Abstract
Background
Sepsis is a life-threatening condition accompanied by organ dysfunction subsequent to a dysregulated host response to infection. Up to 60% of patients with sepsis develop acute kidney injury (AKI), which is associated with a poor clinical outcome. The pathophysiology of sepsis-associated AKI (sepsis-AKI) remains incompletely understood, but mitochondria have emerged as key players in the pathogenesis. Therefore, our aim was to identify mitochondrial damage in patients with sepsis-AKI.
Methods
We conducted a clinical laboratory study using “warm” postmortem biopsies from sepsis-associated AKI patients from a university teaching hospital. Biopsies were taken from adult patients (n = 14) who died of sepsis with AKI at the intensive care unit (ICU) and control patients (n = 12) undergoing tumor nephrectomy. To define the mechanisms of the mitochondrial contribution to the pathogenesis of sepsis-AKI, we explored mRNA and DNA expression of mitochondrial quality mechanism pathways, DNA oxidation and mitochondrial DNA (mtDNA) integrity in renal biopsies from sepsis-AKI patients and control subjects. Next, we induced human umbilical vein endothelial cells (HUVECs) with lipopolysaccharide (LPS) for 48 h to mimic sepsis and validate our results in vitro.
Results
Compared to control subjects, sepsis-AKI patients had upregulated mRNA expression of oxidative damage markers, excess mitochondrial DNA damage and lower mitochondrial mass. Sepsis-AKI patients had lower mRNA expression of mitochondrial quality markers TFAM, PINK1 and PARKIN, but not of MFN2 and DRP1. Oxidative DNA damage was present in the cytosol of tubular epithelial cells in the kidney of sepsis-AKI patients, whereas it was almost absent in biopsies from control subjects. Oxidative DNA damage co-localized with both the nuclei and mitochondria. Accordingly, HUVECs induced with LPS for 48 h showed an increased mnSOD expression, a decreased TFAM expression and higher mtDNA damage levels.
Conclusion
Sepsis-AKI induces mitochondrial DNA damage in the human kidney, without upregulation of mitochondrial quality control mechanisms, which likely resulted in a reduction in mitochondrial mass.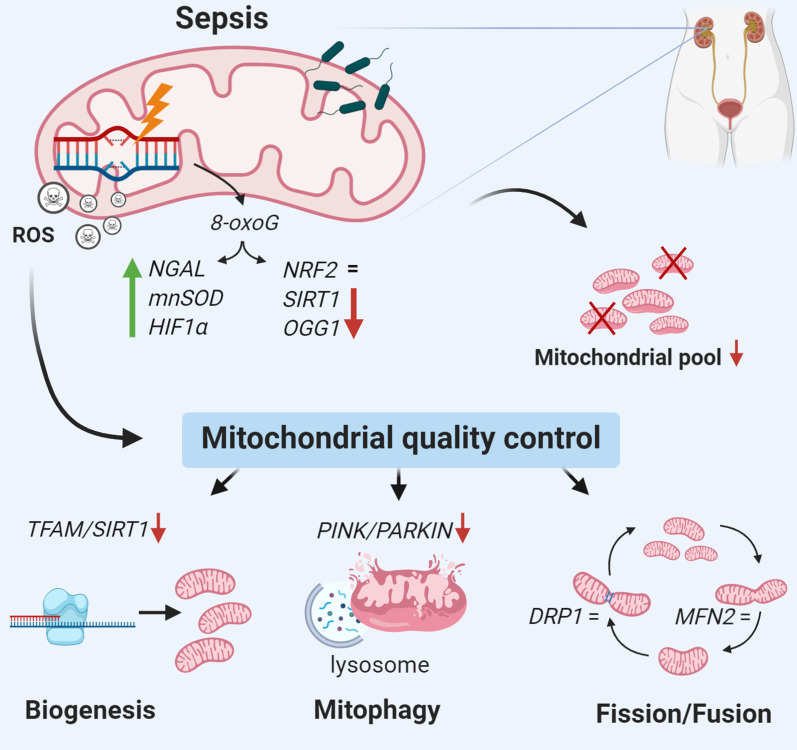
Keywords: Sepsis, Acute kidney injury, Reactive oxygen species, Mitochondria
Introduction
Sepsis is defined as a dysregulated host response to infection, which can lead to loss of organ homeostasis, multiple organ failure, and ultimately, death in patients [1]. Mortality and morbidity associated with sepsis and septic shock are high. In-hospital mortality rate amounts to 20–30% for sepsis and 40–60% for septic shock [1, 2]. Despite extensive research, the pathophysiology of sepsis remains incompletely understood. As a result, therapeutic options directed at the molecular cause of organ dysfunction are lacking, and the current therapy is limited to source control (i.e., antibiotics, drainage) and organ support [1–4]. Up to 60% of patients with sepsis develop acute kidney injury (AKI) and sepsis is the most common cause of AKI in the intensive care unit (ICU) [5, 6]. The occurrence of AKI in sepsis is associated with failure of other organs and increased mortality [5, 6]. Thus, sepsis-AKI constitutes a serious medical health problem.
Mitochondria have emerged as key players in the pathogenesis of sepsis-AKI [7–9]. Healthy mitochondria are essential for maintaining renal homeostasis and are important in supporting the metabolic challenge during sepsis [2, 7, 8]. Mitochondrial failure during sepsis results in renal ATP-depletion and increased levels of reactive oxygen species (ROS), which progresses into loss of cellular homeostasis and organ dysfunction [10]. Sepsis lowers ATP levels, increases biomarkers for mitochondrial dysfunction and reduces antioxidant defense, which are associated with poor survival, as demonstrated in muscle biopsies in 16 critically ill patients compared to 10 healthy, age-matched patients undergoing elective hip surgery [11]. It is known that accumulating levels of ROS during sepsis can cause damage to mitochondrial proteins and DNA in liver and heart from rats [12, 13], further impairing mitochondrial function and leading to a vicious circle in which ROS production continues to increase [12–14]. Conversely, biogenesis of mitochondria is associated with an increased survival of septic shock [11], and experimental models of sepsis-AKI confirm the importance of maintaining healthy mitochondria for renal recovery and survival [15–19].
To date, the precise molecular mechanisms of mitochondria in the pathogenesis of sepsis-AKI are incompletely understood, which hampers the development of a molecular targeted therapy to prevent or treat sepsis-AKI and improve outcome. The high metabolic burden during sepsis leads to increased ROS production by mitochondria. We hypothesized that damage to mitochondrial DNA during sepsis causes mitochondrial dysfunction with relevance to both short-term outcome and potentially also for long-term outcome in sepsis-AKI. We therefore explored the mRNA expression of mitochondrial quality mechanism pathways and studied mitochondrial DNA (mtDNA) oxidation and integrity in renal biopsies from sepsis-AKI patients and control subjects. Next, we validated these results in vitro, by inducing human umbilical vein endothelial cells (HUVECs) with lipopolysaccharide (LPS) for 48 h to mimic sepsis.
Material and methods
Patients
We collected postmortem renal biopsies from patients as quickly as possible after death [24–150-min postmortem] from fourteen patients who died of sepsis with acute kidney injury at the ICU of the University Medical Center Groningen (UMCG). The biopsy was taken from the renal cortex, and once removed from the patient, the biopsies were immediately snap frozen using liquid nitrogen and stored in the − 80 °C until analysis. No in vitro perfusion took place. Due to the rapid sampling time and immediate freezing, we eliminated unwanted tissue necrosis and autolysis, which would disturb the analysis. Patients were defined as having septic shock according to the internal sepsis definitions, comprising sepsis with atrial hypotension despite adequate fluid resuscitation [20], while AKI was classified according to the RIFLE criteria [21]. Severity of illness was defined upon admission to the ICU using the Acute Physiology and Chronic Health Evaluation (APACHE) IV score and the Simplified Acute Physiology Score (SAPS) II score [22, 23]. Informed written consent for performing biopsies was obtained during the family meeting, before or just after death. In control subjects, following preoperative consent, non-septic kidney biopsies were obtained from twelve patients who underwent complete nephrectomy due to renal cell carcinoma. In this procedure, a healthy section of the kidney cortex was taken, as far away as possible from the carcinoma. Nephrectomy biopsies were analyzed by a pathologist and considered to be normal healthy controls. Additional details of the collection of kidney biopsies are described elsewhere [24]. Both septic patients and controls were 18 years or older. Patients with preexisting chronic kidney disease (CKD), active autoimmune disorders with renal involvement and treatment with immune-suppressive medication were excluded from this study. The Medical Ethics Review Committee (METC) of the UMCG reviewed and approved this study (METC 2011/372). Patient characteristics and clinical and laboratory details can be found in Table 1.
