Abstract
Identifying changes in functional connectivity in Attention Deficit Hyperactivity Disorder (ADHD) using functional magnetic resonance imaging (fMRI) can help us understand the neural substrates of this brain disorder. Many studies of ADHD using resting state fMRI (rs-fMRI) data have been conducted in the past decade with either manually crafted features that do not yield satisfactory performance, or automatically learned features that often lack interpretability. In this work, we present a tensor-based approach to identify brain networks and extract features from rs-fMRI data. Results show the identified networks are interpretable and consistent with our current understanding of ADHD conditions. The extracted features are not only predictive of ADHD score but also discriminative for classification of ADHD subjects from typically developed children.
Index Terms—: resting-state fMRI, tensor decomposition, ADHD, brain network identification
1. INTRODUCTION
Attention Deficit Hyperactivity Disorder (ADHD) is a neuropsychological disorder characterized by inappropriate levels of inattention, hyperactivity and impulsivity, which is most typically seen in children with an approximately worldwide prevalence of 5.3% [1]. Many functional magnetic resonance imaging (fMRI) studies on ADHD subjects have been conducted in the past decade, the majority of which are aimed at understanding its impact on cognitive development and function using task-based fMRI. Several meta-analyses of fMRI studies showed reduced brain activity in ADHD subjects, relative to typically developed children (TDC), in regions related to executive functions [2]. In addition, impaired large-scale functional connectivity has been observed indicating dysfunction of entire networks rather than isolated task-related regions [3].
Most resting-state fMRI (rs-fMRI) studies of ADHD use one of the following approaches to analysis [4]: (i) Seeded-correlation [5] in which we place a seed point in a region of interest (ROI), then examine spatial correlations between the seed and all other voxels/vertices. These results can be very sensitive to the selection of the seed point; (ii) Regional homogeneity and amplitude of low frequency fluctuations are measure of the local properties of fMRI signals. Conflicting results have been reported [5]; (iii) Graph-theoretic analysis in which the vertices represent cortical/subcortical ROIs and edge strengths are based on the Pearson correlation between signals in each ROI. Graphic metrics such as modularity or centrality are then computed and statistically tested to contrast ADHD subjects to TDC [6]. While these graph metrics can characterize network topology well, they do not provide insights into low level changes of particular brain networks; (iv) Independent component analysis (ICA) has been widely used for brain network studies in ADHD data [7]. The independence assumption between brain networks, however, does not seem to be physiologically plausible as brain networks can be correlated both spatially and temporally. (v) Finally, methods based on machine-learning [8] or more recently deep-learning [9] have been proposed mainly for the problem of classification of ADHD. While so-called “handcrafted” features (such as ROI-wise correlations) have interpretability, they often show unsatisfactory performance. On the other hand, automatically learned features show improved performance but often lack interpretability [8].
Here we present a tensor-based framework to extract meaningful features from rs-fMRI data. Tensor decomposition is a generalization of matrix factorization in higher dimensional space. In contrast to traditional 2D (space × time) methods (e.g. ICA) where higher dimensional low-rank structure inherent in the data is lost by concatenating multi-subject data either spatially or temporally, this structure can be captured naturally using a tensor decomposition. Also, tensor models do not impose any unrealistic constraints, such as independence (as in ICA) or orthogonality (as in PCA). Moreover, the third (subject) dimension in tensor models provides extra information (features) indicating the strength of extracted components (brain networks), which are not available using 2D methods. We show that the extracted features not only enable us to predict ADHD score and perform classification of ADHD subjects against TDC with satisfactory performance, but also are interpretable in the sense that they facilitate exploration of the differences in brain network activity between ADHD and TDC.
2. MATERIALS AND METHODS
We used all 259 subjects recruited from Peking University in the ADHD-200 dataset available in http://fcon_1000.projects.nitrc.org/indi/adhd200/. By examination of the phenotypic information, subjects with incomplete evaluations or measures, such as ADHD score or any other personal characteristic data, were removed. The final total number of subjects that satisfy the requirements was S = 221, where 95 of them were ADHD and the remaining 126 were TDC. We did not differentiate sub-types of ADHD in this study. The rs-fMRI data was recorded using a Siemens Trio 3T scanner with a TR=2000 ms. Other scan parameters vary across 3 sub-datasets and can be found on their project website.
We preprocessed the rs-fMRI data using our previously developed BrainSuite fMRI Pipeline (BFP) [10]. BFP is a dedicated workflow for processing T1 and fMRI data using a combination of software and toolboxes including BrainSuite (https://brainsuite.org), AFNI (https://afni.nimh.nih.gov), FSL (https://www.fmrib.ox.ac.uk/fsl), Global PDF-based non-local means filtering (GPDF) [11, 12] (https://neuroimageusc.github.io/GPDF) and the BrainSync transform [13].
