates digitalis Block Androgen Receptor Transcriptional Activity in Prostate Cancer Cells: Structure Elucidation, Synthesis, and Biological Activity. J. Med. Chem. 2012;55:503–514. doi: 10.1021/jm2014056. [DOI] [PubMed] [Google Scholar]
- 177.Hsu J.C., Zhang J., Dev A., Wing A., Bjeldanes L.F., Firestone G.L. Indole-3-carbinol inhibition of androgen receptor expression and downregulation of androgen responsiveness in human prostate cancer cells. Carcinogenesis. 2005;26:1896–1904. doi: 10.1093/carcin/bgi155. [DOI] [PubMed] [Google Scholar]
- 178.Qiao Y., Wang X.-M., Mannan R., Pitchiaya S., Zhang Y., Wotring J.W., Xiao L., Robinson D.R., Wu Y.-M., Tien J.C.-Y., et al. Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2021450118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang Z.-R., Zhang Y.-N., Li X.-D., Zhang H.-Q., Xiao S.-Q., Deng F., Yuan Z.-M., Ye H.-Q., Zhang B. A cell-based large-scale screening of natural compounds for inhibitors of SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5 doi: 10.1038/s41392-020-00343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kanjanasirirat P., Suksatu A., Manopwisedjaroen S., Munyoo B., Tuchinda P., Jearawuttanakul K., Seemakhan S., Charoensutthivarakul S., Wongtrakoongate P., Rangkasenee N., et al. High-Content Screening of Thai Medicinal Plants Reveals Boesenbergia rotunda Extract and its Component Panduratin A as Anti-SARS-CoV-2 Agents. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-77003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J., Xie H., Ke C.Q., Hu H.C., Gao M.N., et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41:1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shi T.-H., Huang Y.-L., Chen C.-C., Pi W.-C., Hsu Y.-L., Lo L.-C., Chen W.-Y., Fu S.-L., Lin C.-H. Andrographolide and its fluorescent derivative inhibit the main proteases of 2019-nCoV and SARS-CoV through covalent linkage. Biochem. Biophys. Res. Commun. 2020;533:467–473. doi: 10.1016/j.bbrc.2020.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Coelho C., Gallo G., Campos C.B., Hardy L., Würtele M. Biochemical screening for SARS-CoV-2 main protease inhibitors. PLoS ONE. 2020;15:e0240079. doi: 10.1371/journal.pone.0240079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T., Rizzuti B., Velázquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sharma P., Shanavas A. Natural derivatives with dual binding potential against SARS-CoV-2 main protease and human ACE2 possess low oral bioavailability: A brief computational analysis. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1794970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Efferth T., Romero M.R., Wolf D.G., Stamminger T., Marin J.J.G., Marschall M. The Antiviral Activities of Artemisinin and Artesunate. Clin. Infect. Dis. 2008;47:804–811. doi: 10.1086/591195. [DOI] [PubMed] [Google Scholar]
- 187.Baglivo M., Baronio M., Natalini G., Beccari T., Chiurazzi P., Fulcheri E., Petralia P.P., Michelini S., Fiorentini G., Miggiano G.A.D., et al. Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: A possible strategy for reducing SARS-COV-2 infectivity? Acta Biomed. 2020;91:161–164. doi: 10.23750/abm.v91i1.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bailly C., Vergoten G. Glycyrrhizin: An alternative drug for the treatment of COVID-19 infection and the associated res-piratory syndrome? Pharmacol. Ther. 2020;214:107618. doi: 10.1016/j.pharmthera.2020.107618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang S., Li W., Hui H., Tiwari S.K., Zhang Q., Croker B.A., Rawlings S., Smith D., Carlin A.F., Rana T.M. Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39 doi: 10.15252/embj.2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Han J.W., Ahn S.H., Park S.H., Wang S.Y., Bae G.U., Seo D.W., Kwon H.K., Hong S., Lee H.Y., Lee Y.W., et al. Apicidin, a histone deacetylase inhibitor, inhibits proliferation of tumor cells via induction of p21WAF1/Cip1 and gelsolin. Cancer Res. 2000;60:6068–6074. [PubMed] [Google Scholar]