Table 1.
Patient characteristics
| Control (n = 12) | Sepsis-AKI (n = 14) | |
|---|---|---|
| Mean age (years) | 62 (20–79) | 76 (53–85) |
| Sex (male:female) | 5:7 | 11:3 |
| Comorbidities/medical history (n) | ||
| Hypertension | 3 | 6 |
| Diabetes mellitus | 1 | 1 |
| COPD or asthma | 4 | 2 |
| Coronary artery disease | 1 | 5 |
| Renal disease | 0 | 0 |
| Auto-immune disease | 0 | 1 |
| Neoplasms (extra-renal) | 4 | 1 |
| RIFLE stage (n) | N/A | |
| Risk | 0 | |
| Injury | 6 | |
| Failure | 8 | |
| Lost renal function | 0 | |
| End-stage kidney failure | 0 | |
| Need for RRT in ICU: yes/no | N/A | 5/14 |
| Serum creatinine at admission (μmol/L) | N/A | 135 (81–355) |
| Serum creatinine before biopsy (μmol/L) | 73 (58–114) | 156 (73–401) |
| APACHE IV score | N/A | 97 (50–175) |
| SAPS II score | N/A | 65 (35–88) |
| Sepsis focus | N/A | |
| Lower respiratory tract | 5 | |
| Skin/soft tissue | 2 | |
| Intra-abdominal (Mesenteric ischemia, necrotizing pancreatitis) | 6 | |
| Endovascular (endocarditis) | 1 |
Data are presented as median with upper and lower range. N/A not applicable, RRT renal replacement therapy, ICU intensive care unit, APACHE Acute Physiology and Chronic Health Evaluation, SAPS Simplified Acute Physiology Score
Cells
Human umbilical vein endothelial cells (HUVECs) and medium were obtained from the UMCG Endothelial Cell Facility. Briefly, primary isolates of umbilical cords were mixed and subsequently cultured on HUVEC culture medium, consisting of RPMI 1640 (Lonza, Breda, Netherlands) supplemented with 20% heat-inactivated fetal calf serum (ThermoFisher, Waltham, MA), 2 mM l-glutamine (Life Technologies, Carlsbad, CA), 5 U/ml heparin (Leo Pharmaceutical Products, The Netherlands), 1% Penicillin/Streptomycin (Sigma-Aldrich, St. Louis, MI) and 50 μg/ml EC growth factor supplement from (Sigma-Aldrich). Cells were plated in 6-well culture plates (Corning, St. Louis, MI), and at 80% confluency cells were stimulated with 10 µg/ml lipopolysaccharide (LPS) E. coli 0111:B4 (Sigma-Aldrich) for 48 h.
DNA isolation
Total DNA was isolated to perform a polymerase chain reaction (PCR) and determine mitochondrial DNA damage. DNA was isolated from renal biopsies of eight controls subjects and twelve sepsis-AKI patients. First, sections of 5 μm thickness were cut from the renal biopsies. Samples were pretreated with 500 µL collagenase V (300 U/mL; Sigma-Aldrich, Darmstadt, Germany), incubated at 37 °C for 3 h and vortexed every 30 min. Subsequently, 500 µL RPMI (ThermoFisher, Paisly, UK) was added, followed by centrifugation for 10 min at 20,000 G at room temperature. Next, Rapid Sample Concentrator (RSC) blood DNA kit (Promega, Madison, USA) was used to isolate DNA from the pellet using Maxwell 16 MDx AS3000 (Promega), according to the manufacturer’s protocol. DNA from HUVECs was isolated with Nucleospin DNA kit (MACHEREY–NAGEL, GmbH & Co. KG, Germany), according to manufacturer’s protocol.
Quantification of mitochondrial mass and DNA damage
Mitochondrial copy number, indicative of mitochondrial mass, and mitochondrial DNA damage were determined using quantitative polymerase chain reaction (qPCR). Oligonucleotide primers (Sigma-Aldrich; Table 2) were designed using Clone Manager 9 software and validated by assessing the efficiency, melting- and temperature curves using CFX384- Real-Time system (Biorad, California, USA). Amplification of the DNA was performed using the following thermal profile: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s. All reactions were carried out in duplicate, and the obtained threshold cycle (Ct) values were averaged. MtDNA copy number was calculated using the average levels of the mitochondrial genes: NADH dehydrogenase 1 (ND1), NADH dehydrogenase 4 (ND4), NADH dehydrogenase 6 (ND6), Cytochrome C Oxidase I (COX1), Cytochrome B (CYTB) and D-loop, divided by the amount of nuclear housekeeping gene Beta 2-microglobulin (B2M), using the 2 − ∆∆Ct method.
Table 2.
List of primers used for quantification of mRNA and DNA expression levels by qPCR and RT-qPCR
| Gene | Primer sequence | |
|---|---|---|
| ND1, ETC complex 1 | Forward: | TGGCTCCTTTAACCTCTCCA |
| Reverse: | GGTTCGGTTGGTCTCTGCTA | |
| ND4, ETC complex 1 | Forward: | GGCGGCTATGGTATAATACG |
| Reverse: | GTAGGCAGATGGAGCTTGTT | |
| ND6, ETC complex 1 | Forward: | TGATTGTTAGCGGTGTGGTC |
| Reverse: | CCTCAATAGCCATCGCTGTA | |
| COX1, ETC complex 4 | Forward: | CTAACAGACCGCAACCTCAA |
| Reverse: | CCGAAGCCTGGTAGGATAA | |
| Cytochrome b, ETC complex 3 | Forward: | TTCCTAGCCATGCACTACTC |
| Reverse: | GAAGGTAGCGGATGATTCAG | |
| D-loop, regulative region | Forward: | AACCTACCCACCCTTAACAG |
| Reverse: | CACTCTTGTGCGGGATATTG | |
| β2M | Forward: | CTGGGTAGCTCTAAACAATGTATTCA |
| Reverse: | CATGTACTAACAAATGTCTAAAATGGT | |
ETC electron transport chain
Mitochondrial DNA damage was assessed by qPCR for determination of a short (~ 200 bp) mtDNA part and long-range PCR, for determination of a long (10 kb) mtDNA part. First, the relative amount of each mitochondrial gene was quantified by qPCR and divided by the amount of nuclear housekeeping gene B2M, using the 2 − ∆∆Ct method. Next, a long-range PCR was performed using the TaKaRa LA Taq DNA polymerase kit (Takara Bio, Kusatsu, Japan) to amplify a 10-kb mtDNA template, stretching from the ND5 to ND1 gene, thereby comprising more than two-third of the mitochondrial genome. A short mtDNA fragment of 222 bp was amplified by qPCR to serve as reference (Table 3). The long fragment was amplified with T100 Thermal Cycler (Biorad, California, USA), using the following thermal profile: 94 °C for 1 min, followed by 18 cycles of 15 s at 94 °C and 12 min at 64 °C, and ending with 10 min at 72 °C. The short fragment was amplified using the following thermal profile: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Both PCR products were separated and visualized on a 1% agarose gel (45 min, 100 V), and their intensity was analyzed using ImageJ [25]. The ratio of the short to long fragment was calculated to quantify mtDNA damage. Due to limited sample material, DNA from six control subjects and twelve sepsis-AKI patients was used for long-range PCR, whereas material from seven control subjects and twelve sepsis-AKI patients was used for qPCR.
Table 3.
List of primers used for amplification of mitochondrial DNA with long-range PCR and qPCR
| Primer | Primer sequence |
|---|---|
| Forward long fragment mtDNA (10 kb) | TCTAAGCCTCCTTATTCGAGCCGA |
| Forward Short fragment mtDNA (222 bp) | CCCCACAAACCCCATTACTAAACCCA |
| Reverse primer mtDNA (short and long) | TTTCATCATGCGGAGATGTTGGATGG |
mtDNA mitochondrial DNA
RNA isolation and quantification of gene expression
First, total RNA was isolated from twelve control subjects and twelve sepsis-AKI patients for reverse transcription by quantitative polymerase chain reaction (RT-qPCR). RNA was isolated from 20 × 5 µm kidney cryosections using the RNeasy Mini Plus Kit (Qiagen, Leusden, The Netherlands), according to the manufacturer's protocol. RNA integrity was determined by gel electrophoresis and consistently found intact. RNA yield and purity were measured by an ND-1000 UV–Vis spectrophotometer (NanoDrop Technologies, Rockland, DE). cDNA was synthesized as previously described [26]. RNA from HUVECs was isolated using Nucleospin RNA kit (MACHEREY–NAGEL). Cells were pretreated with TRIzol, followed by RNA isolation according to manufacturer’s protocol. Next, oligonucleotide primers (Sigma-Aldrich; Table 4) were designed using Clone Manager 9 software and validated by assessing the efficiency, melting- and temperature curve using RT-qPCR. RT-qPCR amplification was performed using the following thermal profile: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C 30 s. Reactions were carried out in duplicate, and the obtained Ct values were averaged. Gene expression (mRNA) was normalized using β-actin as a housekeeping gene, and 2 − ∆∆Ct was used to obtain relative quantity.
