Abstract
Background
Laboratory testing is commonly performed in patients with COVID-19. Each of the laboratory parameters has potential value for risk stratification and prediction of COVID-19 outcomes. This systematic review and meta-analysis aimed to evaluate the difference between these parameters in severe and nonsevere disease and to provide the optimal cutoff value for predicting severe disease.
Method
We performed a systematic literature search through electronic databases. The variables of interest were serum procalcitonin, albumin, C-reactive protein (CRP), D-dimer, and lactate dehydrogenase (LDH) levels in each group of severity outcomes from COVID-19.
Results
There were a total of 4848 patients from 23 studies. Our meta-analysis suggest that patients with severe COVID-19 infections have higher procalcitonin, (mean difference 0.07; 95% CI 0.05–0.10; p < 0.00001), CRP (mean difference 36.88; 95% CI 29.10–44.65; p < 0.00001), D-Dimer (mean difference 0.43; 95% CI 0.31–0.56; p < 0.00001), and LDH (mean difference 102.79; 95% CI 79.10–126.49; p < 0.00001) but lower levels of albumin (mean difference −4.58; 95% CI −5.76 to −3.39; p < 0.00001) than those with nonsevere COVID-19 infections. The cutoff values for the parameters were 0.065 ng/mL for procalcitonin, 38.85 g/L for albumin, 33.55 mg/L for CRP, 0.635 μ/L for D-dimer, and 263.5 U/L for LDH, each with high sensitivity and specificity.
Conclusion
This meta-analysis suggests elevated procalcitonin, CRP, D-dimer, and LDH and decreased albumin can be used for predicting severe outcomes in COVID-19.
Keywords: COVID-19, SARS-CoV-2, Coronavirus, Laboratory, Biomarker
1. Introduction
In December 2019, new emerging cases of atypical pneumonia were first reported in Wuhan, Hubei Province, China. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the pathogen of this atypical pneumonia outbreak, called coronavirus disease 2019 (COVID-19), which targets the lower respiratory tract and other organs expressing the ACE2 receptor. Respiratory and airborne droplets are the main routes of transmission of this disease. The World Health Organization (WHO) declared this disease a public health emergency [1]. As of July 13th, 2020, a total of 13,082,304 cases of COVID-19 were recorded worldwide, with a total number of deaths reaching 572,551 [2]. Based on its features, this disease can be divided into ordinary, mild, severe, and critically ill types [3]. The initial symptoms and signs in COVID-19 patients are usually very mild, and the infection can even be asymptomatic. However, the disease can deteriorate over a short period of time (between 7 and 10 days) into acute respiratory distress syndrome (ARDS) and other multiorgan complications due to rapid viral replication and cytokine storms [4,5]. This abrupt onset of disease progression has contributed to an increase in the mortality rate of the disease. Several comorbidities have also been demonstrated to be associated with the development of severe COVID-19, such as hypertension, diabetes mellitus, dyslipidemia, thyroid disease, cardiovascular disease, dementia, and pulmonary disease [[6], [7], [8], [9]]. Therefore, prompt identification and containment, which are achievable through strict surveillance and early diagnosis, are very important.
One of the tests that physicians perform most often in the setting of COVID-19 is laboratory testing. During the detection, treatment, and follow-up of COVID-19, physicians frequently check various laboratory parameters to see the dynamic changes in each. These laboratory parameters have been suggested for risk stratifications in COVID-19, as timely detection of disease progression is crucial for appropriate management and intervention. Currently, combinations of several laboratory tests have been used in some settings to show the hyperinflammatory state and prognosis. These combinations include the neutrophil to lymphocyte ratio (NLR) and the lymphocyte to C-reactive protein ratio (LCR) [10]. Based on the pathophysiology of severe COVID-19, which involves a hyperinflammatory state, coagulation cascade, and multiorgan dysfunction [11], several biomarkers that represent each of those conditions, such as CRP, procalcitonin, D-dimer, LDH, and albumin, might be helpful to predict the outcome of COVID-19. Unfortunately, there are still missing puzzle pieces regarding the most significant laboratory markers and the cutoff values that can differentiate severe and nonsevere outcomes. This systematic review and meta-analysis aimed to analyze the differences in several biomarkers, including serum procalcitonin, CRP, D-dimer, LDH, and albumin, in severe and nonsevere disease, as well as the cutoff value for each biomarker to predict severe outcomes in COVID-19 infection.
2. Material and methods
2.1. Eligibility criteria
We conducted a systematic review and meta-analysis study. Studies were included in this review if they met the following inclusion criteria: representation for clinical questions (P: positive/confirmed cases of COVID-19; I: a group of patients with severe COVID-19; C: a group of patients with nonsevere COVID-19; O: information on laboratory parameters such as procalcitonin, albumin, C-reactive protein (CRP), D-dimer, and lactate dehydrogenase (LDH) levels), the type of study was a randomized control trial, cohort, clinical trial, case cohort, and crossover design, and if the full-text article was available. The following types of articles were excluded: articles other than original research (e.g., review articles or commentaries); case reports; articles not in the English language; articles on research in pediatric populations (17 years of age or younger); and articles on research in pregnant women.
2.2. Search strategy and study selection
We performed a systematic literature search from PubMed, PubMed Central, and Google Scholar with the search terms: ‘Characteristics’ OR ‘Laboratory parameters’ AND ‘COVID-19’ OR ‘Coronavirus disease 2019’. Duplicate results were removed. The remaining articles were independently screened for relevance by their abstracts with two authors. The full texts of residual articles were assessed according to the inclusion and exclusion criteria. The search started on July 6, 2020 and was finalized on July 13, 2020. The study was carried out according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.
2.3. Data extraction and quality assessment
Data extraction was performed independently by two authors, and we used standardized forms that included author, year, study design, number of participants, age, sex, serum procalcitonin, albumin, CRP, D-dimer, LDH, and severe COVID-19.
The variables of interest in our meta-analysis were serum procalcitonin, serum albumin, serum CRP, serum D-dimer, and serum LDH concentrations in mean ± SD or median (interquartile range) from each group of severe COVID-19 and nonsevere COVID-19 infections. Severe COVID-19 infection was defined as patients who had any of the following features at the time of or after admission: (1) respiratory distress (≥30 breaths per min); (2) oxygen saturation at rest ≤ 93%; (3) ratio of the partial pressure of arterial oxygen (PaO2) to a fractional concentration of oxygen inspired air (FiO2) ≤ 300 mmHg; or (4) critical complications (respiratory failure, septic shock, and/or multiple organ dysfunction/failure) or admission to the ICU [12].
Two investigators independently evaluated the quality of the included cohort and case-control studies using the Newcastle–Ottawa Scale (NOS) [13]. The selection, comparability, and exposure of each study were broadly assessed, and studies were assigned a score from zero to nine. Studies with scores ≥ 7 were considered of good quality. They also independently evaluated the quality of the included case-series studies using the Joanna Briggs Institute Critical Appraisal Checklist for Case Series [14].
