Abstract
COVID-19 is a pandemic that began to spread worldwide caused by SARS-CoV-2. Lung cancer patients are more susceptible to SARS-CoV-2 infection. The SARS-CoV-2 enters into the host by the ACE2 receptor. Thus, ACE2 is the key to understand the mechanism of SARS-CoV-2 infection. However, the lack of knowledge about the biomarker of COVID-19 warrants the development of ACE2 biomarkers. The analysis of ACE2 expression in lung cancer was performed using The Cancer Genome Atlas (TCGA). Therefore, we investigated the prognosis, clinical characteristics, and mutational analysis of lung cancer. We also analyzed the shared proteins between the COVID-19 and lung cancer, protein-protein interactions, gene-miRNAs, gene-transcription factors (TFs), and the signaling pathway. Finally, we compared the mRNA expression of ACE2 and its co-expressed proteins using the TCGA. The up-regulation of ACE2 in lung adenocarcinoma (LUAD) and lung squamous carcinoma (LUSC) was found irrespective of gender and age. We found the low survival rate in high expression of ACE2 in lung cancer patients and 16 mutational positions. The functional assessment of targeted 12,671, 3107, and 29 positive genes were found in COVID-19 disease, LUAD, and LUSC, respectively. Then, we identified eight common genes that interact with 20 genes, 219 miRNAs, and 16 TFs. The common genes performed the mRNA expression in lung cancer, which proved the ACE2 is the best potential biomarker compared to co-expressed genes. This study uncovers the relationship between COVID-19 disease and lung cancer. We identified ACE2 and also its co-expressed proteins are the potential biomarker and therapy as the current COVID-19 disease and lung cancer.
Keywords: COVID-19, Lung cancer, ACE2, mRNA expression analysis, Relationship of lung cancer and COVID-19, Potential biomarker
Graphical abstract
Highlights
-
•
High transcriptional expression of ACE2 due to COVID-19 may result in poor lung cancer outcome.
-
•
The investigation of clinical profiles and mutational positions of ACE2 in lung cancer.
-
•
The gene ontologies and pathways of ACE2 and their co-expressed genes were identified.
-
•
The protein-protein, gene-miRNA, and gene-TF interactions were identified.
-
•
ACE2 and also its co-expressed proteins are the potential biomarker and therapy as the current COVID-19 disease and lung cancer.
1. Introduction
A respiratory tract disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become pandemic around the world. The first incidence of this SARS-CoV-2 was found in Wuhan, China, in December 2019 [1]. Among four coronavirus genera α, β, γ, and δ, SARS-CoV-2 belongs to β-coronavirus. Additionally, the other two β coronaviruses are middle east respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV, but SARS-CoV-2 is more identical to SARS-CoV [2,3].
From the day of first reporting, above 4.5 million cases were confirmed globally with more than 2.47 lacs deaths [4,5]. Although immune-compromised patient, older people and people with disease history may develop dangerous outcome like acute respiratory distress syndrome (ARDS) [2], many research suggested that cancer patients are more susceptible to SARS-CoV-2 infection than individuals with COVID-19 alone because of their systemic immunosuppressive state caused by the malignancy [6,7]. A retrospective study in a hospital in Wuhan China, a group of 28 patients with COVID-19 and cancer was taken for observation. In that study, 10 patients (35.7%) were in stage IV where lung cancer was the most frequent (7; 25%). As of February 28th, 15 (53.6%) patients have developed severe conditions, 10 (35.7%) patients have developed the life-threatening condition and 8 (28.6%) patients died [7]. Metastasis in the lung makes patients seriously vulnerable to COVID-19 that might result in poor prognosis, pneumonia, and various lung problem [8]. Several signs and symptoms were found in COVID-19 patients, including invasive lung lesions, fatigue, fever, breathlessness, cough, and many others [5,9,10]. Due to elevated cytokine levels such as TNFα, MIP1A, GCSF, IL (2, 7, 10), which may develop into worse outcomes like pneumonia, respiratory failure, and death have been described [11]. Lung cancer patients are profoundly affected by SARS-CoV-2 [6]. Till March 2020, the lung cancer patients who had SARS-CoV-2 infection have a mortality rate of 5.6% in China and 15.2% in outside of China [12].
Angiotensin-converting enzyme 2 (ACE2) is a vital regulator of the renin-angiotensin system (RAS) expressed in the lung alveolar cell type II [13,14]. The primary function of ACE2 is to convert angiotensin II into angiotensin-(1–7). The blocking of the angiogenesis system and the growth of tumor cells are other roles of ACE2 [15]. SARS-CoV-2 occupies the ACE2 receptor for its entry by serving as attaching sites for spike (S) protein [16]. The pro-inflammatory response is an immunoreaction of the lung to defend against SARS-CoV-2 viral infection [17]. This response comprises the cytokine storm, including tumor necrosis factor-alpha (TNF-α) and interleukin six (IL-6) that play an essential role in toll-like receptor (TLR) signaling pathway. By activating T helper cells, these elevated levels of cytokine cause acute respiratory distress syndrome (ARDS) and many organ failures. Besides, TLR4 is also responsible for immune-pathological exposure of COVID-19 as it is the most effective innate immune receptor and showed a strong binding affinity toward surface protein. Some recent investigations also recorded that TLR1, TLR4, and TLR6 exhibit efficient binding affinity toward S glycoprotein of SARS-CoV-2 [17,18].
ACE2 is also expressed in other organs like heart, kidney, endothelium, and intestine, but due to the nature of COVID-19 infection, we choose lung as our target organ. Moreover, the COVID-19 virus has its primary infection site in the lung and it also shows the worst severity in the lung [19,20]. As the COVID-19 virus demonstrates its primary symptom in lung and the infection increase the expression of ACE2, there is a significant possibility of lung cancer progression due to COVID-19 infection. In lung cancer angiotensin-(1–7) show an elevated level of concentration in serum from lung and ACE2 inhibitors showed an increase in the apoptosis of lung cancer cell in human and rat alveolar epithelial cells [21]. Thus, build up the relation between lung cancer and SARS-CoV-2 infection. Scarcity of ACE2 expression is a guide to abnormal lung function due to decayed vascular permeability, neutrophil infiltration, and enhanced lung edema that make the lung more prone to SARS-CoV-2 infection [22,23]. Maintained level of ACE2 improves patient's survival in lung cancer; enzymatic activity leads to an inflammatory storm that will help to eliminate SARS-CoV-2 from the lung by interrupting in gas exchange between alveoli and capillaries. Elevated or uncontrolled levels of ACE2 expression with S protein enhance the chance of SARS-CoV-2 infection [22,23]. Finally, ACE2 could be identified as a potential biomarker and therapy target in SARS-CoV-2 infection.