After applying the BFP pipeline, the rs-fMRI data was co-registered across subjects and re-sampled onto a common mid-cortical tessellated surface with 11K vertices per hemisphere. Each subject’s rs-fMRI data can then be represented using a V × T matrix, where V ≈ 22K is the number of vertices combined from two hemispheres and T = 235 the number of time points. Because rs-fMRI data across multiple subjects are not directly comparable, we applied the BrainSync transform [13] to temporally synchronize data from each of subjects 2 to N to the first subject. They were then arranged into a third-order tensor as illustrated in Fig. 1. We modeled our 3D data using a canonical polyadic (CP) form, where we assumed that the data tensor has a low rank structure and can be approximated by a sum of rank-1 tensors, Fig. 1. Mathematically,
| (1) |
where , , have unit norm; λi is the scale of the ith component; ○ is the vector outer product and R is the rank or the number of components.
Fig. 1:
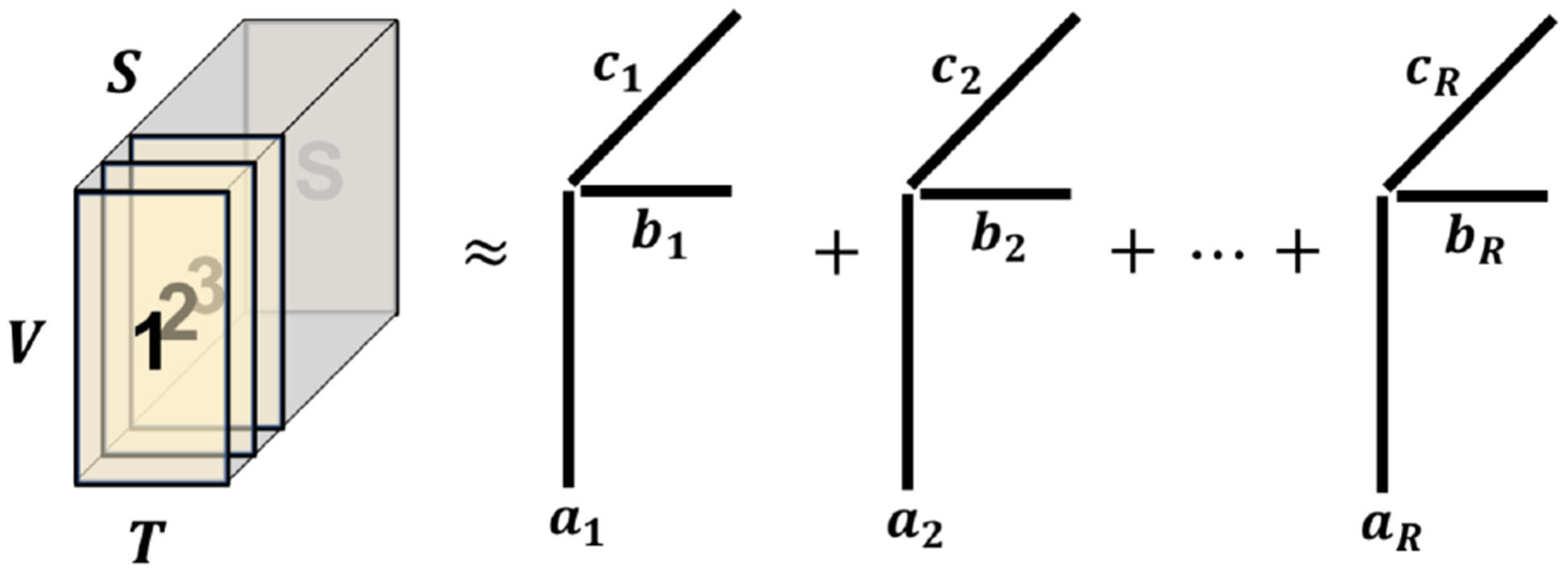
Data tensor formulation and canonical polyadic model
Each rank-1 solution represents a distinct brain network with a common spatial map ai and a temporal dynamics bi and the third subject mode ci indicating the contribution from each subject to the ith network. This model has been applied to task-based fMRI data and meaningful common brain networks had been successfully identified [14]. This tensor decomposition problem can be efficiently and robustly solved using the rank-recursive scalable and robust sequential CP decomposition (SRSCPD) algorithm [15, 16]. In this study, we applied a gradient-based accelerated version of SRSCPD described in [14] to further improve the robustness of solutions.