2.4. Statistical analysis
The software programs Review Manager 5.4 (Cochrane Collaboration) and Comprehensive Meta-Analysis version 3 were used for meta-analysis. Continuous variables were calculated using the inverse-variance formula with random effects models regardless of heterogeneity. The effect estimate was reported as the mean difference (MD) and its standard deviation (SD) along with its 95% confidence interval (CI). Heterogeneity was assessed by using the I2 statistic, and values of <25%, 26%–50%, and >50% were considered low, moderate, and high degrees of heterogeneity, respectively. The p-value was two-tailed, and the statistical significance was set at ≤0.05. When data were reported as medians and interquartile ranges, we converted them to means and standard deviations for meta-analysis pooling using the formula by Wan et al. [15] The mean values of parameters that were found to be significant in the meta-analysis were then used to generate receiver operating characteristic (ROC) curves using SPSS ver. 24 to determine the area under the curve (AUC). The optimal cutoff for parameters with a significant p-value (two-tailed) was determined using Youden's index [16], and the corresponding sensitivity and specificity for the cutoff were also calculated. Subgroup analysis comparing prospective and retrospective studies was performed for each component of variable of interest. A funnel plot, Begg's rank correlation method [17], and Egger's weighted regression method [18] were adopted to statistically assess publication bias (p < 0.05 was considered statistically significant).
3. Results
3.1. Study selection and characteristics
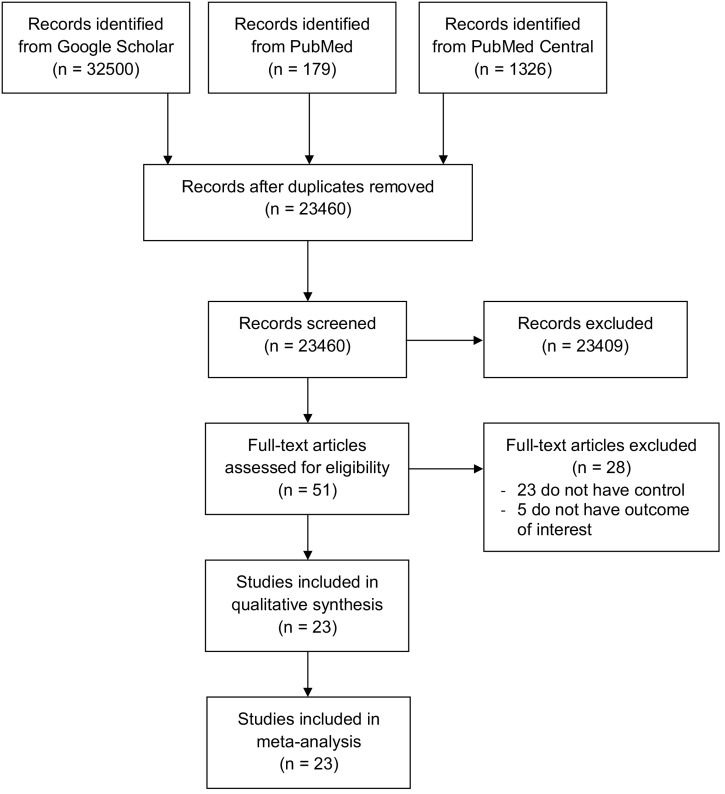
An initial search yielded 34,005 records, and 23,460 records remained after the removal of duplicates. A total of 23,408 records were excluded after screening the titles/abstracts. After evaluating 52 full texts for eligibility, 23 full-text articles were excluded because they did not have a control/comparison group, and 5 were excluded because they had no outcome of interest. Twenty-four studies were included in the qualitative synthesis and meta-analysis (Fig. 1 ) [[19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41]]. There were a total of 4848 patients from 23 studies. Of a total of 23 included studies, 19 were retrospective cohorts, 3 were prospective cohorts and 1 was a case-series study. All studies involved adult patients over 18 years old and used RT-PCR from respiratory tract samples as a confirmatory test for COVID-19 infections. The clinical characteristics of the included studies are summarized in Table 1 .
Fig. 1.
PRISMA diagram of the detailed process of selection of studies for inclusion in the systematic review and meta analysis.
Table 1.
Characteristics of included studies
| Study | Number participants | Type of study | Laboratory parameter | Severe disease |
Non-Severe disease |
||
|---|---|---|---|---|---|---|---|
| n (%) | Age (years) | n (%) | Age (years) | ||||
| Almazeedi et al. [19] | 1096 | Retrospective cohort | Procalcitonin, Albumin, CRP, D-Dimer | 42 (3.8%) | 54.8 ± 11 | 1054 (96.2%) | 37.1 ± 16 |
| Alshukry et al. [20] | 193 | Retrospective cohort | Procalcitonin, Albumin, CRP, D-Dimer, LDH | 22 (11.4%) | 52.3 ± 13.5 | 171 (88.6%) | 44.6 ± 15.7 |
| Cheng et al. [21] | 456 | Retrospective cohort | Procalcitonin, Albumin, CRP, D-Dimer, LDH | 251 (55%) | 59.8 ± 17.4 | 205 (45%) | 48.9 ± 18.1 |
| Dreher et al. [22] | 50 | Prospective cohort | Procalcitonin, CRP, D-Dimer, LDH | 24 (48%) | 63.3 ± 8.8 | 26 (52%) | 69.3 ± 16.2 |
| Duan et al. [23] | 348 | Retrospective cohort | Procalcitonin, Albumin, CRP, D-Dimer | 20 (5.7%) | 58 ± 15 | 328 (94.3%) | 44 ± 15 |
| Feng et al. [24] | 406 | Retrospective cohort | Procalcitonin, Albumin, CRP, D-Dimer, LDH | 54 (13.3%) | 57.6 ± 14 | 352 (86.7%) | 50.3 ± 19.2 |