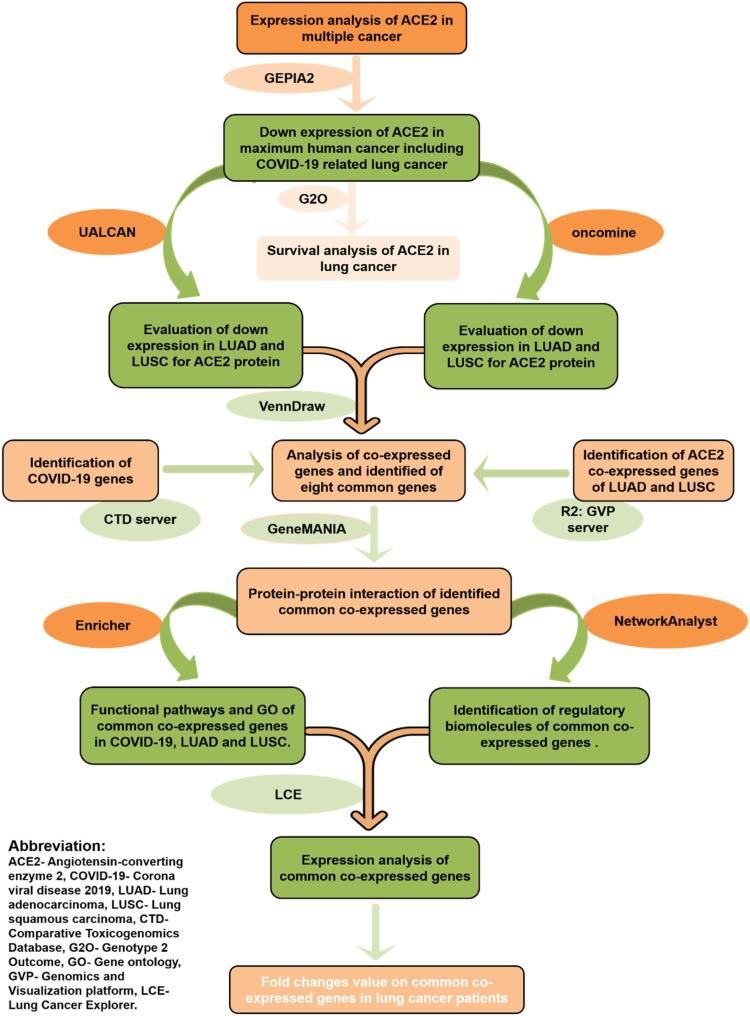
Due to a lack of a specific vaccine or drug, it is challenging to face such a wide-spreading pandemic. In our study, we presented a bioinformatics analysis using various web-servers to identify the expression, clinical characteristics, mutational positions, the survival rate of patients, and co-expression of ACE2. We also revealed the co-expressed genes of ACE2 and their possible protein-protein and transcriptional interaction. Our result uncovered that ACE2 is vitally involved with both COVID-19 and lung cancer. Our aim is to improve patient survival and early diagnosis by using ACE2 as a biomarker. A flow chart representing the overall procedure for the target identification and analysis has been illustrated in Fig. 1 .
Fig. 1.
Schematic representation of the overall workflow of this research.
2. Methods and materials
2.1. Conducting expression analysis of ACE2
To explore the finding behind the transcriptional expression of ACE2 in multiple cancer, we applied a publicly available online tool Gene Expression Profiling Interactive Analysis 2 (GEPIA2) server (http://gepia2.cancer-pku.cn/#index) using the TCGA (The Cancer Genome Atlas) datasets [24]. The GEPIA2 server contains data from 9667 tumors and 602 healthy tissues. Furthermore, we studied the mRNA expression data of ACE2 in lung cancer through the UALCAN website (http://ualcan.path.uab.edu/index.html) using the TCGA dataset [25,26]. The control samples were 59 and cancer samples were 515 in lung adenocarcinoma (LUAD) while lung squamous carcinoma (LUSC) control samples were 52 and cancer samples were 552. Finally, the mRNA expression of ACE2 was analyzed using the Oncomine tool (https://www.oncomine.org/resource/main.html) and TCGA datasets (control samples = 50 and LUAD samples = 226, control samples = 65 and LUSC samples = 27) [27,28]. Oncomine and UALCAN both are user-friendly web-platforms used to reveal the gene expression of TCGA data and relative expression of a query gene. We set the threshold of p-value <0.05.
2.2. Relation of ACE2 expression with clinicopathological parameters in lung cancer patients
UALCAN is a user-friendly publicly available web-portal that is used to explore the expression of a query gene and measure the influence of clinical features on gene expression level. To observe the association of ACE2 mRNA expression with the patient's gender and age in LUAD and LUSC, we used the UALCAN web portal (http://ualcan.path.uab.edu/index.html) [25,26].
2.3. Survival analysis and mutational analysis of ACE2 in lung cancer
The survival analysis of ACE2 in lung cancer was performed using the Genotype 2 Outcome server. The Genotype 2 Outcome server provides the Kaplan-Meier plots using the TCGA data. The number of patient samples was 555. The threshold for statistical significance in the survival analysis was set at p-value < 0.01 and average HR >1.4 (average HR is based on the mean of HR in the cohort having a higher HR and 1/HR of the cohort having a lower HR value). In the entire analysis pathway, none of the samples involved in the training (ROC analysis) are included in the test (survival analysis) as well [29]. Therefore, we performed the analysis of the mutational position by cBioPortal (http://cbioportal.org/) in lung cancer through the TCGA PanCancer Atlas data [30]. The total number of samples was 1053 and threshold parameters were set to default.
2.4. Identification of COVID-19 genes and co-expression profile of ACE2
To collect the co-related genes of LUAD and LUSC in R2 Genomics Analysis and Visualization Platform using TCGA datasets [31]. The COVID-19 genes were downloaded from the Comparative Toxicogenomics Database (CTD) [32] and all data was downloaded in text format. The gathered genes were visualized using FunRich software (http://funrich.org/index.html) for the Venn diagram [33].
2.5. Protein-protein interaction of commonly co-expressed genes
GeneMANIA provides a very flexible interface for the query of genomic, proteomic, and gene function data. It is an extensive database of functional association data, including protein and gene interactions, pathways, and co-expression [34,35]. For uncovering the vital proteins and pathways in the gene set, the PPI network of the signature extracted from GeneMANIA (https://genemania.org/). The figure was visualized using Cytoscape 3.7.2 software (https://cytoscape.org/) [36].
2.6. Network analysis of miRNAs and TFs with target genes
The regulatory Network (miRNAs and transcription factors) analysis with target genes was done by using the Network analyst web tool (https://www.networkanalyst.ca/) [37]. We employed a comprehensive human transcriptional and post-transcriptional regulatory network in the identification of transcription factors (TFs) and miRNAs. Changes in TF and miRNA show a significant change in transcription level and post-transcription level of the gene ACE2.
2.7. Functional pathway and gene ontology analysis of common genes with COVID-19 and lung cancer
Gene ontology was predicted for the computational presentation of the biological system, mainly of those biological products that are involved in the cellular process. It was also designed to view the functional annotation of genes for various model organisms. That has three different sectors, such as molecular function, cellular function, biological function [38]. Kyoto encyclopedia of genomes and gene (KEGG) is a database for utilities of the biological systems and high-level role, through molecular-level information of genome sequencing and other high-throughput experimental technologies [18]. Common co-related genes of COVID-19 and lung cancer were analyzed for their functional enrichment pathway and gene ontology performed by the Enrichr web portal [39]. The study of pathway databases was used for pathway enrichment investigation to assess the potential association of the signature with pathways.
2.8. Examination of co-expression of common genes in COVID-19 and lung cancer
The expression analysis of common genes in lung cancer was conducted by the Lung Cancer Explorer Platform (http://lce.biohpc.swmed.edu/lungcancer/index.php#about) [40]. The Lung Cancer Explorer is a comprehensive analysis tool for lung cancer microarray data from the TCGA. The MORPHEUS server (https://software.broadinstitute.org/morpheus/) was used for the visualization of mRNA expression via heatmap [41]. Therefore, we also investigated the mRNA expression of common genes in the Oncomine database through the TCGA dataset [27,28]. The parameters were set to p-value <0.05 and fold change = 1.