Table 4.
List of primers for quantification of mRNA expression of mitochondrial quality mechanisms, mitochondrial complexes and oxidation pathways by RT-qPCR
| Gene | Primer sequence | Process | |
|---|---|---|---|
| PINK1 | Forward: | GACGCTGTTCCTCGTTATGA | Mitophagy |
| Reverse: | TCCAGCTCCACAAGGATGTT | ||
| PARKIN | Forward: | GACCCTCAACTTGGCTACTC | Mitophagy |
| Reverse: | CTTCGCAGGTGACTTTCCTC | ||
| MFN2 | Forward: | CCGCCACATAGAGGAAGGAC | Fusion |
| Reverse: | CGCACAGACACAGGAAGGAG | ||
| DRP1 | Forward: | AAGCTGCTGCCATAGTCCTC | Fission |
| Reverse: | ACCACAGCCATGTCAGTGTC | ||
| NRF2 | Forward: | GCTACTAATCAGGCTCAGTC | Mitochondrial biogenesis |
| Reverse: | GTAGTCTCAACCAGCTTGTC | ||
| TFAM | Forward: | CATGGACTTCTGCCAGCATA | Mitochondrial biogenesis |
| Reverse: | AGAACACCGTGGCTTCTACA | ||
| mnSOD | Forward: | ACGCGGCCTACGTGAACAAC | Antioxidant enzyme |
| Reverse: | CAACAGATGCAGCCGTCAGC | ||
| OGG1 | Forward: | GTGTGCGACTGCTGCGACAA | mtDNA damage repair (excision of 8-oxoguanine) |
| Reverse: | CTGGATGAGCCGAGGTCCAA | ||
| HIF1α | Forward: | TGAGGGGACAGGAGGATCAG | Master regulator of cellular and systemic homeostatic response to hypoxia |
| Reverse: | CACGCGGAGAAGAGAAGGAA | ||
| SIRT1 | Forward: | ATGCTGGCCTAATAGAGTGG | Regulates epigenetic gene silencing, biogenese and antioxidant mechanisms |
| Reverse: | TCTGGAACATCAGGCTCATC | ||
| NGAL | Forward: | GGTGAGCACCAACTACAACC | Early biomarker of acute kidney injury |
| Reverse: | GCCCAGAGATTTGGAGAAGC | ||
| NDUFA1 | Forward; | ACTGGCTACTGCGTACATCC | ETC complex 1 (nDNA) |
| Reverse: | AGATGCGCCTATCTCTTTCC | ||
| COX5b | Forward: | CATTGGCTCCTTCTCCCATA | ETC complex 4 (nDNA) |
| Reverse: | CATACCAGGTGGTCCCATTC | ||
| B-actin | Forward: | AGGATGCAGAAGGAGATCAC | Housekeeping gene |
| Reverse: | AGTCATAGTCCGCCTAGAAG | ||
mtDNA mitochondrial DNA, ETC electron transport chain, nDNA nuclear DNA
Immunohistochemical analysis
DNA oxidation was assessed by immunohistochemical analysis of 8-oxoguanine (8-oxoG) on formalin-fixed paraffin-embedded kidney sections. Therefore, sections were deparaffinized in xylene and rehydrated in graded ethanol series (70–100%) and distilled water. After deparaffination, heat-induced epitope retrieval was performed using citrate buffer (pH 6) antigen retrieval. Endogenous peroxidase activity was blocked by incubating the slides with 3% hydrogen peroxide. After washing, the slides were incubated with anti-human 8-oxoG Antibody, (Abcam, Cambridge, UK) diluted 1:400 in antibody solution (5% fetal calf serum in PBS) for 1 h at room temperature. After washing, slides were incubated with rabbit anti-mouse IgG secondary antibody (Southern Biotech, Birmingham, USA) diluted 1:100 in antibody solution with 2% normal human serum (NHS) for 45 min at room temperature. Slides were washed and incubated with anti-rabbit horse radish peroxidase-labeled antibody (EnVision kit, DAKO Cytomation, Glostrup, Denmark). Peroxidase activity was detected using 3-amino-9-ethylcarbazole (AEC) complex, and the sections were subsequently counterstained with Mayer’s hematoxylin (Merck, Darmstadt, Germany) before mounting in Aquatex mounting agent (Merck). 8-oxoG staining was visualized with a Leica DCF295 color camera (Leica, Heerbrugg, Switzerland), and images taken with the Leica software application suite (LAS version 4, Leica).
Immunofluorescent analysis
Co-localization of DNA oxidation and mitochondria was assessed by immunofluorescent analysis of Translocase Of Outer Mitochondrial Membrane (TOM20) (mitochondrial immunolabeling) and 8-oxoG on kidney cryosections (9 µm). In short, the slides were air-dried, fixated with ice-cold acetone for 10 min, washed with PBS and permeabilized with 0.25% Triton-X-100 for 10 min. Then, sections were incubated for 1 h at room temperature with the primary 8-oxoG antibody (Novus Biologicals, Abingdon, Oxon), diluted 1:75 in 1% bovine serum albumin (BSA) in PBS, washed with PBS and incubated for 30 min with Donkey anti-Goat secondary antibody (ThermoFisher), diluted 1:100 in 1% PBS/BSA. Next, slides were incubated for 1 h at room temperature with TOM20 primary antibody (Santa Cruz, Dallas, USA), diluted 1:20 in 1% PBS/BSA, followed by washing with PBS and incubation for 30 min with Donkey anti-Rabbit IgG secondary antibody (ThermoFisher), diluted 1:100 in 1% PBS/BSA. Slides were mounted in Vectashield with DAPI (Vector Laboratories Inc., Burlingame, CA, USA) and visualized with a Leica DM2000 microscope (Leica, Amsterdam, Netherlands). Co-localization was identified using ImageJ software.
Statistical analysis and data presentation
Statistical analysis was performed with IBM SPSS 23.0 for Windows (IBM Corp., Armonk, N.Y., USA) and GraphPad Prism Software version 7.02 (GraphPad Prism software Inc., San Diego, CA, USA). Two-tailed Mann–Whitney U tests were used to calculate statistical differences between groups (using GraphPad Prism software), whereas correlations between selected groups were assessed by Spearman’s Rho tests in IBM SPSS. P < 0.05 was considered significant different. Data are expressed as median with upper and lower ranges. Figures were made with GraphPad Prism (GraphPad Prism software Inc.), bars represent the median and each dot represents an individual.
Results
Study population
Renal biopsies from fourteen patients with sepsis-AKI and twelve control subjects were examined. Six sepsis-AKI patients had an intra-abdominal infection, five patients suffered from a lower respiratory tract infection, two patients had a skin/soft tissue infection and one patient suffered from endocarditis. The median age of patients with sepsis-AKI was 76 (range 53–85) years, and for control subjects the median age was 62 (range 20–79) (Table 1). The male-to-female ratio was 11:3 and 5:7 for sepsis-AKI and control subjects, respectively. Six sepsis-AKI patients were categorized in the Injury stage of the RIFLE criteria and eight in the Failure stage. Five patients needed renal replacement therapy in the ICU. Clinical characteristics are listed in Table 1, and serum creatinine before nephrectomy in control subjects or before the biopsy was taken in sepsis-AKI patients is shown in Fig. 1.
Fig. 1.