In total, 20 networks that explain the most variance in the data were extracted using the method as described in [14]. We then grouped the subject modes ci, i = 1, 2, …, R, together, forming matrix , where S = 221 is the number of subjects and R = 20 is the number of networks. F can then be viewed as a feature matrix, where each subject has a feature vector with length of 20, indicating the strength of brain activity for that subject corresponding to each of the 20 networks. In the following section we use this feature matrix F to predict ADHD score, to perform classification of ADHD and most importantly to explore network activity differences between ADHD subjects and TDC.
3. EXPERIMENTS AND RESULTS
3.1. ADHD Index Prediction Using Linear Regression
In the first experiment, we treated the feature matrix F as a set of bases and fitted a multiple linear regression model to ADHD scores. The ADHD scores used in this dataset were ADHD Rating Scale IV, a combined measure of inattention and hyperactivity-impulsivity. The higher this measure, the higher the severity of ADHD. Let yi be the ADHD score for subject i, then we estimated the regression coefficients by
| (2) |
Then the predicted ADHD score . Fig. 2 (a) shows the scatter plot between the predicted ADHD scores ŷ and the clinically measured ADHD scores y. The statistical significance of the regression model was established using a standard F-test with 200 degrees of freedom and a corresponding p-value of 4.3 × 10−4.
Fig. 2:
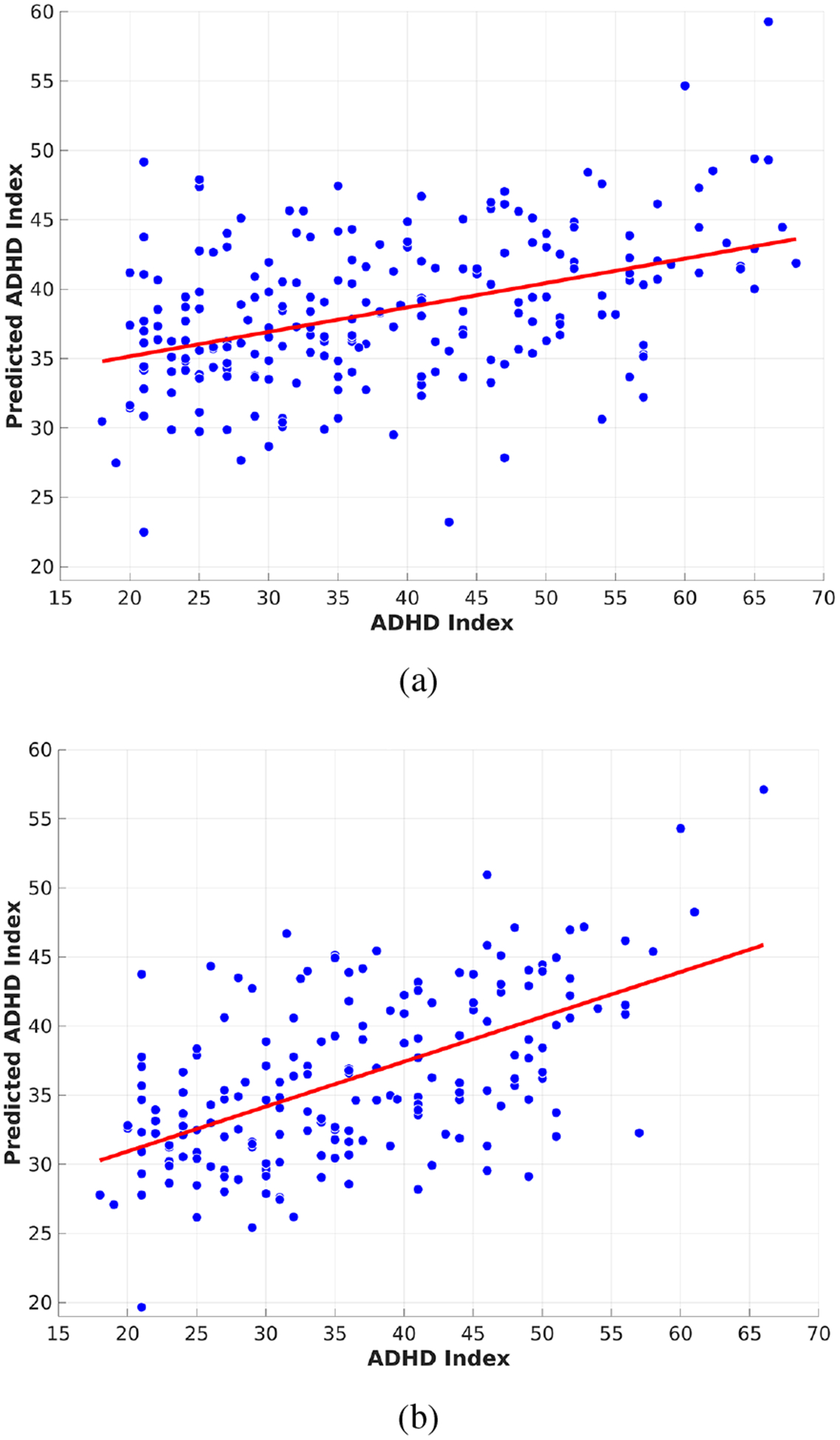
Scatter plot of the predicted ADHD scores vs the clinically measured ADHD scores using linear regression on (a) the entire dataset and (b) the outlier-removed dataset.