3. Results
3.1. Conducting expression analysis of ACE2
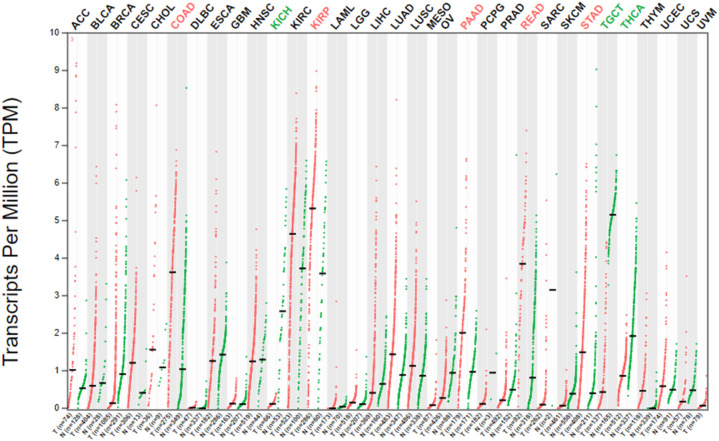
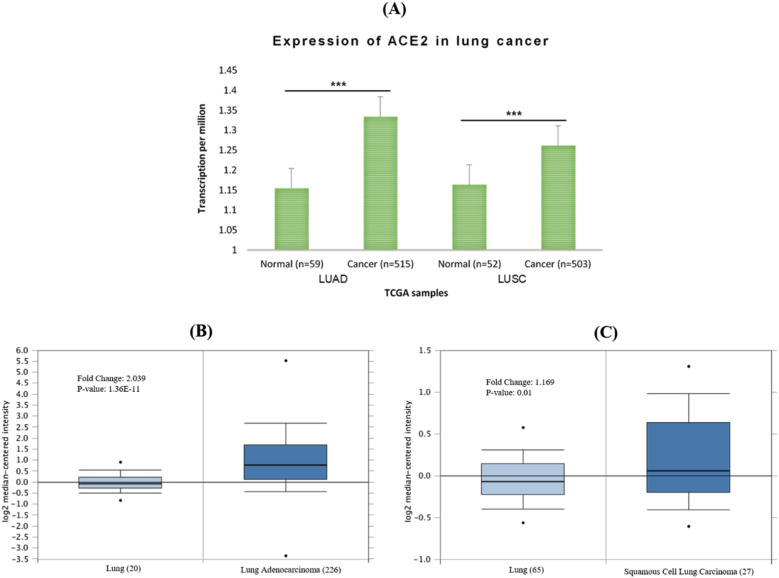
Using the GEPIA2 server, we observed the ACE2 expression in multiple human cancers. The result was illustrated that ACE2 is expressed in both LUAD and LUSC, which suggests that ACE2 is also involved with lung cancer (Fig. 2 ). For further examination, interactive web resources Oncomine and UALCAN were used for mRNA expression of ACE2 in lung cancer (LUAD and LUSC). Significant up-regulation of the ACE2 mRNA level was noticed in LUAD and LUSC when contrasted with healthy control in the TCGA dataset (Fig. 3A). Further, investigation of ACE2 mRNA data also showed indicatory expression in LUAD and LUSC as compared with normal cells (Fig. 3B-C). The results suggested strong and significant evidence regarding the higher expression of ACE2 in LUAD and LUSC tissues compared to their corresponding normal tissues (Fig. 2, Fig. 3).
Fig. 2.
The transcriptional expression of ACE2 in COVID-19 infected human cancer. The red color was representing the cancer cells and the green color was representing the healthy cells. Abbreviation: LUAD- Lung adenocarcinoma, LUSC- lung squamous carcinoma. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
The mRNA expression of ACE2 in lung cancer. (A) ACE2 expression in lung cancer obtained by using UALCAN web, (B) ACE2 expression in LUAD, and (C) LUSC using Oncomine online tools compared to healthy cells. The threshold p-value: * < 0.05, ** < 0.01, and *** < 0.001. Abbreviation: LUAD- Lung adenocarcinoma, LUSC- lung squamous carcinoma. TCGA- The cancer genome atlas.
3.2. Association of clinical parameter and ACE2 expression in LUAD and LUSC
The relation between ACE2 mRNA expression and the clinical characteristics of LUAD and LUSC was done using the TCGA dataset. Compared with normal tissues, ACE2 expression was augmented by the patient's age and gender. Both gender and age's people identified with up-regulation in both LUAD and LUSC (Table 1 ). Although more male, female, young, and older ages samples were identified with high mRNA expression compared with normal samples in both LUAD and LUSC. The results also suggest strong and significant evidence regarding the higher expression of ACE2 in lung cancer.
Table 1.
The ACE2 mRNA expression levels of LUAD and LUSC in human gender and age patients.
| Cancer typea | Type of samples | Expression of mRNA | Number of samples | p-value |
|---|---|---|---|---|
| LUAD | Patient's gender | |||
| Normal | ↓ | 59 | ||
| Male | ↑ | 238 | 6.10E-04 | |
| Female | ↑ | 276 | 1.45E-09 | |
| Patient's age | ||||
| Normal | ↓ | 59 | ||
| 21–40 Yrs | ↑ | 12 | 8.32E-01 | |
| 41–60 Yrs | ↑ | 90 | 3.74E-05 | |
| 61–80 Yrs | ↑ | 149 | 2.29E-06 | |
| 81–100 Yrs | ↑ | 32 | 9.72E-03 | |
| LUSC | Patient's gender | |||
| Normal | ↓ | 52 | ||
| Male | ↑ | 366 | 4.06E-06 | |
| Female | ↑ | 128 | 6.17E-04 | |
| Patient's age | ||||
| Normal | ↓ | 52 | ||
| 21–40 Yrs | ↑ | 2 | 5.71E-02 | |
| 41–60 Yrs | ↑ | 103 | 7.70E-03 | |
| 61–80 Yrs | ↑ | 361 | 2.56E-06 | |
| 81–100 Yrs | ↑ | 20 | 1.51E-01 |
Down-arrow (↓) indicates the under-expression while up-arrow (↑) indicates the over-expression.
3.3. Survival-analysis and identification of mutational positions of ACE2 in human LUAD and LUSC
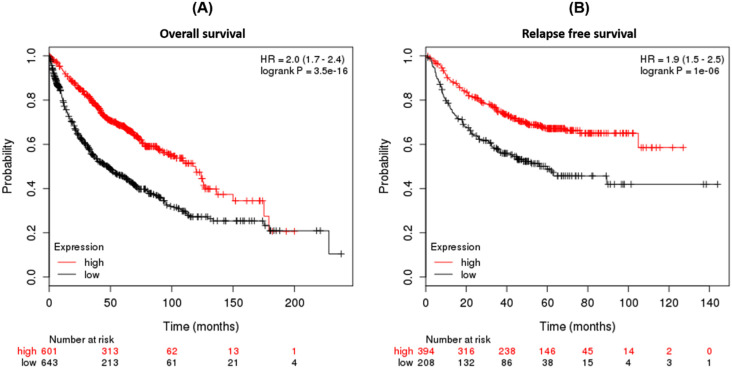
We investigated the ACE2 survival rate in lung cancer. We found both overall (OS) and relapse-free survival (RFS) had a low survival rate in high mRNA expression of ACE2 and higher HR value in lung cancer patients. In the entire analysis pathway, none of the samples involved in the training (ROC analysis) are included in the test (survival analysis) as well (Fig. 4 ). Therefore, we also investigated the mutational analysis of ACE2 protein, where in total 16 mutations in lung cancer were observed - 9 missense mutations in LUAD, while 4 missenses, 2 splice, and 1 frameshift mutations in LUSC (Table 2 ).
Fig. 4.
Survival plot of lung cancer patients associated with differential ACE2 expression. (A) Overall survival and (B) relapse-free survival in lung cancer. The red color was indicating the high expression and black color was indicating the low expression. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
The ACE2 mutational types and positions in human LUAD and LUSC obtained from the TCGA dataset.