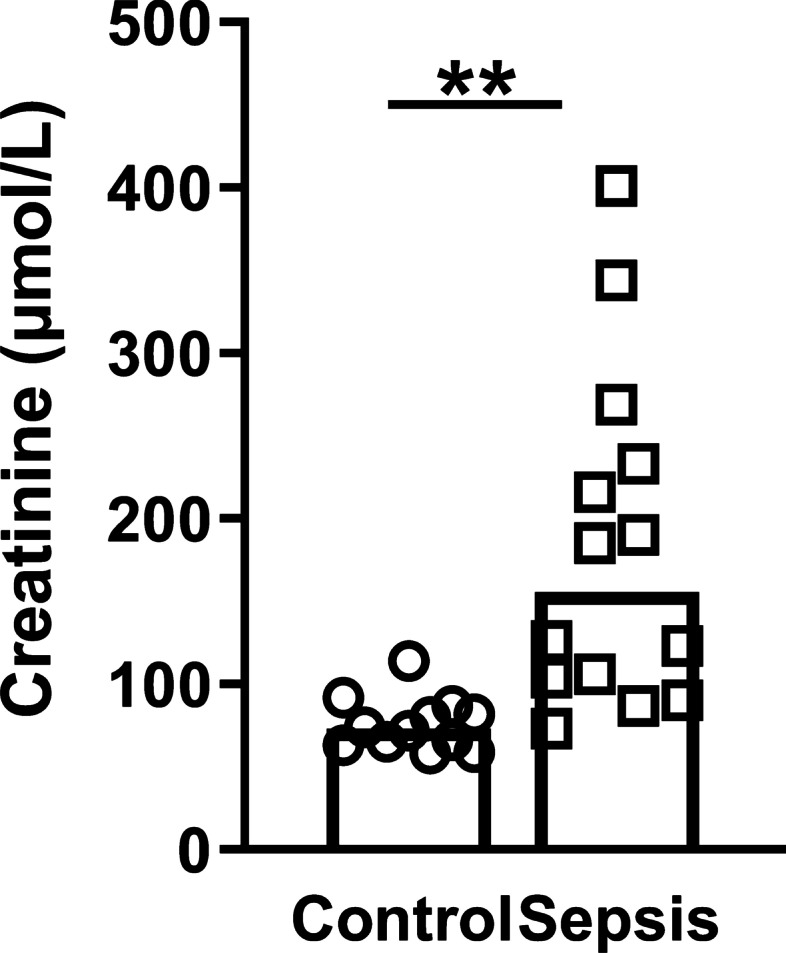
Serum creatinine concentrations. Serum concentrations of creatinine (µmol/L) in control subjects and sepsis-AKI patients before nephrectomy or prior to extirpation, respectively
Sepsis-AKI is associated with renal oxidative stress
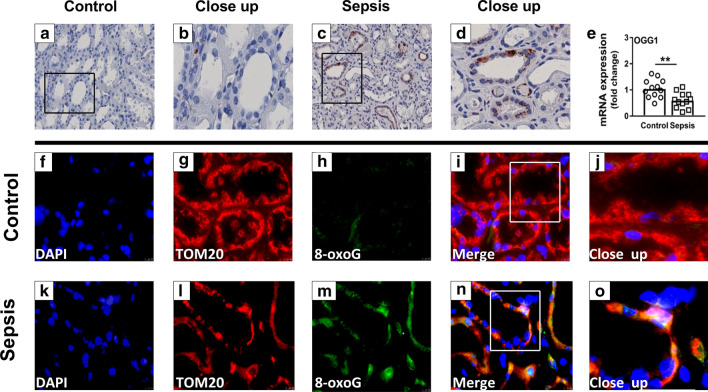
To explore whether sepsis-AKI leads to oxidation of DNA, we performed an 8-oxoG staining and quantified the expression of the base-excision repair enzyme 8-oxoguanine DNA glycosylase (OGG1) in kidney biopsy material. 8-oxoG staining, a mutagenic base byproduct that occurs as a result of exposure to reactive oxygen species, was present in some of the tubular epithelial cells of all sepsis-AKI patients, but almost absent in biopsies from control subjects (Fig. 2a–d). Expression of OGG1, the primary enzyme responsible for the excision of 8-oxoG, was downregulated in patients with sepsis (p < 0.005; Fig. 2e). Fluorescent staining of the mitochondrial marker TOM20 and 8-oxoG demonstrated their colocalization, implicating oxidation of the mitochondrial DNA (Fig. 2j, o).
Fig. 2.
Oxidative DNA damage in controls and sepsis-AKI subjects. a–d Immunohistochemistry of 8-oxoG (DNA oxidation marker) e mRNA expression of OGG1 as quantified by RT-qPCR f–o immunohistochemistry of TOM20 (mitochondrial marker) and 8-oxoG (DNA oxidation marker). Bars represent median, dots represent individual levels; ** means p < 0.005
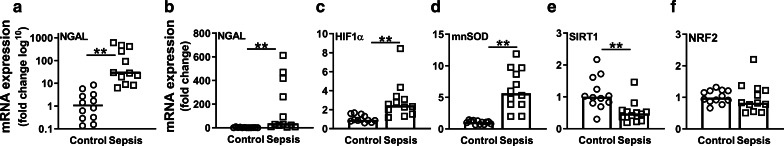
To confirm the presence of kidney damage in sepsis-AKI patients, we quantified mRNA expression of neutrophil gelatinase-associated lipocalin (NGAL), a marker for kidney damage and oxidative stress [26]. Septic patients showed a significantly upregulation of NGAL (p < 0.001; Fig. 3a, b). In addition, sepsis-AKI patients showed an increased expression of hypoxia-inducible factor 1-alpha (HIF1α), the master oxygen sensor within cells and manganese superoxide dismutase (mnSOD), a key antioxidant enzyme, suggesting an active response to oxidative stress (both p < 0.001; Fig. 3c, d). mnSOD positively correlated with NGAL and HIFα (R = 0.858 and R = 0.921, respectively, both p < 0.001). In contrast, sepsis-AKI patients had lowered mRNA expression of Sirtuin 1 (SIRT1), involved in inhibiting oxidative stress and in biogenesis, as compared to control subjects (p < 0.01; Fig. 3e), while NRF2 important in regulation of antioxidant protein expression was not different between both groups (Fig. 3f). Also in HUVECs the mnSOD expression was significantly increased after 48 h of LPS induction compared to controls (p < 0.001, Fig 4a), while NRF2 was not different (Fig. 4b). Taken together, patients with sepsis-AKI likely display increased levels of oxidative stress, as demonstrated by increased levels of 8-oxoG in tubular cells and increased mRNA expression of oxidative stress defense pathways.
Fig. 3.
Gene expression of oxidative damage- and antioxidant markers. mRNA expression of a NGAL (log10), b NGAL, c HIF1α, d mnSOD, e SIRT1, and f NRF2 as quantified by RT-qPCR. Bars represent median, dots represent individual levels; ** means p < 0.005
Fig. 4.