Further, due to the inherent low signal-to-noise ratio of fMRI data as well as variance in the ADHD score measure, we computed the Cook’s distance [17] for each point during the regression above to identify potential outliers. We examined the histogram of the Cook’s distances and observed a small peak with large Cook’s distance which we used to select a threshold to remove outliers. 187 out of 221 (~85%) subjects survived after the outlier removal. Then we re-fitted the linear regression model using the outlier removed dataset and the result shows a substantial improvement with a p-value of 3.17 × 10−7 using a F-test with 166 degrees of freedom, Fig. 2 (b).
To the best of our knowledge, most fMRI-based ADHD studies are focused on either the classification problem or studies of differences in brain networks in ADHD subjects relative to TDC as discussed in the introduction. We are unaware of any previous study in the literature that predicts ADHD scores, except for our previously reported correlation of 0.32 in [18] based on a deep neural network.
3.2. Classification of ADHD
In the second experiment, we input the feature matrix F into a support vector machine (SVM) to identify ADHD subjects from TDC. The ground truth diagnostic/label was given by the ADHD-200 official website. We used the MATLAB (the Mathworks, Inc., Natick, MA) implementation “fitcsvm” with a Gaussian kernel. Based on our experiments, the classification result shown below is not sensitive to choice of the regularization parameter for penalizing falsely classified samples. We chose the regularization parameter to be 1 and the kernel scale was automatically estimated to be 3.94. The extracted features (the rows of F) had been standardized to have zero mean and unit variance for each subject before inputting to the SVM classifier. A standard 10-fold cross validation strategy was employed to avoid overfitting.
Table 1 top row shows the classification results and associated precision, recall and accuracy. We were able to achieve 65.6% cross validated prediction accuracy, which is substantially higher than that reported in the literature using a dataset with a comparable size to ours [8]. For example, the ADHD-200 competition (http://fcon_1000.projects.nitrc.org/indi/adhd200/results.html) reported less than 61% accuracy from all participating teams. The prediction accuracy was even lower, 51%, when using subjects from Peking University only, which is the dataset we used in this work. Higher performance was achieved using deep-learning-based methods. For example, Han et al. [19] reported 65% accuracy using an unsupervised hierarchical convolutional auto-encoder on the ADHD-200 dataset. However, it has been shown that high performance of deep learning methods in neuroimaging studies can often be attributed to overfitting [9] due to the limited number of samples in the dataset relative to the huge number of parameters used in those models. In contrast, only 20 features were extracted from the rs-fMRI data in this work.
Table 1:
ADHD Classification Results
| Predicted ADHD | Predicted TDC | Statistics | ||
|---|---|---|---|---|
| rfMRI only | ADHD | 45 | 50 | 0.4742 |
| TDC | 26 | 100 | ||
| Statistics | 0.6343 | 0.6564 | ||
| rfMRI + PC1 | ADHD | 72 | 23 | 0.7582 |
| TDC | 22 | 104 | ||
| Statistics | 0.7663 | 0.7964 |
Personal Characteristic Data;
Recall;
Precision;
Accuracy.
An interesting report was published by Brown et al. [20] where only non-imaging Personal Characteristic (PC) data were used for classification of ADHD and the result outperformed any of the imaging-based methods in the ADHD-200 competition, achieving an overall 62.5% classification accuracy. Recently, Riaz et al. [21] showed that using a fusion of rs-fMRI data and non-imaging data achieved a higher classification accuracy of 64.7% on the ADHD-200 dataset from Peking University only, indicating that rs-fMRI data and non-imaging data might provide complementary information regarding brain activity in ADHD subjects. To explore this question using tensor-based feature extraction method, we appended five additional personal characteristic attributes: gender, age, verbal IQ, performance IQ, and full scale IQ, to our feature matrix F, forming a new feature matrix and repeated the previous classification procedure. We were able to achieve 79.6% prediction accuracy as shown in Table 1 bottom row, which outperforms all other classification methods (on large datasets) that we are aware of.
