Abstract
Such testing and trying time probably never seen before in the human history. The novel coronavirus disease abbreviated as COVID-19 is the ongoing health crisis which entered into human life in late December 2019. The ease of transmission between humans and the undetectability in early stage makes COVID-19 frightening and unprecedented. The disease is characterised by pneumonia progressing to breathing difficulty, acute respiratory distress syndrome (ARDS) and multi-organ failure. Clinical studies suggest excessive release of inflammatory mediators leads to cytokine storm, a phenomenon which appears to be potentially life-threatening in COVID-19. Across the globe, when the world authorities are grappling to contain the virus, our review provides a glimpse on structure, pathophysiology of the virus and further sheds light on various clinical complications associated with the disease in order to open up/raise new horizons to explore various possible theoretical targets for COVID-19. The review also portrays a question and debates: Can targeting cytokine storm can be a feasible approach to combat COVID-19?
Keywords: COVID-19, SARS-COV-2, Acute respiratory distress syndrome, Cytokine storm, Interleukins, TNF-α, KV1.3
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acquired respiratory syndrome coronavirus 2 (SARS-CoV-2) is an ongoing global health crisis. The first case of COVID-19 was announced in Wuhan, Hubei Province, China in late December 2019 [1] and since then it has shaken the whole world. As of 9th December2020, almost 68 million people have been confirmed to be infected, and around 1.6 million people have already succumbed to this deadly viral disease. The previous virus attacks such as SARS CoV infection of 2003 killed fewer than 800 people [2], whereas middle east respiratory syndrome coronavirus (MERS-CoV) infection of 2012 killed less than 700 people [3]. However, previous virus attacks appear mild as compared to what the world has witnessed so far this year due to high transmissibility of SARS-CoV-2. World Health Organization (WHO) brought the outbreak to global attention and declared it pandemic on March 11, 2020 (https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/novel-coronavirus-2019-ncov).
Coronavirus (CoV) is a member of family Coronaviridae, order Nidovirales. It is an enveloped positive-sense single-stranded RNA (ssRNA) virus. Genotypically and serologically they are divided into four genera namely α, β, γ and δ- coronaviruses. Human coronavirus infections are caused by α- and β-CoVs [4]. The SARS-CoV-2 viruses were isolated in bronchoalveolar lavage fluid (BALF) samples of the patients and upon sequence analysis, it was confirmed to be the member of β-coronaviruses [5]. Genome wide studies suggest SARS-CoV-2 shares around 79% and 50% sequence similarity with SARS-CoV and MERS-CoV, respectively [[5], [6], [7]]. However, the amino acid sequence of SARS-CoV-2 differs from other coronaviruses specifically in the regions of 1ab polyprotein and surface glycoprotein [8]. Respiratory droplets and contact transmission are considered to be the major transmission routes [9]. Studies suggest that SARS-CoV-2 have been detected in the urine and stools of laboratory confirmed patients as well [10]. Recently, WHO has also acknowledged the possibility of airborne transmission (https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions). The ease of transmission between humans and the mildness or undetectability of its symptoms makes COVID-19 frightening and unprecedented. Patients infected have reported no or little flu like symptoms including dry cough, fever, runny nose, throat pain and anosmia. In severe forms of the disease, marked inflammation and pneumonia have been identified, progressing to breathing difficulty, acute respiratory distress syndrome (ARDS) and multi-organ failure. Older as well as comorbid patients suffering from diabetes, hypertension and lung diseases are more susceptible to COVID-19 complications [6,11]. A study suggests that smokers are 1.4 times more prone to COVID-19 infection and approximately possess 2.4 times more chances of getting admitted to intensive care unit (ICU) or die as compared to non-smokers [12]. Clinical evidences indicate that there is an involvement of wide variety of cytokines and the situation is worsened by the excessive release of pro-inflammatory cytokines including Interleukin (IL)-1, IL-6, IL-12, Interferon (IFN) γ and Tumour Necrosis Factor (TNF) α which mainly targets the lung tissue [11]. The dysregulated release of signalling mediators is broadly known as “cytokine storm” and is sometimes, also used interchangeably, with cytokine release syndrome (CRS). Although, the concept of cytokine storm is not new; it has garnered the attention primarily due to its striking role in COVID-19. It is a race against time with containment the only option since there are no specific therapeutics and vaccines available for the prophylaxis and treatment of this infection. However, a better understanding of pathophysiology is highly recommended for the development of vaccine and suitable therapeutic agents against the virus. In order to provide a summary to public health authorities and potential readers across the planet, we provide a detailed review summarising the structure, pathogenesis, immunopathological progression, pathological complications and the potential agents available which can be explored to control the cytokine storm observed in COVID-19.