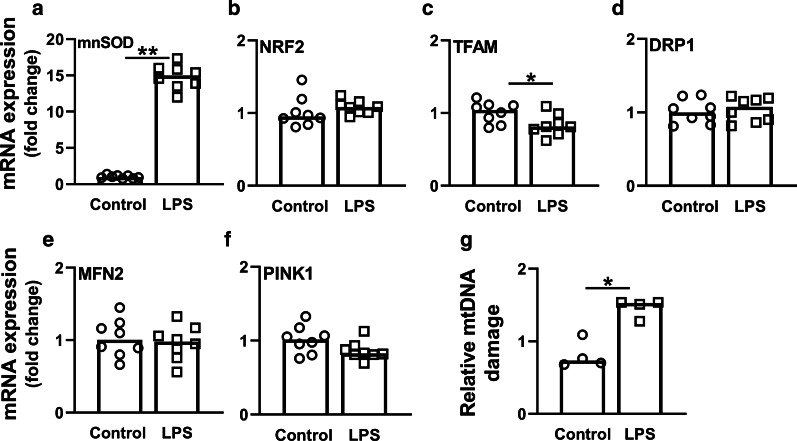
Changes in mitochondrial biogenesis, fusion, fission and mitophagy after 48-h LPS induction in HUVECs. Cells were induced with 10 µg/mL LPS for 48 h. mRNA expression of a mnSOD, b NRF2, c TFAM, d DRP1, e MFN2, and f PINK1; g Relative mtDNA damage levels of control and LPS-induced HUVECs. Bars represent median, dots represent individual levels; * means p < 0.05 and ** p <0.005
Sepsis-AKI is associated with mitochondrial DNA damage in the kidney
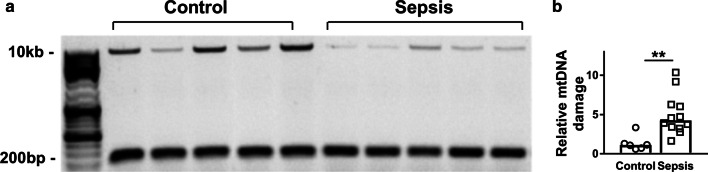
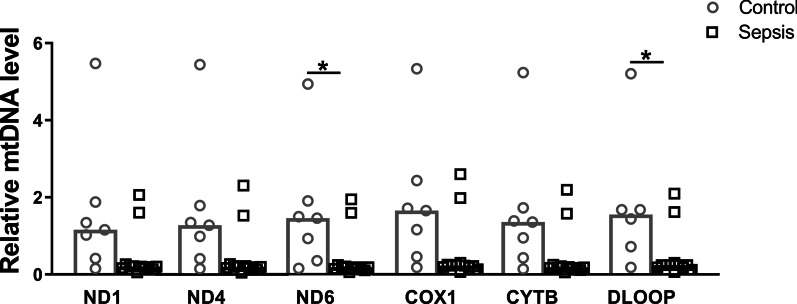
The integrity of mtDNA is critical to maintain normal mitochondrial function [27]. To investigate whether sepsis-AKI alters the integrity of mtDNA, we measured mtDNA damage by long-range PCR and quantified the ratio between a long and short mtDNA fragment, with higher ratios denoting more mtDNA damage (Fig. 5a). Kidney mtDNA damage was higher in sepsis-AKI patients as compared to patients without sepsis-AKI (p < 0.01, Fig. 5b). Levels of mtDNA damage positively correlated with expression of mnSOD and HIF1α, markers for oxidative stress, and renal damage marker NGAL (R = 0.77, R = 0.68 and R = 0.89, respectively, all p < 0.01) and negatively correlated with base-excision repair enzyme OGG1 and biogenesis marker mitochondrial transcription factor A (TFAM) (R = − 0.51 and R = − 0.65, respectively, both p < 0.05). However, mtDNA damage did not correlate with the severity of critical illness as defined by the APACHE IV and SAPSII scores. Furthermore, sepsis-AKI patients had lower DNA levels of mitochondrial genes ND6, and D-loop, whereas DNA levels of ND1, ND4, COX1 and CYTB were not different between control subjects and patients with sepsis-AKI, probably due to the small sample size and thus a type 2 error (Fig. 6). Next, we explored the mtDNA damage in 48-h LPS-induced HUVECs. In keeping, we found an increase in mtDNA damage after 48 h of LPS induction (Fig. 4g). Collectively, these findings show that sepsis is associated with mtDNA damage.
Fig. 5.
Mitochondrial DNA damage in control and sepsis-AKI subjects. A 10-kb-long mtDNA sequence was amplified by long-range PCR, while a 222-bp short mtDNA sequence was amplified with qPCR. a Amplified mtDNA sequences were loaded on agarose gel from representative samples of control subjects and sepsis-AKI subjects; b relative mtDNA damage of the control subjects compared to the sepsis-AKI patients. Bars represent median, dots represent individual levels; ** means p < 0.005
Fig. 6.
Relative mitochondrial DNA levels. Relative mtDNA levels of ND1, ND4, ND6, COX1, CYTB, and D-loop as quantified by qPCR. Bars represent the median, dots represent individual levels; * means p < 0.05
Mitochondrial quality control does not compensate for mtDNA damage
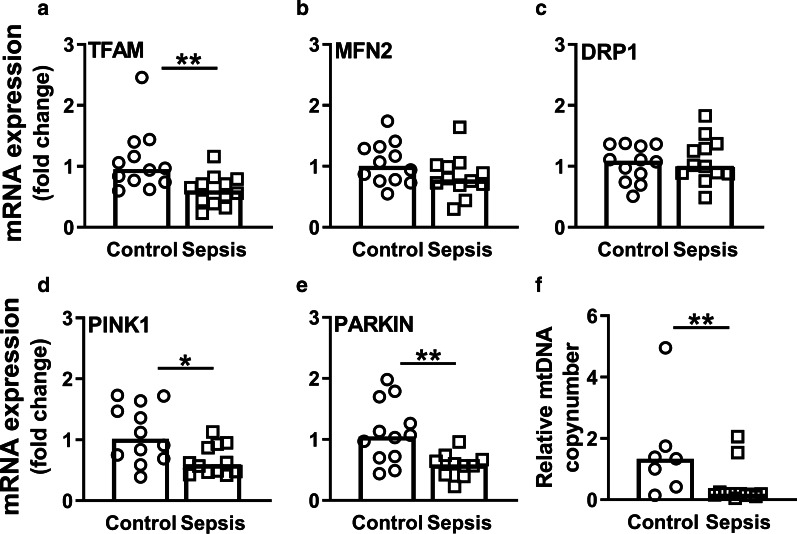
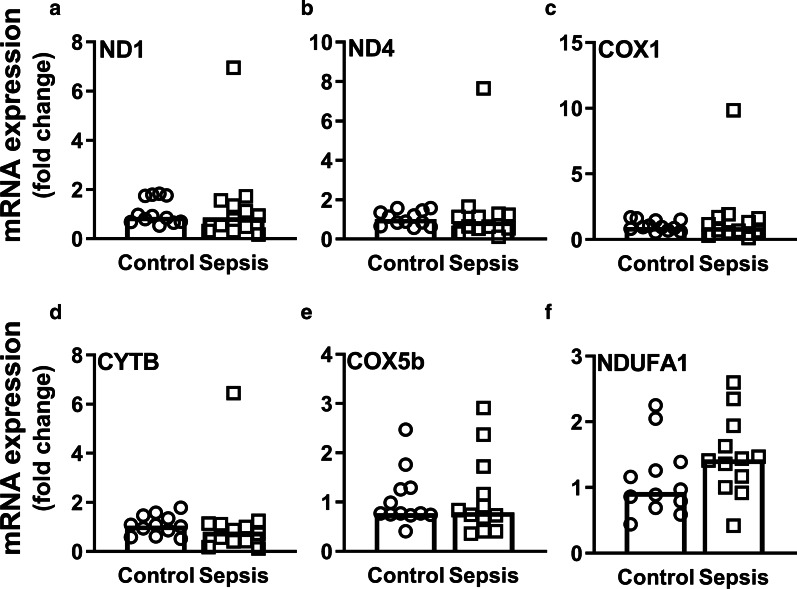
To assess mitochondrial quality control, which is important to maintain a healthy mitochondrial pool and consist of biogenesis, mitophagy, fission and fusion [28], we measured mRNA expression of genes involved in these processes. Expression of the biogenesis marker TFAM was lower in sepsis-AKI patients than in control subjects (p < 0.01; Fig. 7a). Although sepsis-AKI led to reduced mtDNA levels and integrity, it did not affect mRNA expression of mitochondrial genes encoding key components of the mitochondrial electron transport chain complexes (i.e., ND1, ND4, COX1, CYTB, COX5b and NDUFA1) (Fig. 8). Further, sepsis-AKI did not affect mRNA expression of the mitochondrial fusion and fission markers mitofusin 2 (MFN2) and dynamin-related protein 1 (DRP1), respectively (Fig. 7b, c). As expected, however, MFN2 and DRP1 did correlate with each other (R = 0.58, p < 0.01). Sepsis-AKI patients had lower mRNA expression of PTEN-induced putative kinase 1 (PINK1) and PARKIN, key components in regulating mitophagy and therefore important in the removal of unhealthy mitochondria, as compared to control subjects (p < 0.05 and p < 0.01, respectively; Fig. 7d, e). Expression of PINK1 correlated with PARKIN expression (R = 0.56, p < 0.01). Accordingly, 48-h LPS induction in HUVECS caused a decrease in TFAM mRNA expression, but did not change MFN2, DRP1 or PINK1 expression (Fig. 4c–f). Collectively, these data imply that sepsis-AKI patients and LPS-induced HUVECs did not have a compensatory increase in mitochondrial quality control mechanisms.
Fig. 7.