- 192.Kindrachuk J., Ork B., Hart B.J., Mazur S., Holbrook M.R., Frieman M.B., Traynor D., Johnson R.F., Dyall J., Kuhn J.H., et al. Antiviral Potential of ERK/MAPK and PI3K/AKT/mTOR Signaling Modulation for Middle East Respiratory Syndrome Coronavirus Infection as Identified by Temporal Kinome Analysis. Antimicrob. Agents Chemother. 2015;59:1088–1099. doi: 10.1128/AAC.03659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Coleman C.M., Sisk J.M., Mingo R.M., Nelson E.A., White J.M., Frieman M.B. Abelson Kinase Inhibitors Are Potent Inhibitors of Severe Acute Respiratory Syndrome Coronavirus and Middle East Respiratory Syndrome Coronavirus Fusion. J. Virol. 2016;90:8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Emadi A., Chua J.V., Talwani R., Bentzen S.M., Baddley J.W. Safety and Efficacy of Imatinib for Hospitalized Adults with COVID-19: A structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21:1–5. doi: 10.1186/s13063-020-04819-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhao H., Mendenhall M., Deininger M.W. Imatinib is not a potent anti-SARS-CoV-2 drug. Leukemia. 2020;34:3085–3087. doi: 10.1038/s41375-020-01045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. Genetic mechanisms of critical illness in Covid-19. Nat. Cell Biol. 2020;1 doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 197.Sigurdsson S., Nordmark G., Göring H.H.H., Lindroos K., Wiman A.-C., Sturfelt G., Jönsen A., Rantapää-Dahlqvist S., Möller B., Kere J., et al. Polymorphisms in the Tyrosine Kinase 2 and Interferon Regulatory Factor 5 Genes Are Associated with Systemic Lupus Erythematosus. Am. J. Hum. Genet. 2005;76:528–537. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dowty M.E., Lin T.H., Jesson M.I., Hegen M., Martin D.A., Katkade V., Menon S., Telliez J. Janus kinase inhibitors for the treatment of rheumatoid arthritis demonstrate similar profiles of in vitro cytokine receptor inhibition. Pharmacol. Res. Perspect. 2019;7:e00537. doi: 10.1002/prp2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.The Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat. Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 200.Forman S.B., Pariser D.M., Poulin Y., Vincent M.S., A Gilbert S., Kieras E.M., Qiu R., Yu D., Papacharalambous J., Tehlirian C., et al. TYK2/JAK1 Inhibitor PF-06700841 in Patients with Plaque Psoriasis: Phase IIa, Randomized, Double-blind, Placebo-controlled Trial. J. Investig. Dermatol. 2020;140:2359–2370.e5. doi: 10.1016/j.jid.2020.03.962. [DOI] [PubMed] [Google Scholar]
- 201.Page K.M., Suarez-Farinas M., Suprun M., Zhang W., Garcet S., Fuentes-Duculan J., Li X., Scaramozza M., Kieras E., Banfield C., et al. Molecular and Cellular Responses to the TYK2/JAK1 Inhibitor PF-06700841 Reveal Reduction of Skin In-flammation in Plaque Psoriasis. J. Investig. Dermatol. 2020;140:1546–1555.e4. doi: 10.1016/j.jid.2019.11.027. [DOI] [PubMed] [Google Scholar]
- 202.Gerstenberger B.S., Ambler C.M., Arnold E.P., Banker M.-E., Brown M.F., Clark J.D., Dermenci A., Dowty M.E., Fensome A., Fish S., et al. Discovery of Tyrosine Kinase 2 (TYK2) Inhibitor (PF-06826647) for the Treatment of Autoimmune Diseases. J. Med. Chem. 2020;63:13561–13577. doi: 10.1021/acs.jmedchem.0c00948. [DOI] [PubMed] [Google Scholar]
- 203.Liu M., Xiao C., Sun M., Tan M., Hu L., Yu Q. Parthenolide Inhibits STAT3 Signaling by Covalently Targeting Janus Kinases. Molecules. 2018;23:1478. doi: 10.3390/molecules23061478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.