Graphical abstract
Keywords: COVID-19, Treatment, Interferons, Chloroquine/hydroxychloroquine, Azithromycin, Vaccine
Highlights
-
•
SARS-CoV-2 infections induce low level production of type I and III IFNs.
-
•
Reduced antiviral immunity and cytokine storm contribute to severe COVID-19.
-
•
Treatment with type I IFN demonstrates encouraging results.
-
•
CQ/HCQ and AZM-based treatments are controversial due to adverse effects.
-
•
COVID-19 candidate vaccines showcase promising results.
Abstract
The ongoing pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has drawn the attention of researchers and clinicians from several disciplines and sectors who are trying to find durable solutions both at preventive and treatment levels. To date, there is no approved effective treatment or vaccine available to control the coronavirus disease-2019 (COVID-19). The preliminary in vitro studies on viral infection models showed potential antiviral activities of type I and III interferons (IFNs), chloroquine (CQ)/hydroxychloroquine (HCQ), and azithromycin (AZM); however, the clinical studies on COVID-19 patients treated with CQ/HCQ and AZM led to controversies in different regions due to their adverse side effects, as well as their combined treatment could prolong the QT interval. Interestingly, the treatment with type I IFNs showed encouraging results. Moreover, the different preliminary reports of COVID-19 candidate vaccines showcase promising results by inducing the production of a high level of neutralizing antibodies (NAbs) and specific T cell-mediated immune response in almost all participants. The present review aims to summarize and analyze the recent progress evidence concerning the use of IFNs, CQ/HCQ, and AZM for the treatment of COVID-19. The available data on immunization options to prevent the COVID-19 are also analyzed with the aim to present the promising options which could be investigated in future for sustainable control of the pandemic.
1. Introduction
In less than one year, the pandemic of coronavirus disease-2019 (COVID-19) has killed more than 1,193,755 out of over 45,899,858 people infected in the world, as of October 31th, 2020 [1]. To date, no antiviral therapy or vaccines are available against COVID-19; nevertheless, scientists around the world are mobilized to identify the treatment candidates and vaccines. To date, several solutions have been proposed, involving the use of existing antivirals, antibiotics, and anti-inflammatory drugs with known features such as optimal dosage and pharmacokinetics in the therapeutic strategies, for the treatment of COVID-19 [[2], [3], [4]]. Most strategies considered in the clinical trials aim to accelerate the viral clearance and inhibit the cytokine storm to minimize the need for mechanical ventilation, long hospital stays, and COVID-19 associated mortality. To date, different anti-inflammatory drugs such as tocilizumab, chloroquine (CQ)/hydroxychloroquine (HCQ), and dexamethasone have been proposed to decrease or suppress the release/production of pro-inflammatory cytokines with the aim to minimize the cytokine storm associated with SARS-CoV-2 [5,6]. Several clinical trials are underway; however, none of the drugs has been consistently proven significantly effective (Fig. 1 ) [[7], [8], [9], [10]]. Among the trials of using different drugs, the use of CQ/HCQ in combination with azithromycin (AZM) has the highest numbers of trials and patients [7]. In vitro studies have shown the effectiveness of CQ/HCQ and AZM as antiviral and anti-inflammatory drugs [4,[11], [12], [13]]. In addition, there are specially approved treatment strategies for urgent use during the COVID-19 pandemic despite the lack of substantial data. These emergency treatment strategies include the use of remdesivir and convalescent plasma in the USA and favipiravir in China [[14], [15], [16]]. Along with trials of different treatment options, several vaccine candidates are under evaluation. An effective vaccine could ultimately reduce the person-to-person transmission, viral shedding, and disease severity [17].
Fig. 1.
Illustration of replication mechanism and potential drug targets of SARS-CoV-2. The SARS-CoV-2 infects the host cell in two ways: either through the plasma membrane or endosomes or through the interaction between its spike protein (S) and cell receptor, ACE2. The SARS-CoV-2 may also enter the host cell through the interaction between its S protein and sialic acid receptors. The TMPRSS2 close to the ACE receptor activates the S protein in the endosome via followed by initiating the fusion of the SARS-CoV-2 membrane with the plasma membrane. The viral RNA is released and translated in the viral polyprotein followed by proteolysis to produce non-structural proteins (NSPs) and form a replication-transcription complex (RTC). The RTC drives the synthesis of (-) RNA. The full length (-) RNA copies of the genome provide the templates for full-length (+) RNA genomes. The transcription further produces a subset of subgenomic RNAs, including those encoding the accessory and structural proteins. The translated structural proteins (M, N, E, and S) and the genomic RNA are assembled into the viral nucleocapsid and envelope in the ER-Golgi intermediate compartment, which is subsequently released via exocytosis. The potential drug targets are shown in red such as inhibitors of viral entry to human cells: chloroquine (CQ)/hydroxychloroquine (HCQ), umifenovir; viral protease inhibitor: lopinavir/ritonavir, ivermectin; RNA-dependent RNA polymerase inhibitor: remdesivir, favipiravir; interleukin (IL)-6 inhibitor; IL-1 inhibitor: anakinra; Janus kinases inhibitor: baricitinib, ruxolitinib, upadacitinib; corticosteroid: dexamethasone; an inhibitor of the cellular serine protease TMPRSS2: camostat mesylate; antimicrobial/antibiotics: azithromycin (AZM); immunoglobulins: convalescent plasma; interferons (IFNs); angiotensin-converting enzyme inhibitor (ACEI); angiotensin II receptor blocker (ARB). Abbreviations: ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane protease serine 2; AT1R, angiotensin II type 1 receptor; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; RNA, ribonucleic acid; (+) RNA, Positive-strand (5′-to-3′) RNA; (-) RNA, negative-strand (3′-to-5′) RNA; N, nucleocapsid protein; M, matrix protein; E, envelope protein; S, spike protein.
Likewise in earlier viral outbreaks, some patients recover without any treatment at account of effective immune system, including the innate immunity, cell-mediated immunity, and humoral immunity [18]. Such patients either have an effective primary immune response or a secondary immune response due to previous contact with a pathogen showing similar epitopes with SARS-CoV-2 [19]. The use of drugs that can restore, strengthen, and modulate the immune system is, therefore, a perfect approach in handling the COVID-19. The immune response against human coronaviruses (HCoVs), including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [6,[20], [21], [22], [23], [24]] and severe acute respiratory syndrome coronavirus (SARS-CoV) [[25], [26], [27], [28]], is characterized in most cases by low levels of type I and III interferons (IFNs) resulting in reduced innate response despite the excessive production of pro-inflammatory cytokines (cytokine storm). Although the IFNs effectors’ pathways are not affected by HCoVs, it makes sense to consider exogenous IFNs as therapeutic options which could increase the body’s response to the infecting virus. This strategy is already in use considerable success in treating viral infections such as hepatitis viruses [29,30]. In fact, the IFNs stimulate the important factors of innate immunity, that is an essential pivot of host defense against viruses and intracellular pathogens [29,30].
This review aims to describe and summarize the recent progress evidence concerning the use of IFNs, CQ/HCQ, and AZM for the treatment of COVID-19. We highlight the importance of IFNs in the treatment of COVID-19 and describe the modalities of the immune response protection against coronaviruses, including SARS-CoV, SARS-CoV-2, and MERS-CoV viruses.
2. Interferons and coronaviruses
Interferons (IFNs) are important cytokines which orchestrate the immune response to pathogens such as fungi, bacteria, and viruses. The production of IFNs is triggered by the recognition of pathogenic components through pattern-recognition receptors (PRR) [31]. The types I (IFN-α, IFN-β, IFN-ω, IFN- ε et IFN-κ) and III (IFN- λ1, IFN- λ2, IFN- λ2, IFN- λ4) IFNs are involved in the innate antiviral response in most cells, while type II INF (IFN-γ) predominantly serves as the communication molecule between the specialized cells of the immune system [32,33]. Essentially produced by the plasmacytoid dendritic cells (pDC), the type I IFNs target the heterodimeric receptor (IFNAR) [[31], [32], [33]]. The IFN-λ molecules are expressed by certain myeloid and epithelial cells and thought to produce similar effects with type I IFNs. The growing evidence suggests that IFN-λ plays a non-redundant role in promoting the innate immunity and stimulating the adaptive immune response to viral pathogens [34]. IFN-λ induces a non-specialized state of antiviral resistance and uses the same transduction pathway as type I IFNs, although through a different receptor (IFNLR) [35].
IFNs activate hundreds of genes known as IFN-stimulated genes (ISGs) through the Janus kinase-signal transducer/activator of transcription (JAK/STAT) signaling pathway, which provides a complete antiviral response targeting all stages of viruses' replicative cycles [32,36]. For example, MX2 (the IFN-induced GTP-binding protein) prevents the replication of human immunodeficiency virus (HIV) after its association with the capsid [37] and IFITMs (IFN-induced transmembrane protein) probably modify the membrane structure and the pH of endosomes by inhibiting the fusion of viral membranes with the cell membrane [38]. In addition to many roles in regulating the immune response [38], it is demonstrated that IFITM proteins prevent the entry of SARS-CoV and MERS-CoV into the cell [39,40]. Similarly, ISG15 binds and inactivates certain viral proteins [41], while tetherin inhibits the release of viral particles by sequestering them on the membrane of infected cells [42]. IFNs also activate other genes that lead to the overexpression of PRRs to increase the alertness of cells to pathogens or inhibit the translation of new proteins, thus reducing the production of new viral particles [32,36]. Moreover, IFNs can induce apoptosis of infected cells and modulate the adaptive immune response [32,36]. Thus, IFN-α and β participate in the differentiation of certain T lymphocytes, while IFN-λ intervenes in the maturation of T and B lymphocytes [32,36]. The response of IFN could be affected by the viral factors as well as the host factors such as the age [43,44]. The antiviral response mediated by the IFNs depends on the viral load. A low viral load is associated with adequate IFN response which leads to early and effective virus clearance whereas a high virus load delays the IFNs production and lead to inflammation and lung damage [26,45].
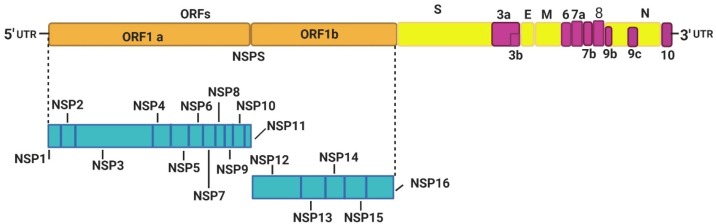
The discovery of the effectiveness of IFNs in antiviral control has made it possible to envisage their therapeutic use [33]. The IFN-α and β are mainly used to treat hepatitis viruses because these induce a state of antiviral resistance used in conjunction with other drugs with direct antiviral action [46,47]. In clinical studies, IFN-λ also showed promising results against chronic hepatitis viruses [[48], [49], [50]]. Many viruses inhibit the production of IFNs by acting on the JAK/STAT signaling pathway [51,52]. As for coronaviruses, these use various strategies to escape the antiviral response of the innate immune system [52,53]. The HCoVs RNA genome (Fig. 2 ) consisted of open-reading frames (ORFs) encodes the nonstructural proteins (NSP1-16), structural proteins (nucleocapsid N, membrane M, envelope E, and spike S), and accessory proteins, which target numerous signaling pathways involved in antiviral response (Table 1 and Fig. 3 ) [54,55]. HCoVs can interfere with the processes of induction of innate immunity, including the recognition of the virus by PRRs, inhibition of IFNs signaling, IFNs production, and ISG effector’s function. SARS-CoV avoids its recognition by the PRR either by protecting its PAMPS (viral RNA) or by inhibiting the activation of PRR [56]. To avoid activation of the RIG-I-like receptor (RLR) by dsRNA replication intermediates, the SARS-CoV can replicate inside the double-membrane vesicles [56].
Fig. 2.
The SARS-CoV-2 genome and encoded proteins. ORF1a and ORF1b encode the nonstructural proteins (NSP1-16), structural proteins (S, E, M, and N), and accessory proteins (ORF3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10). The 5′-UTR and 3′-UTR are untranslated extremities of the genome. NSP, non-structural protein; ORF, open reading frame; UTR, untranslated region.
Table 1.
Coronavirus antagonism upstream of interferon induction.
| Coronavirus | Mechanism of evasion upstream of IFN signaling | References |
|---|---|---|
| SARS-CoV | Viral protein PLP (NSP3) inhibits IFNs production by blocking the nuclear translocation and phosphorylation of IRF3. PLP adversely controls the antiviral immune response by inhibiting the STING signaling. | [84,85,86] |
| Viral NP, ORF3b, and ORF6 prevent the TRIM25 activation of RIG-I and inhibit the transcription of ISGs. Viral NP inhibits the IRF3 function. | [87,88,89,90] | |
| ORF6 antagonizes the STAT1 function. | [91] | |
| Viral protein NSP14 mimics the 5ꞌcap structure on the viral RNA by its guanine -N7-methyltransferase activity. | [92] | |
| Viral protein NSP15 acts on negative-sense viral RNA by cleaving its 50-polyuridines. | [93] | |
| ORF3a induces the degradation of IFNAR1. | [94] | |
| Viral EP, ORF3a, and ORF8b induce the pro-inflammatory cytokines through NF‐κB activation after triggering the NLRP3 inflammasome. | [95,96,97,98] | |
| SARS-CoV-2 | ORF3b suppresses the induction of type I IFNs by acting on IRF3 or MAVS. | [99] |
| Viral protein NSP15 inhibits the induction of IFNs production by interacting and closing the signaling RIG-I/MAVS and IRF3. | [55] | |
| Viral protein NSP15 interacts with RNF41, an activator of IRF3 and TBK1. The viral protein NSP13 interacts with the signaling transitional TBK1 that mediates the production of IRF3. NSP13 antagonizes the NF-kB pathway. | [55] | |
| Viral protein NSP10 induces the production of cytokine storm by facilitating the IL-8 induction after interacting with NKRF. | [100] | |
| ORF6 inhibits nuclear export of host mRNA by targeting the NUP98-RAE1 complex. | [55] | |
| ORF9c antagonizes the NF-kB pathway. | [55] | |
| MERS-CoV | ORF, 4a, 4b, and 5 inhibit the nuclear translocation of IRF3 and induce the suppression of IFNs production. | [101] |
| Viral protein NS4a is a type I and type III IFNs antagonist, which itself binds to the dsRNA, impedes PKR activation, and inhibits the PACT, an activator of RLRs. | [102,103,104] | |
| Viral protein NS4b prevents the host RNase L activation, another activator of RLRs. | [105] | |
| ORF4b protein inhibits the type I IFN production through nuclear and cytoplasmic targets. ORF4b impedes the innate immune response by inhibiting the NF-kB nuclear transport. | [106,107] | |
| HCoVs | Viral proteins NSP10 and NSP16 protect the viral RNA from MDA5 recognition by modifying the 5ꞌ cap structure with its 2ꞌ-0-methyltransferase activity. | [108,109] |
| Viral protein NSP1 inhibits the phosphorylation of STAT1 and host mRNA translation by binding to 40S ribosomal subunit. | [87,88,110,111,112,113] | |
| ORF9b (except for MERS-CoV) interacts indirectly with the RIG-I/MAVS signaling adapter via its association with Tom70, suggesting a preserved mechanism of IFNs escape. ORF9b interacts with the IRF3 signaling pathway. | [55,114] | |
| Viral MP inhibits the TBK1 signaling and nuclear translocation of IRF3. Viral MP interacts with the innate receptors and signaling molecules to impound them in the cytoplasmic compartments. | [101,115] | |
| ORF6 (except for MERS-CoV) inhibits the MAVS, TBK1, and IRF3. | [116] |
NSP, nonstructural protein; IRF, IFNs regulatory factor; STAT1, signal transducer and activator of transcription 1; ORF, open reading frame; PLP, papain-like protease; ISG, IFN-stimulated genes; TRIM 25, tripartite motif-containing 25; NP, nucleocapsid protein; MP, matrix protein; EP, envelope protein; RIG-I, retinoic acid-inducible gene I; MAVS, mitochondrial antiviral signaling protein; TBK1, tank-binding kinase-1; Tom70, translocase of the mitochondrial outer membrane 70; RNF41, ring finger protein 41; NKRF, NF-Kβ repressing factor; PKR, protein kinase R; RLRs, RIG-I-like receptors; STING, stimulator of interferon genes; PACT, protein activator; RAE1, ribonucleic acid export 1; NUP98, Nucleoporin 98.
Fig. 3.
Schematic illustration of the innate immune response to human coronavirus infection. The HCoVs infect the host cell through interaction between its spike protein (S) and the cell receptor, ACE2. Upon viral RNA recognition, the PRRs activate the innate cellular response reactions that normally lead to the production of pro-inflammatory cytokines and IFNs through NF-kB and IRF3/IRF7 (A). IFNs, secreted in an autocrine and paracrine behavior, induce the expression of ISGs via the JAK-STAT signaling pathway involved in antiviral response (B). Some viral proteins such as SARS-CoV’s NSP1, PLP, NSP10, NSP13, NSP14, NSP15, ORF3b, and ORF9b proteins and the MERS-CoV’s NS4a, NS4b, ORF4a, ORF4b, and ORF5 proteins inhibit this natural response at several levels. See Table 1 for detailed mechanisms at each level.