| Cancer type | Sample ID | Protein change | Mutation type | Sample |
|---|---|---|---|---|
| LUAD | TCGA-95-7948-01 | I256M | Missense | 142 |
| TCGA-44-2659-01 | R219P | Missense | 396 | |
| TCGA-44-7670-01 | G211W | Missense | 1017 | |
| TCGA-69-7980-01 | L320F | Missense | 737 | |
| TCGA-73-4658-01 | D693N | Missense | 323 | |
| TCGA-99-7458-01 | H34N | Missense | 437 | |
| TCGA-55-8205-01 | T798P | Missense | 473 | |
| TCGA-55-8506-01 | V670L | Missense | 1583 | |
| TCGA-62-A46R-01 | A99S | Missense | 170 | |
| LUSC | TCGA-22-1016-01 | V491L | Missense | 315 |
| TCGA-39-5035-01 | G147V | Missense | 137 | |
| TCGA-43-6770-01 | X233_splice | Splice | 202 | |
| TCGA-33-4587-01 | Y633Lfs*2 | Frame Shift | 206 | |
| TCGA-56-7731-01 | G395V | Missense | 325 | |
| TCGA-92-7341-01 | W477R | Missense | 154 | |
| TCGA-L3-A524-01 | X195_splice | Splice | 299 |
3.4. Employment of COVID-19 and lung cancer-infected genes
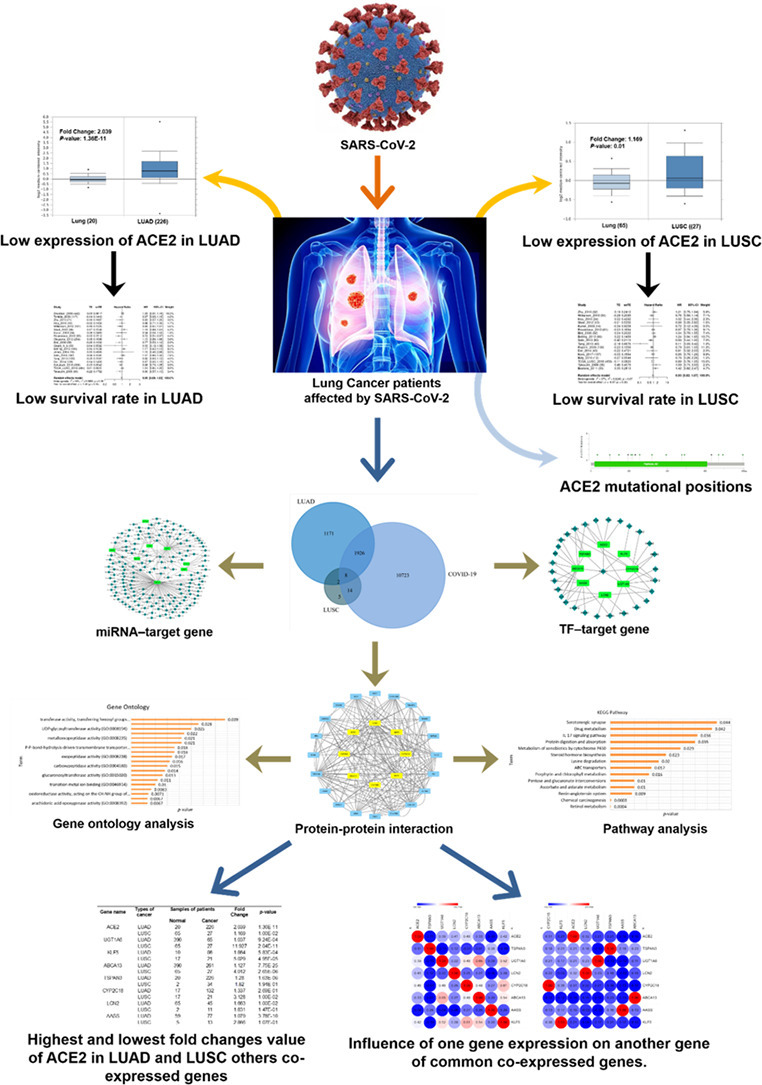
Using the comparative toxicogenomics database, R2 genomics analysis and visualization platform, we collected the genes of COVID-19 and used the R2 genomics analysis and visualization platform for identification of co-expressed genes of ACE2 in LUAD and LUSC. Total 3107 genes of LUAD, 29 genes of LUSC, and COVID-19 genes of 12,671 were found to be co-expressed (Supplementary file 1). To elucidate the overlapping between COVID-19, LUAD, and LUSC genes, we applied FunRich analysis tool (Supplementary file 2). The graphical visualization depicted eight genes that were marked as co-expressed genes of COVID-19, LUAD, and LUSC from the Venn diagram (Fig. 5 , Supplementary file 2). The 8 genes are angiotensin-converting enzyme 2 (ACE2), human ATP binding cassette gene subtype (ABCA13), uridine diphosphoglucuronosyltransferase 1A6 (UGT1A6), krüppel-like factors 5 (KLF5), lipocalin-2 (LCN2), tetraspanins-3 (TSPN3), α-aminoadipic semialdehyde synthase (AASS), and hepatic cytochrome 450 superfamily gene (CYP2C18). The overlapped region points out the commonly co-expressed genes in LUAD, LUSC, and COVID-19.
Fig. 5.
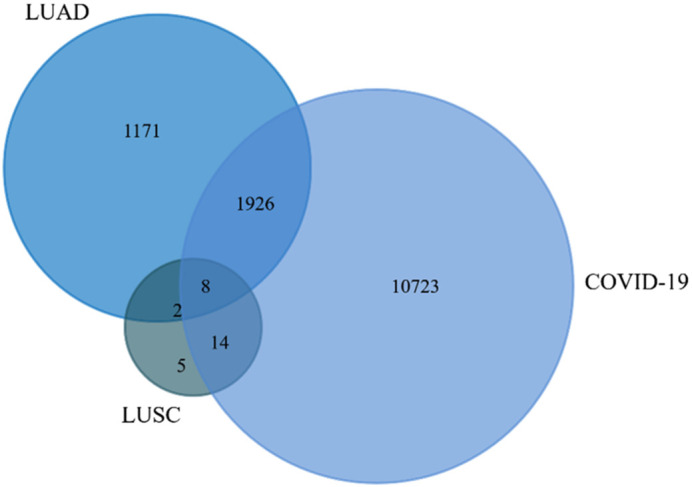
Identification of commonly co-expressed genes with lung cancer and COVID-19 performed by the FunRich software. Abbreviation: LUAD- Lung adenocarcinoma, LUSC- lung squamous carcinoma.
3.5. Construction of protein-protein interaction (PPI) network
To know the depth of the disease and to predict phenotypic-genotypic association, sufficient resources were used to construct protein networks. The protein-protein interaction of ACE2, ABCA13, UGT1A6, KLF5, LCN2, TSPN3, AASS, and CYP2C18 form a protein network was consisting themselves and keratin 7 (KRT7), CEA cell adhesion molecule 5 (CEACAM5), transmembrane 4 six family member 4 (TM4SF4), tripartite motif-containing 31 (TRIM31), glutathione S-transferase Mu 3 (GSTM3), E74 Like ETS transcription factor 3 (ELF3), peptidase inhibitor 3 (PI3), olfactomedin 4 (OLFM4), galectin 4 (LGALS4), chromosome 1 open reading frame 106 (C1orf106), Cbl proto-oncogene C (CBLC), argininosuccinate synthase 1 (ASS1), serine peptidase inhibitor kazal type 1 (SPINK1), S100 calcium-binding protein P (S100P), tetraspanin 8 (TSPAN8), paraoxonase 1 (PON1), fyn related src family tyrosine kinase (FRK), ATPase secretory pathway Ca2+ transporting 2 (ATP2C2), suppressor of cytokine signaling 6 (SOCS6), and secretory leukocyte peptidase inhibitor (SLPI). Other genes that have a relation with these eight genes are also presented (Fig. 6 ). The result suggested that the identified ACE2 and co-expressed genes might be potentially associated with COVID-19 and lung cancer.
Fig. 6.