| Gao et al. [25] | 43 | Retrospective cohort | Procalcitonin, CRP, D-Dimer | 15 (34.8%) | 45.2 ± 7.6 | 28 (65.2%) | 42.9 ± 14 |
| Gong et al. [26] | 189 | Retrospective cohort | Procalcitonin, CRP, Albumin, D-Dimer, LDH | 28 (14.8%) | 63.3 ± 12.9 | 161 (86.2%) | 46.6 ± 21.4 |
| Huang et al. [27] | 41 | Prospective cohort | Procalcitonin, Albumin, D-Dimer, LDH | 13 (31.7%) | 50.3 ± 14.8 | 28 (68.3%) | 49.1 ± 12.2 |
| Jiang et al. [28] | 60 | Retrospective cohort | D-Dimer | 8 (13.3%) | 56.3 ± 27.4 | 52 (86.7%) | 40.3 ± 42.2 |
| Khamis et al. [29] | 63 | Retrospective cohort | CRP, D-Dimer, LDH | 24 (38%) | 50 ± 17 | 39 (62%) | 47 ± 16 |
| Lv et al. [30] | 270 | Retrospective cohort | Procalcitonin, CRP, D-Dimer | 155 (57.4%) | 58.6 ± 47.4 | 115 (42.6%) | 54.3 ± 41.4 |
| Shang et al. [31] | 443 | Retrospective cohort | Procalctionin, Albumin, CRP, D-Dimer, LDH | 139 (31.3%) | 63.6 ± 14 | 304 (68.7%) | 57.3 (14.8) |
| Shi et al. [32] | 134 | Retrospective cohort | Procalcitonin, Albumin, CRP, D-Dimer, LDH | 46 (34.3%) | 56 ± 14.8 | 88 (65.7%) | 40 ± 15.5 |
| Sun et al. [33] | 18 | Prospective cohort | Albumin, CRP, D-Dimer, LDH | 10 (55.5%) | 59 ± 38.5 | 8 (44.5%) | 24.6 ± 33.3 |
| Wan et al. [34] | 135 | Retrospective case series | Procalcitonin, Albumin, CRP, D-Dimer, LDH | 40 (29.6%) | 60.3 ± 15.5 | 95 (70.4%) | 42 ± 11.8 |
| Wang et al. [35] | 45 | Retrospective cohort | Albumin, D-Dimer, LDH | 10 (22.2%) | 44.3 ± 25.1 | 35 (77.8%) | 38.6 ± 34 |
| Wang et al. [36] | 138 | Retrospective cohort | Procalcitonin, D-Dimer, LDH | 36 (26%) | 67 ± 15.5 | 102 (74%) | 50 ± 18.5 |
| Wei et al. [37] | 167 | Retrospective cohort | Procalcitonin, Albumin, CRP, D-Dimer, LDH | 30 (17.9%) | 49 ± 12.6 | 137 (82.1%) | 40.8 ± 15.4 |
| Yang et al. [38] | 200 | Retrospective cohort | Procalcitonin, Albumin, CRP, D-Dimer, LDH | 29 (14.5%) | 71 ± 13.4 | 171 (85.5%) | 52 ± 16.2 |
| Yi et al. [39] | 100 | Retrospective cohort | Procalcitonin, CRP, D-Dimer | 49 (49%) | 60.6 ± 14 | 51 (51%) | 48 ± 16.2 |
| Zhang et al. [40] | 140 | Retrospective cohort | Procalcitonin, CRP, D-Dimer | 56 (40.5%) | 58.6 ± 45.9 | 82 (59.5%) | 51.8 ± 38.5 |
| Zhang et al. [41] | 113 | Retrospective cohort | Procalcitonin, Albumin, CRP, LDH | 61 (53.9%) | 53.6 ± 13.3 | 52 (46.1%) | 34.2 ± 19.6 |
3.2. Quality of study assessment
Studies with various study designs, including cohort and case series, were included in this review and were assessed accordingly with the appropriate scale or tool. The Newcastle Ottawa Scale (NOS) was used to assess the cohort and case-control studies (Table 2 ), while the Joanna Briggs Institute Critical Appraisal checklist was used for case series studies (Table 3 ). All included studies were rated ‘good’ based on the criteria used in the Newcastle Ottawa Scale (NOS) and the Joanna Briggs Institute Critical Appraisal checklist. In conclusion, all studies were deemed fit to be included in the meta-analysis.
Table 2.
Newcastle-Ottawa quality assessment of observational trials.
| First author, year | Study design | Selection | Comparability | Outcome | Total score | Result |
|---|---|---|---|---|---|---|
| Almazeedi et al. [19] | Cohort | **** | ** | *** | 9 | Good |
| Alshukry et al. [20] | Cohort | *** | ** | *** | 8 | Good |
| Cheng et al. [21] | Cohort | *** | ** | *** | 8 | Good |
| Dreher et al. [22] | Cohort | ** | ** | *** | 7 | Good |
| Duan et al. [23] | Cohort | **** | ** | *** | 9 | Good |
| Feng et al. [24] | Cohort | *** | ** | *** | 8 | Good |
| Gao et al. [25] | Cohort | ** | ** | *** | 7 | Good |
| Gong et al. [26] | Cohort | *** | ** | *** | 8 | Good |
| Huang et al. [27] | Cohort | *** | ** | *** | 8 | Good |
| Jiang et al. [28] | Cohort | *** | ** | *** | 8 | Good |
| Khamis et al. [29] | Cohort | *** | ** | *** | 8 | Good |
| Lv et al. [30] | Cohort | *** | ** | *** | 8 | Good |
| Shang et al. [31] | Cohort | *** | ** | *** | 8 | Good |
| Shi et al. [32] | Cohort | *** | ** | *** | 8 | Good |