HCoVs, human coronavirus; PRRs, pattern-recognition receptors; TLRs, toll-like receptors; RIG-I, retinoic acid-inducible gene I; MAVS, mitochondrial antiviral signaling protein; NSP, non-structural protein; IRF, IFNs regulatory factor; STAT, signal transducer and activator of transcription; ORF, open reading frame; PLP, papain-like protease; ISG, IFN-stimulated genes; N, nucleocapsid protein; M, matrix protein; E, envelope protein; TBK1, TANK-binding kinase-1; MyD88, myeloid differentiation primary response 88; TRIF, TIR domain-containing adapter-inducing IFN-β; PKR, protein kinase R; RIG-I, retinoic acid-inducible gene 1; RLRs, RIG-I-like receptors; STING, stimulator of interferon genes; MDA-5, melanoma differentiation-associated protein 5; IkB, inhibitor of nuclear factor kB; IKK-ε, IκB kinase-ε; NLRP3, NOD-like receptor family pyrin domain containing 3; IFNAR, interferon alpha and beta receptor; IFNLR, interferon lambda receptor; TRIMs, tripartate motif proteins; P, phosphate; GBPs, guanylate binding proteins; OASs, oligo adenylate synthases; NOS2, nitric oxide synthase 2; IFITMs, IFN-induced transmembrane proteins.
In vitro observations and clinical studies of SARS-CoV patients have shown a deficiency of cytokines, including IFN-α/β and IFN-γ, compared to influenza patients [25]. Specifically, decreased amounts of IFN-α/β and IFN-λ is also reported in COVID-19 patients [22]. Similarly, in mice, SARS-CoV infection induces a low level of IFN-α/β and IFN-λ [27]. Moreover, a study in ferrets has shown that SARS-CoV-2 significantly induces a lower production of IFNs than the Influenza A virus (IAV) [22]. Additionally, the production of IFN-β and ISGs in IAV infection arises more rapidly than in SARS-CoV and MERS-CoV (Middle East respiratory syndrome coronavirus) [57]. Thus, a cohort clinical study of Covid-19 patients tested 8–12 days subsequent first symptoms showed a low level of blood type I IFN associated with higher viral load, chemokines, and pro-inflammatory cytokines resulting in decreased innate immunity and COVID-19 progression. The ISGs expression was lower in severe than in mild COVID-19 [20]. In contrast, a study carried out using bronchoalveolar lavage fluid from 8 cases of COVID-19 reported a higher expression of ISGs and pro-inflammatory genes, including chemokines, compared to controls [58]. Clinical studies of SARS-CoV and MERS patients reported that the severity of the disease leads to a high increase in IFNs with the viral load remaining high [59,60].
In vitro studies involving Vero cells showed that SARS-COV-2 exhibits a greater sensitivity to IFN-α/β than SARS-COV; where the viral titers were significantly reduced by over 4 log after IFN-α or IFN-β treatment at 50 IU/Ml [61,62]. Similar results were found with MERS-CoV infected cells [63]. Better viral outcomes were observed in SARS-COV infected macaques after receiving pegylated IFN-α [64]. During IAV infection in mice, IFNs-λ was the predominant IFN type with viral replication control capability without induction of inflammation [65]. IFN-λ plays a role by providing protection against IAV respiratory tract infection. Similarly, in vitro studies showed that IFN-λ4 inhibits the replication of MERS-COV in the respiratory epithelial cells, with higher induction of ISG [66]. Moreover, the use of IFN-β1b decreased the viral load and lung pathology in marmosets infected with MERS-CoV [67]. Similar results were found with MERS-CoV infected macaques after receiving IFN-α2b and ribavirin [68]. Similarly, mycophenolic acid and IFN-β reduced viral load in MERS-CoV infected cells [69]. In contrast, IFN-β associated with lopinavir/ritonavir (LV/RTV) ameliorated the pulmonary function but did not heal the lung pathology in mice infected with MERS-CoV [70].
Vapor inhalation of IFN-α combined with ribavirin was recommended in China as guidelines for the treatment of COVID-19 [71,72]. In Hubei province, 2,944 healthcare workers categorized in high-risk and low-risk groups received daily nasal drops of the recombinant human interferon-gamma (rhIFN-α) during 28 days for COVID-19 prevention. The rhIFN-α was combined with thymosin-α1 for the high-risk group. The findings of the study showed the potential of IFN for prevention of COVID-19 [73]. In Hong Kong, the administration of LV/RTV and ribavirin (RV) in addition to subcutaneous injection of IFN β-1b significantly reduced the virus load, symptoms, and the recovery time in COVID-19 patients in comparison to the antiviral drugs used without IFN [74]. In another clinical trial in Wuhan, China on 77 patients with moderate COVID-19, who were administered either oral umifenovir (N = 24), IFN-α2b (N = 7) or a combination of both compounds (N = 46), showed that patients treated with IFN present rapid viral clearance and reduced inflammatory signs compared to others [74]. A retrospective cohort study in Shenzhen city and Hubei province (China) on 141 patients with COVID-19 who inhaled IFN-α2b (N = 70) or a combination of IFN-α2b and umifenovir (N = 71) showed that patients treated with IFN- α2b present rapid SARS-CoV-2 clearance and reduced hospital stay [75]. In a study made in Turkey, the type I IFN associated with HCQ, AZM, and enoxaparin sodium was used to treat COVID-19 patients with multiple sclerosis, which demonstrated a rapid recovered of patients. It was difficult to evaluate the effect of IFN because the patients were treated with other drugs [76]. In the United Arab Emirates, the Pegylated IFN-α-2a was used to treat three patients with severe COVID-19 (2 patients with no significant medical history and one patient was known as types II diabetes mellitus), where the patients recovered with progress in clinical status within 36 h [77]; however, the reliability of the findings of this study is limited because of the small sample size (i.e., n = 3). In a clinical study conducted in Iran, 20 COVID-19 patients received IFN-β-1a in combination with HCQ, and LV/RTV as the treatment [78]. Within 10 days, the viral clearance results showed a significant decrease with the recovery of all patients in 14 days. In a randomized clinical trial (RCT) in Iran, the IFN β-1b reduced the mortality by significantly raising the discharge rate of 33 patients on day 14 compared to the control group comprising of 33 patients treated with LV/RTV or atazanavir/ RTV in combination with HCQ [98]. Furthermore, the inhaled IFN-β-1a reduced the risk of developing severe COVID-19 among 101 patients by 79 % in phase 2 clinical trial (NCT04385095) in the United Kingdom [79].
Overall, these studies demonstrate that despite the inhibition of endogenous IFNs production by HCoVs, the supply of exogenous IFN represents a promising therapeutic approach. An early and suitable administration of IFN-α/β and IFN-λ to COVID-19 patients could be of great value in controlling the viral infection while avoiding inflammation and tissue damage. In animal models, an early IFN administration better protects animals in comparison to the late treatments [28,45,80]. Moreover, IFN-α/β signaling avoids excessive IFN-γ-mediated responses and therefore prevents lung injury and chronic inflammation [81]. IFN-λ is essential for controlling viral replication in the upper respiratory tract [82]. It decreases the viral spread in the lower respiratory tract as well as the transmission to the new hosts. When the virus reaches the lungs due to failed initial viral control, the lungs are protected by IFNs-α/β [65,82,83]. Additionally, the unique quality of the IFN-λ response is long-lasting without inflammatory effects [61]. These features make IFN-λ attractive for COVID-19 prevention and treatment strategies.
3. Chloroquine/hydroxychloroquine, azithromycin, and coronaviruses
Chloroquine (CQ) is a drug indicated in the treatment and prevention of malaria as well as in rheumatology and dermatology for rheumatoid arthritis and certain lupus [117,118]. It induces the production of reactive oxygen species (ROS)-mediated toxicity in inner glial cells after prolonged treatment requiring the use of vitamin C to prevent the ROS production [119]. High dose of vitamin C inhibits cytokines storm in COVID-19 patients by exerting immunomodulatory properties [120,121] and induces the production of IFNs in patients with viral infections [122]. Furthermore, vitamin C reduces pulmonary lung inflammation and lung injury due to ROS release by phagocytes (Fig. 4 ) [26,123]. Hydroxychloroquine (HCQ) is a less toxic drug derived from CQ with similar properties to the latter [124] and used in the management of certain autoimmune diseases, including rheumatoid arthritis and lupus erythematosus [118]. In vitro and in vivo studies have certified that CQ can inhibit the replication of numerous intracellular microorganisms, including HCoVs, Zika virus, and HIV [4,11,12]. The action of CQ and HCQ may be different depending on the type of microorganisms, but it is shown that it targets the interaction between the SARS-CoV and its receptor by hampering the glycosylation of its cellular receptors blocking the SARS-CoV infections [125]. Besides, CQ/HCQ blocks the sialic acid receptors, thus preventing the entry of SARS-CoV-2 into the upper respiratory tract [126]. Furthermore, CQ has an immunomodulatory ability on activated immune cells [11,127]. It downregulates the expression of TLRs (Toll-like receptors) and TLR-mediated signal transduction and decreases or suppresses the release and production of TNF-α, IL-1, and IL-6, which facilitate the inflammation (cytokines storm) in COVID-19. In fact, CQ inhibits the interactions between TLRs of antigen-presenting cells (APC) and viral nucleic acid, and subsequently prevent the production of IFNs [11,[127], [128], [129]]. CQ (lysosomotropic drug) is also a powerful inhibitor of endosome-lysosome acidification, which allows the development of SARS-CoV and then its processing in the cytosol [11,127]. CQ inactivates all endosomal proteinases and results in APC reduced function; hence, impairing the adaptive immunity [130]. Moreover, CQ inhibits the viral RNA and DNA polymerase, virion assembly, and viral shedding (Fig. 1) [16]. CQ can interact with the LV/RTV in COVID-19 patients, and cause the prolongation of the QT interval [131]. HCQ being less toxic with potent antiviral activity against SARS-CoV-2 compared to the CQ, is preferred for the treatment of COVID-19 [131,132].
Fig. 4.
The immunological response to SARS-CoV-2 infection showing the release of ROS by phagocytes.
pDS, plasmacytoid dendritic cells; IL, interleukin; TNF-α, tumor necrosis factor-alpha; IFN-γ, interferon-gamma; Inflamm: inflammatory; Eosino, eosinophils; Mono, monocytes; Macs, macrophages; Neutro, neutrophils; NK cell, Natural killer cell; Abs, antibodies; ADCC, Antibody-dependent cell-mediated cytotoxicity; ROS, reactive oxygen species.
Azithromycin (AZM) is the first macrolide antibiotic in the azalide group, which inhibits the synthesis of bacterial proteins, quorum-sensing, and biofilm formation [133,134]. It has an antibacterial mechanism related to that of other macrolides but accumulates more effectively in the cells, mainly phagocytes [133]. Recently, AZM has received increasing attention because of additional properties on host-defense reactions by exerting the immunomodulation effects in chronic inflammatory diseases [133,135]. The modulation of immune responses enables the lasting therapeutic advantage of AZM in chronic obstructive pulmonary disease, cystic fibrosis, non-eosinophilic asthma, and non-fibrous cystic bronchiectasis [133]. Macrolides such as AZM present the antiviral properties on bronchial epithelial cells by decreasing the rate of rhinovirus replication in the isolated bronchial cells of cystic fibrosis patients using a mechanism related to IFN induction [136]. AZM is associated with increased expression of PRR, IFN β, IFN λ, and IFN-stimulated genes [136]. It also has antiviral properties against rhinoviruses by decreasing the synthesis of intercellular adhesion molecules such as ICAM-1, which are determinant routes for virus integration [137]. In vitro studies have also demonstrated an inhibitory effect of macrolides on the respiratory syncytial virus (RSV) in bronchial epithelial cells [138]. This would be linked to the prevention of interaction with the cellular viral target [138]. AZM also seems to inhibit the Zika virus replication in vitro on Vero cells; however, the antiviral action mechanism, in this case, has not been studied [139]. Similarly, AZM has shown in vitro activity against the Ebola virus on Vero cells [140]. In in vitro model for the influenza virus (H1N1), AZM interferes with the cellular internalization process of the virus [141]; however, the drug must be present before the infection for its maximum efficacy in this model. Macrolides such as AZM and spiramycin have also shown some activity on Enterovirus species in animal models of Enterovirus A71 infection by relieving the symptoms and increasing the animal survival [142]. As a senolytic drug, AZM has the ability to accelerate the phagocytic power of macrophages and selectively kills the senescent cells [143,144]. In a retrospective study on Saudi patients with severe MERS-CoV infection, macrolides used according to different regimens showed a weak (but not statistically significant) protective effect on mortality or viral clearance compared to the macrolide-free approach [145]. In a clinical trial carried out in 40 children with RSV infection randomized between AZM (10 mg/kg once a day for 7 days, then 5 mg/kg once a day for an additional 7 days) and placebo treatment for 14 days showed a significant statistical reduction in IL-8 and wheezing were put forward in the AZM group compared to placebo. An anti-inflammatory effect has been advanced to explain the decrease in the release of cytokines [146]. In another clinical trial on 50 patients hospitalized for influenza, randomized between oseltamivir versus oseltamivir and AZM, the effect on viral clearance was not significant, on the other hand, the decrease in pro-inflammatory cytokines (IL-6, IL-18, IL-17, CXCL-8, CXCL-9, sTNFR-1) and C-reactive protein was faster in the group receiving AZM and oseltamivir compared to the oseltamivir alone [13]. AZM increases the pH of organelles such as trans-Golgi network and endosome in primary bronchial epithelial cells and then alters the intracellular activities of virus [147]. These organelles play a significant role in packaging the proteins into vesicles for secretion, in a process exploited by the viruses to facilitate their replication [147]. The increase in pH of these organelles may alter the glycosylation of ACE2 (Angiotensin-Converting Enzyme 2) and therefore inhibit SARS-CoV-2 and host-cell interaction [147]. AZM reduces the level of furin in the spike protein of SARS-CoV-2, and therefore prevents the viral entry into the cells (Fig. 1) [147].
In summary, several in vitro studies showed a possible antiviral and anti-inflammatory effect of CQ/HCQ and AZM in preclinical models of viral infections. Different hypotheses on the mechanism of antiviral and anti-inflammatory activities have been proposed. CQ or its derivative HCQ and AZM have been used in treating COVID-19 patients [4,125,131]. In vitro evaluation indicated that HCQ and AZM exhibit a synergistic effect on SARS-CoV-2 at concentrations similar to that achieved in humans [148]. Additionally, AZM increases the effectiveness of HCQ in decreasing the viral load [149]. Several studies published at the start of the COVID-19 pandemic have confirmed the effectiveness of CQ on COVID-19. An in vitro study conducted in China demonstrated that CQ prevents the entry steps as well as the release of SARS-CoV-2, thus preventing its replication and propagation in the host [150]. Another study reported an apparent efficacy and an acceptable tolerance of a treatment based on CQ in view of the data from 10 hospitals in Guangzhou, Shanghai, Beijing, Jingzhou, Chongqing, Wuhan, and Ningbo in China [125]. HCQ also presented encouraging results in clinical studies showing the high rates of patients’ recovery in comparison to the untreated COVID-19 patients in France, China, and Italy [18,[151], [152], [153], [154], [155]]. Despite the small number of patients (i.e., sample size), these studies show that these drugs deserve deeper investigations. It was also shown in several studies that the association of HCQ with AZM increases the treatment effectiveness in comparison to HCQ alone [18,153,[152], [153], [154], [155]].