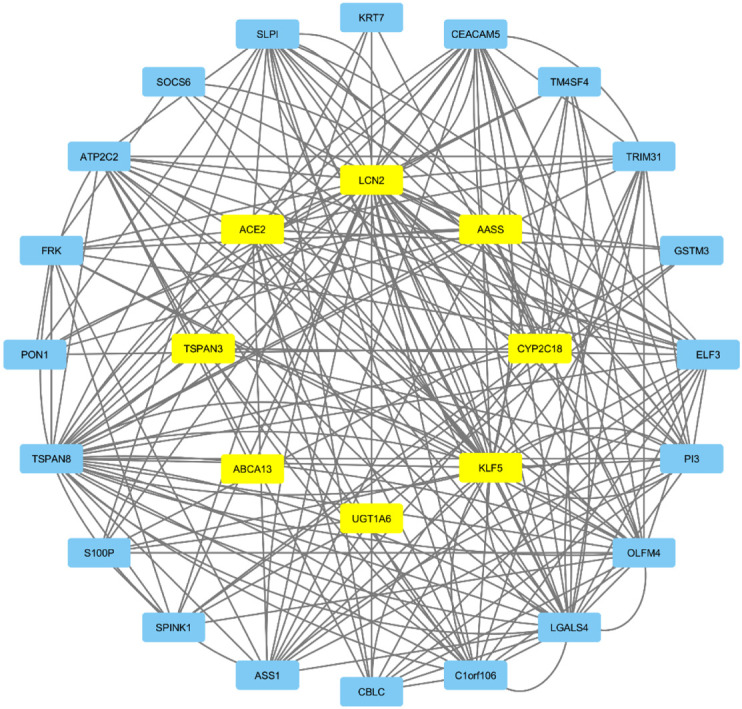
Protein-protein interaction of commonly co-expressed genes in lung cancer and COVID-19 performed by the GeneMANIA server and visualized in Cytoscape 3.7.2 software. The yellow color was representing the commonly co-expressed genes and blue color representing the interacting genes with commonly co-expressed genes. Abbreviation: LUAD- Lung adenocarcinoma, LUSC- lung squamous carcinoma. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
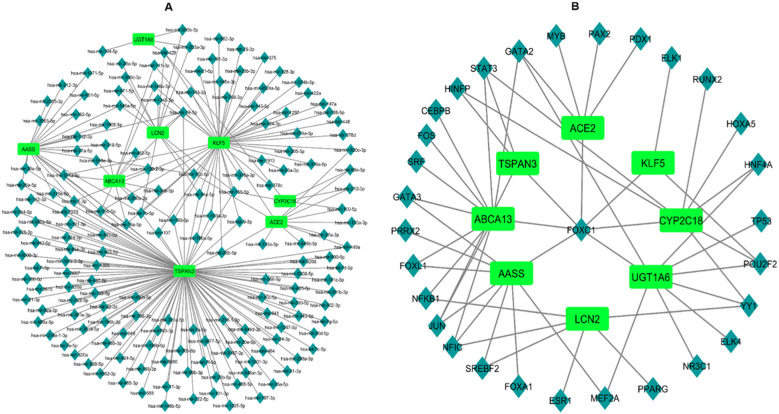
3.6. miRNAs and TFs target genes network investigation
After the construction of miRNAs and TFs target genes network, we observed that there are a total of 219 miRNAs shared between the 8 co-expressed genes (Fig. 7A). On the other hand, there are 30 TFs between the 8 co-expressed genes (Fig. 7B). From the 219 miRNAs ACE2 shares has-mir-26b-5p with TSPN3 and KLF5, has-mir-10b-5p with KLF5 and AASS, has-mir-499a with TSPN3, has-mir-100-5p with CYP2C18, has-mir-520c-3p with KLF5, has-mir-99a-5p with CYP2C18, has-mir-210-3p with CYP2C18 and has-mir-449a with TSPN3. The correlating TFs of ACE2 with the other 7 genes are GATA2, MYB, PAX2, PDX1, and FOXC1. In TFs, we can see that FOXC1 has a relation with almost all the 8 genes except LCN2. TFs and miRNAs relation might indicate the expression of the genes and disease progression.
Fig. 7.
(A) Regulatory interacting network of the miRNAs-target genes. Herein, the square shape indicates the gene symbols while diamond shape indicates the miRNA symbols. (B) The interacting regulatory network of the TFs-target genes. Herein, the square shape indicates the gene symbols while diamond shape indicates the TF symbols.
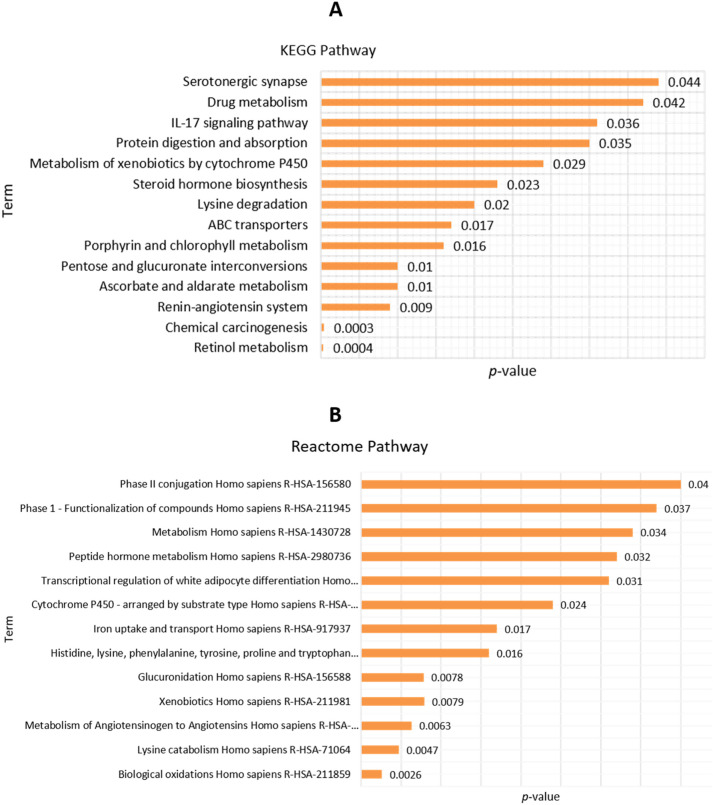
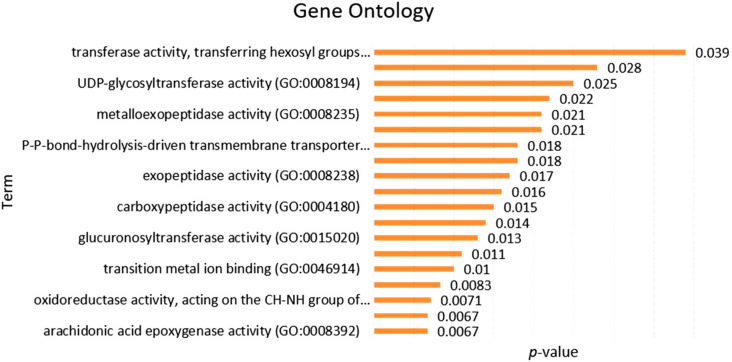
3.7. Functional pathway and gene ontology
For integrative analysis of enrichment pathways, we used the Enrichr web tool. At first, eight co-expressed genes that were shared between COVID-19 and lung cancer used for functional pathway analysis. The KEGG pathways are involved in the serotonergic synapse, drug metabolism, IL-17 signaling pathway, protein digestion, and absorption, metabolism of xenobiotic, steroid hormone biosynthesis, ABC transporter, and renin-angiotensin system (Fig. 8A, Supplementary file 3). The Reactome pathway involved in phase II conjugation, metabolism of Homo sapiens, peptide hormone metabolism, iron uptake, and transport (Fig. 8B, Supplementary file 4). Gene ontology was also performed by Enrichr that involved transferase activity, transferring hexosyl, UDP-glycosyltransferase, and metalloexopeptide activity (Fig. 9 Supplementary file 5). Our data claim that the common co-expressed genes of ACE2 are significantly involved with many biological processes of the cell.
Fig. 8.
The functional pathways of common co-expressed genes with lung cancer and COVID-19 performed by the Enricher online tool: (A) KEGG pathway, and (B) Reactome pathway.
Fig. 9.
The functional gene ontology analysis of commonly co-expressed genes in lung cancer and COVID-19 was performed by the Enricher online tool.
3.8. Importance of ACE2 selection for LUAD and LUSC
Through the Oncomine server, we collected the fold change value of ACE2 for LUAD and LUSC. The result showed that high fold change value in LUAD and low at LUSC compared to other co-expressed genes. The fold change value indicates the ACE2 is the best potential biomarker and other co-expressed genes also potentials biomarkers in COVID-19 infected lung cancer (Table 3 ).
Table 3.