| Sun et al. [33] | Cohort | ** | ** | *** | 7 | Good |
| Wang et al. [35] | Cohort | ** | ** | *** | 7 | Good |
| Wang et al. [36] | Cohort | *** | ** | *** | 8 | Good |
| Wei et al. [37] | Cohort | *** | ** | ** | 7 | Good |
| Yang et al. [38] | Cohort | *** | ** | *** | 8 | Good |
| Yi et al. [39] | Cohort | ** | ** | *** | 7 | Good |
| Zhang et al. [40] | Cohort | **** | ** | *** | 9 | Good |
| Zhang et al. [41] | Cohort | *** | ** | *** | 8 | Good |
Each (*) means one score given to that criteria, so *** means the score of the study under that criteria is 3.
Table 3.
Joanna Briggs Institute Critical Appraisal tool for case series.
| Wan et al. [34] | |
|---|---|
| 1. Were there clear criteria for inclusion in the case series? | Yes |
| 2. Were the conditions measured in a standard, reliable way for all participants included in the case series? | Yes |
| 3. Were valid methods used for identification of the condition for all participants included in the case series? | Yes |
| 4. Did the case series have consecutive inclusion of participants? | Yes |
| 5. Did the case series have complete inclusion of participants? | Yes |
| 6. Was there clear reporting of the demographics of the participants in the study? | Yes |
| 7. Was there clear reporting of the clinical information of the participants? | Yes |
| 8. Were the outcomes or the follow-up results of the cases clearly reported? | Yes |
| 9. Was there clear reporting of the presenting site(s)/clinic(s) demographic information? | Yes |
| 10. Was the statistical analysis appropriate? | Yes |
| Quality | Include study |
3.3. Outcomes
3.3.1. Albumin levels
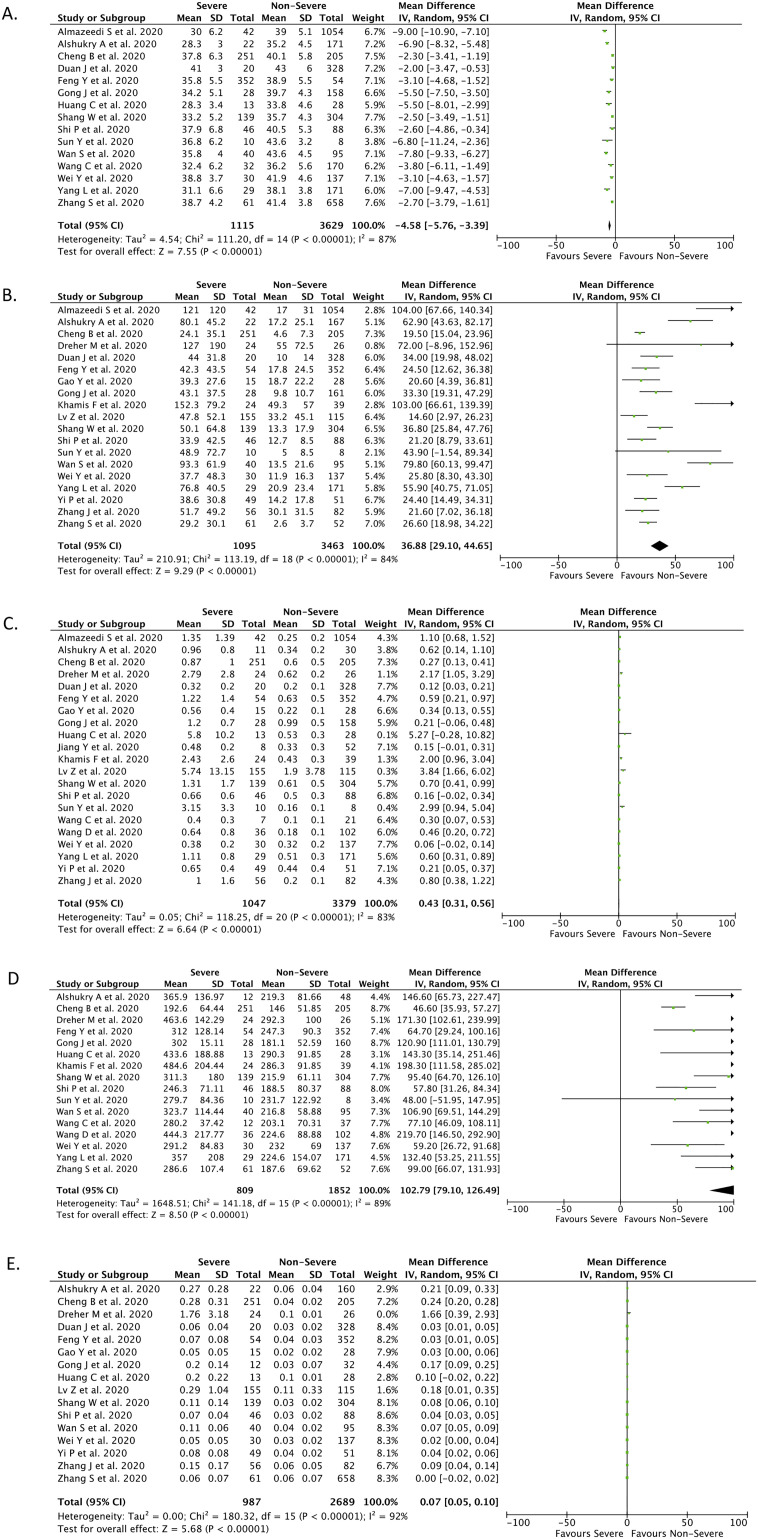
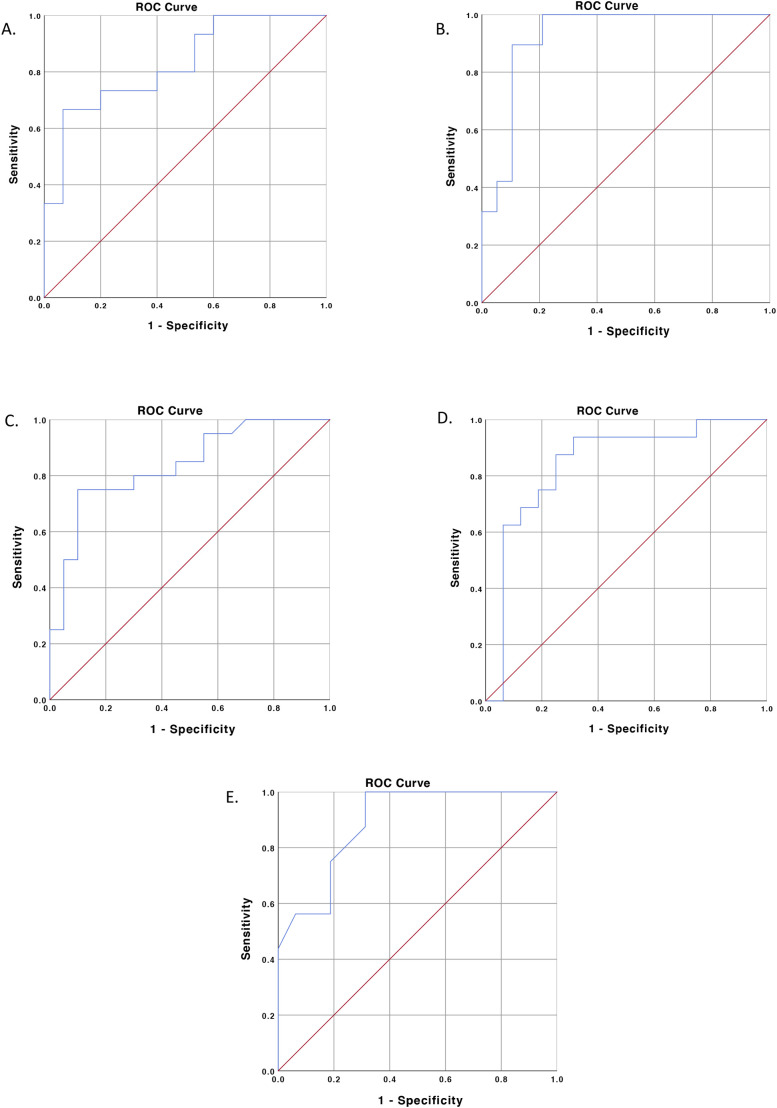
Fifteen studies (n = 4744) reported the levels of serum albumin in each group of outcomes. Decreased albumin was associated with severe disease based on our meta-analysis, with high heterogeneity (mean difference (MD) −4.58; 95% CI −5.76 to −3.39; p < 0.00001; I 2 = 87%; random-effects modeling) (Fig. 2A). The ROC curve for albumin parameters is shown in Fig. 3A, demonstrating that serum albumin provides good discrimination (AUC = 0.827, p = 0.002) between severe COVID-19 and nonsevere COVID-19 infections, with an optimal cutoff of 38.85 g/L, yielding a sensitivity of 66.7% and a specificity of 93.3% (Table 4 ).
Fig. 2.
Forest-plot analysis for serum albumin (A), CRP (B), D-Dimer (C), LDH (D), and procalcitonin (E) in severe and non-severe COVID-19.
Fig. 3.
ROC-curve analysis for serum albumin (A), CRP (B), D-Dimer (C), LDH (D), and procalcitonin (E) for predicting severe COVID-19 infection.
Table 4.
The summary of cut-off value for predicting severe outcome of COVID-19 from each laboratory parameter, their sensitivity and specificity, and their Begg's and Egger's test results
| Laboratory parameter | AUC | p-Value | Cut-off value | Sensitivity | Specificity | Begg's test | Egger's test |
|---|---|---|---|---|---|---|---|
| Albumin | 0.827 | 0.002 | ≤38.85 g/L | 66.7% | 93.3% | 0.367 | 0.11 |
| CRP | 0.922 | <0.001 | ≥33.55 mg/L | 89.5% | 89.5% | 0.119 | 0.07 |
| d-Dimer | 0.836 | <0.001 | ≥0.635 μg/L | 75% | 90% | 0.029 | 0.012 |
| LDH | 0.844 | 0.001 | ≥263.5 U/L | 87.5% | 75% | 0.138 | 0.003 |
| Procalcitonin | 0.891 | <0.001 | ≥0.065 ng/mL | 75% | 81.2% | 0.04 | 0.008 |
3.3.2. C-reactive protein (CRP) levels
Nineteen studies (n = 4558) reported the levels of serum CRP in each group of outcomes. Elevated CRP was associated with severe disease based on our meta-analysis, with high heterogeneity (mean difference (MD) 36.88; 95% CI 29.10–44.65; p < 0.00001; I 2 = 84%; random-effects modeling) (Fig. 2B). The ROC curve for CRP parameters is shown in Fig. 3B, demonstrating that serum CRP provides good discrimination (AUC = 0.922, p < 0.001) between severe COVID-19 and nonsevere COVID-19 infections with an optimal cutoff of 33.55 mg/L, yielding a sensitivity of 89.5% and a specificity of 89.5% (Table 4).