3.3. Brain Network Comparison and Exploration
As we discussed previously, in ADHD studies methods designed for studying brain networks (e.g. seeded correlation, ICA, graph theory, etc.) in general do not directly provide useful features for accurate classification of ADHD. On the other hand, methods designed for classification (e.g. handcrafted feature extraction with traditional machine learning or deep learning) lack interpretability of extracted features due to the high dimensionality of the feature space and black-box operations. In this work, our tensor-based feature extraction strategy bridged these two issues: the subject mode not only provided a set of informative bases for the prediction of ADHD scores (Sec. 3.1) as well as a discriminative feature matrix for the superior performance of ADHD classification (Sec. 3.2), but also allows us to investigate differences in brain activity between ADHD subjects and TDC in a variety of brain networks concurrently, hence potentially facilitating the exploration of neurological differences associated with ADHD.
In this section, we examine the common (across ADHD and TDC) resting-state brain networks identified by the tensor decomposition and exploratively interpret how those networks in ADHD subjects differ from those in TDC. Of the 20 extracted networks, we identified and recognized the 6 networks which explained the most variance (according to the λi in Eq. (1)) in the data. The spatial maps (ai in Eq. (1)) of these 6 networks are shown in Fig. 3: (a) default mode network (DMN); (b) ventral attention network (VAN); (c) executive control network (ECN); (d) fronto-parietal network (FPN); (e) visual network (VN); and (f) sensorimotor network (SMN) around the face region. For each of the 6 networks, we took the subject mode (ci in Eq. (1)) and separated them into two sets, one for ADHD and the other for TDC, based on the ground truth labels. We then computed the median statistic of that subject mode for each group separately, resulting in 6 median values for the ADHD group and 6 median values for the TDC group. Finally these 12 median values were normalized jointly to have a maximum of 1 and a minimum of 0.5 and displayed using a radar plot for easy visualization and comparison as shown in Fig. 4. Results, consistent with literature, illustrate that ADHD subjects present a decreased activity level compared to TDC in DMN [5] as well as networks related to executive functions, such as FPN, ECN, and VAN [3, 4]. On the other hand, SMN and VN show an increased level of activity [22] in ADHD subjects relative to TDC. This is as we expected, since ADHD subjects tend to be more inattentive and physically hyperactive than TDC.