Changes in mitochondrial biogenesis, fusion, fission, mitophagy and copy number in response to sepsis-AKI. mRNA expression of a TFAM, b MFN2, c DRP1, d PINK1, and e PARKIN as quantified by RT-qPCR; f mtDNA levels were quantified by RT-qPCR to calculate the mitochondrial copy number. Bars represent median, dots represent individual levels; */** means p < 0.05/0.005; mtDNA mitochondrial DNA
Fig. 8.
mRNA expression of mitochondrial electron transport chain complexes. mRNA expression of a ND1, b ND4, c COX1, d CYTB, e COX5b, and f NDUFA1 as quantified by RT-qPCR. Bars represent median, dots represent individual levels
Sepsis-AKI causes a decrease in mitochondrial mass
Based on the increase in antioxidant markers and decrease in mitochondrial biogenesis markers, we hypothesized that patients with sepsis-AKI had a reduction in the mitochondrial mass, due to increased oxidative stress while having reduced mitochondrial quality control. To estimate the mitochondrial mass, we determined the mitochondrial copy number by calculating the ratio between the expression of mitochondrial genes and the nuclear housekeeping gene B2M. In keeping with the elevated mitochondrial DNA damage ratio, mitochondrial DNA levels were reduced in sepsis-AKI compared to control subjects, denoting a decrease in mitochondrial mass (p < 0.05; Fig. 7f). Hence, sepsis-AKI shows increased mitochondrial DNA damage ratio, a decreased mitochondrial mass, in the absence of compensation of mitochondrial quality control markers.
Discussion
Sepsis leads to increased levels of ROS, which is in part due to the overwhelming immune response to facilitate bacterial killing [1, 27, 29]. In turn, ROS can cause collateral damage by oxidizing proteins, lipids and DNA [12, 13]. Subsequent damage to mtDNA can impair mitochondrial function and further increase ROS generation [27, 29]. Whether this process underlies the pathophysiology in sepsis-AKI was not yet known. Here, we demonstrate that sepsis-AKI patients have an upregulated mRNA expression of markers important in the renal oxidative stress response, leading to higher levels of mtDNA oxidation, mtDNA damage and a reduced number of mitochondria in the kidney. Despite the mitochondrial damage, we did not find evidence of compensatory upregulation of mitochondrial quality control mechanisms, which further substantiates mitochondrial dysfunction in sepsis-AKI. Together, these data demonstrate key mechanisms leading to mitochondrial failure in the pathophysiology of sepsis-AKI.
The way cells deal with ROS under inflammatory circumstances is important for the distinction between survival, long-term complications, or mortality of patients with sepsis [30]. Induction of NGAL, mnSOD and HIF1α is indicative of renal oxidative stress and injury [31–33]. One of the first markers of renal injury is upregulation of NGAL expression [34], as demonstrated in our biopsies derived from patients with sepsis-AKI. Increased NGAL expression is regulated by ROS to suppress bacterial growth and modulate the inflammatory response [31]. In turn, NGAL activates antioxidant defense mechanisms, leading to the upregulation of mnSOD, as demonstrated in vitro [31]. Accordingly, we found a positive correlation between NGAL expression and mnSOD expression in the renal biopsies. The importance of upregulated mnSOD in sepsis is confirmed in mouse models of sepsis (i.e., lipopolysaccharide [LPS] injection and cecal ligature and puncture [CLP]), where higher mnSOD expression prevented ATP depletion and subsequent mortality [33, 35, 36]. Similarly, we found an increased gene expression of mnSOD expression in patients with sepsis-AKI as compared to control subjects. Upregulation of HIF1α switches mitochondrial aerobic respiration to glycolysis metabolism, thereby bypassing the dysfunctional ROS-producing mitochondria and lowering oxidative stress [32]. Higher HIF1α expression levels were detected in whole blood cells from patients with septic shock [37], in line with the increased expression in the kidney as demonstrated here in sepsis-AKI. In contrast to NGAL, mnSOD and HIF1α, the expression of SIRT1, involved in inhibiting oxidative stress and the suppression of biogenesis and mitophagy [38, 39], was downregulated in sepsis-AKI patients. Taken together, sepsis-AKI is associated with upregulation of genes encoding molecules involved in inhibiting inflammation and antioxidant defense in the kidney.
Compared to control subjects, sepsis-AKI patients had upregulated mRNA expression of oxidative damage markers and high levels of mitochondrial DNA damage. Also 48 h of LPS induction in HUVECs caused an increase in mnSOD expression and mtDNA damage. Although several studies demonstrated increased biomarkers suggestive of mitochondrial dysfunction in sepsis [10, 11, 15], to our knowledge only one other study so far demonstrated the occurrence of mtDNA damage in patients with sepsis, as illustrated by the depletion of mtDNA defined by RT-qPCR in monocytes and lymphocytes [40]. Extending on the observations of these previous studies, we now directly demonstrate the presence of mtDNA damage in the septic kidney. Whereas mtDNA damage in monocytes and lymphocytes correlated with the APACHE score, we did not find such a correlation between mtDNA damage and APACHE score in sepsis-AKI patients, which might be due to the small sample size (n = 147 vs n = 12, respectively). The observed mtDNA damage in the kidney in patients with sepsis-AKI is in line with findings from experimental murine sepsis models, which showed profound damage to mtDNA in skeletal muscle and 50% mtDNA depletion in the liver [9, 41]. mtDNA damage was associated with increased DNA oxidation (i.e., 8-oxoG) in sepsis-AKI as compared to control subjects, while immunofluorescent staining showed co-localization of DNA oxidation with both nuclei and mitochondria. In line with this observation, sepsis in mice also leads to accumulation of 8-oxoG in mitochondria and mitochondrial dysfunction, as demonstrated in the brain [42]. Hence, mitochondrial dysfunction leading to increased oxidative stress, oxidation of mtDNA and damage that further impairs mitochondrial function might play a key role in the pathophysiology sepsis-AKI.
Mitochondrial quality control mechanisms, consisting of biogenesis (making new mitochondria), fission/fusion of mitochondria and mitophagy (removal of damaged mitochondria), can counteract mitochondrial damage and subsequent dysfunction [7, 8, 43]. Sepsis-AKI patients had lower mRNA expression of TFAM (marker for biogenesis) and PINK and PARKIN (markers for mitophagy), but no change in mRNA expression of MFN2 and DRP1 (markers for fusion and fission). HUVECs induced with LPS for 48 h showed a similar pattern, where TFAM was decreased, but no change in PINK, MFN2 or DRP1. Similar to our findings, sepsis is associated with increased mRNA expression and protein levels of TFAM in muscle, suggestive of biogenesis and a lowered mitochondrial mass [11]. Adequate compensation of mitochondrial damage seems to play an important role in determining the outcome of sepsis, as mRNA expression of Peroxisome Proliferator-activated Receptor Gamma Coactivator 1-alpha (PGC1α) (marker of biogenesis upstream of TFAM) is associated with survival from sepsis [11]. Also, LPS-induced mice with AKI suffer from decreased mRNA expression of PGC1α in the renal cortex, whereas overexpression of PGC1α shows recovery from LPS-induced AKI [15]. The protective role of mitophagy is illustrated by PARK2-deficient mice, which exhibited degradation of mitochondrial functions and impaired recovery of cardiac contractility in sepsis [44]. Additionally, inhibition of mitophagy in mice increases the sensitivity of multiple organ failure and death from sepsis [44, 45]. Here, we found a decreased expression of PINK1 and PARKIN, the main mitophagy regulators, in the septic kidney, which suggests that removal of damaged mitochondria by mitophagy is impaired. Lastly, we found no changes in mRNA expression DRP1 or MFN2 (markers for fission/fusion) in renal biopsies or LPS treated HUVECs. Accordingly, fission and fusion did not differ between septic patients and controls in human PBMCs [46]. However, since PINK1 and PARKIN mediate degradation of MFN2 and activation of DRP1 to prevent fusion while promoting fission [47, 48], we cannot conclude that protein levels or activity of PINK1 and PARKIN and hence fusion/fission are unaltered in sepsis. Together, sepsis is associated with mtDNA damage in sepsis without upregulation of genes encoding for mitochondrial quality control processes to safeguard the mitochondria.
A strength of our study is the use of fresh direct postmortem kidney biopsies from sepsis-AKI patients and control subjects, which allowed us to directly investigate the effect of sepsis on mitochondria in the kidney in association with immunohistochemical analysis, albeit in a small cohort of patients, while other studies estimate kidney and mitochondrial damage indirect using surrogate markers in urine or blood. However, we could only analyze non-survivors, which represent the most severe critically ill patients, as it would be ethically unacceptable to obtain renal biopsies from living patients with sepsis-AKI. Postmortem changes in mRNA expression should be taken into consideration when interpreting our data, but are unlikely to have been of major relevance, as samples were collected as quickly as possible after death (24–150-min postmortem), and the kidney is among the less susceptible organs to changes in RNA integrity and gene expression as demonstrated up to 24-h postmortem [49]. Lastly, there is uncertainty as to whether renal cell carcinoma might have affected mitochondrial mass or DNA levels in surrounding healthy tissue. However, mtDNA copy number, DNA content and activities of mitochondrial enzymes are shown to be reduced within RCC tissue [50, 51]. Furthermore, impairment of mitochondrial DNA levels and mass depends on RCC aggressiveness, a trend that is not found in healthy tissue within the same kidney [52, 53]. Thus, in contrast to effects of cancer on mitochondrial mass and DNA within the tumor, this does not seem to be the case for surrounding tissue. However, even if mitochondrial mass and DNA in surrounding tissue would have been affected by the tumor, this would have led to an underestimation of the effect of sepsis and thus not compromise our findings.