3.3.3. D-dimer levels
Twenty-one studies (n = 4426) reported the levels of serum D-dimer in each group of outcomes. Elevated D-dimer was associated with severe disease based on our meta-analysis, with high heterogeneity (mean difference (MD) 0.43; 95% CI 0.31–0.56; p < 0.00001; I 2 = 83%; random-effects modeling) (Fig. 2C). The ROC curve for D-dimer parameters is shown in Fig. 3C, demonstrating that serum D-dimer provides good discrimination (AUC = 0.836, p < 0.001) between severe COVID-19 and nonsevere COVID-19 infections with an optimal cutoff of 0.635 μg/L, yielding a sensitivity of 75% and a specificity of 90% (Table 4).
3.3.4. Lactate dehydrogenase (LDH) levels
Sixteen studies (n = 2661) reported the levels of serum LDH in each group of outcomes. Elevated LDH was associated with severe disease based on our meta-analysis, with high heterogeneity (mean difference (MD) 102.79; 95% CI 79.10–126.49; p < 0.00001; I 2 = 89%; random-effects modeling) (Fig. 2D). The ROC curve for LDH parameters is shown in Fig. 3D, demonstrating that serum LDH provides good discrimination (AUC = 0.844, p = 0.001) between severe COVID-19 and nonsevere COVID-19 infections with an optimal cutoff of 263.5 μg/L, yielding a sensitivity of 87.5% and a specificity of 75% (Table 4).
3.3.5. Procalcitonin levels
Sixteen studies (n = 3676) reported the levels of serum procalcitonin in each group of outcomes. Elevated procalcitonin was associated with severe disease based on our meta-analysis, with high heterogeneity (mean difference (MD) 0.07; 95% CI 0.05–0.10; p < 0.00001; I 2 = 92%; random-effects modeling) (Fig. 2E). The ROC curve for procalcitonin parameters is shown in Fig. 3E, demonstrating that serum procalcitonin provides good discrimination (AUC = 0.891, p ≤ 0.001) between severe COVID-19 and nonsevere COVID-19 infections with an optimal cutoff of 0.065 ng/mL, yielding a sensitivity of 75% and a specificity of 81.2% (Table 4).
3.3.6. Subgroup analysis
Subgroup analysis for retrospective studies revealed lower mean differences (MD) for the serum levels of albumin (MD = −4.42; 95% CI −5.69 to −3.16; p < 0.00001; I 2 = 89%; random-effect modeling), CRP (MD = 36.45; 95% CI 28.53–44.38; p < 0.00001; I 2 = 86%; random-effect modeling), D-dimer (MD = 0.39; 95% CI 0.27–0.51; p < 0.00001; I 2 = 82%; random-effect modeling), and LDH (MD = 99.50; 95% CI 74.40–124.61; p < 0.00001; I 2 = 91%; random-effect modeling) in non severe COVID-19 showed lower MD when compared to their prospective studies (MD for prospective studies of Albumin, CRP, D-dimer, and LDH consecutively: MD = −5.82 (95% CI −8.00 to −3.63), p < 0.00001, I 2 = 0%, random-effect modeling; MD = 50.63; (95% CI 11.00–90.26), p = 0.01; I 2 = 0%; random-effect modeling; MD = 2.45; 95% CI 1.48–3.42; p < 0.00001; I 2 = 0%; random-effect modeling; MD = 126.43; 95% CI 52.15–200.71; p = 0.0009; I 2 = 50%; random-effect modeling.) On the other hand, subgroup analysis for retrospective studies (mean difference (MD) 0.07; 95% CI 0.04–0.09; p < 0.00001; I 2 = 92%; random-effect modeling) showed a lower but more significant mean difference (MD) for procalcitonin levels between severe and nonsevere COVID-19 compared with prospective studies (mean difference (MD) 0.75 (95% CI −0.76–2.25), p = 0.33, I 2 = 83%, random-effect modeling).
3.3.7. Publication bias
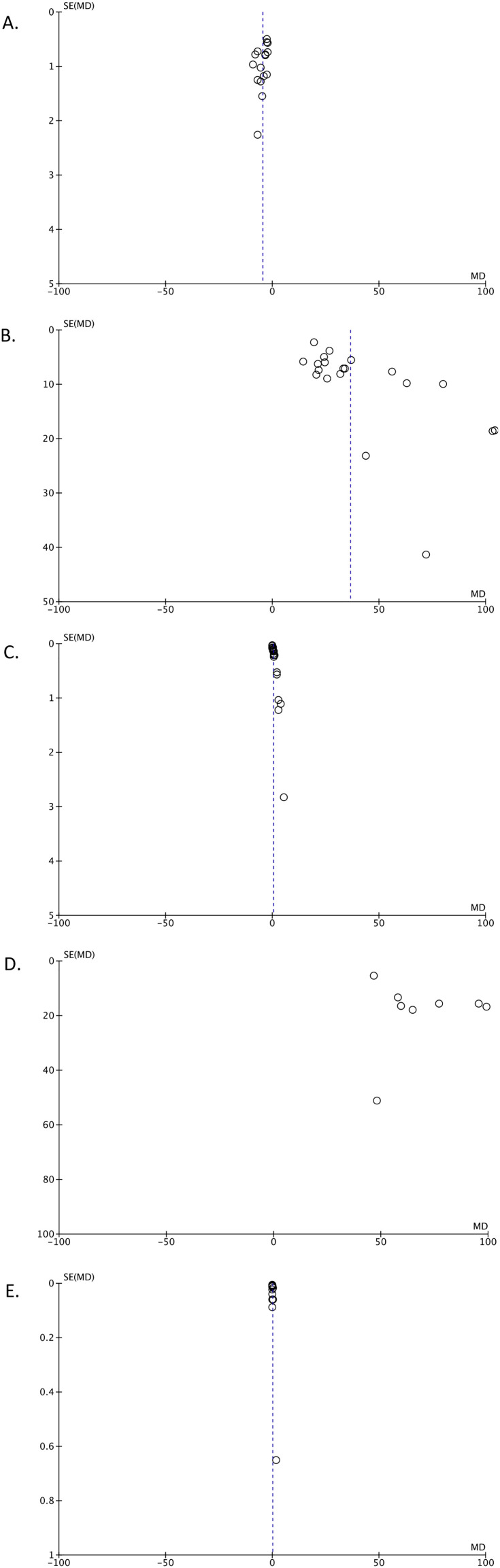
The funnel-plot analysis showed a relatively symmetrical inverted funnel plot for the albumin (Fig. 4 A), CRP (Fig. 4B), D-dimer (Fig. 4C), and procalcitonin (Fig. 4E), an parameters but showed an asymmetrical shape for the LDH (Fig. 4D) parameter, indicating possible publication bias. Meanwhile, rank-correlation Begg's test and regression-based Egger's test were not statistically significant for albumin and CRP parameters, showing no indication of publication bias, but were statistically significant for procalcitonin, D-dimer, and LDH parameters, showing a possible indication of publication bias (Table 4).