Besides the studies showing the effectiveness of HCQ associated with AZM, some studies revealed adverse effects, which limit the administration of this treatment to COVID-19 patients [[156], [157], [158]]. These adverse effects include prolonged QT interval, death, and transfer to intensive care with the persistence of virus after 6 days of treatment in more than 80 % of the patients [159,160]. Moreover, an RCT (ChiCTR2000029868) revealed no significant difference in the rate of viral elimination between HCQ and standard of care [160]. A retrospective study on 807 COVID-19 patients in the USA concluded that HCQ used alone or in combination with AZM did not reduce the risk of mechanical ventilation but increased the mortality [161]. In another retrospective study conducted in the USA on 1376 patients showed that HCQ does not reduce or increase the risk of a composite endpoint for intubation or death [162]. In a randomized trial (NCT04308668) of HCQ as the post-exposure prophylaxis for COVID-19 conducted across the USA and Canada, HCQ did not show significant results between the participants receiving HCQ (49/414) and those receiving placebo (58/407) [163]. In this study, no significant adverse effects were reported in the HCQ group. Moreover, an early administration of HCQ did not reduce the severity of symptoms in non-hospitalized patients with early COVID-19 in another randomized trial (NCT04308668) [164]. In Spain, an early administration of HCQ to mild COVID-19 patients did not improve the viral clearance, disease regression, and symptoms reduction. In this study, the HCQ-treated patients experienced adverse events during the 28 days of follow up [165]. In another RCT (NCT04381936) conducted in 176 hospitals across the United Kingdom on 7,513 participants, HCQ did not reduce the mortality rate after 28 days compared to the standard of care. Moreover, the patients who received HCQ had a longer hospital stay associated with a higher risk of undergoing mechanical ventilation [166]. Additionally, HCQ provided no clinical benefit in 11,000 hospitalized patients with COVID-19 in phase 2/3 clinical trials (NCT04381936) across the United Kingdom [167]. Among the 504 COVID-19 hospitalized patients, HCQ, as well as HCQ/AZM regiments, did not improve the clinical outcomes after 15 days in an RCT (NCT04322123) performed in Brazil. In this study, patients treated with HCQ/AZM (39.3 %) or HCQ alone (33.7 %) presented adverse events, whereas the standard of care group showed 22.6 % of adverse outcomes [168]. In another RCT (NCT04321278) conducted in 57 centers in Brazil, the patients treated with AZM/HCQ presented adverse events with no special improvement regarding healing [169]. Similarly, The European discovery trial (NCT04315948) and the World Health Organization (WHO)/Solidarity trial have stopped their clinical trials, declaring that the CQ/HCQ did not reduce the death rate in hospitalized COVID-19 patients compared to the standard care [170,171]. Based on scientific data associated with serious adverse effects, the US Food and Drug Administration (US FDA) revoked the authorization for emergency use of CQ and HCQ, stating that these two drugs are not effective in treating COVID-19 [172]. Similarly, the US National Institutes of Health (NIH), Agenzia Italiana del Farmaco (AIFA), European Medicines Agency (EMA), and Infectious Disease Society of America (IDSA) did not recommend the use of CQ/HCQ or CQ/HCQ/AZM for COVID-19 treatment in hospitalized and non-hospitalized patients [[173], [174], [175]]. Among the adverse effects associated with CQ/HCQ and AZM, the QT prolongation was the most common [[176], [177], [178]]. It was recorded in several countries on COVID-19 patients treated with HCQ/AZM combination with prolongations exceeding 500 ms in some studies [158,176]. The rate of HCQ/AZM treated patients presenting QT prolongation ranges from 0.77 % to 61 %, according to some studies [158,176,179]. The polymorphic ventricular tachycardia or fibrillation, acute kidney failure, and resuscitated cardiac arrest were also recorded in the patients under HCQ/AZM treatment [169,177]. In a randomized, double-blind, and phase 2b study (NCT04323527), a high dose of CQ (600 mg x 2/day for 10 days) was associated with an increased risk of the prolonged QT interval and mortality in severe COVID-19 patients, mainly when used in combination with AZM compared to low dose (450 mg x 2/1day followed by 450 mg x 1/day for 4 days) [180]. A reduced dose of CQ/HCQ results in a normal QT interval [178].
Overall, elevated doses of CQ/HCQ or their combination with AZM are associated with serious adverse effects and do not provide post-exposure prophylaxis nor reduce the infection rate. However, at this time, no significant antiviral results regarding the impacts of CQ/HCQ on the first viral phase of COVID-19 have been reported. In contrast, the inhibition of IFNs production, an essential pivot of host defense against SARS-CoV-2, by CQ/HCQ may exclude the use of the latter in the viral phase of COVID-19. Besides, CQ/HCQ administration may not control the entire inflammatory process (i.e., cytokine storm) in the COVID-19 course compared to other anti-inflammatory drugs [181,182].
4. Vaccine for coronaviruses
Effective SARS-CoV-2 vaccines are required to control the COVID-19 pandemic [183]. Presently, no vaccine is approved to prevent the SARS-CoV-2 infection. It was shown that coronavirus S protein, that binds to ACE2 during the infection process, is highly conserved between SARS-CoV-2, SARS-CoV, and MERS-CoV, indicating this protein a good vaccination target [184]. It was shown that SARS-CoV-2 infection induces the production of protective antibodies [185]. Two monkeys were protected from re-infection during the convalescence period after recovering from SARS-CoV-2 infection [186]. In a study carried out in Shanghai on 175 COVID-19 patients, 30 % showed a low titer of neutralizing antibodies (NAbs), whereas undetectable titers were registered in 10 patients [187]. These results demonstrate that a robust antibody response necessary for lasting the immunity is not systematically induced by mild COVID-19. Epidemiological analyses have projected that infections with other coronaviruses causing mild cold symptoms only confer immunity for approximately one year [188]. The NAb titers decreased 2–3 years post-infection in individuals infected with SARS-CoV or MERS-CoV [189,190], indicating that immunity may not last a lifetime even with more serious coronaviral infections. Sub-neutralizing or non-NAbs against certain epitopes of SARS-CoV may induce the Antibody Disease Enhancement (ADE), which results in negating the main objective of vaccination. These antibodies can improve the entry of the virus into the human cells either by interacting with the conformational epitopes in the ACE2-binding domain [191] or via the Fcγ II receptor [192]. Although the in vivo significance of these results remains to be determined, we can add the ADE to the list of concerns that need to be addressed in the COVID-19 vaccine development. Three methods have been proposed to alleviate the harmful effects of ADE, including the shielding of the non-neutralizing epitopes of the S proteins, immunofocusing marking only the critical neutralizing epitope, and removing the epitope sequences which promote the ADE [193,194]. The protein sequences responsible for ADE have been recognized at S597-603 of the SARS-CoV S protein, a section that is also preserved in SARS-CoV-2. Therefore, the COVID-19 vaccines could be developed to decrease the ADE via the removal of the epitope [193,194]. It was shown that the COVID-19 vaccine comprising residues of S-RBD319-545 (S-Receptor-Binding Domain) induce a strong protective antibody immune response in non-human primates, rabbits, and mice [185]. ADE is of clinical impact in some viral infections, including MERS-CoV, SARS-CoV [195,196], influenza [197], Dengue virus [198,199], RSV [200], HIV [201,202], and Ebola virus (EBV) [197,203]. The vaccine PiCoVacc Sinovac Biotech candidate was administered to non-human primates, rats, and mice [204], where the immune system induced in animals was able to combat SARS-CoV-2 strains without observable ADE of infection. Similarly, partial or whole protection in animals against SARS-CoV-2 was achieved after three immunizations of two doses (3 μg or 6 μg per dose) each. Recently, Chinese macaques infected with SARS-CoV after being vaccinated with a modified vaccinia virus Ankara (MVA) coding for the glycoprotein SARS-CoV S produced a high titer of antibodies associated with reduced viral load [205]. However, these macaques showed severe acute lung damage associated with diffuse alveolar damage. Immunization of mice with an inactivated SARS-CoV with or without adjuvant or a recombinant baculovirus expressing an S protein or particles of coronavirus (VLP) reduced the occurrence of morbidity and mortality related to the virus by decreasing the viral load [206,207]. However, histopathological findings revealed the type 2 inflammatory response at the pulmonary level characterized by the eosinophilic pulmonary infiltrates. The vaccine against the RSV [208] and recombinant vaccinia virus (VV) [209] expressing the SARS-CoV S and N proteins have also shown pneumonia or pulmonary manifestations in pediatric subjects and mice, respectively. Similarly, the inactive MERS-CoV vaccine candidate has also induced NAbs in mice associated with type 2 pulmonary pathology as depicted by the eosinophilic infiltration and elevated concentrations of IL-13 and IL-5 [210]. Interestingly, TLR agonists [211] or Inulin Delta [212] could be used as an adjuvant to improve the healing of type 2 lung disease. An immunization study involving ferrets, naturally susceptible to SARS-CoV, revealed that the recombinant vaccinia virus Ankara (rMVA) expressing the SARS-CoV S induces a rapid and high titer of NAbs. However, the rMVA-S immunized ferrets developed severe hepatitis [213]. Several attempts to develop vaccines against the coronavirus (FIPV) which causes feline infectious peritonitis (FIP) in cats have failed. In fact, the vaccination induced low levels of NAbs, and the immune response induced by the FIPV vaccine was turned out harmful to the animals [214,215]. When the FIPV virus was injected into animals, the mortality of the treated group was further elevated compared to the control group [214]. The hypothesis was that the virus itself, in the presence of antibodies and macrophages, induced the formation of immune complexes that led to the mortality in immunized animals [216]. These data, on previous vaccine experiments with coronaviruses, suggest that the histopathological pulmonary and clinical evaluation should be carried out during the preclinical development of SARS-Cov2 vaccines.
The vaccine approach often chosen is the induction of antibody-based immunity [217]. Unfortunately, the immune system cannot be reduced to antibody-based immunity alone but works in a coordinated and complementary manner, especially concerning the antiviral immunity [218]. Furthermore, the main difficulty in implementing a candidate vaccine against the viral infections is the ability of a vaccine to produce a high level of NAbs, hence the importance of acting on innate immunity, then on cell-mediated immunity as a vaccine supplement. Additionally, the problem of setting up a good design comes mainly from the extreme genetic variability of viruses. HIV is the perfect example, where during the progression of natural infection, only 1% of the individuals produced NAbs which can neutralize most HIV-1 subtypes [219,220]. Genetic analyses of 86 SARS-CoV-2 genomes have provided evidence of genetic diversity by revealing numerous mutations and deletions on coding and non-coding regions [221]. A study reported that 198 sites in the SARS-CoV-2 genome seem to have already undergone mutations [222], but the genetic variability of SARS-CoV remains limited compared to other RNA viruses such as HIV [223]. SARS-CoV-2 displays, like other RNA viruses, a relatively high potential of mutation that can cause the evolution of potential epitopes. Additionally, a mutation in SARS-CoV-2 spike protein such as D614 G may increase its infectivity and make the vaccine less effective [224]. In parallel, the B cells, which produce antibodies, are also subject to the phenomena of somatic mutations at the origin of antibody variants exhibiting increasingly optimized activities [225]. There is, therefore, competition between the immune system and the virus. Thus, after several years of infection, this phenomenon of optimization of recognition sometimes leads to the appearance of more active antibodies capable of accessing very conserved areas of the virus [225]. This results in a broad-spectrum neutralization capacity, effective against many viral isolates; called broadly NAbs, or bNAbs.
Numerous COVID-19 vaccine technologies are in the process of development, including the DNA, RNA, protein subunit, live attenuated virus, inactivated virus, virus-like particles, non-replicating, and replicating viral vectors vaccines [226]. To date, 201 COVID-19 candidate vaccines have been listed by WHO landscape summary, out of which 45 are under clinical evaluation, including 21 in phase 1 (safety and dosage evaluation), 12 in phase 1/2, 2 in phase 2 (safety, immunogenicity and immunization process evaluation), 1 in phase 2/3, and 9 in phase 3 (safety and effectiveness evaluation) clinical trials (Table 2 ) and 156 in preclinical evaluation (efficacy and safety) [226]. A preliminary report of phase 1 clinical trial (NCT04283461) showed that candidate vaccine mRNA-1273 induced an immune response in 45 healthy participants (18–55 years old) with elevated levels of antibodies against the SARS-CoV-2 on day 28 without significant local or systemic adverse events [227]. The induction of antibodies was dose-dependent, and the titers of NAbs increased after the second vaccination. Moreover, this candidate vaccine induced T cells response with more T helper (Th) 1 cytokine (IFN-γ, TNF-α, IL-2) than the Th2 (IL-4, IL-13). The Th1 cell is often involved in antiviral immune responses by inducing the cell-mediated immunity, while the primary role of Th2 is to initiate an antibody response cooperating with B cell [228]. The Th1/Th2 cytokine profile constitutes an essential basis for the development of viral vaccines [228]. In the final study comprised of 40 aged adults (≥ 56 years old), the vaccine mRNA-1273 induced the NAbs and T cell immune response [229]. The side effects such as headache, pain at injection, chills, and myalgia were dose-dependent and increased after the second vaccination. In phase 1/2 clinical trial (NCT04324606), the ChAdOx1-S candidate vaccine induced a strong antibodies response in 1077 participants on day 28 and cellular immune response on day 14 [230]. All participants showed production of higher NAbs after receiving a second dose of the vaccine. The side effects such as headache, malaise, fever, chills, muscle ache, and pain were reduced by administering the prophylactic paracetamol to participants. Similarly, the Ad5 vectored COVID-19 candidate vaccine induced a robust NAbs response at day 28 and specific T cellular response at day 14 in most of the 108 participants included in the phase 1 clinical trial (NCT04313127) [231]. The side effects were associated with a higher dose of vaccine. In the phase 2 trial (NCT04341389), the Ad5 vectored COVID-19 candidate vaccine induced a strong immune response in the majority of 508 participants without side effects [232]. Moreover, the COVID-19 RNA vaccine BNT162b1 in phase 1/2 (NCT04368728) induced a strong NAbs in 45 participants. The NAbs titers were significantly higher than those of a panel of COVID-19 convalescent human plasma [233]. Most participants developed a Th1 response associated with IFN-γ production without severe adverse events. The robust NAbs associated with favorable IFN response is beneficial for protection against COVID-19 [234]. In two RCTs comprised of 96 adults in phase 1 and 224 adults in phase 2 (ChiCTR2000031809), the interim analysis of the inactivated vaccine against SARS-CoV-2 from Wuhan Institute of Biological Products/Sinopharm reported that the vaccine induced a NAbs response in all participants after 14 days without severe adverse reactions [235]. The NAbs increased significantly after the second and third immunization suggesting the necessity for at least two immunizations. Furthermore, analysis from two open and non-randomized phase 1/2 (NCT04436471/NCT04437875) trials showed that the rAd26 and rAd5 vector-based vaccine induced a strong NAbs at day 14 and cell-mediated immunity with favorable median CD4+ and CD8 + T cells proliferation associated with IFN-γ production at day 28 in 76 participants. The minor adverse events such as asthenia (21 %), headache (42 %), hyperthermia (50 %), and pain at the injection site (24 %) were reported [236]. In this trial, participants who received the vaccine had the same titer of NAbs against SARS-CoV-2 as recovering COVID-19 patients suggesting a capability of natural SARS-CoV-2 infection to induce NAbs. In phase 1/2 trial (NCT04368988), 83 healthy participants received the rSARS-CoV-2 vaccine with adjuvant (Matrix-M), 25 without Matrix-M, and 23 healthy participants received placebo [237]. The rSARS-CoV-2 vaccine adjuvanted with Matrix M1 (NVX-CoV2373) induced strong antibody and Th1 immune response compared to the vaccine without adjuvant. The titers of NAbs induced by the adjuvanted dose was significantly higher than those of convalescent human plasma. More Th1 cytokines (IFN-γ, TNF-α, IL-2) were induced than Th2 (IL-5, IL-13), suggesting the importance of cell-mediated immunity in viral clearance. All participants reported no severe adverse events. At this stage of clinical trials, the direct comparisons between different candidate vaccines should be interpreted with caution due to the small size of the trials, the different doses of vaccine used, and the variety of analytical methods.
Table 2.
Vaccines under clinical evaluation for COVID-19 according to the WHO as of October 29, 2020. The information provided here are based on the analysis from official website of WHO [226].