The fold changes value on common co-expressed genes in lung cancer patients from the Oncomine database.
| Gene name | Types of cancer | Samples of patients |
Fold change | p-value | |
|---|---|---|---|---|---|
| Normal | Cancer | ||||
| ACE2 | LUAD | 20 | 226 | 2.039 | 1.36E-11 |
| LUSC | 65 | 27 | 1.169 | 1.00E-02 | |
| UGT1A6 | LUAD | 390 | 65 | 1.037 | 9.24E-04 |
| LUSC | 65 | 27 | 11.927 | 2.04E-11 | |
| KLF5 | LUAD | 10 | 86 | 1.864 | 5.83E-04 |
| LUSC | 17 | 21 | 5.029 | 4.95E-05 | |
| ABCA13 | LUAD | 390 | 261 | 1.127 | 7.75E-25 |
| LUSC | 65 | 27 | 4.012 | 2.65E-06 | |
| TSPAN3 | LUAD | 20 | 226 | 1.28 | 1.63E-06 |
| LUSC | 2 | 34 | 1.82 | 1.94E-01 | |
| CYP2C18 | LUAD | 17 | 132 | 1.337 | 2.69E-01 |
| LUSC | 17 | 21 | 3.128 | 1.00E-02 | |
| LCN2 | LUAD | 65 | 45 | 1.663 | 1.00E-02 |
| LUSC | 2 | 11 | 1.831 | 1.47E-01 | |
| AASS | LUAD | 59 | 77 | 1.079 | 3.78E-10 |
| LUSC | 5 | 13 | 2.866 | 1.07E-01 | |
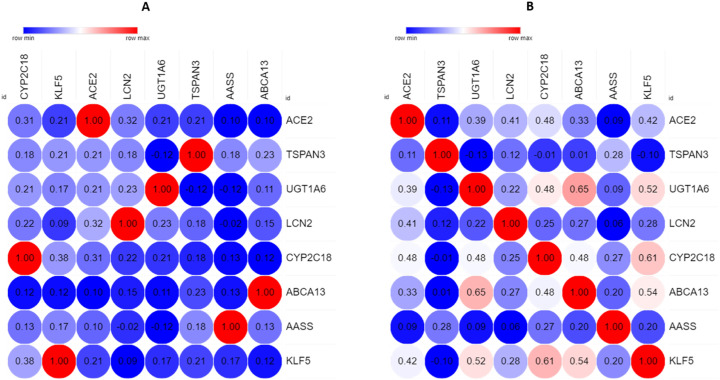
3.9. Investigation of co-expression of ACE2 associated genes
To address the impact of each gene expression on another expression, we used the Lung Cancer Explorer server. The heatmap depicted that the eight co-expressed genes were expressed. There is the influence of the one gene expression on another gene expression (Fig. 10 ). The result suggested that the identified co-expressed genes of ACE2 might be potentially associated with lung cancer.
Fig. 10.
The co-expression analysis of common co-expressed genes in COVID-19 infected lung cancer was performed by the Lung Cancer Explorer server: (A) LUAD, and (C) LUSC. The correlation result was visualized by the MORPHEUS server.
4. Discussion
COVID-19 is now the number one public health issue in the world as it was declared a pandemic by the World Health Organization [42]. This research aims to possibly establish a relationship between lung cancer and COVID-19 using a commonly expressed gene ACE2. The relation between lung cancer and COVID-19 patients would indicate that ACE2 can be a biomarker for COVID-19 cases. It will deescalate the risk factor of lung cancer patients with SARS-CoV-2 infection and improve the patient's survival rate.
In our study, we observed that ACE2 mRNA expression in human cancer and COVID-19 infected lung cancers by analyzing in GEPIA2, Oncomine, and UALCAN servers. The interesting fact about this is, in COVID-19 infected cases, the ACE2 gene is up-regulated, where in both LUAD and LUSC cases, the gene was also up-regulated. The spike protein of COVID-19 uses ACE2 as a host receptor for entry into a host cell. It has been proven in rat or mouse models [[43], [44], [45]]. The upregulation of ACE2 is common in LUAD and LUSC; the tumor growth observation in various studies correlated with ACE2 overexpression [46,47]. The up-regulation of ACE2 can be explained as COVID-19 inhibits RAS, which is the main pathway of ACE2 peptide [48]. There has been a correlation between smoking and the upregulation of ACE2 in LUAD and LUSC patients. It puts those patients at high risk of SARS-CoV-2 infection and possibly deadly outcome [13,49,50]. Our study showed an increased rate of expression of ACE2 in an elderly male patient in the age group of 60–80 who has LUAD or LUSC. Recent research on COVID-19 patients in Italy and United States (US) suggested that patients in the age group of 60–80 are highly susceptible to this disease and their mortality rate is also high [[51], [52], [53]]. So, it might correlate COVID-19 patients and lung cancer patients via the overexpression of ACE2. We also conducted a prognostic analysis of ACE2 in lung cancer. In both OS and RFS, we found that ACE2 mRNA is highly expressed; this correlates with studies that indicate poor survival of COVID-19 patients with lung malignancy [54,55]. ACE2 has been proven to be an essential regulator in tumorigenesis. For instance, ACE2 inhibits breast cancer angiogenesis through suppressing VEGFa/VEGFR2/ERK pathway [56] and reduces cell invasion and migration in NSCLC cells [57]. Furthermore, we also found 16 mutations in the ACE2 gene in lung cancer patients that may aid in COVID-19 virus entry or disease severity depending on the mutations type and position [58].
This study also associates eight genes in positive co-expressed in LUAD, LUSC, and COVID-19 patients via the Venn diagram. The eight genes protein-protein interaction analysis showed 20 other genes that interact with these eight co-expressed genes; this was done by GeneMANIA server. Human ATP binding cassette gene subtype (ABCA13) up-regulates when exposed to cigarette smoke, which increases the risk factor of LUAD and LUSC [59]. Another peptide Uridine diphosphoglucuronosyltransferase 1A6 (UGT1A6) is responsible for the detoxification of carcinogenic agents from lung cells [60]. Nevertheless, in our finding, we observed that UGT1A6 is significantly less expressed when ACE2 expression is high. Krüppel-like factors 5 (KLF5) is another gene that is expressed along with ACE2, KLF5 inhibits non-small cell lung cancer cell by HIF-1α [61]. Our co-expression study indicates low expression of KLF5 in LUAD when ACE2 expression is high, but for LUSC the expression is significantly higher. Lipocalin-2 (LCN2) showed significant radioresistance in other studies for oral and lung carcinoma [62]. So, in our study, the moderated co-expression of LCN2 still bears significance in the clinical outcome of LUAD and LUSC because of non-responsiveness to radiotherapy. The other three genes Tetraspanins-3 (TSPN3), α-Aminoadipic Semialdehyde Synthase (AASS), and hepatic cytochrome 450 superfamily gene (CYP2C18) show little relation to lung carcinoma and their co-expression with ACE2 is also low. There is a significant lack of study considering these 3 genes concerning lung cancer, but they might also correlate with lung cancer. So, 5 out of 8 genes (ACE2, ABCA13, UGT1A6, KLF5, LCN2) are significantly related to COVID-19 and Lung cancer. That further correlates these co-expressed genes with ACE2. It also up-regulates in LUAD and LUSC development. So, ACE2 helping in the entry of the COVID-19 virus into the cell may lead to lung cancer, as evident in the co-expressed genes contributing to lung cancer progression and its upregulation in LUAD and LUSC. The co-expressed genes also aid in the progression of lung cancer and COVID-19 by suppressing the immune system and resisting therapy. Moreover, transcription factor (TF) controls the rate of transcription [63] and miRNA plays a role in RNA silencing and regulation of gene expression at the post transcription level [64]. In this regard, this study uncovers the relationship shared between the co-expressed genes and their respective TFs and regulatory miRNAs. The functional pathway analysis of commonly co-expressed genes was done by Enricher online tool, where the KEGG pathway indicates IL-17 signaling pathway significant for the commonly expressed genes. That concludes the hyper-inflammatory state of COVID-19 patients upon infection compared with normal inflammation in viral infection [65]. This cytokine storm leads to prolonged alveolar damage and excessive neutrophil migration. The damage caused in severe condition effect gas exchange, as a result, there is difficulty in breathing among patients [66]. The selection of ACE2 is important for LUAD and LUSC as a biomarker. We observed high fold change value in LUAD and low at LUSC compared to other co-expressed gene. The fold change value indicates the ACE2 is the best potential biomarker and others co-expressed genes also potential biomarker in COVID-19 infected lung cancer.
Finally, the ACE2 overexpression, enforcing the viral entry, and cytokine storm is the reason for the higher susceptibility and fatality rate in lung cancer patients with COVID-19. So, this research shines a light on the fact that ACE2 can be a biomarker for COVID-19 in lung cancer patients.