Fig. 4.
Funnel-plot analysis for serum albumin (A), CRP (B), D-Dimer (C), LDH (D), and procalcitonin (E).
4. Discussion
This systematic review and meta-analysis included 24 articles with quite large samples of COVID-19 patients. Our meta-analysis suggests that patients with severe COVID-19 infections have higher procalcitonin, CRP, D-dimer, and LDH levels but lower levels of albumin than those with nonsevere COVID-19 infections.
Albumin is a natural colloid that is abundant in plasma. It is exclusively synthesized in the liver and serves several purposes, such as maintaining intravascular oncotic pressure, acting as a carrier of several different endogenous and exogenous compounds, and maintaining the acid-base balance, and is often used as a marker of nutritional status and particular disease severity (e.g., liver cirrhosis) [42]. Several conditions can decrease the levels of serum albumin, such as decreased albumin production (e.g., advanced stage of hepatic cirrhosis and increased catabolism due to systemic illness), nutritional deficiencies (e.g., kwashiorkor), and increased loss of albumin (e.g., renal loss, gut loss, and extravascular loss). Systemic inflammation, such as what happens in sepsis, will increase systemic vascular permeability and capillary leakage, causing albumin to shift toward the extravascular space. In such patients, the synthesis of albumin is also disturbed, making hypoalbuminemia more profound [43]. In the case of COVID-19, the low levels of albumin in severe disease may be caused by malfunctioning system organs (vascular permeability, renal, and gastrointestinal), contributing to a greater extent of albumin excretion. Moreover, albumin has been found to have the ability to downregulate ACE2 receptors [44], which is crucial for modulating COVID-19 infection; therefore, low levels of albumin will possibly result in upregulation of ACE2 receptors and an increase in COVID-19 infectivity.
C-reactive protein (CRP) is a sensitive biomarker for inflammation. As an acute-phase inflammatory mediator, CRP is synthesized and released by the liver to the bloodstream, contributing to the host's resistance against invading pathogens [45]. The elevation of CRP may also be caused by bacterial coinfection that occurs in severe COVID-19. Moreover, a robust inflammatory response that occurs in severe disease may cause the levels of CRP to increase significantly.
D-dimer is one marker associated with thrombotic events and may also be elevated in
infections such as influenza, SARS, and CAP [46,47]. In COVID-19 patients, elevation of the D-dimer level is common and is associated with disease severity and mortality. D-dimers are fragments produced when plasmin cleaves fibrin to break down clots. Therefore, every process that increases fibrin production or breakdown will elevate plasma D-dimer levels. It is assumed that in severe COVID-19 (critically ill) patients, proinflammatory cytokines and the coagulation cascade, including D-dimer, are activated [48]. Studies also suggest that under inflammatory conditions, dysregulation of the coagulation cascade in SARS-CoV-2 infection results in diffuse alveolar damage with cellular fibromyxoid exudates, pneumocyte desquamation, and formation of a hyaline membrane, pulmonary edema with hyaline membrane formation, and interstitial mononuclear inflammatory infiltrates dominated by lymphocytes [49,50]. This event shifts the alveolar hemostatic balance toward prothrombotic activity. Proinflammatory cytokines also contribute to endothelial injury, which could activate coagulation and inhibit fibrinolysis in patients with severe COVID-19 [51]. This elevation of D-dimer suggests that there is a hypercoagulable state contributing to the severity of the disease and increased mortality.
Lactate dehydrogenase (LDH) is a cytoplasmic glycolytic enzyme that catalyzes the reversible conversion of L-lactate and pyruvate with concomitant interconversion of NADH and NAD+. It is present in the cytoplasm of all human tissues, with higher concentrations in the liver, heart, and skeletal muscle [52]. It has been reported that elevated LDH levels are one of the most common findings in patients infected with MERS-CoV [53] and H5N1 [54] and are also one of the biomarkers most strongly associated with ARDS mortality [55]. Multiple organ injury and decreased oxygenation with upregulation of the glycolytic pathway can result in abnormal values because the acidic extracellular pH resulting from infection and tissue injury will activate metalloproteases and enhance macrophage-mediated angiogenesis [56]. Increased LDH in severe COVID-19 suggested possible subclinical tissue damage. LDH itself is released when cellular necrosis happens; therefore, in severe infections in which a large number of cells typically undergo necrosis, high serum LDH levels can be observed. In particular, severe COVID-19 infections that mainly affect the lungs will release a high amount of LDH isozyme commonly found in lung tissue (LHD isoenzyme 3) [57,58].
A peptide precursor of the hormone calcitonin, PCT, has been widely investigated as a promising biomarker for the initial investigation of a bacterial infection [59]. An elevated serum PCT is often found in patients with sepsis and septic shock [60]. While it is still controversial whether PCT can accurately distinguish bacterial or viral pneumonia [61], PCT-guided therapy in acute respiratory infections may reduce antibiotic exposure [62]. Bacterial infections trigger extrathyroidal synthesis of PCT, which is actively maintained by elevated values of IL-6, IL-1β, and TNF-α, while viral infections hinder PCT production due to interferon-γ [63]. In this meta-analysis we found that an elevated serum PCT was associated with severe COVID-19.
Our systematic review and meta-analysis have several strengths. First, we included all published studies, therefore, the risk of publication bias was minimized. Second, the pooled effect estimates of our meta-analysis were very precise. Third, we also provided a cutoff value for each parameter with high sensitivity and specificity based on ROC curve analysis to help physicians predict the severe outcome of COVID-19.
Nonetheless, this review has its limitations. Given the observational design of the included studies and the retrospective data collection, the possibility that the observed association between each laboratory parameter and the severity of COVID-19 was affected by bias or confounding factors should still be considered. The asymmetrical inverted funnel plot for serum LDH and the significant p-values from Begg's and Egger's tests for serum procalcitonin, D-dimer, and LDH imply the presence of publication bias. Another limitation is that the total sample size of our meta-analysis was not very high due to the limited number of studies that provided data regarding laboratory parameters that matched our inclusion criteria. We also combined the results from prospective and retrospective cohort studies. Finally, there is an unclear association between the time at which biomarkers were tested and the determination of severe or nonsevere COVID-19; however, we believe that early laboratory testing since the patients were first admitted to the emergency unit can still be used to monitor and anticipate the progression of nonsevere to severe COVID-19; therefore, timely and suitable management can be performed.
Further research can explore another potential laboratory parameter to predict the severity or mortality of COVID-19. A cutoff value for new potential parameters should also be included to help physicians predict severe COVID-19.
5. Conclusion
This meta-analysis suggests serum procalcitonin, CRP, D-dimer, and LDH are elevated in patients with severe COVID-19 infection, while serum albumin levels are lower in severe illness compared with nonsevere COVID-19 infections. Further study is needed concerning specific threshold levels of these laboratory markers. Finally, COVID-19 patients with high levels of serum procalcitonin, CRP, D-dimer, and LDH and low levels of serum albumin should be monitored closely to minimize the risk of progression to severe disease.
Authorship
All authors whose names appear on the submission:
-
1)
made substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data; or the creation of new software used in the work;
-
2)
drafted the work or revised it critically for important intellectual content;
-
3)
approved the version to be published; and
-
4)
agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
None.