| Platform | Candidate vaccine (Developer) |
Current stage (participants) |
Status (completion date) |
Subject | Study location |
|---|---|---|---|---|---|
| Inactivated | Inactivated + alum (Sinovac) |
Phase 3 NCT04456595 (8,870) Phase 1/2 NCT04383574 (422) NCT04352608 (744) |
Recruiting (October 2021) Recruiting (December 2020) |
18−59 years 18−59 years |
Brazil China |
| Inactivated (Wuhan Institute of Biological Products/Sinopharm) |
Phase 3 ChiCTR2000034780 (15,000) Phase 1/2 ChiCTR2000031809 809 (1,120) |
Recruiting (July 2021) Completed |
≥18 years 18−59 years |
United Arab Emirates China |
|
| Inactivated (Beijing Institute of Biological Products/Sinopharm) |
Phase 3 ChiCTR2000034780 (15,000) Phase 1/2 ChiCTR2000032 459 (1,904) |
Recruiting (July 2021) Recruiting (-) |
≥ 18 years ≥ 3 years |
China China |
|
| Inactivated (Research Institute for Biological Safety Problems, Rep of Kazakhstan | Phase 1/2 NCT04530357 (244) |
Recruiting (April 2021) |
18−100 years |
Kazakhstan | |
| Inactivated (Beijing M Inhai Biotechnology Co., Ltd.,) |
Phase 1 ChiCTR2000038804 (180) |
Not yet recruiting (-) |
≥ 18 years | China | |
| Inactivated (Institute of Medical Biology Chinese Academy of Medical Sciences) |
Phase 1/2 NCT04412538 (942) NCT04470609 (471) |
Recruiting (September 2021) (November 2021) |
18−59 years ≥ 60 years |
China China |
|
| Whole-Virion inactivated (Bharat Biotech) | Phase 1/2 NCT04471519 (755) |
Active, not recruit ing (June 2021) |
12−65 years |
India | |
| Non-replicating viral |
ChAdOx1-S (University of Oxford/ Astrazeneca) |
Phase 3 ISRCTN8995424 (2000) Phase 2b/3 2020-001228-32 (-) Phase 1/2 PACTR20200692216 5132 (2000) NCT04324606 (1090) |
Recruiting (July 2021) Recruiting (-) Recruiting (December 2021) Completed |
18−55 years ≥ 18 years 18−65 years with or wit hout HIV 18−55 years |
Brazil UK South Africa UK |
| Adenovirus Type 5 Vector (Cansino Biological Inc./Beijing Institute of Biotechnology) |
Phase 3 NCT04526990 (40,000) |
Recruiting (January 2022) |
≥ 18 years | Pakistan | |
| Phase 2 ChiCTR2000031781 (500) Phase 1 ChiCTR2000030906 (108) |
Completed Completed |
≥ 18 years 18−60 years |
China China |
||
| Replication defective Simian Adenovirus (GRAd) encoding S (ReiThera/LEUKOCA RE/Univercells) |
Phase 1 NCT04528641 (90) |
Recruiting (July 2021) |
18−85 years |
Italy | |
| Ad5-nCoV(Institute of Biotechnology, Academy of Military Medical Sciences, PLA of China |
Phase 1 NCT04552366 (144) |
Recruiting (June 2021) |
≥ 18 years | China | |
| Adeno-based (Gamaleya Research institute) |
Phase 3 NCT04530396 (40,000) |
Recruiting (May 2021) |
≥ 18 years | Russia | |
| Phase 1/2 NCT04436471 NCT04437875 (38) |
Active, not recruit ing (August 2020) |
18−60 years |
Russia | ||
| Ad26COVS1(Janssen Pharmaceutical Companies) |
Phase 3 NCT04505722 (60,000) |
Recruiting (March 2023) |
≥ 18 years | USA, Peru Argentina Brazil, Chile Mexico, South Africa, Columbia |
|
| Phase 1/2 NCT04436276 (1045) |
Recruiting (November 2023) |
≥ 18 years | USA Belgium |
||
| Ad5 adjuvanted Oral Vaccine platform (Vaxart ) |
Phase 1 NCT04563702 (48) |
Recruiting (October 2021) |
18−54 years |
USA | |
| MVA-SARS-2-S (Ludwing-Maximillians- -University of Munich) |
Phase1 NCT04569383 (30) |
Not yet recruiting (May 2021) | 18−55 years |
Germany | |
| RNA | LNP-encapsulated mR NA (Moderna/ NIAID) |
Phase 3 NCT04470427 (30,000) Phase 2 NCT04405076 (600) Phase 1 NCT04283461 (155) |
Recruiting (October 2022) Active, not recruit ing (August 2021) Completed |
≥ 18 years ≥ 18 years 18−99 years |
USA USA USA |
| 3LNP-mRNAs (BioNTech/Fosun Pharma/Pfizer) |
Phase 2/3 NCT04368728 (43,998) Phase 1/2 2020-001038-36 (444) Phase 1 ChiCTR2000034825 (144) |
Recruiting (November 2022) Recruiting (-) Recruiting (December 2020) |
18−85 Years ≥ 18 years ≥ 18 years |
USA Argentina Brazil Germany China |
|
| LNP-nCoVsaRNA (Imperial College London ) |
Phase 1 ISRCTN17072692 (320) |
Recruiting (July 2021) |
18−75 years |
UK | |
| mRNA (Curevac) | Phase 2 NCT04515 147 (691) |
Not yet recruiting (November 2021) |
≥ 18 years | Germany Belgium |
|
| Phase 1 NCT04449276 (168) |
Recruiting (August 2021) |
18−60 years |
Germany Belgium |
||
| mRNA (People’s Liberation Army (PLA) Academy of Military Sciences/Walvax Biotech) |
Phase 1 ChiCTR2000034112 (168) |
Not yet recruiting (December 2021) |
18−80 years |
China | |
| mRNA(Arcturus/Duke -Nus) |
Phase 1/2 NCT04480957 (92) |
Recruiting (January 2021) |
21−80 years |
Singapore | |
| DNA | DNA plasmid vaccine with electroporation (Inovio Pharmaceuticals/International Vaccine Institute) |
Phase 1/2 NCT04447781(160) Phase 1 NCT04336410 (120) |
Recruiting (February 2022) Recruting (July 2021) |
19−64 Years ≥ 18 years |
Republic of Korea USA |
| DNA plasmid + Adjuvant (Osaka University/AnGes/TakaraBio) | Phase 1/2 NCT04463472 (30) |
Recruiting (July 2021) |
20−55 years |
Japan | |
| DNA plasmid vaccine (Cadila Healthcare Limited) |
Phase 1/2 CTRI/2020/07/0 263552 (1048) |
Recruiting (July 2021) |
18−55 years |
India | |
| DNAVaccine (GX-19) (Genexine Consortium ) |
Phase 1 NCT04445389 (210) |
Recruiting (June 2022) |
18−50 years |
Republic of Korea |
|
| Protein Subunit |
Full length recombinant SARS-CoV-2 glycoprotein nanoparticle vaccine adjuvanted with Matrix M (Novavax) |
Phase 3 (2020-004123-16) (-) Phase 2b NCT04533399 (2,900) Phase 1/2 NCT04368988 (131) |
Recruiting (-) Recruiting (November 2021) Recruiting (July 2021) |
≥ 18 years 18−64 Years 18−59 years |
UK South Africa USA |
| Native-like Trimeric subunit spike protein vaccine (Clover Bioph Pharmaceuticals Inc. /GSK/Dynavax) |
Phase 1 NCT0440590 (150) |
Recruiting (March 2021) |
18−75 years |
Australia | |
| RBD (Baculovirus production expressed in Sf9 cells) (West China Hospital, Sichuan University |
Phase 1 ChiCTR2000037518 (168) |
Recruiting (August 2021) |
≥ 18 years | China | |
| Adjuvanted recombinat protein (RBD Di mer) (Anhui Zhifei Longcom Biopharmaceutical /Institute of Microbiology , Chinese Academy of Sciences) |
Phase 2 NCT04466085 (900) Phase 1 NCT04445194 (50) |
Recruiting (December 2021) Recruiting (September 2021) |
18−59 Years 18−59 years |
China China |
|
| S protein (Baculovirus Production) Sanofi Pasteur/GSK |
Phase 1/2 NCT04537208 (440) |
Recruiting (October 2021) |
≥ 18 years | USA | |
| Recombinant spike protein with AdvaxTM Adjuvant (Vaxine Pty Ltd./Medytox) |
Phase 1 NCT04453852 (40) |
Recruiting (July 2021) |
18−65 years |
Australia | |
| SARS-CoV-2 HLA- DR peptides (University Hospital Tuebingen) |
Phase 1 NCT04546841 (36) |
Not yet recruiting (December 2021) |
≥ 18 years | Germany | |
| Molecular clamp stabilized spike protein (University of Queensland /GSK/Dynavax) |
Phase 1 ACTRN1262000067 4932P (120) |
Recruiting (-) |
18−55 years |
Australia | |
| RBD-based (Kentucky Bioprocessing, Inc) |
Phase 1/2 NCT04473690 (180) |
Not yet recruiting (November 2021) |
18−70 years |
USA | |
| S1-RBD-protein (COVAXX) | Phase 1 NCT04545749 (60) |
Recruiting (August 2021) |
20−55 years |
Taiwan | |
| S-2 P protein + CpG101 8(Medigen Vaccine Biologics corporation/NIAID /Dynavax |
Phase 1 NCT04487210 (45) |
Not yet recruiting (December 2021) |
20−50 years |
Taiwan | |
| RBD + Adjuvant (Instituto Finaly de Vacunas, Cuba |
Phase 1/2 IFV/COR/04 (676) |
Recruiting (Febuary 2021) |
19−80 years |
Cuba | |
| Peptide (FBRI SRC VB B VECTOR, Rospotrebnadzor , Koltsovo) |
Phase 1/2 NCT04527575 (100) |
Active, not recruit Ing (October 2020) |
18−60 Years |
Russia | |
| VLP | Plant-derived VLP (Medicago Inc. ./Universite’ Lava) |
Phase 1 NCT04450004 (180) |
Recruiting (April 2021) |
18−55 years |
Canada |
| RBD-HBsAg VLPs (SpyBiotech/serum Institute of India |
Phase 1/2 ACTRN126200000 817943 (280) |
Recruiting (-) |
18−79 years |
Australia | |
| Replicating viral vector | Intranasal flu-based- RBD (Beijing Wantai Biological Pharmacy/ Xiamen University |
Phase 1 ChiCTR2000037782 (60) |
Not yet recruiting (October 2021) |
≥ 18 years | China |
| Meascles-vector based (Institute Pasteur/Thermis/University of Pittsburg CVR/Merck Sharp & Dohme) |
Phase 1 NCT04497298 (90) |
Recruiting (October 2021) |
18−55 years |
France Belgium |
Overall, most of the COVID-19 vaccines with the published report have induced the production of NAbs and T cell immune response after the activation of SARS-CoV-2-specific CD4+ and CD8 + T cells. The CD4 + T cell is at the center of the initiation of the adaptive immune response, which can be either an antibody immune response or a cellular response or both responses depending on the type of pathogen [238]. The production of antibodies is an essential part of the immune response following the infection or vaccination. The antibodies are produced by the B cells and are specific for the antigens that trigger the immune response. The term neutralization relates to the formation of the immune complex which prevents the biological activity of the antigen [239]. This neutralization function is the main correlate of protection of many vaccines and convalescent plasma. The antibodies are also able to interact with many cells of the immune system through the receptors which recognize the constant region of immunoglobulins called the fragment crystallizable (Fc). These Fc receptors (FcR) are necessary for the proper functioning of the humoral response, for example for the clearance of immune complexes. Different types of cells of the immune system express these FcRs, allowing the induction of effector functions complementary to neutralization [240]. Among these, the antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent phagocytosis (ADP) target the cells or the antigenic elements that have been recognized and are covered by antibodies and lead to their destruction (Fig. 5 ) [240]. The cellular effectors of ADCC are mainly NK (natural killer) cells, which express the CD16 molecule (or FcγRIII), an FcR that recognizes the isotype G immunoglobulin. Moreover, monocytes, neutrophils, and macrophages are also involved in ADCC.
Fig. 5.
Schematic representation of ADCC and ADP induced by neutralizing antibodies (NAbs). A cell infected with SARS-CoV-2 expresses viral antigens on its surface, that allows certain NAbs to attach to the plasma membrane. The NK (natural killer) cells (A) and phagocytes (B) are then activated via the receptor for the Fc fragment of immunoglobulins. The NK cells then release cytolytic granules (granzymes and perforins), and phagocytes internalize the virus-infected cell in the phagolysosome. Both cytolytic granules and phagolysosome destroy the infected cell recognized by the NAbs.
ADCC, antibody-dependent cellular cytotoxicity; ADP, antibody-dependent phagocytosis (ADP).
The CD8 + T cells are involved in the adaptive cell-mediated immune response, which is an effective immune response against viruses and other intracellular parasites [241]. The effective control of viral infection requires the elimination of the infected cells in order to limit the production and the propagation of the virus as well as the establishment of an immune memory specifically directed against the viral antigens. The latter relies on the expansion of cytotoxic CD8 + T lymphocytes (CTL) [242]. In SARS-CoV survivors, the CD8 + T cells conferred lasting immune memory that persisted up to 11 years [243]. The candidate vaccine such as COVID-19 RNA vaccine BNT162b induced a robust release of IFN-γ, which participate in several antiviral responses in particular by inhibiting the replication of SARS-COV-2. The robust CD8+ producing IFN-γ with antiviral properties could complement the strong response of NAbs. Furthermore, the SARS-COV-2 specific cellular immune response could be effective in controlling the COVID-19, even in the absence of NAbs [244]. Therefore, the vaccine inducing strong NAbs and cell-mediated immune response could be effective in controlling the COVID-19 pandemic.
The BCG (Bacillus Calmette–Guérin) is an example of a vaccine that induces innate immunity [245]. It is a live vaccine obtained from the attenuation of Mycobacterium bovis strain that is injected into the infants usually in the first year after their birth [245]. The BCG vaccine has other long-standing effects, including the protection against other types of respiratory infections such as RSV infection [[246], [247], [248]]. BCG, as a live vaccine, induces the expression of genes which are involved in the innate immunity (trained immunity) as well as long-term cellular immunity [249,250]. The BCG vaccine offers protection up to 60 years of immunization, and therefore the old population must be revaccinated [249]. This very broad spectrum immunity presents a form of memory which would somehow be boosted by these live vaccines since they are real pathogens, even weakened. This practice would help the body to better regulate its overall immune response [250]. COVID-19, in the last phase of the infection, disrupts the inflammatory system (cytokine storm), which ends up in catastrophic chain reactions in the patient [5,6]. However, the aforementioned trained innate immunity would prevent such unwanted chain reactions, and therefore partly avoid the severe cases of the disease. A recent publication shows that the nationwide differences in the impact of COVID-19 could be explained, in part, by the national policies concerning the vaccination of children with BCG [251]. Thus, countries without a BCG vaccination policy (United States, Italy, the Netherlands) were more seriously affected than those with long-standing universal BCG policies. In European countries, each 10 % increase in the BCG index was associated with a 10.4 % drop in the mortality from COVID-19 [252]. Clinical trials are underway in the Netherlands (NCT04328441) [253], South Africa (NCT04379336) [254], and Australia (NCT04327206) [255] to confirm this association.
5. Concluding remarks
More effective COVID-19 patient management protocols to ensure the reduced need for prolonged mechanical ventilation, short hospital stays, and to reduce the morbidity and mortality associated with COVID-19 are still lacking. Despite the controversial results of CQ/HCQ and AZM in the treatment of COVID-19 in recent days, several countries continue to allow their use in the treatment of COVID-19 patients. However, the use of CQ/HCQ and AZM should remain restrained because the results of new studies do not validate these treatments in the management of COVID-19. Nevertheless, in the situation of a supposed bacterial super-infection that can complicate the COVID-19, clinicians may wish to prescribe AZM. The lung injury in COVID-19 patients is associated with ROS release by phagocytes, and thus the use of antioxidants is necessary for the management of COVID-19. Besides, type I and type III IFNs have shown antiviral activities both in vitro and in vivo. Type I IFN was effective in treating COVID-19 patients than other forms of treatment without IFN; thus, we hypothesize that the use of type I and III IFNs in the early stage of COVID-19 combined with an adapted antioxidant and anti-inflammatory drug in the advanced stage could help improve the clinical outcomes. The vaccine remains an efficient agent for COVID-19 protection, and the preliminary reports of the candidate vaccines showcase some encouraging results.
Author contribution
The manuscript was written through the contribution of all authors.
Funding Source
This work was supported by the National Natural Science Foundation of China (Grant No. 21774039) and the National Key Research and Development Program of China (2018YFE0123700).
Declaration of Competing Interest
The authors declare that there is no conflict of interest associated with the publication of this manuscript.