Fig. 3:
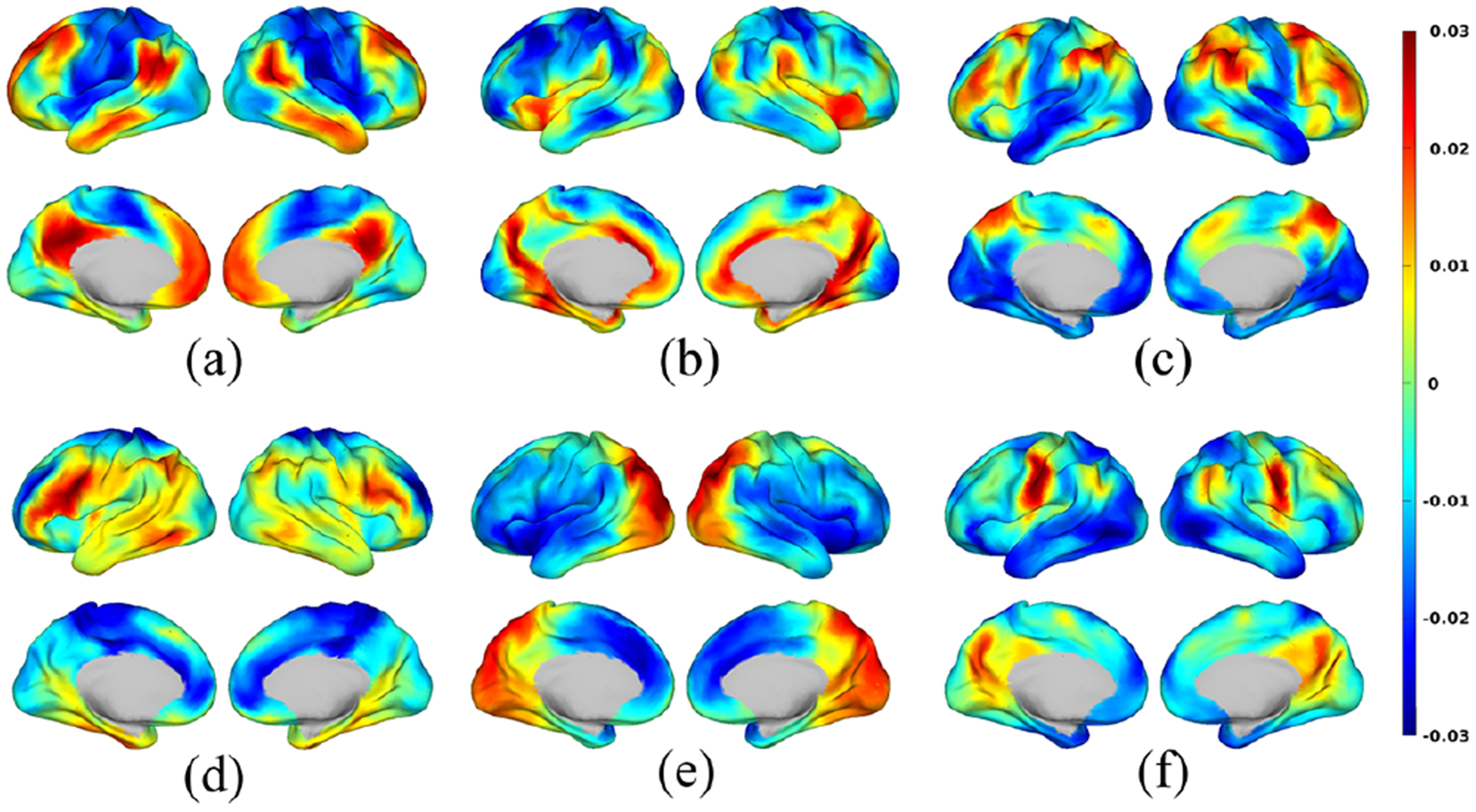
Spatial maps of six identified brain networks: (a) Default mode; (b) Ventral attention; (c) Executive control; (d) Fronto-parietal; (e) Visual; and (f) Sensorimotor. The spatial maps were normalized to have zero mean and unit norm, illustrating relative level of brain activity across different regions.
Fig. 4:
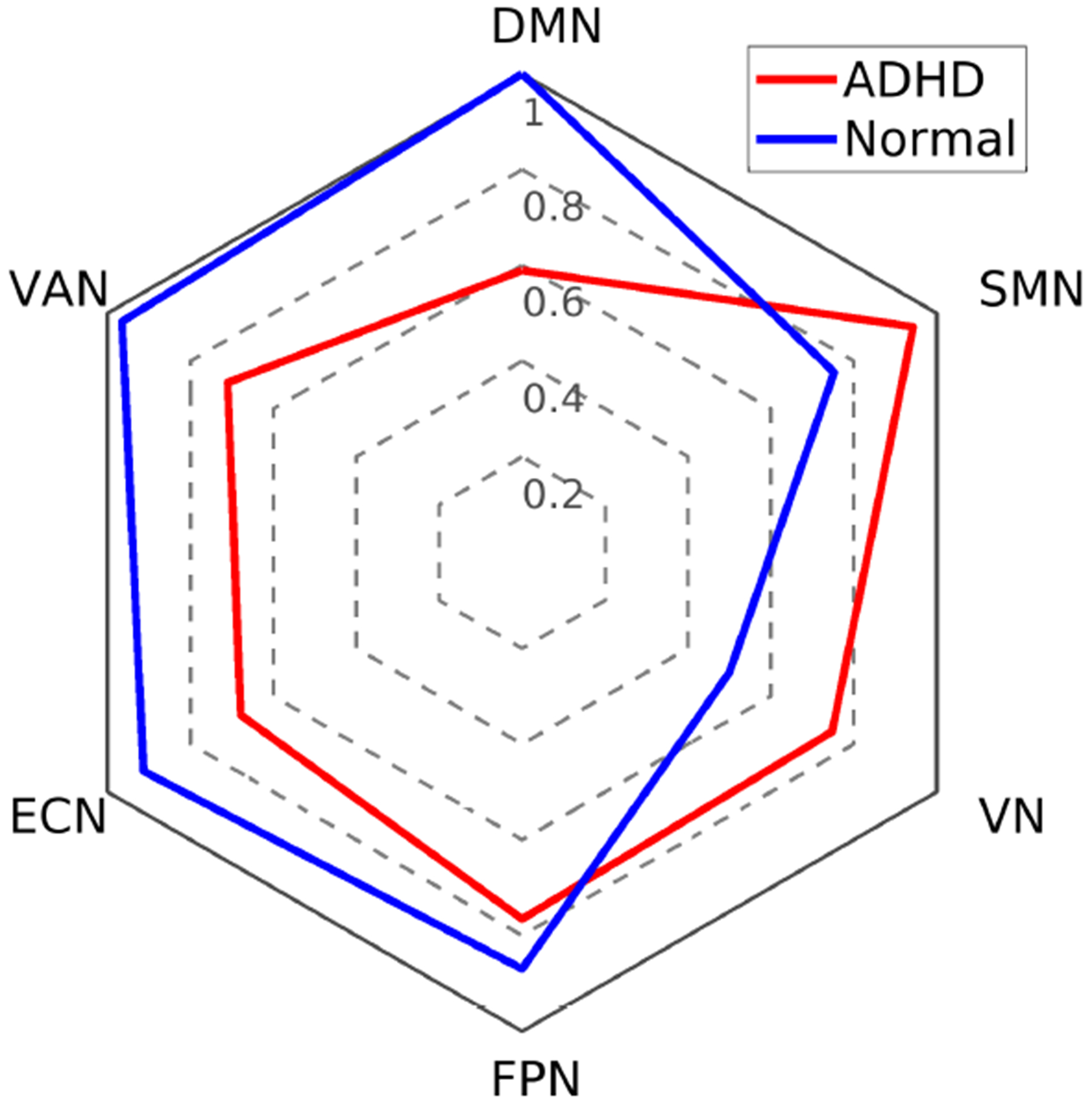
Radar plot of the normalized median subject mode for the 6 identified brain networks.
4. CONCLUSION
In this paper, we presented a feature extraction framework based on tensor decomposition of rs-fMRI data, which generates interpretable features that allow us to study ADHD in several ways: the predicted ADHD scores are significantly correlated with the clinically measured scores; The SVM-based classification achieved state-of-the-art accuracy even without using personal characteristic data; A further brain network study showed decreased activity level in DMN and other executive related networks such as FPN, ECN, VAN, but increased activity in SMN and VN in ADHD relative to TDC controls.
Acknowledgments
This work was supported by the National Institutes of Health with grant R01-NS089212, R01-NS074980, and R01-EB026299
5. REFERENCES
- [1].Polanczyk Guilherme, De Lima Mauricio Silva, Horta Bernardo Lessa, Biederman Joseph, and Rohde Luis Au-gusto, “The worldwide prevalence of ADHD: A systematic review and metaregression analysis,” American Journal of Psychiatry, vol. 164, no. 6, pp. 942–948, 2007. [DOI] [PubMed] [Google Scholar]
- [2].McCarthy H, Skokauskas N, and Frodl T, “Identifying a consistent pattern of neural function in attention deficit hyperactivity disorder: A meta-analysis,” Psychological Medicine, vol. 44, no. 4, pp. 869–880, 2014. [DOI] [PubMed] [Google Scholar]
- [3].Rubia Katya, “Cognitive neuroscience of attention deficit hyperactivity disorder (ADHD) and its clinical translation,” Frontiers in Human Neuroscience, vol. 12, pp. 1–23, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Castellanos Francisco X. and Aoki Yuta, “Intrinsic functional connectivity in attention-deficit/hyperactivity disorder: A science in developments,” Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, vol. 1, no. 3, pp. 253–261, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Castellanos Francisco X., Margulies Daniel S., Kelly Clare, Uddin Lucina Q., Ghaffari Manely, Kirsch Andrew, Shaw David, Shehzad Zarrar, Di Martino Adriana, Biswal Bharat, Sonuga-Barke Edmund J.S., Rotrosen John, Adler Lenard A., and Milham Michael