2. Structure and pathogenesis of SARS-COV-2
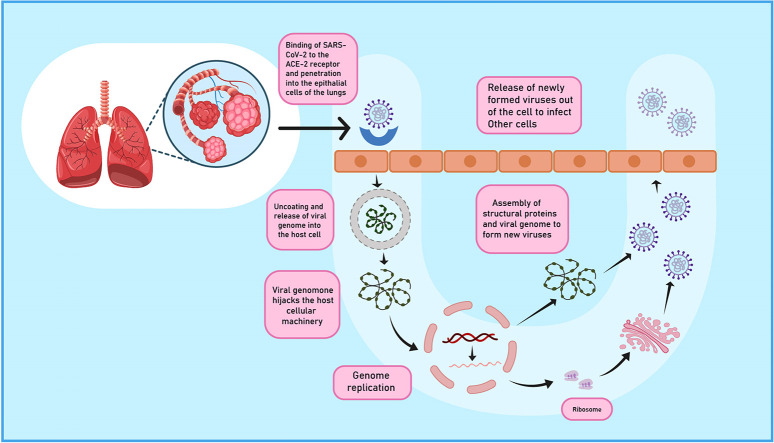
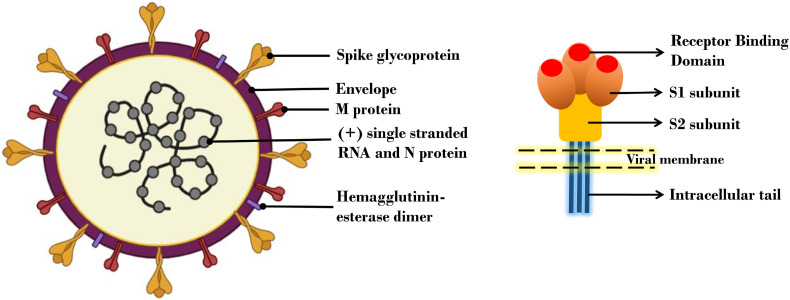
SARS-CoV-2 is an enveloped, pleomorphic or spherical shaped positive sense RNA virus having varying genome length of around 30 kb with a 5′- cap structure and 3′- poly A tail with an arrangement as 5′-replicase open reading frame (ORF) 1ab-spike(S)-envelope(E)-membrane(M)-N-3′ [13,14]. Population genetics analysis of 103 SARS-CoV-2 genomes indicate that these viruses have evolved into two major lineages namely the more prevalent CT haplotype or the L lineage and the TC haplotype also called the S lineage [15]. Two-third of the genomic RNA is used as a template to directly translate two large polyproteins namely pp1a and pp1ab which encode for non-structural proteins (nsps). Remaining one-third of the genome encodes for four structural proteins namely Spike (S), Envelope (E), Membrane (M) and Nucleocapsid (N) [16]. The genes encoding S, E, M and N proteins are around 3822, 228, 669 and 1260 nucleotides in length [13]. The replication and transcription of genome of SARS-CoV-2 occurs via RNA dependent RNA polymerase (RdRp). Structurally, RdRp resembles that of right-hand comprising fingers, thumb and palm subdomain. The cryo-electron microscopic structure of the SARS-CoV-2 reveals that the structure of RdRp consists of nsp7, nsp8 and nsp12 subunits with over two turns of RNA template-product duplex. The nsp12 is composed of three domains: N-terminal nidovirus RdRp associated nucleotidyltransferase (NiRAN) domain, an interface domain and a C-terminal RdRp domain. The subunit nsp7 binds to the thumb whereas two copies of nsp8 which adopt different structure in RdRp binds to the finger and thumb subdomains [17]. The densely glycosylated S proteins, similar to SARS-CoV S protein, is a clove shaped, trimeric class I fusion protein on the virus surface consists of a large ectodomain, a single-pass transmembrane anchor and a short intracellular tail. The ectodomain segment consists of a receptor-binding subunit S1 and a membrane-fusion subunit S2. The S1 component having a receptor binding domain shows conformational movements and can hide or expose the determinants for receptor binding. In each spike, three S1 heads attach at the top of a trimeric S2 stalk. The C-terminal domain of S1 binds to the host receptor surface whereas the N-terminal of S1 is in contact with the S2 [18]. The S2 protein has showed much higher sequence similarity (around 93%) with bat-SL-CoVZC45 and bat-SL-CoVZXC21 in comparison to S1 domain which had only around 68% identity with these bats derived viruses [7]. The E-protein localised in the ER-Golgi Intermediate Compartment (ERGIC) is involved in protein-protein interaction and is a determinant of SARS-CoV-2 virulence. E-protein PDZ-binding motif activates p38 MAPK which is involved in over expression of inflammatory cytokines [19]. The M proteins are the most abundant structural proteins which define the shape of the virion and act as an organiser of coronavirus activity [20]. A simplified pictorial representation of virus and its spike protein is shown in Fig. 1 .
Fig. 1.
A simplified structure of SARS COV-2 virus and spike protein. SARS-CoV-2 is an enveloped, pleomorphic or spherical shaped positive sense RNA virus. The genome of the virus encodes for four structural proteins namely Spike (S), Envelope (E), Membrane (M) and Nucleocapsid (N). S protein is clove shaped and consists of a receptor-binding subunit S1 and a membrane-fusion subunit S2.