References
- 1.2020. Coronavirus Cases.https://www.worldometers.info/coronavirus/ (accessed October 31, 2020) [Google Scholar]
- 2.Salvi R., Patankar P. Emerging pharmacotherapies for COVID-19. Biomed. Pharmacother. 2020;128:110267. doi: 10.1016/j.biopha.2020.110267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez M.A. Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob. Agents Chemother. 2020;64:1–7. doi: 10.1128/AAC.00399-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colson P., Rolain J.M., Lagier J.C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao M. Cytokine storm and immunomodulatory therapy in COVID-19: role of chloroquine and anti-IL-6 monoclonal antibodies. Int. J. Antimicrob. Agents. 2020;55:105982. doi: 10.1016/j.ijantimicag.2020.105982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jamilloux Y., Henry T., Belot A., Viel S., Fauter M., El Jammal T., Walzer T., François B., Sève P. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020;19:102567. doi: 10.1016/j.autrev.2020.102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu T., Luo S., Libby P., Shi G.-P. Cathepsin L-selective inhibitors: a potentially promising treatment for COVID-19 patients. Pharmacol. Ther. 2020;213:107587. doi: 10.1016/j.pharmthera.2020.107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.US National Library of Medicine . 2020. Camostat Mesylate in COVID-19 Outpatients, ClinicalTrials.Gov.https://clinicaltrials.gov/ct2/show/NCT04353284 (accessed September 28, 2020) [Google Scholar]
- 9.Ullah M.W., Manan S., Guo Z., Yang G. Therapeutic options for treating COVID-19. Eng. Sci. 2020 doi: 10.30919/es8d765. [DOI] [Google Scholar]
- 10.Luo W., Li Y.-X., Jiang L.-J., Chen Q., Wang T., Ye D.-W. Targeting JAK-STAT signaling to control cytokine release syndrome in COVID-19. Trends Pharmacol. Sci. 2020;41:531–543. doi: 10.1016/j.tips.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today’s diseases? Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keyaerts E., Li S., Vijgen L., Rysman E., Verbeeck J., Van Ranst M., Maes P. Antiviral activity of chloroquine against human coronavirus OC43 infection in newborn mice. Antimicrob. Agents Chemother. 2009;53:3416–3421. doi: 10.1128/AAC.01509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee N., Wong C.K., Chan M.C.W., Yeung E.S.L., Tam W.W.S., Tsang O.T.Y., Choi K.W., Chan P.K.S., Kwok A., Lui G.C.Y., Leung W.S., Yung I.M.H., Wong R.Y.K., Cheung C.S.K., Hui D.S.C. Anti-inflammatory effects of adjunctive macrolide treatment in adults hospitalized with influenza: a randomized controlled trial. Antiviral Res. 2017;144:48–56. doi: 10.1016/j.antiviral.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 14.U.S. FOOD and DRUG Administration . 2020. Coronavirus (COVID-19) Update: FDA Warns of Newly Discovered Potential Drug Interaction That May Reduce Effectiveness of a COVID-19 Treatment Authorized for Emergency Use.https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-warns-newly-discovered-potential-drug-interaction-may-reduce (accessed September 26, 2020) [Google Scholar]
- 15.U.S. FOOD and DRUG Administration . 2020. FDA Issues Emergency Use Authorization for Convalescent Plasma As Potential Promising COVID–19 Treatment, Another Achievement in Administration’s Fight Against Pandemic | FDA.https://www.fda.gov/news-events/press-announcements/fda-issues-emergency-use-authorization-convalescent-plasma-potential-promising-covid-19-treatment (accessed September 26, 2020) [Google Scholar]
- 16.Hossen M.S., Barek M.A., Jahan N., Safiqul Islam M. A review on current repurposing drugs for the treatment of COVID-19: reality and challenges. SN Compr. Clin. Med. 2020 doi: 10.1007/s42399-020-00485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartsch S.M., O’Shea K.J., Ferguson M.C., Bottazzi M.E., Wedlock P.T., Strych U., McKinnell J.A., Siegmund S.S., Cox S.N., Hotez P.J., Lee B.Y. Vaccine efficacy needed for a COVID-19 coronavirus vaccine to prevent or stop an epidemic as the sole intervention. Am. J. Prev. Med. 2020;59:493–503. doi: 10.1016/j.amepre.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Gioia C., Horejsh D., Agrati C., Martini F., Capobianchi M.R., Ippolito G., Poccia F. T-cell response profiling to biological threat agents including the SARS coronavirus. Int. J. Immunopathol. Pharmacol. 2005;18:525–530. doi: 10.1177/039463200501800312. [DOI] [PubMed] [Google Scholar]
- 20.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pene F., Marin N., Roche N., Szwebel T.-A., Smith N., Merkling S., Treluyer J.-M., Veyer D., Mouthon L., Blanc C., Tharaux P.-L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kerneis S., Terrier B. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Brien T.R., Thomas D.L., Jackson S.S., Prokunina-Olsson L., Donnelly R.P., Hartmann R. Weak induction of interferon expression by severe acute respiratory syndrome coronavirus 2 supports clinical trials of Interferon-λ to treat early coronavirus disease 2019. Clini. Infect. Dis. 2020 doi: 10.1093/cid/ciaa453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., TenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020:1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassoun A., Thottacherry E.D., Muklewicz J., Ul ain Aziz Q., Edwards J. Utilizing tocilizumab for the treatment of cytokine release syndrome in COVID-19. J. Clin. Virol. 2020;128:104443. doi: 10.1016/j.jcv.2020.104443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reghunathan R., Jayapal M., Hsu L.Y., Chng H.H., Tai D., Leung B.P., Melendez A.J. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2172. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Channappanavar S., Fehr R., Vijay A.R., Mack R., Zhao M., Meyerholz J., Perlman D.K. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-Infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin F., Young H.A. Interferons : success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 2014;25:369–376. doi: 10.1016/j.cytogfr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vicenzi E., Poli G. The interferon-stimulated gene TRIM22: a double-edged sword in HIV-1 infection. Cytokine Growth Factor Rev. 2018;40:40–47. doi: 10.1016/j.cytogfr.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y.-J. IPC: Professional Type 1 Interferon-Producing Cells and Plasmacytoid Dendritic Cell Precursors. Annu. Rev. Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 32.Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye L., Schnepf D., Staeheli P. Interferon-λ orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 2019;19:614–625. doi: 10.1038/s41577-019-0182-z. [DOI] [PubMed] [Google Scholar]
- 35.Lazear H.M., Schoggins J.W., Diamond M.S. Shared and distinct functions of type I and type III interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Betancor G., Dicks M.D.J., Jimenez-Guardeño J.M., Ali N.H., Apolonia L., Malim M.H. The GTPase domain of MX2 interacts with the HIV-1 capsid, enabling its short isoform to moderate antiviral restriction. Cell Rep. 2019;29:1923–1933. doi: 10.1016/j.celrep.2019.10.009. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suddala K.C., Lee C.C., Meraner P., Marin M., Markosyan R.M., Desai T.M., Cohen F.S., Brass A.L., Melikyan G.B. Interferon-induced transmembrane protein 3 blocks fusion of sensitive but not resistant viruses by partitioning into virus-carrying endosomes. PLoS Pathog. 2019;15:6347298. doi: 10.1371/journal.ppat.1007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrensch F., Winkler M., Pöhlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6:3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., Longobardi L.E., Boltz D., Kuhn J.H., Elledge S.J., Bavari S., Denison M.R., Choe H., Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skaug B., Chen Z.J. Emerging role of ISG15 in antiviral immunity. Cell. 2010;143:187–190. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Tortorec A., Willey S., Neil S.J.D. Antiviral inhibition of enveloped virus release by tetherin/BST-2: action and counteraction. Viruses. 2011;3:520–540. doi: 10.3390/v3050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knolle P.A., Kremp S., Hohler T., Krummenauer F., Schirmacher P., Gerken G. Viral and host factors in the prediction of response to interferon-alpha therapy in chronic hepatitis C after long-term follow-up. J. Viral Hepat. 1998;5:399–406. doi: 10.1046/j.1365-2893.1998.00127.x. [DOI] [PubMed] [Google Scholar]
- 44.Agrawal A. Mechanisms and implications of age-associated impaired innate interferon secretion by dendritic cells: a mini-review. Gerontology. 2013;59:421–426. doi: 10.1159/000350536. [DOI] [PubMed] [Google Scholar]
- 45.Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manns M.P., Wedemeyer H., Cornberg M. Treating viral hepatitis C: efficacy, side effects, and complications. Gut. 2006;55:1350–1359. doi: 10.1136/gut.2005.076646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S.F., Gong M.J., Zhao F.R., Shao J.J., Xie Y.L., Zhang Y.G., Chang H.Y. Type i interferons: distinct biological activities and current applications for viral infection. Cell. Physiol. Biochem. 2018;51:2377–2396. doi: 10.1159/000495897. [DOI] [PubMed] [Google Scholar]
- 48.Phillips S., Mistry S., Riva A., Cooksley H., Hadzhiolova-Lebeau T., Plavova S., Katzarov K., Simonova M., Zeuzem S., Woffendin C., Chen P.J., Peng C.Y., Chang T.T., Lueth S., De Knegt R., Choi M.S., Wedemeyer H., Dao M., Kim C.W., Chu H.C., Wind-Rotolo M., Williams R., Cooney E., Chokshi S. Peg-interferon lambda treatment induces robust innate and adaptive immunity in chronic hepatitis B patients. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muir A.J., Shiffman M.L., Zaman A., Yoffe B., De La Torre A., Flamm S., Gordon S.C., Marotta P., Vierling J.M., Lopez-Talavera J.C., Byrnes-Blake K., Fontana D., Freeman J., Gray T., Hausman D., Hunder N.N., Lawitz E. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010;52:822–832. doi: 10.1002/hep.23743. [DOI] [PubMed] [Google Scholar]
- 50.Makjaroen J., Somparn P., Hodge K., Poomipak W., Hirankarn N., Pisitkun T. Comprehensive proteomics identification of IFNλ3-regulated antiviral proteins in HBV-transfected cells. Mol. Cell Proteomics. 2018;17:2197–2215. doi: 10.1074/mcp.RA118.000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.García-Sastre A. Ten strategies of interferon evasion by viruses. Cell Host Microbe. 2017;22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim Y., Ng Y., Tam J., Liu D. Human Coronaviruses: A Review of Virus–Host Interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., Pia L., Risson E., Saffern M., Salomé B., Esai Selvan M., Spindler M.P., Tan J., van der Heide V., Gregory J.K., Alexandropoulos K., Bhardwaj N., Brown B.D., Greenbaum B., Gümüş Z.H., Homann D., Horowitz A., Kamphorst A.O., Curotto de Lafaille M.A., Mehandru S., Merad M., Samstein R.M., Agrawal M., Aleynick M., Belabed M., Brown M., Casanova-Acebes M., Catalan J., Centa M., Charap A., Chan A., Chen S.T., Chung J., Bozkus C.C., Cody E., Cossarini F., Dalla E., Fernandez N., Grout J., Ruan D.F., Hamon P., Humblin E., Jha D., Kodysh J., Leader A., Lin M., Lindblad K., Lozano-Ojalvo D., Lubitz G., Magen A., Mahmood Z., Martinez-Delgado G., Mateus-Tique J., Meritt E., Moon C., Noel J., O’Donnell T., Ota M., Plitt T., Pothula V., Redes J., Reyes Torres I., Roberto M., Sanchez-Paulete A.R., Shang J., Schanoski A.S., Suprun M., Tran M., Vaninov N., Wilk C.M., Aguirre-Ghiso J., Bogunovic D., Cho J., Faith J., Grasset E., Heeger P., Kenigsberg E., Krammer F., Laserson U. Immunology of COVID-19: current state of the science. Immunity. 2020;1:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z., Shen L., Gu X. Evolutionary dynamics of MERS-CoV: potential recombination, positive selection and transmission. Sci. Rep. 2016;6:25049. doi: 10.1038/srep25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y.F., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., D’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menachery V.D., Eisfeld A.J., Schäfer A., Josset L., Sims A.C., Proll S., Fan S., Li C., Neumann G., Tilton S.C., Chang J., Gralinski L.E., Long C., Green R., Williams C.M., Weiss J., Matzke M.M., Webb-Robertson B.-J., Schepmoes A.A., Shukla A.K., Metz T.O., Smith R.D., Waters K.M., Katze M.G., Kawaoka Y., Baric R.S. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5:e01174–14. doi: 10.1128/mBio.01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Yang J., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890. doi: 10.1016/j.chom.2020.04.017. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., Willey B.M., DeVries M.E., Fang Y., Seneviratne C., Bosinger S.E., Persad D., Wilkinson P., Greller L.D., Somogyi R., Humar A., Keshavjee S., Louie M., Loeb M.B., Brunton J., McGeer A.J., Kelvin D.J. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81:8692–8706. doi: 10.1128/jvi.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim E.S., Choe P.G., Park W.B., Oh H.S., Kim E.J., Nam E.Y., Na S.H., Kim M., Song K.H., Bang J.H., Park S.W., Bin Kim H., Kim N.J., Oh M.D. Clinical progression and cytokine profiles of middle east respiratory syndrome coronavirus infection. J. Korean Med. Sci. 2016;31:1717–1725. doi: 10.3346/jkms.2016.31.11.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lokugamage K.G., Hage A., Schindewolf C., Rajsbaum R., Menachery V.D. SARS-CoV-2 is sensitive to type I interferon pretreatment. bioRxiv. 2020;21:1–9. doi: 10.1101/2020.03.07.982264. [DOI] [Google Scholar]
- 62.Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179:7188648. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W.A.L., Posthuma C.C., van der Meer Y., Bárcena M., Haagmans B.L., Snijder E.J., van den Hoogen B.G. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haagmans B.L., Kuiken T., Martina B.E., Fouchier R.A.M., Rimmelzwaan G.F., Van Amerongen G., Van Riel D., De Jong T., Itamura S., Chan K.H., Tashiro M., Osterhaus A.D.M.E. Pegylated interferon-α protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 2004;10:290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galani I.E., Triantafyllia V., Eleminiadou E.E., Koltsida O., Stavropoulos A., Manioudaki M., Thanos D., Doyle S.E., Kotenko S.V., Thanopoulou K., Andreakos E. Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46:875–890. doi: 10.1016/j.immuni.2017.04.025. e6. [DOI] [PubMed] [Google Scholar]
- 66.Jeon Y.J., Gil C.H., Jo A., Won J., Kim S., Kim H.J. The influence of interferon-lambda on restricting Middle East Respiratory Syndrome Coronavirus replication in the respiratory epithelium. Antivir. Res. 2020;180:104860. doi: 10.1016/j.antiviral.2020.104860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jasper Fuk-Woo Chan K.-Y.Y., Yao Yanfeng, Yeung Man-Lung, Deng Wei, Bao Linlin, Jia Lilong, Li Fengdi, Xiao Chong, Gao Hong, Pin Yu, Cai Jian-Piao, Chu Hin, Zhou Jie, Chen Honglin, Qin Chuan. Treatment with Lopinavir / ritonavir or Interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Falzarano D., De Wit E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P., Brining D., Bushmaker T., Martellaro C., Baseler L., Benecke A.G., Katze M.G., Munster V.J., Feldmann H. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat. Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hart B.J., Dyall J., Postnikova E., Zhou H., Kindrachuk J., Johnson R.F., Olinger G.G., Frieman M.B., Holbrook M.R., Jahrling P.B., Hensley L. Interferon-β and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 2014;95:571–577. doi: 10.1099/vir.0.061911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:41467. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 72.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci. Trends. 2020;14:69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 73.Meng Z., Wang T., Li C., Chen X., Li L., Qin X., Li H., Luo J. An experimental trial of recombinant human interferon alpha nasal drops to prevent coronavirus disease 2019 in medical staff in an epidemic area. medRxiv. 2020 doi: 10.1101/2020.04.11.20061473. 2020.04.11.20061473. [DOI] [Google Scholar]
- 74.Hung I.F.N., Lung K.C., Tso E.Y.K., Liu R., Chung T.W.H., Chu M.Y., Ng Y.Y., Lo J., Chan J., Tam A.R., Shum H.P., Chan V., Wu A.K.L., Sin K.M., Leung W.S., Law W.L., Lung D.C., Sin S., Yeung P., Yip C.C.Y., Zhang R.R., Fung A.Y.F., Yan E.Y.W., Leung K.H., Ip J.D., Chu A.W.H., Chan W.M., Ng A.C.K., Lee R., Fung K., Yeung A., Wu T.C., Chan J.W.M., Yan W.W., Chan W.M., Chan J.F.W., Lie A.K.W., Tsang O.T.Y., Cheng V.C.C., Que T.L., Lau C.S., Chan K.H., To K.K.W., Yuen K.Y. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu P., Huang J., Fan Z., Huang W., Qi M., Lin X., Song W., Yi L. Arbidol/IFN-α2b therapy for patients with corona virus disease 2019: a retrospective multicenter cohort study. Microbes Infect. 2020;22:200–205. doi: 10.1016/j.micinf.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gemcioglu E., Davutoglu M., Ozdemir E.E., Erden A. Are type 1 interferons treatment in Multiple Sclerosis as a potential therapy against COVID-19? Mult. Scler. Relat. Disord. 2020;42:102196. doi: 10.1016/j.msard.2020.102196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.El-Lababidi R.M., Mooty M., Bonilla M.-F., Salem N.M. Treatment of severe pneumonia due to COVID-19 with peginterferon alfa 2a. IDCases. 2020;21:e00837. doi: 10.1016/j.idcr.2020.e00837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dastan F., Nadji S.A., Saffaei A., Marjani M., Moniri A., Jamaati H., Hashemian S.M.R., Baghaei P., Abedini A., Varahram M., Yousefian S., Tabarsi P. Subcutaneous administration of interferon beta-1a for COVID-19: a non-controlled prospective trial. Int. Immunopharmacol. 2020;85:7275997. doi: 10.1016/j.intimp.2020.106688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.https://www.synairgen.com/ (accessed August 1, 2020).