Declaration of Competing Interest
The authors declare no conflict of interest regarding this article.
Acknowledgment
None.
References
- 1.World Health Organization . World Health Organization; 2020. World Health Organization. Statement on the second meeting of the international health regulations (2005) emergency committee regarding the outbreak of novel coronavirus (2019-nCoV)https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov Available from: [Google Scholar]
- 2.World Health Organization . World Health Organization; 2020. Coronavirus disease 2019 (COVID-19) situation report −138.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200606-covid-19-sitrep-138.pdf?sfvrsn=c8abfb17_4 Available from: [Google Scholar]
- 3.World Health Organization . World Health Organization; 2020. Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected: interim guidance, 25 January 2020.https://apps.who.int/iris/handle/10665/330854 Available from: [Google Scholar]
- 4.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwenandar F., Japar K.V., Damay V., Hariyanto T.I., Tanaka M., Lugito N.P.H., et al. Coronavirus disease 2019 and cardiovascular system: a narrative review. Int J Cardiol Heart Vasc. 2020 Jun 3;29:100557. doi: 10.1016/j.ijcha.2020.100557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hariyanto T.I., Kurniawan A. Thyroid disease is associated with severe coronavirus disease 2019 (COVID-19) infection. Diabetes Metab Syndr. 2020 Sep-Oct;14(5):1429–1430. doi: 10.1016/j.dsx.2020.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hariyanto T.I., Kurniawan A. Metformin use is associated with reduced mortality rate from coronavirus disease 2019 (COVID-19) infection. Obes Med. 2020 Sep;19:100290. doi: 10.1016/j.obmed.2020.100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hariyanto T.I., Kurniawan A. Statin therapy did not improve the in-hospital outcome of coronavirus disease 2019 (COVID-19) infection. Diabetes Metab Syndr. 2020 Aug 26;14(6):1613–1615. doi: 10.1016/j.dsx.2020.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hariyanto T.I., Putri C., Situmeang R.F.V., Kurniawan A. Dementia is a predictor for mortality outcome from coronavirus disease 2019 (COVID-19) infection. Eur Arch Psychiatry Clin Neurosci. 2020 Oct;26:1–3. doi: 10.1007/s00406-020-01205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y., Liu A., Liang L., Jiang J., Luo H., Deng W., et al. Diagnostic value of blood parameters for community-acquired pneumonia. Int Immunopharmacol. 2018 Nov;64:10–15. doi: 10.1016/j.intimp.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 11.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020 Aug 25;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization . World Health Organization; 2020. Report of the WHO-China joint mission on coronavirus disease 2019 COVID-19 report.https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19---final-report-1100hr-28feb2020-11mar-update.pdf?sfvrsn=1a13fda0_2&download=true Available from: [Google Scholar]
- 13.Margulis A.V., Pladevall M., Riera-Guardia N., Varas-Lorenzo C., Hazell L., Berkman N.D., et al. Quality assessment of observational studies in a drug-safety systematic review, comparison of two tools: the Newcastle-Ottawa Scale and the RTI item bank. Clin Epidemiol. 2014;6:359–368. doi: 10.2147/CLEP.S66677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moola S., Munn Z., Tufanaru C., Aromataris E., Sears K., Sfetcu R., et al. In: Joanna Briggs Institute reviewer’s manual. Aromataris E., Munn Z., editors. The Joanna Briggs Institute; 2017. Chapter 7: systematic reviews of etiology and risk.https://reviewersmanual.joannabriggs.org/ Available from: [Google Scholar]
- 15.Wan X., Wang W., Liu J., Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14:135. doi: 10.1186/1471-2288-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fluss R., Faraggi D., Reiser B. Estimation of the Youden index and its associated cutoff point. Biom J. 2005;47(4):458–472. doi: 10.1002/bimj.200410135. [DOI] [PubMed] [Google Scholar]
- 17.Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 18.Egger M., Davey S.G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almazeedi S., Al-Youha S., Jamal M.H., Al-Haddad M., Al-Muhaini A., Al-Ghimlas F., et al. Characteristics, risk factors and outcomes among the first consecutive 1096 patients diagnosed with COVID-19 in Kuwait. EClinicalMedicine. 2020 doi: 10.1016/j.eclinm.2020.100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alshukry A., Ali H., Ali Y., Al-Taweel T., Abu-Farha M., AbuBaker J., et al. Clinical characteristics of coronavirus disease 2019 (COVID-19) patients in Kuwait. PLoS One. 2020 Nov 20;15(11) doi: 10.1371/journal.pone.0242768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng B., Hu J., Zuo X., Chen J., Li X., Chen Y., et al. Predictors of progression from moderate to severe coronavirus disease 2019: a retrospective cohort. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.06.033. (S1198-743X(20)30379-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dreher M., Kersten A., Bickenbach J., Balfanz P., Hartmann B., Cornelissen C., et al. The characteristics of 50 hospitalized COVID-19 patients with and without ARDS. Dtsch Arztebl Int. 2020;117(16):271–278. doi: 10.3238/arztebl.2020.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan J., Wang X., Chi J., Chen H., Bai L., Hu Q., et al. Correlation between the variables collected at admission and progression to severe cases during hospitalization among patients with COVID-19 in Chongqing. J Med Virol. 2020 doi: 10.1002/jmv.26082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y., Ling Y., Bai T., Xie Y., Huang J., Li J., et al. COVID-19 with different severities: a multicenter study of clinical features. Am J Respir Crit Care Med. 2020;201(11):1380–1388. doi: 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Y., Li T., Han M., Li X., Wu D., Xu Y., et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol. 2020;92(7):791–796. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong J., Ou J., Qiu X., Jie Y., Chen Y., Yuan L., et al. A tool to early predict severe corona virus disease 2019 (COVID-19): a multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y., He S., Zhang C., Wang X., Chen X., Jin Y., et al. Clinical characteristics of 60 discharged cases of 2019 novel coronavirus-infected pneumonia in Taizhou, China. Ann Transl Med. 2020;8(8):547. doi: 10.21037/atm.2020.04.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khamis F., Al-Zakwani I., Al Naamani H., Al Lawati S., Pandak N., Omar M.B., et al. Clinical characteristics and outcomes of the first 63 adult patients hospitalized with COVID-19: an experience from Oman. J Infect Public Health. 2020;13(7):906–913. doi: 10.1016/j.jiph.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lv Z., Cheng S., Le J., Huang J., Feng L., Zhang B., et al. Clinical characteristics and co-infections of 354 hospitalized patients with COVID-19 in Wuhan, China: a retrospective cohort study. Microbes Infect. 2020;22(4–5):195–199. doi: 10.1016/j.micinf.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shang W., Dong J., Ren Y., Tian M., Li W., Hu J., et al. The value of clinical parameters in predicting the severity of COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi P., Ren G., Yang J., Li Z., Deng S., Li M., et al. Clinical characteristics of imported and second-generation coronavirus disease 2019 (COVID-19) cases in Shaanxi outside Wuhan, China: a multicentre retrospective study. Epidemiol Infect. 