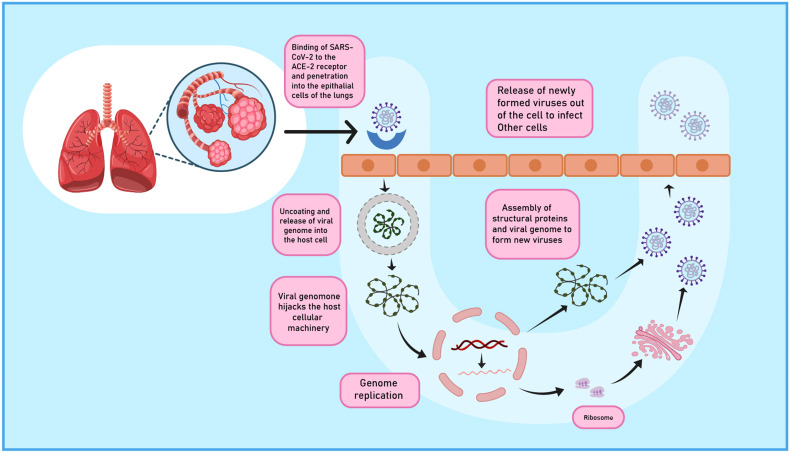
The virus may gain entry through various routes such as droplets released in the environment by the infected patient via coughing, sneezing, saliva or through faecal-oral route [9,10]. Upon exposure, S protein of virus binds to the Angiotensin converting enzyme-2 (ACE-2) receptor [6] unlike MERS-CoV which binds to dipeptidyl peptidase 4 (DPP 4) [3]. The receptor is expressed on the type II pneumocytes in the respiratory tract as well as on vital organs including kidney, liver, heart and intestine. [21]. The entry of virus into the lung epithelium occurs by cleavage of S protein into S1 domain (responsible for receptor binding) and S2 domain (helps in membrane fusion) similar to influenza or by endocytosis [18]. Studies demonstrate that 3CL like protease (3CLpro), papain like protease (PLpro) [16] and RdRp are the major enzymes responsible for proteolysis, replication and production of new virions [17]. Upon membrane fusion to the host surface, viral RNA genome is released into the cytoplasm and hijacks the host's translational machinery. The RNA is uncoated to allow translation of two polyproteins, the newly formed envelop glycoproteins are inserted into the Rough endoplasmic reticulum (RER) or Golgi membranes. Viral particles germinate in ERGIC and which fuse with plasma membrane to release the virus out of the cell to transfect other host cells [16]. Fig. 2 gives a bird eye view of the pathogenesis of SARS-CoV-2 virus.
Fig. 2.
Pathogenesis of SARS-COV-2 virus. SARS-CoV-2 infects the host lungs epithelium by binding to ACE-2 receptor and releases its genome into host cell. The viral genome hijacks host cellular machinery. Viral particles germinate in ERGIC and fuses with plasma membrane to release more copy of viruses.
3. The cytokine storm
The pathophysiology of SARS-CoV-2 involves a particular phenomenon associated with aberrant release of several mediators such as interleukins, interferons, chemokines etc., as a result of hyperactive immune response. Pathological findings of patients admitted to ICU reported increased levels of cytokines including IL-2, IL-7, Macrophage-Colony Stimulating Factor (M-CSF), Granulocyte CSF (G-CSF), interferon γ -induced protein (IP-10), Monocyte Chemoattractant Protein-1 (MCP-1), Macrophage Inflammatory Protein 1-α (MIP1-α) and TNF-α. This vicious cycle of cytokines overproduction and excessive release is termed as “cytokine storm” or “hypercytokinemia” [11,22]. Although, these mediators are primarily involved in physiological signalling and are part of strictly regulated immune system for surveillance and clearance of foreign objects from the body; the unfettered release of these agents, in response to infection, may result in exaggerated immune response which negatively affects the host cells. In context of SARS-CoV-2 infection, cytokine storm has caught the attention of research scientists, health workers and general public due to its possible close association with pathogenesis of the virus. The release of aforementioned mediators, notably TNF-α show a good correlation with severity of SARS-CoV-2 based on the results of clinical studies. Adding to this fact, the monoclonal antibodies such as Tocilizumab and Sarilumab (although, intended for rheumatoid arthritis) have shown their effectiveness in SARS-CoV-2 and have entered the clinical trials highlighting the striking role of TNF-α in disease.