- 80.Kindler E., Thiel V. SARS-CoV and IFN: Too Little, Too Late. Cell Host Microbe. 2016;19:139–141. doi: 10.1016/j.chom.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meissner N., Swain S., McInnerney K., Han S., Harmsen A.G. Type-I IFN signaling suppresses an excessive IFN-γ response and thus prevents lung damage and chronic inflammation during Pneumocystis (PC) clearance in CD4 T cell-competent mice. Am. J. Pathol. 2010;176:2806–2818. doi: 10.2353/ajpath.2010.091158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klinkhammer J., Schnepf D., Ye L., Schwaderlapp M., Gad H.H., Hartmann R., Garcin D., Mahlakõiv T., Staeheli P. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife. 2018;7:e33354. doi: 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davidson S., McCabe T.M., Crotta S., Gad H.H., Hessel E.M., Beinke S., Hartmann R., Wack A. IFN λ is a potent anti‐influenza therapeutic without the inflammatory side effects of IFN α treatment. EMBO Mol. Med. 2016;8:1099–1112. doi: 10.15252/emmm.201606413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T.K., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D.B., Clementz M.A., Banach B.S., Li K., Baker S.C., Chen Z. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012;7:22312431. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.-T.K., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/jvi.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/jvi.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frieman M., Baric R. Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiol. Mol. Biol. Rev. 2008;72:672–685. doi: 10.1128/mmbr.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu Y., Li W., Gao T., Cui Y., Jin Y., Li P., Ma Q., Liu X., Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-Mediated RIG-I ubiquitination. J. Virol. 2017;91:28148787. doi: 10.1128/jvi.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic Reticulum/Golgi membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/jvi.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen I.Y., Moriyama M., Chang M.F., Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10:3389. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nieto-Torres J.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shi C.S., Nabar N.R., Huang N.N., Kehrl J.H. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:41420. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Siu K.L., Yuen K.S., Castano-Rodriguez C., Ye Z.W., Yeung M.L., Fung S.Y., Yuan S., Chan C.P., Yuen K.Y., Enjuanes L., Jin D.Y. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., Nakagawa S., Sato K. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., Wang C., Zhang Y., Zhu Z., Liu Z., Peng C., Zhu T., Liang Q. Virus-host interactome and proteomic survey of PBMCs from COVID-19 patients reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. 2020 doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang Y., Zhang L., Geng H., Deng Y., Huang B., Guo Y., Zhao Z., Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Comar C.E., Goldstein S.A., Li Y., Yount B., Baric R.S., Weiss S.R. Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. mBio. 2019;10:e00319–19. doi: 10.1128/mBio.00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Niemeyer D., Zillinger T., Muth D., Zielecki F., Horvath G., Suliman T., Barchet W., Weber F., Drosten C., Muller M.A. Middle east respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 2013;87:12489–12495. doi: 10.1128/jvi.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siu K.-L., Yeung M.L., Kok K.-H., Yuen K.-S., Kew C., Lui P.-Y., Chan C.-P., Tse H., Woo P.C.Y., Yuen K.-Y., Jin D.-Y. Middle East Respiratory Syndrome Coronavirus 4a Protein Is a Double-Stranded RNA-Binding Protein That Suppresses PACT-Induced Activation of RIG-I and MDA5 in the Innate Antiviral Response. J. Virol. 2014;88:4866–4876. doi: 10.1128/jvi.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Thornbrough J.M., Jha B.K., Yount B., Goldstein S.A., Li Y., Elliott R., Sims A.C., Baric R.S., Silverman R.H., Weissa S.R. Middle east respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio. 2016;7:4817253. doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z., Tan W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type i interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015;5:26631542. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Canton J., Fehr A.R., Fernandez-Delgado R., Gutierrez-Alvarez F.J., Sanchez-Aparicio M.T., García-Sastre A., Perlman S., Enjuanes L., Sola I. MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection. PLoS Pathog. 2018;14:29370303. doi: 10.1371/journal.ppat.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Menachery V.D., Debbink K., Baric R.S. Coronavirus non-structural protein 16: evasion, attenuation, and possible treatments. Virus Res. 2014;194:191–199. doi: 10.1016/j.virusres.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tazikeh-Lemeski E., Moradi S., Raoufi R., Shahlaei M., Janlou M.A.M., Zolghadri S. Targeting SARS-COV-2 non-structural protein 16: a virtual drug repurposing study. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1779133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7:22174690. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lokugamage K.G., Narayanan K., Nakagawa K., Terasaki K., Ramirez S.I., Tseng C.-T.K., Makino S. Middle East Respiratory Syndrome Coronavirus nsp1 Inhibits Host Gene Expression by Selectively Targeting mRNAs Transcribed in the Nucleus while Sparing mRNAs of Cytoplasmic Origin. J. Virol. 2015;89:10970–10981. doi: 10.1128/jvi.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thoms B.R., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Stürzel C.M., Fröhlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.05.18.102467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shi C.-S., Qi H.-Y., Boularan C., Huang N.-N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-coronavirus open reading Frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Siu K.L., Kok K.H., Ng M.H.J., Poon V.K.M., Yuen K.Y., Zheng B.J., Jin D.Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding theformation of TRAF3·TANK·TBK1/IKKε complex. J. Biol. Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yuen C.-K., Lam J.-Y., Wong W.-M., Mak L.-F., Wang X., Chu H., Cai J.-P., Jin D.-Y., To K.K.-W., Chan J.F.-W., Yuen K.-Y., Kok K.-H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schrezenmeier E., Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 118.Aguiar A.C.C., Murce E., Cortopassi W.A., Pimentel A.S., Almeida M.M.F.S., Barros D.C.S., Guedes J.S., Meneghetti M.R., Krettli A.U. Chloroquine analogs as antimalarial candidates with potent in vitro and in vivo activity. Int. J. Parasitol. Drugs Drug Resist. 2018;8:459–464. doi: 10.1016/j.ijpddr.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oliveira K.R.H.M., dos Anjos L.M., Araújo A.P.S., Luz W.L., Kauffmann N., Braga D.V., da Conceição Fonseca Passos A., de Moraes S.A.S., de Jesus Oliveira Batista E., Herculano A.M. Ascorbic acid prevents chloroquine-induced toxicity in inner glial cells. Toxicol. In Vitro. 2019;56:150–155. doi: 10.1016/j.tiv.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 120.Carr A.C. A new clinical trial to test high-dose vitamin C in patients with COVID-19. Crit. Care. 2020;24:13054. doi: 10.1186/s13054-020-02851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cheng R.Z. Can early and high intravenous dose of vitamin C prevent and treat coronavirus disease 2019 (COVID-19)? Med. Drug Discov. 2020;5:100028. doi: 10.1016/j.medidd.2020.100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim Y., Kim H., Bae S., Choi J., Lim S.Y., Lee N., Kong J.M., Hwang Y., Kang J.S., Lee W.J. Vitamin C is an essential factor on the anti-viral immune responses through the production of Interferon-α/β at the initial stage of influenza a virus (H3N2) infection. Immune Netw. 2013;13:70. doi: 10.4110/in.2013.13.2.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boretti A., Banik B.K. Intravenous vitamin C for reduction of cytokines storm in acute respiratory distress syndrome. PharmaNutrition. 2020;12:7172861. doi: 10.1016/j.phanu.2020.100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lim H.S., Im J.S., Cho J.Y., Bae K.S., Klein T.A., Yeom J.S., Kim T.S., Choi J.S., Jang I.J., Park J.W. Pharmacokinetics of hydroxychloroquine and its clinical implications in chemoprophylaxis against malaria caused by plasmodium vivax. Antimicrob. Agents Chemother. 2009;53:1468–1475. doi: 10.1128/AAC.00339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:32074550. doi: 10.5582/BST.2020.01047. [DOI] [PubMed] [Google Scholar]
- 126.Satarker S., Ahuja T., Banerjee M., V.B. E, Dogra S., Agarwal T., Nampoothiri M. Hydroxychloroquine in COVID-19: potential mechanism of action against SARS-CoV-2. Curr. Pharmacol. Rep. 2020;6:203–211. doi: 10.1007/s40495-020-00231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rainsford K.D., Parke A.L., Clifford-Rashotte M., Kean W.F. Therapy and pharmacological properties of hydroxychloroquine and chloroquine in treatment of systemic lupus erythematosus, rheumatoid arthritis and related diseases. Inflammopharmacology. 2015;23:231–269. doi: 10.1007/s10787-015-0239-y. [DOI] [PubMed] [Google Scholar]
- 128.An J., Woodward J.J., Sasaki T., Minie M., Elkon K.B. Cutting Edge: Antimalarial Drugs Inhibit IFN-β Production through Blockade of Cyclic GMP-AMP Synthase–DNA Interaction. J. Immunol. 2015;194:4089–4093. doi: 10.4049/jimmunol.1402793. [DOI] [PubMed] [Google Scholar]
- 129.Martinson J.A., Montoya C.J., Usuga X., Ronquillo R., Landay A.L., Desai S.N. Chloroquine modulates HIV-1-Induced plasmacytoid dendritic cell alpha interferon: implication for T-Cell activation. Antimicrob. Agents Chemother. 2010;54:871–881. doi: 10.1128/AAC.01246-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu T., Luo S., Libby P., Shi G.P. Cathepsin L-selective inhibitors: a potentially promising treatment for COVID-19 patients. Pharmacol. Ther. 2020;213:107587. doi: 10.1016/j.pharmthera.2020.107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sahraei Z., Shabani M., Shokouhi S., Saffaei A. Aminoquinolines against coronavirus disease 2019 (COVID-19): chloroquine or hydroxychloroquine. Int. J. Antimicrob. Agents. 2020;55:105945. doi: 10.1016/j.ijantimicag.2020.105945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Parnham M.J., Haber V.E., Giamarellos-Bourboulis E.J., Perletti G., Verleden G.M., Vos R. Azithromycin: Mechanisms of action and their relevance for clinical applications. Pharmacol. Ther. 2014;143:225–245. doi: 10.1016/j.pharmthera.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 134.Schönfeld W., Mutak S. Macrolide Antibiot. Birkhäuser Basel; Basel: 2002. Azithromycin and novel azalides; pp. 73–95. [DOI] [Google Scholar]
- 135.Kuo C.-H., Lee M.-S., Kuo H.-F., Lin Y.-C., Hung C.-H. Azithromycin suppresses Th1- and Th2-related chemokines IP-10/MDC in human monocytic cell line. J. Microbiol. Immunol. Infect. 2019;52:872–879. doi: 10.1016/j.jmii.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 136.Schögler A., Kopf B.S., Edwards M.R., Johnston S.L., Casaulta C., Kieninger E., Jung A., Moeller A., Geiser T., Regamey N., Alves M.P. Novel antiviral properties of azithromycin in cystic fibrosis airway epithelial cells. Eur. Respir. J. 2015;45:428–439. doi: 10.1183/09031936.00102014. [DOI] [PubMed] [Google Scholar]
- 137.Gielen V., Johnston S.L., Edwards M.R. Azithromycin induces anti-viral responses in bronchial epithelial cells. Eur. Respir. J. 2010;36:646–654. doi: 10.1183/09031936.00095809. [DOI] [PubMed] [Google Scholar]
- 138.Asada M., Yoshida M., Suzuki T., Hatachi Y., Sasaki T., Yasuda H., Nakayama K., Nishimura H., Nagatomi R., Kubo H., Yamaya M. Macrolide antibiotics inhibit respiratory syncytial virus infection in human airway epithelial cells. Antiviral Res. 2009;83:191–200. doi: 10.1016/j.antiviral.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 139.Retallack H., Di Lullo E., Arias C., Knopp K.A., Laurie M.T., Sandoval-Espinosa C., Mancia Leon W.R., Krencik R., Ullian E.M., Spatazza J., Pollen A.A., Mandel-Brehm C., Nowakowski T.J., Kriegstein A.R., DeRisi J.L. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc. Natl. Acad. Sci. U. S. A. 2016;113:14408–14413. doi: 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A., Davey R., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. Evaluation of ebola virus inhibitors for drug repurposing. ACS Infect. Dis. 2016;1:317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- 141.Tran D.H., Sugamata R., Hirose T., Suzuki S., Noguchi Y., Sugawara A., Ito F., Yamamoto T., Kawachi S., Akagawa K.S., Ōmura S., Sunazuka T., Ito N., Mimaki M., Suzuki K. Azithromycin, a 15-membered macrolide antibiotic, inhibits influenza A(H1N1)pdm09 virus infection by interfering with virus internalization process. J. Antibiot. 2019;72:759–768. doi: 10.1038/s41429-019-0204-x. [DOI] [PubMed] [Google Scholar]
- 142.Zeng S., Meng X., Huang Q., Lei N., Zeng L., Jiang X., Guo X. Spiramycin and azithromycin, safe for administration to children, exert antiviral activity against enterovirus A71 in vitro and in vivo. Int. J. Antimicrob. Agents. 2019;53:362–369. doi: 10.1016/j.ijantimicag.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 143.Sargiacomo C., Sotgia F., Lisanti M.P. COVID-19 and chronological aging: senolytics and other anti-aging drugs for the treatment or prevention of corona virus infection? Aging (Albany. NY) 2020;12:6511–6517. doi: 10.18632/aging.103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kagkelaris K.A., Makri O.E., Georgakopoulos C.D., Panayiotakopoulos G.D. An eye for azithromycin: review of the literature. Ther. Adv. Ophthalmol. 2018;10 doi: 10.1177/2515841418783622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Arabi Y.M., Deeb A.M., Al-Hameed F., Mandourah Y., Almekhlafi G.A., Sindi A.A., Al-Omari A., Shalhoub S., Mady A., Alraddadi B., Almotairi A., Al Khatib K., Abdulmomen A., Qushmaq I., Solaiman O., Al-Aithan A.M., Al-Raddadi R., Ragab A., Al Harthy A., Kharaba A., Jose J., Dabbagh T., Fowler R.A., Balkhy H.H., Merson L., Hayden F.G. Saudi Critical Care Trials group, macrolides in critically ill patients with middle east respiratory syndrome. Int. J. Infect. Dis. 2019;81:184–190. doi: 10.1016/j.ijid.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Beigelman A., Isaacson-Schmid M., Sajol G., Baty J., Rodriguez O.M., Leege E., Lyons K., Schweiger T.L., Zheng J., Schechtman K.B., Castro M., Bacharier L.B. Randomized trial to evaluate azithromycin’s effects on serum and upper airway IL-8 levels and recurrent wheezing in infants with respiratory syncytial virus bronchiolitis. J. Allergy Clin. Immunol. 2015;135:1171–1178. doi: 10.1016/j.jaci.2014.10.001. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Deretic V., Timmins G.S. Azithromycin and ciprofloxacin have a chloroquine-like effect on respiratory epithelial cells. bioRxiv. 2020 doi: 10.1101/2020.03.29.008631. 2020.03.29.008631. [DOI] [Google Scholar]