2020 Sep 30;148 doi: 10.1017/S0950268820002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Y., Dong Y., Wang L., Xie H., Li B., Chang C., et al. Characteristics and prognostic factors of disease severity in patients with COVID-19: the Beijing experience. J Autoimmun. 2020 doi: 10.1016/j.jaut.2020.102473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan S., Xiang Y., Fang W., Zheng Y., Li B., Hu Y., et al. Clinical features and treatment of COVID-19 patients in Northeast Chongqing. J Med Virol. 2020;92(7):797–806. doi: 10.1002/jmv.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C., Deng R., Gou L., Fu Z., Zhang X., Shao F., et al. Preliminary study to identify severe from moderate cases of COVID-19 using combined hematology parameters. Ann Transl Med. 2020;8(9):593. doi: 10.21037/atm-20-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y.Y., Wang R.R., Zhang D.W., Tu Y.H., Chen C.S., Ji S., et al. Risk factors for severe COVID-19: evidence from 167 hospitalized patients in Anhui, China. J Infect. 2020;81(1):e89–e92. doi: 10.1016/j.jinf.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L., Liu J., Zhang R., Li M., Li Z., Zhou X., et al. Epidemiological and clinical features of 200 hospitalized patients with corona virus disease 2019 outside Wuhan, China: a descriptive study. J Clin Virol. 2020;129:104475. doi: 10.1016/j.jcv.2020.104475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi P., Yang X., Ding C., Chen Y., Xu K., Ni Q., et al. Risk factors and clinical features of deterioration in COVID-19 patients in Zhejiang, China: a single-centre, retrospective study. BMC Infect Dis. 2020 Dec 10;20(1):943. doi: 10.1186/s12879-020-05682-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q., et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75(7):1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S.Y., Lian J.S., Hu J.H., Zhang X.L., Lu Y.F., Cai H., et al. Clinical characteristics of different subtypes and risk factors for the severity of illness in patients with COVID-19 in Zhejiang, China. Infect Dis Poverty. 2020;9(1):85. doi: 10.1186/s40249-020-00710-6. (Version 2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cabrerizo S., Cuadras D., Gomez-Busto F., Artaza-Artabe I., Marín-Ciancas F., Malafarina V. Serum albumin and health in older people: review and meta-analysis. Maturitas. 2015;81(1):17–27. doi: 10.1016/j.maturitas.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 43.Levitt D.G., Levitt M.D. Human serum albumin homeostasis: a new look at the roles of synthesis, catabolism, renal and gastrointestinal excretion, and the clinical value of serum albumin measurements. Int J Gen Med. 2016;9:229–255. doi: 10.2147/IJGM.S102819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu B.C., Gao J., Li Q., Xu L.M. Albumin caused the increasing production of angiotensin II due to the dysregulation of ACE/ACE2 expression in HK2 cells. Clin Chim Acta. 2009;403(1–2):23–30. doi: 10.1016/j.cca.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 45.Sproston N.R., Ashworth J.J., et al. Role of C-Reactive Protein at Sites of Inflammation and Infection. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Wissen M., Keller T.T., van Gorp E.C., Gerdes V.E., Meijers J.C., van Doornum G.J., et al. Acute respiratory tract infection leads to procoagulant changes in human subjects. J Thromb Haemost. 2011;9(7):1432–1434. doi: 10.1111/j.1538-7836.2011.04340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agapakis D.I., Tsantilas D., Psarris P., Massa E.V., Kotsaftis P., Tziomalos K., et al. Coagulation and inflammation biomarkers may help predict the severity of community-acquired pneumonia. Respirology. 2010;15(5):796–803. doi: 10.1111/j.1440-1843.2010.01773.x. [DOI] [PubMed] [Google Scholar]
- 48.Shorr A.F., Thomas S.J., Alkins S.A., Fitzpatrick T.M., Ling G.S. D-dimer correlates with proinflammatory cytokine levels and outcomes in critically ill patients. Chest. 2002;121(4):1262–1268. doi: 10.1378/chest.121.4.1262. [DOI] [PubMed] [Google Scholar]
- 49.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lippi G., Favaloro E.J. D-dimer is associated with severity of coronavirus disease 2019: a pooled analysis. Thromb Haemost. 2020;120(5):876–878. doi: 10.1055/s-0040-1709650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu P.P., Sabatini D.M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 53.Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013;13(9):752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oner A.F., Bay A., Arslan S., Akdeniz H., Sahin H.A., Cesur Y., et al. Avian influenza a (H5N1) infection in eastern Turkey in 2006. New Engl J Med. 2006;355(21):2179–2185. doi: 10.1056/NEJMoa060601. [DOI] [PubMed] [Google Scholar]
- 55.Hoeboer S.H., Oudemans-van Straaten H.M., Groeneveld A.J. Albumin rather than C-reactive protein may be valuable in predicting and monitoring the severity and course of acute respiratory distress syndrome in critically ill patients with or at risk for the syndrome after new onset fever. BMC Pulm Med. 2015;15(1):22. doi: 10.1186/s12890-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez-Outschoorn U.E., Prisco M., Ertel A. Ketones and lactate increase cancer cell “stemness,” driving recurrence, metastasis and poor clinical outcome in breast cancer: achieving personalized medicine via metabolo-genomics. Cell Cycle. 2011;10(8):1271–1286. doi: 10.4161/cc.10.8.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hariyanto T.I., Kurniawan A., et al. Tocilizumab administration is associated with the reduction in biomarkers of coronavirus disease 2019 infection. J Med Virol. 2020 doi: 10.1002/jmv.26698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henry B.M., Aggarwal G., Wong J., Benoit S., Vikse J., Plebani M., et al. Lactate dehydrogenase levels predict coronavirus disease 2019 (COVID-19) severity and mortality: A pooled analysis. Am J Emerg Med. 2020;38(9):1722–1726. doi: 10.1016/j.ajem.2020.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Creamer A.W., Kent A.E., Albur M. Procalcitonin in respiratory disease: use as a biomarker for diagnosis and guiding antibiotic therapy. Breathe (Sheff) 2019 Dec;15(4):296–304. doi: 10.1183/20734735.0258-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song J., Park D.W., Moon S., Cho H.J., Park J.H., Seok H., et al. Diagnostic and prognostic value of interleukin-6, pentraxin 3, and procalcitonin levels among sepsis and septic shock patients: a prospective controlled study according to the Sepsis-3 definitions. BMC Infect Dis. 2019 Nov 12;19(1):968. doi: 10.1186/s12879-019-4618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kamat I.S., Ramachandran V., Eswaran H., Guffey D., Musher D.M. Procalcitonin to distinguish viral from bacterial pneumonia: a systematic review and meta-analysis. Clin Infect Dis. 2020 Jan 16;70(3):538–542. doi: 10.1093/cid/ciz545. [DOI] [PubMed] [Google Scholar]
- 62.Schuetz P., Wirz Y., Sager R., Christ-Crain M., Stolz D., Tamm M., et al. Effect of procalcitonin-guided antibiotic treatment on mortality in acute respiratory infections: a patient level meta-analysis. Lancet Infect Dis. 2018;18:95–107. doi: 10.1016/S1473-3099(17)30592-3. [DOI] [PubMed] [Google Scholar]
- 63.Schuetz P., Albrich W., Mueller B. Procalcitonin for diagnosis of infection and guide to antibiotic decisions: past, present and future. BMC Med. 2011 Sep 22;9:107. doi: 10.1186/1741-7015-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]