4. Immunopathological basis of cytokine storm in SARS-CoV-2
Once SARS-CoV-2 encounters the host cells, the immune system of host gets activated and triggers adaptive as well as innate immune responses [23]. The early activation of innate immune response is an incumbent reaction of the body to eliminate the infection. Cellular component of innate immunity such as natural killer (NK) cells, macrophages, mast cells work in concert with soluble components including cytokines, chemokines, complement system etc. to provide a first line of defence mechanism to the body. The details of immune response activation are beyond the scope of this review and the interested readers are requested to refer certain excellent reviews on the topic [24,25]. However, the activation of cytokine storm as a result of hyperactive immune response leads to progressive inflammation in the organs. Although, our knowledge of SARS-CoV-2 pathogenesis is quite limited; the in vitro as well as clinical studies have shown a definite role of cytokine storm in SARS-CoV-2. Within hours of infection with virus, the body starts producing and releasing various inflammatory cytokines followed by a delayed production of IFN. The sudden spurt in the levels of proinflammatory mediators dominates the relatively slower defence mechanism of the body. The released cytokines attract a number of inflammatory cells such as neutrophils, mast cells etc. further fuelling the inflammatory response. Inflammation begins when the virus replicates within the local macrophages resulting in apoptosis. Antigen presenting cells (APCs) engulf the foreign antigen and digests them into small peptide fragments further releasing pro-inflammatory cytokines, mainly IL-1β, IL-12, IL-6, TNF-α and chemokines including MCP-1 and IP-10 to attract other immune cells to the site of infection [11,26]. Macrophages bind to Toll like receptors present on CD4+ T-cells and activate them. Activation promotes the secretion of IL-2 which acts in an autocrine manner and binds to specific receptor on itself causing proliferation and differentiation, which in turn generates the CD4+ T-effector and memory cells. The effector T-cells promote the production of T-helper cells which stimulate the release of IL-4, IL-5 and IL-6, which is manifested as inflammation [27]. Patients infected with COVID-19 have presented high amounts of IL-1β, IFNγ, IP-10, and MCP-1, leading to activated T-helper-1 (Th1) cell responses. However, evidences suggest infected patients also showed increased secretion of T-helper-2 (Th2) cytokines namely IL-4 and IL-10 that suppress inflammation, which is opposite to what was encountered in SARS-CoV infection [28]. CD8+ T-effector cells secrete cytotoxic substances like granzyme and perforin in order to kill the virus infected cells resulting in local tissue damage [29]. Activated forms of B-lymphocytes undergo differentiation to generate memory B-cells and plasma cells in order to protect the body from invading exogenous antigens. Plasma cells liberate IL-5 and IL-6 to protect the host cells by neutralization and lysing the infected cells. The cytokine storm syndrome has been associated with infectious diseases including influenza, SARS, COVID 19 and non-infectious diseases like multiple sclerosis and pancreatitis [30]. Hence, immune response induction may not always be good as it is used to remove the infected cells by committing suicide of host cells to clear the virus. Pattern recognition receptors (PRRs) such as retinoic acid - inducible gene I protein (RIG) or melanoma differentiation associated protein 5 (MDA 5) trigger complex signalling cascades involving MYD88 leading to production of type I Interferons (IFNs) and activation of transcription factor nuclear factor - κB (NF-κB) [31]. Type I IFN signals through downstream molecules such a signal transducer and activator of transcription (STAT) proteins to stimulate the production of antiviral proteins. Hence, establishing the antiviral response to limit the virus replication in neighbouring cells [32]. Healthy serum IL-6 levels are 4 pg/ml but this level rises to several folds in case of novel coronavirus infected patients [11]. High levels of IL-6 bind to its receptor CD126 (also called IL-6R) and forms a complex. IL-6-IL-6R complex further binds to CD130 (also called gp130) and forms a hexameric structure. This hexameric structure having two molecules each of IL-6, CD126 and CD130 formed activates and phosphorylates JAK followed by phosphorylation of tyrosine residues on gp130. Furthermore, STAT1, STAT3 and SH2 domain containing protein tyrosine phosphatase (SHP2) recruitment occur on gp130. STAT1 and STAT3 are phosphorylated by JAKs and translocate into the nucleus causing transcription of target genes [33]. On the other hand, SHP2 acts by activating the Ras-MAP pathway which causes inflammation [34]. It can also be expected that cAMP hydrolysing enzymes i.e.; phosphodiesterase which is expressed in monocytes, lymphocytes and neutrophils might also contribute to inflammation upon stimulation by cytokines such as TNFα, IL-1β, IL-6 and matrix metalloproteases (MMPs) [35,36]. Retrospective longitudinal studies of recovered patients when compared to died patients demonstrated early expression of IFNα, IFNγ, Chemokine C—C motif ligand 2 (CCL2) and proteins that are encoded by IFN-stimulated genes in all patients but only the patients who survived had expression profiles, which indicates development of adaptive immunity. Also the newly recruited immune cells recognise the viral pathogen and release more anti-inflammatory cytokines such as IL-10 to recruit more immune cells thus amplifying the cytokine response [37]. It has been well understood that when there is an over-exuberant release of cytokines and severe inflammation, the tissue repair process is unable to keep up resulting in a major damage to the lung tissue. In severe cases these inflammatory cytokines may enter the systemic circulation resulting into shock like condition and ultimately causing multi organ failure. Table 1 summarises the physiological role of various cytokines and evidences which suggests their possible involvement in COVID-19.
Table 1.