5. Conclusion
Currently, COVID-19 is an ongoing pandemic in the world, and it is spreading rapidly. Intense research into the virus receptor ACE2's molecular mechanism may aid in the diagnosis, prevention, and therapy for this disease. Form this study; we have gained knowledge about transcriptional expression, survival rate, mutational positions, PPI, miRNAs, TFs, gene ontology, and signaling pathways in human lung cancer. We also conclude that the overexpression of ACE2 may lead to lung carcinoma progression. Therefore, we can assume that the high expression of ACE2 may lead to higher susceptibility of lung cancer patients toward COVID-19, and ACE2 can serve as the best potential biomarker for this disease compared to co-expressed genes. However, a more in-depth research is needed to confirm our findings through wet-lab experiments.
Author contributions
AS designed the project; AS performed the experiments; AS and TJ evaluated and interpreted the data; AS, TJ and JHR prepared the draft manuscript; AS finalized the manuscript. All authors approved the final version of the manuscript.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
Not applicable.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
We thank the Biological Solution Centre (BioSol Centre) for their technical support. We are also thankful to the reviewers for their thoughtful comments and suggestions, which were truly helpful in revising the manuscript.
References
- 1.Passaro A., Peters S., Mok T.S.K., Attili I., Mitsudomi T., de Marinis F. Testing for COVID-19 in lung cancer patients. Ann. Oncol. 2020 doi: 10.1016/j.annonc.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Sen Tan K., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak- a n update on the status. Mil. Med. Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nain Z., Rana H.K., Liò P., Islam S.M.S., Summers M.A., Moni M.A. Pathogenetic profiling of COVID-19 and SARS-like viruses. Brief. Bioinform. 2020 doi: 10.1093/bib/bbaa173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engineering G., Science N. 2020. Assessment of ACE2, CXCL10 and their Co-Expressed Genes: An in-Silico Approach to Evaluate the Susceptibility and Fatality of Lung Cancer Patients towards COVID-19 Infection. [Google Scholar]
- 5.Mirza J., Ganguly A., Ostrovskaya A., Ph D., Ph D., Viswanathan R., Sc D.M. Command suicidal hallucination as initial presentation of coronavirus disease 2019 (COVID-19): a case report. Psychosomatics. 2019;2020 doi: 10.1016/j.psym.2020.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang W., Guan W., Chen R., Wang W., Li J., Xu K., Li C., Ai Q., Lu W., Liang H., Li S., He J. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020 doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sidaway P. COVID-19 and cancer: what we know so far. Nat. Rev. Clin. Oncol. 2020;17:2020. doi: 10.1038/s41571-020-0366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adashek J., Hajjar J., Chemaly R., Kurzrock R. Are cancer patients at higher risk of death with COVID-19? J. Immunother. Precis. Oncol. 2020;0:0. doi: 10.4103/2666-2345.280883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samad A., Ahammad F., Nain Z., Alam R., Imon R.R., Hasan M., Rahman M.S. Designing a multi-epitope vaccine against SARS-CoV-2: an immunoinformatics approach. J. Biomol. Struct. Dyn. 2020:1–17. doi: 10.1080/07391102.2020.1792347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baud D., Qi X., Nielsen-Saines K., Musso D., Pomar L., Favre G. Real estimates of mortality following COVID-19 infection. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai G. Bulk and single-cell transcriptomics identify tobacco-use disparity in lung gene expression of ACE2, the receptor of 2019-nCov. MedRxiv. 2020 doi: 10.1101/2020.02.05.20020107. 2020.02.05.20020107. [DOI] [Google Scholar]
- 14.Burrell L.M., Johnston C.I., Tikellis C., Cooper M.E. ACE2, a new regulator of the renin-angiotensin system. Trends Endocrinol. Metab. 2004 doi: 10.1016/j.tem.2004.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J., Li H., Hu S., Zhou Y. Vol. 12. 2020. NSfMQRDyZ3r4ijG6S; pp. 6518–6535. [Google Scholar]
- 16.Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W., Duan G. Virology, epidemiology, pathogenesis, and control of covid-19. Viruses. 2020 doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol. 2020 doi: 10.1002/jmv.25987. jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection. Radiol. Cardiothorac. Imag. 2020;2 doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24 doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher P.E., Cook K., Soto-Pantoja D., Menon J., Tallant E.A. Angiotensin peptides and lung cancer. Curr. Cancer Drug Targets. 2011;11:394–404. doi: 10.2174/156800911795538048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L., Zhong L. Lung Adenocarcinoma Patients Own Higher Risk of SARS- CoV-2 Infection. 2020. www.preprints.org [DOI] [PMC free article] [PubMed]
- 23.Cheng H., Wang Y., Wang G.Q. Organ-protective effect of angiotensin-converting enzyme 2 and its effect on the prognosis of COVID-19. J. Med. Virol. 2020 doi: 10.1002/jmv.25785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.V.S.K., Varambally S. 2017. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses, Neoplasia (United States) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha S.K., Kader M.A., Samad K.A., Biswas K.C., Rahman M.A., Parvez M.A.K., Rahman M.S. Prognostic and clinico-pathological significance of BIN1 in breast cancer. Informatics Med. Unlocked. 2020;19:100327. doi: 10.1016/j.imu.2020.100327. [DOI] [Google Scholar]
- 27.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia (New York, NY) 2007;9:166. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barman U.D., Saha S.K., Kader M.A., Jamal M.A.H.M., Sharma S.P., Samad A., Rahman M.S. Clinicopathological and prognostic significance of GPC3 in human breast cancer and its 3D structure prediction. Netw. Model. Anal. Heal. Informatics Bioinforma. 2020;9:1–18. [Google Scholar]
- 29.Pongor L., Kormos M., Hatzis C., Pusztai L., Szabó A., Győrffy B. A genome-wide approach to link genotype to clinical outcome by utilizing next generation sequencing and gene chip data of 6,697 breast cancer patients. Genome Med. 2015;7:104. doi: 10.1186/s13073-015-0228-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio Cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koster J., Volckmann R., Zwijnenburg D., Molenaar P., Versteeg R. 2019. R2: Genomics Analysis and Visualization Platform. [Google Scholar]
- 32.Mattingly C.J., Colby G.T., Forrest J.N., Boyer J.L. The comparative toxicogenomics database (CTD) Environ. Health Perspect. 2003;111:793–795. doi: 10.1289/ehp.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathan M., Keerthikumar S., Ang C., Gangoda L., Quek C.Y.J., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 34.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., Maitland A., Mostafavi S., Montojo J., Shao Q., Wright G., Bader G.D., Morris Q. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karim M., Samad A., Adhikari U.K., Kader M., Kabir M., Islam M., Hasan M. A multi-omics analysis of bone morphogenetic protein 5 (BMP5) mRNA expression and clinical prognostic outcomes in different cancers using bioinformatics approaches. Biomedicines. 2020;8:19. doi: 10.3390/biomedicines8020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia J., Benner M.J., Hancock R.E.W. NetworkAnalyst - integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42:167–174. doi: 10.1093/nar/gku443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin D., Brun C., Remy E., Mouren P., Thieffry D., Jacq B. GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai L., Lin S., Girard L., Zhou Y., Yang L., Ci B., Zhou Q., Luo D., Yao B., Tang H. LCE: an open web portal to explore gene expression and clinical associations in lung cancer. Oncogene. 2019;38:2551–2564. doi: 10.1038/s41388-018-0588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khomtchouk B.B., Hennessy J.R., Wahlestedt C. shinyheatmap: Ultra fast low memory heatmap web interface for big data genomics. PLoS One. 2017;12 doi: 10.1371/journal.pone.0176334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Bio-Medica Atenei Parm. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L., Fang Q., Deng F., Wang H., Christopher E.Y., Ba L., Yu W., Lin R.D., Li T., Hu Z. Natural mutations in the receptor binding domain of spike glycoprotein determine the reactivity of cross-neutralization between palm civet coronavirus and severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:4694–4700. doi: 10.1128/JVI.02389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020:1–5. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian Y.-R., Guo Y., Wan H.-Y., Fan L., Feng Y., Ni L., Xiang Y., Li Q.-Y. Angiotensin-converting enzyme 2 attenuates the metastasis of non-small cell lung cancer through inhibition of epithelial-mesenchymal transition. Oncol. Rep. 2013;29:2408–2414. doi: 10.3892/or.2013.2370. [DOI] [PubMed] [Google Scholar]