- 148.Andreani J., Le Bideau M., Duflot I., Jardot P., Rolland C., Boxberger M., Wurtz N., Rolain J.M., Colson P., La Scola B., Raoult D. In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect. Microb. Pathog. 2020;145:104228. doi: 10.1016/j.micpath.2020.104228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Tissot Dupont H., Honoré S., Colson P., Chabrière E., La Scola B., Rolain J.-M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 150.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang M., Tang T., Pang P., Li M., Ma R., Lu J., Shu J., You Y., Chen B., Liang J., Hong Z., Chen H., Kong L., Qin D., Pei D., Xia J., Jiang S., Shan H. Treating COVID-19 with chloroquine. J. Mol. Cell Biol. 2020;12:322–325. doi: 10.1093/jmcb/mjaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen J., Liu D., Liu L., Liu P., Xu Q., Xia L., Ling Y., Huang D., Song S., Zhang D., Qian Z., Li T., Shen Y., Lu H. [A pilot study of hydroxychloroquine in treatment of patients with moderate COVID-19] Zhejiang Da Xue Xue Bao Yi Xue Ban. 2020;49:215–219. doi: 10.3785/j.issn.1008-9292.2020.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Sevestre J., Mailhe M., Doudier B., Aubry C., Amrane S., Seng P., Hocquart M., Eldin C., Finance J., Vieira V.E., Tissot-Dupont H.T., Honoré S., Stein A., Million M., Colson P., La Scola B., Veit V., Jacquier A., Deharo J.-C., Drancourt M., Fournier P.E., Rolain J.-M., Brouqui P., Raoult D. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: a pilot observational study. Travel Med. Infect. Dis. 2020;34:101663. doi: 10.1016/j.tmaid.2020.101663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Million M., Lagier J.-C., Gautret P., Colson P., Fournier P.-E., Amrane S., Hocquart M., Mailhe M., Esteves-Vieira V., Doudier B., Aubry C., Correard F., Giraud-Gatineau A., Roussel Y., Berenger C., Cassir N., Seng P., Zandotti C., Dhiver C., Ravaux I., Tomei C., Eldin C., Tissot-Dupont H., Honoré S., Stein A., Jacquier A., Deharo J.-C., Chabrière E., Levasseur A., Fenollar F., Rolain J.-M., Obadia Y., Brouqui P., Drancourt M., La Scola B., Parola P., Raoult D. Early treatment of COVID-19 patients with hydroxychloroquine and azithromycin: a retrospective analysis of 1061 cases in Marseille, France. Travel Med. Infect. Dis. 2020;35:101738. doi: 10.1016/j.tmaid.2020.101738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lagier J.-C., Million M., Gautret P., Colson P., Cortaredona S., Giraud-Gatineau A., Honoré S., Gaubert J.-Y., Fournier P.-E., Tissot-Dupont H., Chabrière E., Stein A., Deharo J.-C., Fenollar F., Rolain J.-M., Obadia Y., Jacquier A., La Scola B., Brouqui P., Drancourt M., Parola P., Raoult D. IHU COVID-19 Task force, Outcomes of 3,737 COVID-19 patients treated with hydroxychloroquine/azithromycin and other regimens in Marseille, France: a retrospective analysis. Travel Med. Infect. Dis. 2020:101791. doi: 10.1016/j.tmaid.2020.101791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tang W., Cao Z., Han M., Wang Z., Chen J., Sun W., Wu Y., Xiao W., Liu S., Chen E., Chen W., Wang X., Yang J., Lin J., Zhao Q., Yan Y., Xie Z., Li D., Yang Y., Liu L., Qu J., Ning G., Shi G., Xie Q. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ. 2020;369 doi: 10.1136/bmj.m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mitra R.L., Greenstein S.A., Epstein L.M. An algorithm for managing QT prolongation in coronavirus disease 2019 (COVID-19) patients treated with either chloroquine or hydroxychloroquine in conjunction with azithromycin: possible benefits of intravenous lidocaine. Hear. Case Rep. 2020;6:244–248. doi: 10.1016/j.hrcr.2020.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gérard A., Romani S., Fresse A., Viard D., Parassol N., Granvuillemin A., Chouchana L., Rocher F., Drici M.-D. French Network of Pharmacovigilance Centers, “Off-label” use of hydroxychloroquine, azithromycin, lopinavir-ritonavir and chloroquine in COVID-19: a survey of cardiac adverse drug reactions by the French Network of Pharmacovigilance Centers. Therapie. 2020;S0040-5957:30091–30093. doi: 10.1016/j.therap.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Molina J.M., Delaugerre C., Le Goff J., Mela-Lima B., Ponscarme D., Goldwirt L., de Castro N. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Mal. Infect. 2020;50:384. doi: 10.1016/j.medmal.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tang W., Cao Z., Han M., Wang Z., Chen J., Sun W., Wu Y., Xiao W., Liu S., Chen E., Chen W., Wang X., Yang J., Lin J., Zhao Q., Yan Y., Xie Z., Li D., Yang Y., Liu L., Qu J., Ning G., Shi G., Xie Q. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ. 2020;369:m1849. doi: 10.1136/bmj.m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Magagnoli J., Narendran S., Pereira F., Cummings T.H., Hardin J.W., Sutton S.S., Ambati J. Outcomes of hydroxychloroquine usage in United States veterans hospitalized with COVID-19. Med. 2020 doi: 10.1016/j.medj.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D.K., Kubin C., Barr R.G., Sobieszczyk M.E., Schluger N.W. Observational study of hydroxychloroquine in hospitalized patients with Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/nejmoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Boulware D.R., Pullen M.F., Bangdiwala A.S., Pastick K.A., Lofgren S.M., Okafor E.C., Skipper C.P., Nascene A.A., Nicol M.R., Abassi M., Engen N.W., Cheng M.P., LaBar D., Lother S.A., MacKenzie L.J., Drobot G., Marten N., Zarychanski R., Kelly L.E., Schwartz I.S., McDonald E.G., Rajasingham R., Lee T.C., Hullsiek K.H. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N. Engl. J. Med. 2020;383:517–525. doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Skipper C.P., Pastick K.A., Engen N.W., Bangdiwala A.S., Abassi M., Lofgren S.M., Williams D.A., Okafor E.C., Pullen M.F., Nicol M.R., Nascene A.A., Hullsiek K.H., Cheng M.P., Luke D., Lother S.A., MacKenzie L.J., Drobot G., Kelly L.E., Schwartz I.S., Zarychanski R., McDonald E.G., Lee T.C., Rajasingham R., Boulware D.R. Hydroxychloroquine in Nonhospitalized Adults With Early COVID-19: A Randomized Trial. Ann. Intern. Med. 2020 doi: 10.7326/M20-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mitjà O., Corbacho-Monné M., Ubals M., Tebe C., Peñafiel J., Tobias A., Ballana E., Alemany A., Riera-Martí N., Pérez C.A., Suñer C., Laporte P., Admella P., Mitjà J., Clua M., Bertran L., Sarquella M., Gavilán S., Ara J., Argimon J.M., Casabona J., Cuatrecasas G., Cañadas P., Elizalde-Torrent A., Fabregat R., Farré M., Forcada A., Flores-Mateo G., Muntada E., Nadal N., Narejos S., Gil-Ortega A.N., Prat N., Puig J., Quiñones C., Reyes-Ureña J., Ramírez-Viaplana F., Ruiz L., Riveira-Muñoz E., Sierra A., Velasco C., Vivanco-Hidalgo R.M., Sentís A., G-Beiras C., Clotet B., Vall-Mayans M. BCN PEP-CoV-2 RESEARCH GROUP, hydroxychloroquine for early treatment of adults with mild Covid-19: a randomized-controlled trial. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Horby P., Mafham M., Linsell L. Effect of hydroxychloroquine in hospitalized patients with COVID-19: preliminary results from a multi-centre, randomized, controlled trial. medRxiv. 2020 doi: 10.1101/2020.07.15.20151852. [DOI] [Google Scholar]
- 167.US National Library of Medicine . 2020. Randomised Evaluation of COVID-19 Therapy (RECOVERY), ClinicalTrials.Gov. [Google Scholar]
- 168.Cavalcanti A.B., Zampieri F.G., Rosa R.G., Azevedo L.C.P., Veiga V.C., Avezum A., Damiani L.P., Marcadenti A., Kawano-Dourado L., Lisboa T., Junqueira D.L.M., de Barros e Silva P.G.M., Tramujas L., Abreu-Silva E.O., Laranjeira L.N., Soares A.T., Echenique L.S., Pereira A.J., Freitas F.G.R., Gebara O.C.E., Dantas V.C.S., Furtado R.H.M., Milan E.P., Golin N.A., Cardoso F.F., Maia I.S., Hoffmann Filho C.R., Kormann A.P.M., Amazonas R.B., Bocchi de Oliveira M.F., Serpa-Neto A., Falavigna M., Lopes R.D., Machado F.R., Berwanger O. Hydroxychloroquine with or without azithromycin in mild-to-Moderate Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2019014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Furtado R.H.M., Berwanger O., Fonseca H.A., Corrêa T.D., Ferraz L.R., Lapa M.G., Zampieri F.G., Veiga V.C., Azevedo L.C.P., Rosa R.G., Lopes R.D., Avezum A., Manoel A.L.O., Piza F.M.T., Martins P.A., Lisboa T.C., Pereira A.J., Olivato G.B., Dantas V.C.S., Milan E.P., Gebara O.C.E., Amazonas R.B., Oliveira M.B., Soares R.V.P., Moia D.D.F., Piano L.P.A., Castilho K., Momesso R.G.R.A.P., Schettino G.P.P., Rizzo L.V., Neto A.S., Machado F.R., Cavalcanti A.B. Azithromycin in addition to standard of care versus standard of care alone in the treatment of patients admitted to the hospital with severe COVID-19 in Brazil (COALITION II): a randomised clinical trial. Lancet. 2020;396:959–967. doi: 10.1016/S0140-6736(20)31862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.US National Library of Medicine . 2020. Trial of Treatments for COVID-19 in Hospitalized Adults (DisCoVeRy), ClinicalTrials.Gov.https://clinicaltrials.gov/ct2/show/NCT04315948 (accessed September 26, 2020) [Google Scholar]
- 171.World Health Organization . 2020. WHO Discontinues Hydroxychloroquine and lopinavir/ritonavir Treatment Arms for COVID-19.https://www.who.int/news/item/04-07-2020-who-discontinues-hydroxychloroquine-and-lopinavir-ritonavir-treatment-arms-for-covid-19 (accessed September 28, 2020) [Google Scholar]
- 172.U.S. FOOD and DRUG Administration . 2020. Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Chloroquine and Hydroxychloroquine | FDA.https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine-and (accessed September 26, 2020) [Google Scholar]
- 173.US NIH . 2020. COVID-19 Treatment guidelines/Chloroquine or Hydroxychloroquine With or Without Azithromycin.https://www.covid19treatmentguidelines.nih.gov/antiviral-therapy/chloroquine-or-hydroxychloroquine-with-or-without-azithromycin/ (accessed September 30, 2020) [Google Scholar]
- 174.D’Acquarica I., Agranat I. Chiral switches of chloroquine and hydroxychloroquine: potential drugs to treat COVID-19. Drug Discov. Today. 2020;25:1121–1123. doi: 10.1016/j.drudis.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cortegiani A., Ippolito M., Ingoglia G., Iozzo P., Giarratano A., Einav S., Update I. A systematic review on the efficacy and safety of chloroquine/hydroxychloroquine for COVID-19. J. Crit. Care. 2020;59:176–190. doi: 10.1016/j.jcrc.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chorin E., Dai M., Shulman E., Wadhwani L., Bar-Cohen R., Barbhaiya C., Aizer A., Holmes D., Bernstein S., Spinelli M., Park D.S., Chinitz L.A., Jankelson L. The QT interval in patients with COVID-19 treated with hydroxychloroquine and azithromycin. Nat. Med. 2020;26:808–809. doi: 10.1038/s41591-020-0888-2. [DOI] [PubMed] [Google Scholar]
- 177.Chorin E., Wadhwani L., Magnani S., Dai M., Shulman E., Nadeau-Routhier C., Knotts R., Bar-Cohen R., Kogan E., Barbhaiya C., Aizer A., Holmes D., Bernstein S., Spinelli M., Park D.S., Stefano C., Chinitz L.A., Jankelson L. QT interval prolongation and torsade de pointes in patients with COVID-19 treated with hydroxychloroquine/azithromycin. Hear. Rhythm. 2020;S1547-5271 doi: 10.1016/j.hrthm.2020.05.014. 30435–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Karalis V., Ismailos G., Karatza E. Chloroquine dosage regimens in patients with COVID-19: safety risks and optimization using simulations. Saf. Sci. 2020;129:104842. doi: 10.1016/j.ssci.2020.104842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bernardini A., Ciconte G., Negro G., Rondine R., Mecarocci V., Viva T., Santini F., de Innocentiis C., Giannelli L., Witkowska E., Locati E.T., Castelvecchio S., Marrocco-Trischitta M.M., Vicedomini G., Menicanti L., Pappone C. Assessing QT interval in COVID-19 patients:safety of hydroxychloroquine-azithromycin combination regimen. Int. J. Cardiol. 2020 doi: 10.1016/j.ijcard.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Borba M.G.S., Val F.F.A., Sampaio V.S., Alexandre M.A.A., Melo G.C., Brito M., Mourão M.P.G., Brito-Sousa J.D., Baía-da-Silva D., Guerra M.V.F., Hajjar L.A., Pinto R.C., Balieiro A.A.S., Pacheco A.G.F., Santos J.D.O., Naveca F.G., Xavier M.S., Siqueira A.M., Schwarzbold A., Croda J., Nogueira M.L., Romero G.A.S., Bassat Q., Fontes C.J., Albuquerque B.C., Daniel-Ribeiro C.-T., Monteiro W.M., Lacerda M.V.G., CloroCovid-19 Team Effect of high vs low doses of chloroquine diphosphate as adjunctive therapy for patients hospitalized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection: a randomized clinical trial. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Taboada M., Caruezo V., Naveira A., Atanassoff P.G. Corticosteroids and the hyper-inflammatory phase of the COVID-19 disease. J. Clin. Anesth. 2020;66:109926. doi: 10.1016/j.jclinane.2020.109926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Banchereau R., Hong S., Cantarel B., Baldwin N., Baisch J., Edens M., Cepika A.-M., Acs P., Turner J., Anguiano E., Vinod P., Khan S., Obermoser G., Blankenship D., Wakeland E., Nassi L., Gotte A., Punaro M., Liu Y.-J., Banchereau J., Rossello-Urgell J., Wright T., Pascual V. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell. 2016;165:551–565. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Andre H.S.F.E., Booy R., Bock H.L., Clemens J., Datta S.K., John T.J., Lee B.W., Lolekha S., Peltola H., Ruff T.A., Santosham M. 2011. WHO | Vaccination Greatly Reduces Disease, Disability, Death and Inequity Worldwide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Salvatori G., Luberto L., Maffei M., Aurisicchio L., Roscilli G., Palombo F., Marra E. SARS-CoV-2 SPIKE PROTEIN: an optimal immunological target for vaccines. J. Transl. Med. 2020;18:222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., Hong W., Yang Y., Zhao Y., Ye F., Lin S., Deng W., Chen H., Lei H., Zhang Z., Luo M., Gao H., Zheng Y., Gong Y., Jiang X., Xu Y., Lv Q., Li D., Wang M., Li F., Wang S., Wang G., Yu P., Qu Y., Yang L., Deng H., Tong A., Li J., Wang Z., Yang J., Shen G., Zhao Z., Li Y., Luo J., Liu H., Yu W., Yang M., Xu J., Wang J., Li H., Wang H., Kuang D., Lin P., Hu Z., Guo W., Cheng W., He Y., Song X., Chen C., Xue Z., Yao S., Chen L., Ma X., Chen S., Gou M., Huang W., Wang Y., Fan C., Tian Z., Shi M., Wang F.-S., Dai L., Wu M., Li G., Wang G., Peng Y., Qian Z., Huang C., Lau J.Y.-N., Yang Z., Wei Y., Cen X., Peng X., Qin C., Zhang K., Lu G., Wei X. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020 doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 186.Bao L., Deng W., Gao H., Xiao C., Liu J., Xue J., Lv Q., Liu J., Yu P., Xu Y., Qi F., Qu Y., Li F., Xiang Z., Yu H., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao W., Han Y., Zhao L., Liu X., Wei Q., Qin C. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. BioRxiv. 2020 doi: 10.1101/2020.03.13.990226. 2020.03.13.990226. [DOI] [Google Scholar]
- 187.Fan Wu J.H., Wang Aojie, Liu Mei, Wang Qimin, Chen Jun, Xia Shuai, Ling Yun, Zhang Yuling, Xun Jingna, Lu Lu, Jiang Shibo, Hongzhou Lu, Wen Yumei. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 doi: 10.1101/2020.03.30.20047365. 2020.03.30.20047365. [DOI] [Google Scholar]
- 188.Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368:860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Payne D.C., Iblan I., Rha B., Alqasrawi S., Haddadin A., Al Nsour M., Alsanouri T., Ali S.S., Harcourt J., Miao C., Tamin A., Gerber S.I., Haynes L.M., Al Abdallat M.M. Persistence of antibodies against middle east respiratory syndrome coronavirus. Emerg. Infect. Dis. 2016;22:1824–1826. doi: 10.3201/eid2210.160706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mo H., Zeng G., Ren X., Li H., Ke C., Tan Y., Cai C., Lai K., Chen R., Chan-Yeung M., Zhong N. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology. 2006;11:49–53. doi: 10.1111/j.1440-1843.2006.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yang Z.Y., Werner H.C., Kong W.P., Leung K., Traggiai E., Lanzavecchia A., Nabel G.J. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc. Natl. Acad. Sci. U. S. A. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jaume M., Yip M.S., Cheung C.Y., Leung H.L., Li P.H., Kien F., Dutry I., Callendret B., Escriou N., Altmeyer R., Nal B., Daeron M., Bruzzone R., Peiris J.S.M. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent fc r pathway. J. Virol. 2011;85:10582–10597. doi: 10.1128/jvi.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., Zhu H., Liu J., Xu Y., Xie J., Morioka H., Sakaguchi N., Qin C., Liu G. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human Primates. ACS Infect. Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang B., He J., Liu C., Chang L. An effective cancer vaccine modality: lentiviral modification of dendritic cells expressing multiple cancer-specific antigens. Vaccine. 2006;24:3477–3489. doi: 10.1016/j.vaccine.2006.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yip M.S., Cheung C.Y., Li P.H., Bruzzone R., Peiris J.M., Jaume M. Investigation of Antibody-Dependent Enhancement (ADE) of SARS coronavirus infection and its role in pathogenesis of SARS. BMC Proc. 2011;5:P80. doi: 10.1186/1753-6561-5-S1-P80. [DOI] [PubMed] [Google Scholar]
- 196.Wang S.F., Tseng S.P., Yen C.H., Yang J.Y., Tsao C.H., Shen C.W., Chen K.H., Liu F.T., Liu W.T., Chen Y.M.A., Huang J.C. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ochiai H., Kurokawa M., Matsui S., Yamamoto T., Kuroki Y., Kishimoto C., Shiraki K. Infection enhancement of influenza A NWS virus in primary murine macrophages by anti‐hemagglutinin monoclonal antibody. J. Med. Virol. 1992;36:217–221. doi: 10.1002/jmv.1890360312. [DOI] [PubMed] [Google Scholar]
- 198.Kim H.W., Canchola J.G., Brandt C.D., Pyles G., Chanock R.M., Jensen K., Parrott R.H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 199.Halstead S.B., O’Rourke E.J. Dengue viruses and mononuclear phagocytes: I. Infection enhancement by non-neutralizing antibody*. J. Exp. Med. 1977;146:201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chin J., Magoffin R.L., Shearer L.A., Schieble J.H., Lennette E.H. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am. J. Epidemiol. 1969;89:449–463. doi: 10.1093/oxfordjournals.aje.a120957. [DOI] [PubMed] [Google Scholar]
- 201.Robinson W.E., Montefiori D.C., Mitchell W.M. Antibody-dependent enhancement of human immunodeficiency virus type 1 infection. Lancet. 1988;331:790–794. doi: 10.1016/S0140-6736(88)91657-1. [DOI] [PubMed] [Google Scholar]