Physiological role of various cytokines and their evidences reported in COVID-19.
| SL. No. | Cytokines | Physiological Role of cytokines | Evidences related to COVID-19 | References |
|---|---|---|---|---|
| 1. | IL-1β | Pyrogenic, pro-inflammatory, proliferation and differentiation | COVID-19 patients with severe symptoms have elevated levels of IL-1β | [38,39] |
| 2. | IL-2 | Proliferation of T cells, generation of effector and memory T cells. It increases glucose metabolism to promote the proliferation and activation of T-cells, B-cells and NK cells. | A direct relationship between elevated IL-2 levels and disease severity. | [11,40,41] |
| 3. | IL-6 | Differentiation into plasma cells, IgG production | IL-6 levels elevated in patients with COVID-19 and related to poor prognosis. Additionally, they were found to be markedly higher in patients who died from COVID-19 than in those who recovered. | [38,42,43] |
| 4. | IL-4 | Proliferation of B and cytotoxic T cells, enhances MHC class II expression. | Various studies of COVID-19 patients have detected elevated IL-4 levels associated with severe respiratory symptoms. | [11,38,44] |
| 5. | IL-10 | Inhibits the production of pro-inflammatory cytokines such as TNF-α, IL-1β, IL-6. Also prevents dendritic cell maturation by blocking IL-12 | IL-10 levels were found to be higher in patients with COVID-19 as compared to those with SARS-CoV or MERS It has also been indicated that IL-10 may be over-expressed in anti-SARS-CoV-2 immunity. |
[11,38,45] |
| 6. | IL-7 | Plays vital role in lymphocyte differentiation to promote the development of B and T cells. | IL-7 levels are elevated in patients with COVID-19. | [11,38,46] |
| 7. | IL-12 | Major role in the development of Th1 and Th17 cells. It activates NK cells. | Elevated serum IL-12 levels have been reported in patients infected with SARS-CoV-2 | [11,38,44,47] |
| 8. | IL-13 | Regulates immune responses mediated by Th2-type cytokines. | Researchers have found no difference in serum IL-13 levels between those requiring ICU admission and those who did not. In contrast, a study has reported direct relationship between IL-13 levels and the viral load of SARS-CoV-2. | [11,48] |
| 9. | IL-17 | It plays a major role in tissue damage, physiological stress, and infection. There functions vary according to the tissue. | Elevated IL-17 levels have been reported in patients with SARS-CoV-2 and have been associated with disease severity. Paradoxically a study, reported normal IL-17 levels in COVID-19 patients. |
[11,49,50] |
| 10. | M-CSF | Regulates the growth, proliferation, and differentiation of hematopoietic cells, including monocytes, macrophages, and osteoclasts. Mechanically they act via type III tyrosine-kinase receptors. | The elevated levels of M-CSF in patients with COVID-19 may be associated the hyper-expression of itself and other cytokines resulting into lung damage. | [11,38,51] |
| 11. | G-CSF | Required for the proliferation and maturation of polymorphonuclear granulocyte cells (PMNs). PMNs act by the release of lysosomal enzymes and other signalling molecules | Studies suggest elevated G-CSF levels in patients with COVID-19 and the level rises even more in ICU patients. | [11,52] |
| 12. | GM-CSF | Maintains the immune homeostasis in lungs and gut. It stimulates the proliferation and activation of monocytes, macrophages, eosinophils, neutrophils, dendritic cells, and microglial cells. | Serum GM-CSF levels have been detected to rise in SARS-CoV-2 infection in comparison to healthy individuals. | [11,53] |
| 13. | IP-10 | It binds to chemokine receptor 3 (CXCR3) and regulates immune system responses by activating and recruiting leukocytes. | Studies have reported elevation in IP-10 levels in patients infected with COVID-19 and the levels raises more in those who required ICU admission. Hence it can be concluded that IP-10 over-expression has significant role in lung damage and disease severity Findings report high levels of IP-10 in the most severe cases of COVID-19 which can be related to disease progression and mortality. |
[11,54] |
| 14. | MCP-1 | It regulates the migration and infiltration of monocytes, memory T cells, and NK cells. | Elevated levels of MCP-1 in the broncho-alveolar lavage fluid of COVID-19 patients. Levels have also been detected to rise in lung tissue of patients infected with SARS-CoV-2. |
[11,55,56] |
| 15. | TNF-α | It is mediated by IL-1β and IL-6. TNF-α is involved in the regulation of inflammatory processes and infectious diseases. | A study of 522 patients with COVID-19 reported an inverse relationship between TNF-α levels and T-cell counts. In contrast, another study reported normal TNF-α level in patients with COVID-19. |