Conclusion
Our findings shed new light on the contribution of mitochondria in the pathophysiology of sepsis-AKI. We reveal that sepsis induces oxidation of nuclear and mitochondrial DNA and mtDNA damage, without signs of upregulation of mitochondrial quality control mechanisms, resulting in a reduced mitochondrial mass in the septic kidney. These findings are of clinical relevance, as sepsis is the major cause of AKI and death in critically ill patients. Dissecting the molecular mechanisms leading to mitochondrial dysfunction in sepsis-AKI is crucial for the development of novel targeted therapies to prevent or treat sepsis-AKI and potentially improve the survival. Since it is not known whether mtDNA damage is repaired after survival from sepsis, our findings might also be of relevance for long-term outcomes of sepsis. Given the role of mitochondria in the pathophysiology of sepsis-AKI, pharmacologic strategies directed at maintaining mitochondrial function, limiting oxidative stress and mtDNA damage, or enhancing mitochondrial quality control to ameliorate mitochondrial damage, might successfully prevent or halt AKI in sepsis.
Acknowledgements
Not applicable.
Abbreviations
- AKI
Acute kidney injury
- APACHE
Acute Physiology and Chronic Health Evaluation
- B2M
Beta 2-microglobulin
- COX1
Cytochrome C oxidase I
- CYTB
Cytochrome B
- DRP1
Dynamin-related protein 1
- HIF1α
Hypoxia-inducible factor 1-alpha
- ICU
Intensive care unit
- METC
Medical Ethics Review Committee
- MFN2
Mitofusin 2
- mnSOD
Manganese superoxide dismutase
- mtDNA
Mitochondrial DNA
- mRNA
Messenger RNA
- ND1
NADH dehydrogenase 1
- NGAL
Neutrophil gelatinase-associated lipocalin
- OGG1
8-Oxoguanine DNA glycosylase 1
- PCR
Polymerase chain reaction
- PGC1α
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PINK
PTEN-induced putative kinase 1
- RIFLE
Risk, Injury, Failure, Loss of kidney function, and End-stage kidney disease
- ROS
Reactive oxygen species
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- SAPS II
Simplified Acute Physiology Score II
- SIRT1
Sirtuin 1
- TFAM
Mitochondrial transcription factor A
- TOM20
Translocase of outer mitochondrial membrane 20
- UMCG
University Medical Center Groningen
Authors’ contributions
MVM designed the study and collected the biomaterials. ECVDS and JM performed the experiments. ECVDS, BS, and HRB conceptualized the manuscript. ECVDS analyzed the data and wrote the manuscript. BS, MVM, RHH, JM, and HRB edited the manuscript. All authors read and approved the final manuscript.
Funding
Our sepsis research is supported by a grant to HRB from the Dutch Kidney Foundation (16OKG06) and by a MD/PhD grant from the Junior Scientific Masterclass (JSM, UMCG) to ECVDS.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The Medical Ethics Review Committee (METC) of the UMCG reviewed and approved this study (METC 2011/372).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369(9):840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 2.Cecconi M, Evans L, Levy M, Rhodes A. Sepsis and septic shock. Lancet. 2018;392(10141):75–87. doi: 10.1016/S0140-6736(18)30696-2. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart K, Daniels R, Kissoon N, Machado FR, Schachter RD, Finfer S. Recognizing sepsis as a global health priority—a WHO resolution. N Engl J Med. 2017;377(5):414–417. doi: 10.1056/NEJMp1707170. [DOI] [PubMed] [Google Scholar]
- 4.Gotts JE, Matthay MA. Sepsis: pathophysiology and clinical management. BMJ. 2016;353:i1585. doi: 10.1136/bmj.i1585. [DOI] [PubMed] [Google Scholar]
- 5.Schrier RW, Wang W. Acute renal failure and sepsis. N Engl J Med. 2004;351(2):159–169. doi: 10.1056/NEJMra032401. [DOI] [PubMed] [Google Scholar]
- 6.Bellomo R, Kellum JA, Ronco C, Wald R, Martensson J, Maiden M, et al. Acute kidney injury in sepsis. Intensive Care Med. 2017;43(6):816–828. doi: 10.1007/s00134-017-4755-7. [DOI] [PubMed] [Google Scholar]
- 7.Emma F, Montini G, Parikh SM, Salviati L. Mitochondrial dysfunction in inherited renal disease and acute kidney injury. Nat Rev Nephrol. 2016;12(5):267–280. doi: 10.1038/nrneph.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parikh SM, Yang Y, He L, Tang C, Zhan M, Dong Z. Mitochondrial function and disturbances in the septic kidney. Semin Nephrol. 2015;35(1):108–119. doi: 10.1016/j.semnephrol.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rocheteau P, Chatre L, Briand D, Mebarki M, Jouvion G, Bardon J, et al. Sepsis induces long-term metabolic and mitochondrial muscle stem cell dysfunction amenable by mesenchymal stem cell therapy. Nat Commun. 2015;6:10145. doi: 10.1038/ncomms10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brealey D, Brand M, Hargreaves I, Heales S, Land J, Smolenski R, et al. Association between mitochondrial dysfunction and severity and outcome of septic shock. Lancet. 2002;360(9328):219–223. doi: 10.1016/S0140-6736(02)09459-X. [DOI] [PubMed] [Google Scholar]
- 11.Carre JE, Orban JC, Re L, Felsmann K, Iffert W, Bauer M, et al. Survival in critical illness is associated with early activation of mitochondrial biogenesis. Am J Respir Crit Care Med. 2010;182(6):745–751. doi: 10.1164/rccm.201003-0326OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Supinski GS, Callahan LA. Polyethylene glycol-superoxide dismutase prevents endotoxin-induced cardiac dysfunction. Am J Respir Crit Care Med. 2006;173(11):1240–1247. doi: 10.1164/rccm.200410-1346OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suliman HB, Carraway MS, Piantadosi CA. Postlipopolysaccharide oxidative damage of mitochondrial DNA. Am J Respir Crit Care Med. 2003;167(4):570–579. doi: 10.1164/rccm.200206-518OC. [DOI] [PubMed] [Google Scholar]
- 14.Hahn A, Zuryn S. Mitochondrial genome (mtDNA) mutations that generate reactive oxygen species. Antioxidants (Basel). 2019;8(9):392. doi: 10.3390/antiox8090392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran M, Tam D, Bardia A, Bhasin M, Rowe GC, Kher A, et al. PGC-1alpha promotes recovery after acute kidney injury during systemic inflammation in mice. J Clin Invest. 2011;121(10):4003–4014. doi: 10.1172/JCI58662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunst J, Derese I, Aertgeerts A, Ververs EJ, Wauters A, Van den Berghe G, et al. Insufficient autophagy contributes to mitochondrial dysfunction, organ failure, and adverse outcome in an animal model of critical illness. Crit Care Med. 2013;41(1):182–194. doi: 10.1097/CCM.0b013e3182676657. [DOI] [PubMed] [Google Scholar]
- 17.Funk JA, Schnellmann RG. Persistent disruption of mitochondrial homeostasis after acute kidney injury. Am J Physiol Renal Physiol. 2012;302(7):F853–F864. doi: 10.1152/ajprenal.00035.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran MT, Zsengeller ZK, Berg AH, Khankin EV, Bhasin MK, Kim W, et al. PGC1alpha drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature. 2016;531(7595):528–532. doi: 10.1038/nature17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stallons LJ, Funk JA, Schnellmann RG. Mitochondrial homeostasis in acute organ failure. Curr Pathobiol Rep. 2013;1(3):169–177. doi: 10.1007/s40139-013-0023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med. 2003;31(4):1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 21.Bellomo R, Ronco C, Kellum JA, Mehta RL, Palevsky P. Acute Dialysis Quality Initiative w. Acute renal failure - definition, outcome measures, animal models, fluid therapy and information technology needs: the Second International Consensus Conference of the Acute Dialysis Quality Initiative (ADQI) Group. Crit Care. 2004;8(4):R204–R212. doi: 10.1186/cc2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knaus WA, Zimmerman JE, Wagner DP, Draper EA, Lawrence DE. APACHE-acute physiology and chronic health evaluation: a physiologically based classification system. Crit Care Med. 1981;9(8):591–597. doi: 10.1097/00003246-198108000-00008. [DOI] [PubMed] [Google Scholar]