- 47.Feng Y., Ni L., Wan H., Fan L., Fei X., Ma Q., Gao B., Xiang Y., Che J., Li Q. Overexpression of ACE2 produces antitumor effects via inhibition of angiogenesis and tumor cell invasion in vivo and in vitro. Oncol. Rep. 2011;26:1157–1164. doi: 10.3892/or.2011.1394. [DOI] [PubMed] [Google Scholar]
- 48.Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin–angiotensin–aldosterone system inhibitors in patients with Covid-19. N. Engl. J. Med. 2020;382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai G., Bossé Y., Xiao F., Kheradmand F., Amos C.I. Tobacco smoking increases the lung gene expression of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;201(12):1557–1559. doi: 10.1164/rccm.202003-0693LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J., Luo Q., Chen R., Chen T., Li J. 2020. Susceptibility Analysis of COVID-19 in Smokers Based on ACE2. [Google Scholar]
- 51.Liu K., Chen Y., Lin R., Han K. Clinical features of COVID-19 in elderly patients: a comparison with young and middle-aged patients. J. Inf. Secur. 2020;80(6):14–18. doi: 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Covid C.D.C., Team R. Severe outcomes among patients with coronavirus disease 2019 (COVID-19)—United States, February 12–march 16, 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. Jama. 2020;323:1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- 54.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma’ayan A., Karim M., Samad A., Adhikari U.K., Kader M.A., Kabir M., Islam M., Hasan M., Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P., Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.V.S.K., Varambally S., Tang Z., Kang B., Li C.C., Chen T.T., Zhang Z., Saha S.K., Kader M.A., Samad K.A., Biswas K.C., Rahman M.A.S., Parvez M.A.K., Rahman M.A.S., Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T.R., Pandey A., Chinnaiyan A.M., Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., Maitland A., Mostafavi S., Montojo J., Shao Q., Wright G., Bader G.D., Morris Q., Burrell L.M., Johnston C.I., Tikellis C., Cooper M.E., Adashek J., Hajjar J., Chemaly R., Kurzrock R., Chen L., Zhong L., Choudhury A., Mukherjee S., Cheng H., Wang Y., Wang G.Q., Liang W., Guan W., Chen R.R., Wang W., Li J.J., Xu K., Li C.C., Ai Q., Lu W., Liang H., Li S., He J., Jneid R., Sidaway P., Baud D., Qi X., Nielsen-Saines K., Musso D., Pomar L., Favre G., Singhal T., Guo Y.Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.S.S.D., Jin H.J., Tan K. Sen, Wang D.Y., Yan Y., Cai G., Jin Y., Yang H., Ji W., Wu W., Chen S.S.S.D., Zhang W., Duan G., Mirza J., Ganguly A., Ostrovskaya A., Ph D., Ph D., Viswanathan R., Sc D.M., Engineering G., Science N., Passaro A., Peters S., Mok T.S.K., Attili I., Mitsudomi T., de Marinis F., Wan Y., Shang J., Graham R., Baric R.S., Li F., Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S., Qian Y.-R., Guo Y.Y.R., Wan H.H.-Y., Fan L., Feng Y., Ni L., Xiang Y., Li Q.Q.-Y., Wan H.H.-Y., Fan L., Fei X., Ma Q., Gao B., Xiang Y., Che J., Li Q.Q.-Y., Liu L., Fang Q., Deng F., Wang H., Christopher E.Y., Ba L., Yu W., Lin R.D., Li T., Hu Z., Cai G., Bossé Y., Xiao F., Kheradmand F., Amos C.I., Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D., Wang J., Luo Q., Chen R.R., Chen T.T., Li J.J., Leung J.M., Yang C.X., Tam A., Shaipanich T., Hackett T.-L., Singhera G.K., Dorscheid D.R., Sin D.D., Aguiar J.A., Tamminga A., Lobb B., Huff R.D., Nguyen J.P., Kim Y., Dvorkin-Gheva A., Stampfli M.R., Doxey A.C., Hirota J.A., Kua L.-F., Ross S., Lee S.-C., Mimura K., Kono K., Goh B.-C., Yong W.-P., Shiiba M., Saito K., Fushimi K., Ishigami T., Shinozuka K., Nakashima D., Kouzu Y., Koike H., Kasamatsu A., Sakamoto Y., Li X., Liu X., Xu Y., Liu J.J., Xie M., Ni W., Chen S.S.S.D., Pacha O., Sallman M.A., Evans S.E., Chan J.F.-W., Yuan S., Kok K.-H., To K.K.W., Chu H., Yang J., Xing F., Liu J.J., Yip C.C.-Y., Poon R.W.-S., Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Cucinotta D., Vanelli M., Mattingly C.J., Colby G.T., Forrest J.N., Boyer J.L., Khomtchouk B.B., Hennessy J.R., Wahlestedt C., Koster J., Volckmann R., Zwijnenburg D., Molenaar P., Versteeg R., Pathan M., Keerthikumar S., Ang C., Gangoda L., Quek C.Y.J., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A., Barman U.D., Saha S.K., Kader M.A., Jamal M.A.H.M., Sharma S.P., Samad A., Rahman M.A.S., Cai L., Lin S., Girard L., Zhou Y., Yang L., Ci B., Zhou Q., Luo D., Yao B., Tang H., H.L.H.A.S. Collaboration Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Neoplasia. 2020;9:281–286. doi: 10.1002/jmv.25785. [DOI] [Google Scholar]
- 55.Yu J., Ouyang W., Chua M.L.K., Xie C. SARS-CoV-2 transmission in patients with cancer at a tertiary care hospital in Wuhan, China. JAMA Oncol. 2020;6(7):1108–1110. doi: 10.1001/jamaoncol.2020.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q., Lu S., Li T., Yu L., Zhang Y., Zeng H., Qian X., Bi J., Lin Y. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J. Exp. Clin. Cancer Res. 2019;38:1–12. doi: 10.1186/s13046-019-1156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu J., Fan J., Wu F., Huang Q., Guo M., Lv Z., Han J., Duan L., Hu G., Chen L. The ACE2/angiotensin-(1–7)/mas receptor axis: pleiotropic roles in cancer. Front. Physiol. 2017;8:276. doi: 10.3389/fphys.2017.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chai P., Yu J., Ge S., Jia R., Fan X. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: a pan-cancer analysis. J. Hematol. Oncol. 2020;13:1–5. doi: 10.1186/s13045-020-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aguiar J.A., Tamminga A., Lobb B., Huff R.D., Nguyen J.P., Kim Y., Dvorkin-Gheva A., Stampfli M.R., Doxey A.C., Hirota J.A. The impact of cigarette smoke exposure, COPD, or asthma status on ABC transporter gene expression in human airway epithelial cells. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-018-36248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kua L.-F., Ross S., Lee S.-C., Mimura K., Kono K., Goh B.-C., Yong W.-P. UGT1A6 polymorphisms modulated lung cancer risk in a Chinese population. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X., Liu X., Xu Y., Liu J., Xie M., Ni W., Chen S. KLF5 promotes hypoxia-induced survival and inhibits apoptosis in non-small cell lung cancer cells via HIF-1α. Int. J. Oncol. 2014;45:1507–1514. doi: 10.3892/ijo.2014.2544. [DOI] [PubMed] [Google Scholar]
- 62.Shiiba M., Saito K., Fushimi K., Ishigami T., Shinozuka K., Nakashima D., Kouzu Y., Koike H., Kasamatsu A., Sakamoto Y. Lipocalin-2 is associated with radioresistance in oral cancer and lung cancer cells. Int. J. Oncol. 2013;42:1197–1204. doi: 10.3892/ijo.2013.1815. [DOI] [PubMed] [Google Scholar]
- 63.Latchman D.S. Transcription factors: an overview. Int. J. Biochem. Cell Biol. 1997;29:1305–1312. doi: 10.1016/s1357-2725(97)00085-x. [DOI] [PubMed] [Google Scholar]
- 64.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 65.Pacha O., Sallman M.A., Evans S.E. COVID-19: a case for inhibiting IL-17? Nat. Rev. Immunol. 2020;20:345–346. doi: 10.1038/s41577-020-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., H.L.H.A.S. Collaboration COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet (Lond., Engl.) 2020;395:1033. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]