[[56], [57], [58]] |
| 16. | IFN-γ | It is associated with macrophage activation, signal transduction, anti-bacterial and antiviral immunity. | Serum IFN-γ levels are found to be higher in patients with COVID-19 than in healthy individuals. Elevated levels may be a result from the activation of Th1 and Th2 cells. | [11,59] |
5. Complications associated with the cytokine storm
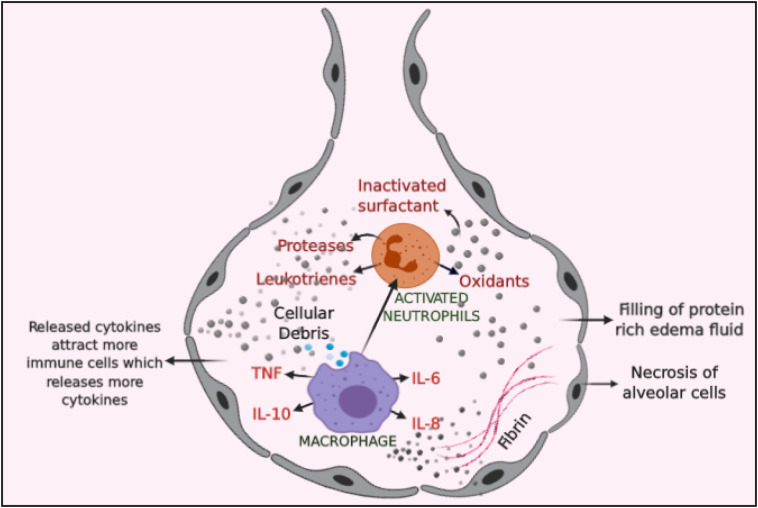
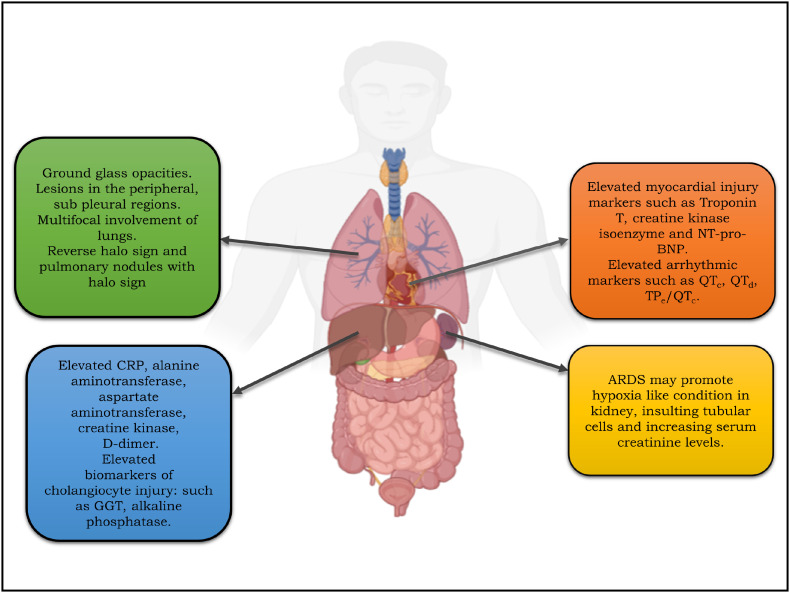
Clinical findings suggest that patients infected with SARS-CoV-2 initially appears stable and the situation deteriorates rapidly with hypoxia finally, inching towards ARDS. The interval from appearance of symptoms to the development of ARDS takes approximately 8–12 days [60]. Comorbid patients are more severely and rapidly affected as compared to patient without any underlying disease [11,59]. Chest scan of patients confers ground glass opacities and consolidation are the noteworthy features in the lungs of patients infected by SARS-CoV-2 and the lesions predominantly appear in the peripheral and sub pleural region [5,11]. The patients showed characteristic reverse halo sign and pulmonary nodules with halo sign which is not been reported in SARS and MERS patients [61,62]. Clinicians have suggested that deformation of bronchus due to fibrosis and strip like lesions may cause irreversible damage to the lung and deteriorate the respiratory functions of the patients [63]. On the other hand, the cytokines entering the circulating blood cause relaxation of the smooth muscles lining the blood vessels and increase the permeability by contracting the epithelial cells. The increased permeability of blood vessels may also promote the accumulation of plasma in the extra cellular spaces decreasing the blood volume which may instigate hypovolemic shock [64]. In response to the cytokines release, the host defence mechanism activates neutrophils generating reactive oxygen species (ROS) and proteases insulting the gaseous exchange by destructing the alveolar lining figuring hypoxia like condition in the patients [65]. Almost 80% of the patients suffering from novel coronavirus have reported elevated body temperature. Data suggest that patients suffering from SARS-CoV-2 infection demonstrate elevation in the heart rate due to stimulation of chemoreceptor by the prevalent hypoxic condition triggering sympathetic nervous system. The exact mechanism of cardiovascular injury in SARS-CoV-2 infection has not been well understood. However, it has been suggested that due to expression of ACE2 on cardiomyocytes, binding of SARS-CoV-2 can occur promoting cardiac insufficiency and poor cardiac outcomes [66]. Further, infected patients have also reported elevated expression of markers of myocardial injury such as Troponin T, creatine kinase isoenzyme and N-terminal of the prohormone brain natriuretic peptide (NT-proBNP) [67]. The excessive release of cytokines can cause potential changes in the ECG arrhythmic markers such as QTc, QTd, TPe/QTc. Elevated cytokines mainly IL-6, IL-1 may result in the electrophysiological changes in cardiomyocytes, which may cause electrical remodelling of the heart prolonging QT interval and ventricular arrhythmia [68]. Patients infected with SARS-CoV-2 virus exhibit elevated C-Reactive Protein, creatine kinase and D-dimer suggesting cardiovascular injury [60]. The exact mechanism of liver injury in COVID-19 infected patients is not known. However, from prevalent knowledge it can be ascertained that, since the biomarkers of cholangiocyte injury namely γ-Glutamyl transferase (GGT), alkaline phosphatase are found to be elevated, there might be a direct binding of SARS-CoV-2 virus to the cholangiocyte expressing ACE-2 receptors [69,70]. Moreover, under hypoxia like conditions due to respiratory distress depletion of adenosine triphosphate, lipid accumulation, glycogen consumption of hepatocyte can inhibit cell survival mechanism inducing death of hepatocytes. In addition, lack of oxygen supply generates oxidative stress contributing to increased ROS and lipid peroxidation products which can promote the transcription of pro-inflammatory cytokines mediated liver damage [70,71]. The prevalence of kidney involvement in novel coronavirus disease is not so high. It can be argued that ARDS may promote to renal medullary causing hypoxia, insulting tubular cells and increasing serum creatinine levels [72]. Fig. 3 gives an overview of complications associated with different organs in COVID-19 disease.