P., “Cingulate-precuneus interactions: A new locus of dysfunction in adult attention-deficit/hyperactivity disorder,” Biological Psychiatry, vol. 63, no. 3, pp. 332–337, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Di Martino Adriana, Zuo Xinian, Kelly Clare, Grzadzinski Rebecca, Mennes Maarten, Schvarcz Ariel, Rodman Jennifer, Lord Catherine, Castellanos F. Xavier, and Milham Michael P., “Shared and distinct intrinsic functional network centrality in autism and attention-deficit/hyperactivity disorder,” Biological Psychiatry, vol. 74, no. 8, pp. 623–632, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hoekzema Elseline, Carmona Susana, Ramos-Quiroga J. Antoni, Fernandez Vanesa Richarte, Bosch Rosa, Soliva Juan Carlos, Rovira Mariana, Bulbena Antonio, Tobeña Adolf, Casas Miguel, and Vilarroya Oscar, “An independent components and functional connectivity analysis of resting state fMRI data points to neural network dysregulation in adult ADHD,” Human Brain Mapping, vol. 35, no. 4, pp. 1261–1272, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wolfers Thomas, Buitelaar Jan K., Beckmann Christian F., Franke Barbara, and Marquand Andre F., “From estimating activation locality to predicting disorder: A review of pattern recognition for neuroimaging-based psychiatric diagnostics,” Neuroscience and Biobehavioral Reviews, vol. 57, pp. 328–349, 2015. [DOI] [PubMed] [Google Scholar]
- [9].Vieira Sandra, Pinaya Walter H.L., and Mechelli Andrea, “Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: Methods and applications,” Neuroscience and Biobehavioral Reviews, vol. 74, pp. 58–75, 2017. [DOI] [PubMed] [Google Scholar]
- [10].Joshi Anand A., McCoy Dakarai, Chong Minqi, Li Jian, Choi Soyoung, Shattuck David, and Leahy Richard M., “BFP: BrainSuite fMRI pipeline,” in 24th Annual Meeting of the OHBM, Singapore, June 2018. [Google Scholar]
- [11].Li Jian, Choi Soyoung, Joshi Anand A., Wisnowski Jessica L., and Leahy Richard M., “Global PDF-based temporal non-local means filtering reveals individual differences in brain connectivity,” in IEEE 15th International Symposium on Biomedical Imaging, April 2018, pp. 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li Jian, Choi Soyoung, Joshi Anand A., Wisnowski Jessica L., and Leahy Richard M., “Temporal non-local means filtering for studies of intrinsic brain connectivity from individual resting fMRI,” Medical Image Analysis, p. 101635, 2020. (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Joshi Anand A., Chong Minqi, Li Jian, Choi Soyoung, and Leahy Richard M., “Are you thinking what I’m thinking? Synchronization of resting fMRI time-series across subjects,” Neuroimage, vol. 172, pp. 740–752, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li Jian, Wisnowski Jessica L., Joshi Anand A., and Leahy Richard M., “Brain network identification in asynchronous task fMRI data using robust and scalable tensor decomposition,” in Proc. SPIE Medical Imaging: Image Processing, San Diego, March 2019, pp. 164–172. [Google Scholar]