- 23.Le Gall JR, Lemeshow S, Saulnier F. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. JAMA. 1993;270(24):2957–2963. doi: 10.1001/jama.1993.03510240069035. [DOI] [PubMed] [Google Scholar]
- 24.Aslan A, van den Heuvel MC, Stegeman CA, Popa ER, Leliveld AM, Molema G, et al. Kidney histopathology in lethal human sepsis. Crit Care. 2018;22(1):359. doi: 10.1186/s13054-018-2287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jou-Valencia D, Molema G, Popa E, Aslan A, van Dijk F, Mencke R, et al. Renal Klotho is reduced in septic patients and pretreatment with recombinant Klotho attenuates organ injury in lipopolysaccharide-challenged mice. Crit Care Med. 2018;46(12):e1196–e1203. doi: 10.1097/CCM.0000000000003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Exline MC, Crouser ED. Mitochondrial mechanisms of sepsis-induced organ failure. Front Biosci. 2008;13:5030–5041. doi: 10.2741/3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Yao YM, Lu ZQ. Mitochondrial quality control mechanisms as potential therapeutic targets in sepsis-induced multiple organ failure. J Mol Med (Berl) 2019;97(4):451–462. doi: 10.1007/s00109-019-01756-2. [DOI] [PubMed] [Google Scholar]
- 29.Martins PS, Kallas EG, Neto MC, Dalboni MA, Blecher S, Salomao R. Upregulation of reactive oxygen species generation and phagocytosis, and increased apoptosis in human neutrophils during severe sepsis and septic shock. Shock. 2003;20(3):208–212. doi: 10.1097/01.shk.0000079425.52617.db. [DOI] [PubMed] [Google Scholar]
- 30.Reitsema VA, Star BS, de Jager VD, van Meurs M, Henning RH, Bouma HR. Metabolic resuscitation strategies to prevent organ dysfunction in sepsis. Antioxid Redox Signal. 2019;31(2):134–152. doi: 10.1089/ars.2018.7537. [DOI] [PubMed] [Google Scholar]
- 31.Bahmani P, Halabian R, Rouhbakhsh M, Roushandeh AM, Masroori N, Ebrahimi M, et al. Neutrophil gelatinase-associated lipocalin induces the expression of heme oxygenase-1 and superoxide dismutase 1, 2. Cell Stress Chaperones. 2010;15(4):395–403. doi: 10.1007/s12192-009-0154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fitzpatrick SF. Immunometabolism and sepsis: a role for HIF? Front Mol Biosci. 2019;6:85. doi: 10.3389/fmolb.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patil NK, Parajuli N, MacMillan-Crow LA, Mayeux PR. Inactivation of renal mitochondrial respiratory complexes and manganese superoxide dismutase during sepsis: mitochondria-targeted antioxidant mitigates injury. Am J Physiol Renal Physiol. 2014;306(7):F734–F743. doi: 10.1152/ajprenal.00643.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagshaw SM, Bennett M, Haase M, Haase-Fielitz A, Egi M, Morimatsu H, et al. Plasma and urine neutrophil gelatinase-associated lipocalin in septic versus non-septic acute kidney injury in critical illness. Intensive Care Med. 2010;36(3):452–461. doi: 10.1007/s00134-009-1724-9. [DOI] [PubMed] [Google Scholar]
- 35.Choumar A, Tarhuni A, Letteron P, Reyl-Desmars F, Dauhoo N, Damasse J, et al. Lipopolysaccharide-induced mitochondrial DNA depletion. Antioxid Redox Signal. 2011;15(11):2837–2854. doi: 10.1089/ars.2010.3713. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Wang X, Zhang L, Zhang R. Alleviation of acute lung injury in rats with sepsis by resveratrol via the phosphatidylinositol 3-kinase/nuclear factor-erythroid 2 related factor 2/heme oxygenase-1 (PI3K/Nrf2/HO-1) pathway. Med Sci Monit. 2018;24:3604–3611. doi: 10.12659/MSM.910245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Textoris J, Beaufils N, Quintana G, Ben Lassoued A, Zieleskiewicz L, Wiramus S, et al. Hypoxia-inducible factor (HIF1alpha) gene expression in human shock states. Crit Care. 2012;16(4):R120. doi: 10.1186/cc11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang BL. Sirt1 and the mitochondria. Mol Cells. 2016;39(2):87–95. doi: 10.14348/molcells.2016.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salminen A, Kaarniranta K, Kauppinen A. Crosstalk between oxidative stress and SIRT1: impact on the aging process. Int J Mol Sci. 2013;14(2):3834–3859. doi: 10.3390/ijms14023834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pyle A, Burn DJ, Gordon C, Swan C, Chinnery PF, Baudouin SV. Fall in circulating mononuclear cell mitochondrial DNA content in human sepsis. Intensive Care Med. 2010;36(6):956–962. doi: 10.1007/s00134-010-1823-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez AS, Elguero ME, Finocchietto P, Holod S, Romorini L, Miriuka SG, et al. Abnormal mitochondrial fusion-fission balance contributes to the progression of experimental sepsis. Free Radic Res. 2014;48(7):769–783. doi: 10.3109/10715762.2014.906592. [DOI] [PubMed] [Google Scholar]
- 42.Leon J, Sakumi K, Castillo E, Sheng Z, Oka S, Nakabeppu Y. 8-Oxoguanine accumulation in mitochondrial DNA causes mitochondrial dysfunction and impairs neuritogenesis in cultured adult mouse cortical neurons under oxidative conditions. Sci Rep. 2016;6:22086. doi: 10.1038/srep22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505(7483):335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piquereau J, Godin R, Deschenes S, Bessi VL, Mofarrahi M, Hussain SN, et al. Protective role of PARK2/Parkin in sepsis-induced cardiac contractile and mitochondrial dysfunction. Autophagy. 2013;9(11):1837–1851. doi: 10.4161/auto.26502. [DOI] [PubMed] [Google Scholar]
- 45.Kang R, Zeng L, Xie Y, Yan Z, Zhou B, Cao L, et al. A novel PINK1- and PARK2-dependent protective neuroimmune pathway in lethal sepsis. Autophagy. 2016;12(12):2374–2385. doi: 10.1080/15548627.2016.1239678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jang DH, Greenwood JC, Owiredu S, Ranganathan A, Eckmann DM. Mitochondrial networking in human blood cells with application in acute care illnesses. Mitochondrion. 2019;44:27–34. doi: 10.1016/j.mito.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci. 2012;125(Pt 4):795–799. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buhlman L, Damiano M, Bertolin G, Ferrando-Miguel R, Lombes A, Brice A, et al. Functional interplay between Parkin and Drp1 in mitochondrial fission and clearance. Biochim Biophys Acta. 2014;1843(9):2012–2026. doi: 10.1016/j.bbamcr.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 49.Ferreira PG, Munoz-Aguirre M, Reverter F, Sa Godinho CP, Sousa A, Amadoz A, et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat Commun. 2018;9(1):490. doi: 10.1038/s41467-017-02772-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin CS, Lee HT, Lee MH, Pan SC, Ke CY, Chiu AW, et al. Role of mitochondrial DNA copy number alteration in human renal cell carcinoma. Int J Mol Sci. 2016;17(6):814. doi: 10.3390/ijms17060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meierhofer D, Mayr JA, Foetschl U, Berger A, Fink K, Schmeller N, et al. Decrease of mitochondrial DNA content and energy metabolism in renal cell carcinoma. Carcinogenesis. 2004;25(6):1005–1010. doi: 10.1093/carcin/bgh104. [DOI] [PubMed] [Google Scholar]
- 52.Simonnet H, Alazard N, Pfeiffer K, Gallou C, Beroud C, Demont J, et al. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23(5):759–768. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- 53.Yusenko MV, Ruppert T, Kovacs G. Analysis of differentially expressed mitochondrial proteins in chromophobe renal cell carcinomas and renal oncocytomas by 2-D gel electrophoresis. Int J Biol Sci. 2010;6(3):213–224. doi: 10.7150/ijbs.6.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.