Fig. 3.
Release of inflammatory cytokines. An overview of excessive release of inflammatory cytokines and their negative impact on the host cell.
6. Potential targets and agents to control cytokine storm
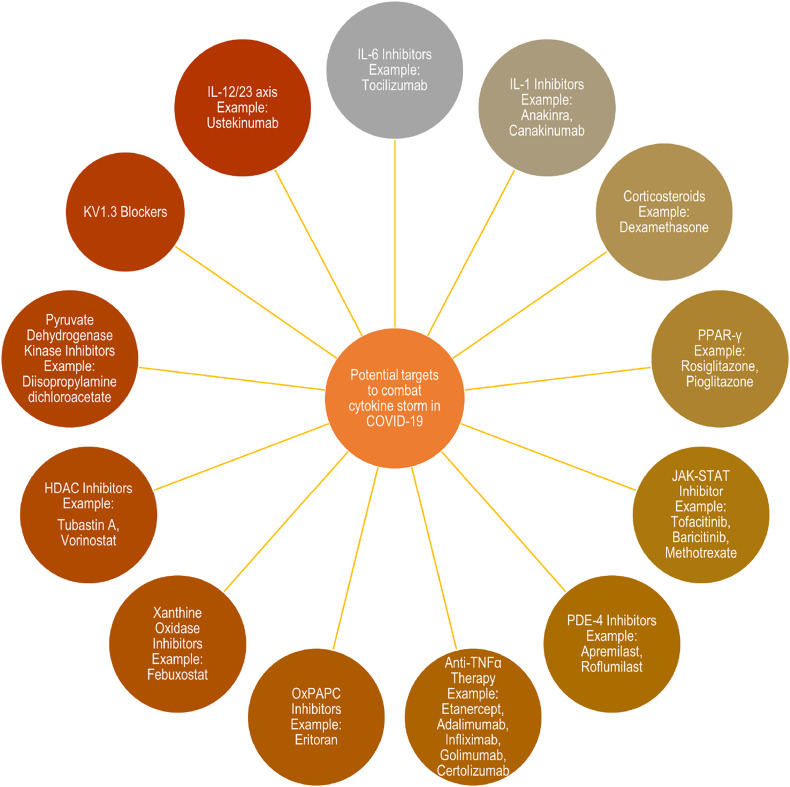
Strong inflammatory cytokine and chemokine response upon viral infection paint a frightening spectre. As the world authorities are grappling to contain the COVID-19 virus, here are a few targets which can be explored to control and manage the over-exuberant release of cytokines. A brief overview of different potential targets has been shown in the Fig. 4 .
Fig. 4.
Multi-organ complications associated with COVID-19. The figure illustrates the evidences and markers which are reported to be elevated in various major organs namely heart, lung, liver and kidneys.
6.1. IL-6 inhibitors
In COVID 19 patients, pathogenic Th1 cells and inflammatory monocytes with high expression of IL-6 exist [11]. They are also responsible for the pathogenesis of autoimmune disorders like Rheumatoid Arthritis. A recent study of patient suffering from COVID-19 reported IL-6 as one of the most robust prognostic markers of survival and is associated with severity and predictive outcome when linked with ventilation and end organ damage [73]. The biopsy samples of autopsy from COVID-19 patients demonstrate pathogenic T cells as well as inflammatory monocytes may enter the pulmonary circulation in large numbers and lead to inflammation [74]. The inflammation occurs via two major signalling pathways namely the mitogen-activated protein kinase (MAPK) pathway [75] and the JAK/STAT pathway [76]. Early detection of increased serum IL-6 levels and its attenuation using suitable approaches can be one of the potential paths to manage SARS-CoV-2 associated complications. A recent study of Tocilizumab (humanized anti-monoclonal antibody) treated COVID-19 patients along with Lopinavir, Methylprednisolone and oxygen therapy demonstrated improved respiratory functions and CRP levels within 3–5 days. In addition, plaque like lesions which appeared before the treatment were significantly reduced. Surprisingly, no adverse drug reactions were reported during the treatment with Tocilizumab at the selected dose [77]. It has also been reported that treatment with Tocilizumab reduced QT prolongation which might be associated with comorbid cases in COVID-19 patients [78]. Though, Tocilizumab has been proved as a promising therapeutic agent in reducing the mortality of severe COVID-19 patients; it should be administered carefully because early administration can increase viral load leading to adverse effects. In addition, the drug is contraindicated in pregnant or lactating women, patients with active pulmonary tuberculosis, patients with mental disorders, those having neutrophil count <0.5 × 109/L, platelet count <50 × 109/L or have undergone organ transplant [77].