- [15].Li Jian, Mosher John C., Nair Dileep R., Gonzalez-Martinez Jorge, and Leahy Richard M., “Robust tensor decomposition of resting brain networks in stereotactic EEG,” in IEEE 51st Asilomar Conference on Signals, Systems and Computers, October 2017, pp. 1544–1548. [Google Scholar]
- [16].Li Jian, Haldar Justin P., Mosher John C., Nair Dileep R., Gonzalez-Martinez Jorge, and Leahy Richard M., “Scalable and robust tensor decomposition of spontaneous stereotactic EEG data,” IEEE Transactions on Biomedical Engineering, vol. 66, no. 6, pp. 1549–1558, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].R Dennis Cook, “Detection of influential observation in linear regression,” Technometrics, vol. 19, no. 1, pp. 15–18, 1977. [Google Scholar]
- [18].Joshi Anand A., Li Jian, Akrami Haleh, and Leahy Richard M., “Predicting cognitive scores from resting fMRI data and geometric features of the brain,” in Proc. SPIE Medical Imaging: Image Processing, March 2019, pp. 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Han Xiaobing, Zhong Yanfei, He Lifang, Yu Philip S., and Zhang Liangpei, “The unsupervised hierarchical convolutional sparse auto-encoder for neuroimaging data classification,” in International Conference on Brain Information and Health, August 2015, pp. 156–166. [Google Scholar]
- [20].Brown Matthew R. G., Sidhu Gagan S., Greiner Russell, Asgarian Nasimeh, Bastani Meysam, Silverstone Peter H., Greenshaw Andrew J., and Dursun Serdar M., “ADHD-200 Global Competition: diagnosing ADHD using personal characteristic data can outperform resting state fMRI measurements,” Frontiers in Systems Neuroscience, vol. 6, pp. 1–22, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Riaz Atif, Asad Muhammad, Alonso Eduardo, and Slabaugh Greg, “Fusion of fMRI and non-imaging data for ADHD classification,” Computerized Medical Imaging and Graphics, vol. 65, pp. 115–128, 2018. [DOI] [PubMed] [Google Scholar]
- [22].Cortese Samuele, Kelly Clare, Chabernaud Camille, Proal Erika, Di Martino Adriana, Milham Michael P, and Castellanos F. Xavier, “Toward systems neuroscience of ADHD: A meta-analysis of 55 fMRI studies,” American Journal of Psychiatry, vol. 169, no. 10, pp. 1038–1055, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]