- 202.Robinson W.E., Montefiori D.C., Mitchell W.M., Prince A.M., Alter H.J., Dreesman G.R., Eichberg J.W. Antibody-dependent enhancement of human immunodeficiency virus type 1 (HIV-1) infection in vitro by serum from HIV-1-infected and passively immunized chimpanzees. Proc. Natl. Acad. Sci. U. S. A. 1989;86:4710–4714. doi: 10.1073/pnas.86.12.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Takada A., Feldmann H., Ksiazek T.G., Kawaoka Y. Antibody-dependent enhancement of ebola virus infection. J. Virol. 2003;77:7539–7544. doi: 10.1128/jvi.77.13.7539-7544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C. Development of an inactivated vaccine candidate for SARS-CoV-2. Science (80-.) 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., Wu T., Cheung K.-W., Chan K.-H., Alvarez X., Qin C., Lackner A., Perlman S., Yuen K.-Y., Chen Z. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lokugamage K.G., Yoshikawa-Iwata N., Ito N., Watts D.M., Wyde P.R., Wang N., Newman P., Te Kent Tseng C., Peters C.J., Makino S. Chimeric coronavirus-like particles carrying severe acute respiratory syndrome coronavirus (SCoV) S protein protect mice against challenge with SCoV. Vaccine. 2008;26:797–808. doi: 10.1016/j.vaccine.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Te Tseng C., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., Peters C.J., Couch R.B. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Castilow E.M., Olson M.R., Varga S.M. Understanding respiratory syncytial virus (RSV) vaccine-enhanced disease. Immunol. Res. 2007;39:225–239. doi: 10.1007/s12026-007-0071-6. [DOI] [PubMed] [Google Scholar]
- 209.Yasui F., Kai C., Kitabatake M., Inoue S., Yoneda M., Yokochi S., Kase R., Sekiguchi S., Morita K., Hishima T., Suzuki H., Karamatsu K., Yasutomi Y., Shida H., Kidokoro M., Mizuno K., Matsushima K., Kohara M. Prior immunization with severe acute respiratory syndrome (SARS)-Associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J. Immunol. 2008;181:6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- 210.Agrawal A.S., Tao X., Algaissi A., Garron T., Narayanan K., Peng B.H., Couch R.B., Tseng C.T.K. Immunization with inactivated Middle East Respiratory Syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum. Vaccines Immunother. 2016;12:2351–2356. doi: 10.1080/21645515.2016.1177688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Iwata-Yoshikawa N., Uda A., Suzuki T., Tsunetsugu-Yokota Y., Sato Y., Morikawa S., Tashiro M., Sata T., Hasegawa H., Nagata N. Effects of toll-like receptor stimulation on eosinophilic infiltration in lungs of BALB/c mice immunized with UV-Inactivated severe acute respiratory syndrome-related coronavirus vaccine. J. Virol. 2014;88:8597–8614. doi: 10.1128/jvi.00983-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Honda-Okubo Y., Barnard D., Ong C.H., Peng B.-H., Tseng C.-T.K., Petrovsky N. Severe Acute Respiratory Syndrome-Associated Coronavirus Vaccines Formulated with Delta Inulin Adjuvants Provide Enhanced Protection while Ameliorating Lung Eosinophilic Immunopathology. J. Virol. 2015;89:2995–3007. doi: 10.1128/jvi.02980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J., Smith G., Jones S., Proulx R., Deschambault Y., Grudeski E., Andonov A., He R., Li Y., Copps J., Grolla A., Dick D., Berry J., Ganske S., Manning L., Cao J. Immunization with modified vaccinia virus ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 2004;78:12672–12676. doi: 10.1128/jvi.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vennema H., de Groot R.J., Harbour D.A., Dalderup M., Gruffydd-Jones T., Horzinek M.C., Spaan W.J.M. Immunogenicity of recombinant feline infectious peritonitis virus spike protein in mice and kittens. Adv. Exp. Med. Biol. 1990;276:217–222. doi: 10.1007/978-1-4684-5823-7_30. [DOI] [PubMed] [Google Scholar]
- 215.Pedersen N.C., Black J.W. Attempted immunization of cats against feline infectious peritonitis, using avirulent live virus or sublethal amounts of virulent virus. Am. J. Vet. Res. 1983;44:229–234. [PubMed] [Google Scholar]
- 216.Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Burton D.R. What are the most powerful immunogen design vaccine strategies? Reverse vaccinology 2.0 shows great promise. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Rouse B.T., Sehrawat S. Immunity and immunopathology to viruses: what decides the outcome? Nat. Rev. Immunol. 2010;10:514–526. doi: 10.1038/nri2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Simek M.D., Rida W., Priddy F.H., Pung P., Carrow E., Laufer D.S., Lehrman J.K., Boaz M., Tarragona-Fiol T., Miiro G., Birungi J., Pozniak A., McPhee D.A., Manigart O., Karita E., Inwoley A., Jaoko W., Dehovitz J., Bekker L.-G., Pitisuttithum P., Paris R., Walker L.M., Poignard P., Wrin T., Fast P.E., Burton D.R., Koff W.C. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kumar R., Qureshi H., Deshpande S., Bhattacharya J. Broadly neutralizing antibodies in HIV-1 treatment and prevention. Ther. Adv. Vaccines Immunother. 2018;6:61–68. doi: 10.1177/2515135518800689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., Ortiz A.T., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Xu D., Zhang Z., Chu F., Li Y., Jin L., Zhang L., Gao G.F., Wang F.-S. Genetic variation of SARS coronavirus in Beijing Hospital. Emerg. Infect. Dis. 2004;10:789–794. doi: 10.3201/eid1005.030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zharko Daniloski N.E.S., Guo Xinyi. The D614G mutation in SARS-CoV-2 Spike increases transduction of multiple human cell types. BioRxiv. 2020 doi: 10.1101/2020.06.14.151357. [DOI] [Google Scholar]
- 225.Mouquet H. Antibody B cell responses in HIV-1 infection. Trends Immunol. 2014;35:549–561. doi: 10.1016/j.it.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 226.World Health Organization . 2020. Draft Landscape of COVID-19 Candidate vaccines-29 October 2020.https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed October 31, 2020) [Google Scholar]
- 227.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Romagnani S. T-cell subsets (Th1 versus Th2) Ann. Allergy Asthma Immunol. 2000;85:9–21. doi: 10.1016/S1081-1206(10)62426-X. [DOI] [PubMed] [Google Scholar]
- 229.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O’Dell S., Schmidt S.D., Corbett K.S., Swanson P.A., Padilla M., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Martinez D.R., Baric R., Buchanan W., Luke C.J., Phadke V.K., Rostad C.A., Ledgerwood J.E., Graham B.S., Beigel J.H. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., Dold C., Faust S.N., Finn A., Flaxman A.L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A.M., Pollock K.M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Aboagye J., Adams K., Ali A., Allen E., Allison J.L., Anslow R., Arbe-Barnes E.H., Babbage G., Baillie K., Baker M., Baker P., Baleanu I., Ballaminut J., Barnes E., Barrett J., Bates L., Batten A., Beadon K., Beckley R., Berrie E., Berry L., Beveridge A., Bewley K.R., Bijker E.M., Bingham T., Blackwell L., Blundell C.L., Bolam E., Boland E., Borthwick N., Bower T., Boyd A., Brenner T., Bright P.D., Brown-O’Sullivan C., Brunt E., Burbage J., Burge S., Buttigieg K.R., Byard N., Cabera Puig I., Calvert A., Camara S., Cao M., Cappuccini F., Carr M., Carroll M.W., Carter V., Cathie K., Challis R.J., Chelysheva I., Cho J.-S., Cicconi P., Cifuentes L., Clark H., Clark E., Cole T., Colin-Jones R., Conlon C.P., Cook A., Coombes N.S., Cooper R., Cosgrove C.A., Coy K., Crocker W.E.M., Cunningham C.J., Damratoski B.E., Dando L., Datoo M.S., Davies H., De Graaf H., Demissie T., Di Maso C., Dietrich I., Dong T., Donnellan F.R., Douglas N., Downing C., Drake J., Drake-Brockman R., Drury R.E., Dunachie S.J., Edwards N.J., Edwards F.D.L., Edwards C.J., Elias S.C., Elmore M.J., Emary K.R.W., English M.R., Fagerbrink S., Felle S., Feng S., Field S., Fixmer C., Fletcher C., Ford K.J., Fowler J., Fox P., Francis E., Frater J., Furze J., Fuskova M., Galiza E., Gbesemete D., Gilbride C., Gorini G., Goulston L., Grabau C., Gracie L., Gray Z., Guthrie L.B., Hackett M., Halwe S., Hamilton E., Hamlyn J., Hanumunthadu B., Harding I., Harris S.A., Harris A., Harrison D., Harrison C., Hart T.C., Haskell L., Hawkins S., Head I., Henry J.A., Hill J., Hodgson S.H.C., Hou M.M., Howe E., Howell N., Hutlin C., Ikram S., Isitt C., Iveson P., Jackson S., Jackson F., James S.W., Jenkins M., Jones E., Jones K., Jones C.E., Jones B., Kailath R., Karampatsas K., Keen J., Kelly S., Kelly D., Kerr D., Kerridge S., Khan L., Khan U., Killen A., Kinch J., King T.B., King L., King J., Kingham-Page L., Klenerman P., Knapper F., Knight J.C., Koleva S., Kupke A., Larkworthy C.W., Larwood J.P.J., Laskey A., Lawrie A.M., Lee A., Ngan Lee K.Y., Lee E.A., Legge H., Lelliott A., Lemm N.-M., Lias A.M., Linder A., Lipworth S., Liu X., Liu S., Lopez Ramon R., Lwin M., Mabesa F., Madhavan M., Mallett G., Mansatta K., Marcal I., Marinou S., Marlow E., Marshall J.L., Martin J., McEwan J., Meddaugh G., Mentzer A.J., Mirtorabi N., Moore M., Moran E., Morey E., Morgan V., Morris S.J., Morrison H., Morshead G., Morter R., Mujadidi Y.F., Muller J., Munera-Huertas T., Munro C., Munro A., Murphy S., Muster V.J., Mweu P., Noé A., Nugent F.L., Nugent E., O’Brien K., O’Connor D., Oguti B., Oliver J.L., Oliveira C., O’Reilly P.J., Osborn M., Osborne P., Owen C., Owens D., Owino N., Pacurar M., Parker K., Parracho H., Patrick-Smith M., Payne V., Pearce J., Peng Y., Peralta Alvarez M.P., Perring J., Pfafferott K., Pipini D., Plested E., Pluess-Hall H., Pollock K., Poulton I., Presland L., Provstgaard-Morys S., Pulido D., Radia K., Ramos Lopez F., Rand J., Ratcliffe H., Rawlinson T., Rhead S., Riddell A., Ritchie A.J., Roberts H., Robson J., Roche S., Rohde C., Rollier C.S., Romani R., Rudiansyah I., Saich S., Sajjad S., Salvador S., Sanchez Riera L., Sanders H., Sanders K., Sapaun S., Sayce C., Schofield E., Screaton G., Selby B., Semple C., Sharpe H.R., Shea A., Shelton H., Silk S., Silva-Reyes L., Skelly D.T., Smee H., Smith C.C., Smith D.J., Song R., Spencer A.J., Stafford E., Steele A., Stefanova E., Stockdale L., Szigeti A., Tahiri-Alaoui A., Tait M., Talbot H., Tanner R., Taylor I.J., Taylor V., Te Water Naude R., Thakur N., Themistocleous Y., Themistocleous A., Thomas M., Thomas T.M., Thompson A., Thomson-Hill S., Tomlins J., Tonks S., Towner J., Tran N., Tree J.A., Truby A., Turkentine K., Turner C., Turner N., Turner S., Tuthill T., Ulaszewska M., Varughese R., Van Doremalen N., Veighey K., Verheul M.K., Vichos I., Vitale E., Walker L., Watson M.E.E., Welham B., Wheat J., White C., White R., Worth A.T., Wright D., Wright S., Yao X.L., Yau Y. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., Wu S.-P., Wang B.-S., Wang Z., Wang L., Jia S.-Y., Jiang H.-D., Wang L., Jiang T., Hu Y., Gou J.-B., Xu S.-B., Xu J.-J., Wang X.-W., Wang W., Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhu F.-C., Guan X.-H., Li Y.-H., Huang J.-Y., Jiang T., Hou L.-H., Li J.-X., Yang B.-F., Wang L., Wang W.-J., Wu S.-P., Wang Z., Wu X.-H., Xu J.-J., Zhang Z., Jia S.-Y., Wang B.-S., Hu Y., Liu J.-J., Zhang J., Qian X.-A., Li Q., Pan H.-X., Jiang H.-D., Deng P., Gou J.-B., Wang X.-W., Wang X.-H., Chen W. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Şahin U., Jansen K.U. Phase 1/2 study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020 doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 234.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., Brachtendorf S., Lörks V., Sikorski J., Hilker R., Becker D., Eller A.-K., Grützner J., Boesler C., Rosenbaum C., Kühnle M.-C., Luxemburger U., Kemmer-Brück A., Langer D., Bexon M., Bolte S., Karikó K., Palanche T., Fischer B., Schultz A., Shi P.-Y., Fontes-Garfias C., Perez J.L., Swanson K.A., Loschko J., Scully I.L., Cutler M., Kalina W., Kyratsous C.A., Cooper D., Dormitzer P.R., Jansen K.U., Türeci Ö. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T-cell responses. Nature. 2020 doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 235.Xia S., Duan K., Zhang Y., Zhao D., Zhang H., Xie Z., Li X., Peng C., Zhang Y., Zhang W., Yang Y., Chen W., Gao X., You W., Wang X., Wang Z., Shi Z., Wang Y., Yang X., Zhang L., Huang L., Wang Q., Lu J., Yang Y., Guo J., Zhou W., Wan X., Wu C., Wang W., Huang S., Du J., Meng Z., Pan A., Yuan Z., Shen S., Guo W., Yang X. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes. JAMA. 2020;324:951. doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Logunov D.Y., Dolzhikova I.V., Zubkova O.V., Tukhvatullin A.I., Shcheblyakov D.V., Dzharullaeva A.S., Grousova D.M., Erokhova A.S., Kovyrshina A.V., Botikov A.G., Izhaeva F.M., Popova O., Ozharovskaya T.A., Esmagambetov I.B., Favorskaya I.A., Zrelkin D.I., Voronina D.V., Shcherbinin D.N., Semikhin A.S., Simakova Y.V., Tokarskaya E.A., Lubenets N.L., Egorova D.A., Shmarov M.M., Nikitenko N.A., Morozova L.F., Smolyarchuk E.A., Kryukov E.V., Babira V.F., Borisevich S.V., Naroditsky B.S., Gintsburg A.L. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396:887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M.B., Haupt R.E., Logue J., McGrath M., Weston S., Piedra P.A., Desai C., Callahan K., Lewis M., Price-Abbott P., Formica N., Shinde V., Fries L., Lickliter J.D., Griffin P., Wilkinson B., Glenn G.M. Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2026920. NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Luckheeram R.V., Zhou R., Verma A.D., Xia B. CD4 + t cells: differentiation and functions. Clin. Dev. Immunol. 2012;2012:1–12. doi: 10.1155/2012/925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lambert P.-H., Liu M., Siegrist C.-A. Can successful vaccines teach us how to induce efficient protective immune responses? Nat. Med. 2005;11:S54–62. doi: 10.1038/nm1216. [DOI] [PubMed] [Google Scholar]
- 240.Bournazos S., DiLillo D.J., Ravetch J.V. The role of Fc-FcγR interactions in IgG-mediated microbial neutralization. J. Exp. Med. 2015;212:1361–1369. doi: 10.1084/jem.20151267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Jansen J.M., Gerlach T., Elbahesh H., Rimmelzwaan G.F., Saletti G. Influenza virus-specific CD4+ and CD8+ T cell-mediated immunity induced by infection and vaccination. J. Clin. Virol. 2019;119:44–52. doi: 10.1016/j.jcv.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 242.Wherry E.J., Ha S.-J., Kaech S.M., Haining W.N., Sarkar S., Kalia V., Subramaniam S., Blattman J.N., Barber D.L., Ahmed R. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 243.Ng O.-W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.-J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Floriane Gallais S.F.-K., Velay Aurelie, Wendling Marie-Josee, Nazon Charlotte, Partisani Marialuisa, Sibilia Jean, Candon Sophie. Intrafamilial exposure to SARS-CoV-2 induces cellular immune response without seroconversion. medRxiv. 2020 doi: 10.1101/2020.06.21.20132449. 2020.06.21.20132449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.O’Neill L.A.J., Netea M.G. BCG-induced trained immunity: can it offer protection against COVID-19? Nat. Rev. Immunol. 2020;20:335–337. doi: 10.1038/s41577-020-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Stensballe L.G., Nante E., Jensen I.P., Kofoed P.E., Poulsen A., Jensen H., Newport M., Marchant A., Aaby P. Acute lower respiratory tract infections and respiratory syncytial virus in infants in Guinea-Bissau: a beneficial effect of BCG vaccination for girls: community based case-control study. Vaccine. 2005;23:1251–1257. doi: 10.1016/j.vaccine.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 247.Wardhana E.A., Datau A., Sultana V.V., Mandang E.Jim. The efficacy of Bacillus calmette-Guerin vaccinations for the prevention of acute upper respiratory tract infection in the elderly. Acta Med. Indones. 2011;43:185–190. [PubMed] [Google Scholar]
- 248.Ohrui T., Nakayama K., Fukushima T., Chiba H., Sasaki H. Prevention of elderly pneumonia by pneumococcal, influenza and BCG vaccinations. Japanese J. Geriatr. 2005;42:34–36. doi: 10.3143/geriatrics.42.34. [DOI] [PubMed] [Google Scholar]
- 249.Sharma A.R., Batra G., Kumar M., Mishra A., Singla R., Singh A., Singh R.S., Medhi B. BCG as a game-changer to prevent the infection and severity of COVID-19 pandemic? Allergol. Immunopathol. 2020 doi: 10.1016/j.aller.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Benn C.S., Netea M.G., Selin L.K., Aaby P. A small jab - a big effect: nonspecific immunomodulation by vaccines. Trends Immunol. 2013;34:431–439. doi: 10.1016/j.it.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 251.Miller A., Reandelar M.J., Fasciglione K., Roumenova V., Li Y., Otazu G.H. Correlation between universal BCG vaccination policy and reduced morbidity and mortality for COVID-19: an epidemiological study. medRxiv. 2020 doi: 10.1101/2020.03.24.20042937. 2020.03.24.20042937. [DOI] [Google Scholar]
- 252.Escobar L.E., Molina-Cruz A., Barillas-Mury C. BCG vaccine protection from severe coronavirus disease 2019 (COVID19) medRxiv. 2020;2019 doi: 10.1101/2020.05.05.20091975. 2020.05.05.20091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.US National Library of Medicine . 2020. Reducing Health Care Workers Absenteeism in Covid-19 Pandemic Through BCG Vaccine (BCG-CORONA), ClinicalTrials.Gov.https://clinicaltrials.gov/ct2/show/NCT04328441 (accessed August 6, 2020) [Google Scholar]
- 254.US National Library of Medicine . 2020. BCG Vaccination for Healthcare Workers in COVID-19 Pandemic, ClinicalTrials.Gov.https://clinicaltrials.gov/ct2/show/NCT04379336 (accessed August 6, 2020) [Google Scholar]
- 255.US National Library of Medicine . 2020. BCG Vaccination to Protect Healthcare Workers Against COVID-19 (BRACE), ClinicalTrials.Gov.https://clinicaltrials.gov/ct2/show/NCT04327206 (accessed August 6, 2020) [Google Scholar]