6.2. IL-1 inhibitors
IL-1β blockade has been found to be effective in the treatment of various inflammatory diseases such as Still's disease, rheumatoid arthritis, osteoarthritis, gout, multiple sclerosis, macrophage activation syndrome and hyper IgD syndrome [79]. IL-1α is released from cells undergoing hypoxic damage, lost membrane integrity and bind to Interleukin-1 receptor type 1 (IL-1RI) receptor and further triggers a cascade of chemokines and inflammatory cytokines. It also promotes the leukocyte infiltration to the site of inflammation. Mechanistically, IL-1binding to its receptor induces phosphorylation dependent pathway such as p38 MAP kinase, and NF-κB pathway which further induces expression of various inflammatory genes. Therefore, blocking IL-1 can limit the production of inflammatory factors such as TNFα, IL-6 and G-CSF [80] and may also prove to be a powerful tool in reversing the damage in SARS-CoV-2 patients. One possible strategy to inhibit inflammatory cytokines could be the use of IL-1 soluble receptors so that the available free IL-1 binds to its specific soluble IL-1 receptors (sIL-1R) and neutralizes the cytokine action. Another possible intervention could be inhibiting the production or release of IL-1β by using IL-1 receptor antagonist (IL-1Ra) such as Anakinra. Several in-vitro and animal studies have shown beneficial effects of Anakinra in disorders with enhanced cytokine levels such as osteoarthritis. In case of osteoarthritis, IL-1RI receptor are increased on the human chondriocytes and synovial fibroblasts due to the biological activation of IL-1β, major cytokine involved in osteoarthritis [79]. The safety and efficacy of Anakinra are well established. It was approved in 2001 for treatment of RA and later for hyper-IgD syndrome and multiple myeloma [79] A multi-centre study involving 472 patients suffering from RA treated with placebo as well as IL-1RI antagonist at 30 mg/kg, 75 mg/kg and 150 mg/kg doses for 24 weeks demonstrated statistically significant results by decreasing the rate of joint damage, inhibiting expression of metalloproteinase, IL-1 induced matrix erosion and reducing CRP value as well as proteoglycan synthesis [81]. In addition, the use of monoclonal antibodies against IL-1 or against IL-1RI can be a good strategy to provide relief to the SARS-CoV-2 suffering patients. Canakinumab, a humanized immunoglobulin monoclonal antibody that binds selectively to the human IL-1β receptor has been found to be more promising than other IL-1β inhibitors because of its selective affinity for IL-1β receptors and long half-life of around 21 to 28 days [82]. In such a testing time, therapeutic agents targeting IL-1 signalling can be a ray of hope for patients suffering from SARS-CoV-2 infection.
6.3. Corticosteroids
Corticosteroids are one of the first line treatments available for the inflammatory disorders. Corticosteroids were found to be primary means of immunomodulation during 2003 SARS outbreak [83]. Historical data suggests treatment with systemic glucocorticoids in SARS and Influenza infected patients were associated with decreased mortality rates [84]. In SARS, treatment with high dosage of Methylprednisolone (up to 1000 mg/kg) was associated with survival benefits, ARDS reduction and early discharge from hospital [85]. A comparison of 2100 Dexamethasone treated patients who received a dose of 6 mg/kg for 10 days against 4300 people who received standard care showed impressive results. Mortality rates were reduced by 20% among patients with O2 therapy. In case of SARS-CoV-2, treatment with Dexamethasone showed statistically significant reduction of hospital stay and earlier chances of discharge from hospital (https://www.sciencemag.org/news/2020/06/cheap-steroid-first-drug-shown-reduce-death-covid-19-patients). Dexamethasone has been reported to reduce the expression of a number of cytokines in various cell lines and animal models. Expression of IL-22 has been reported to be impaired in-vitro in human peripheral blood mononuclear cells (PBMC) as well as in rat sepsis model. It causes a robust reduction in the expression of IL-22 and IL-8. IL-5 gene expression assessed in unfractioned PBMC obtained from normal human volunteers stimulated by various agents [86]. On the contrary, the administration of glucocorticoids in MERS was associated with adverse outcomes [87]. The use of corticosteroids also brings certain concerns such as decreased immunity and increased chances of infections. Though, recovery using corticosteroids has been quite promising; the immunosuppressive effects may aggravate the viral load and increase the risk of secondary infections [84]. Timing of treatment and blood levels should be taken into consideration while treating with corticosteroids as it may have both stimulatory as well as inhibitory effects on immune system [84,88]. Hence it can be strived upon that corticosteroid therapy has been found to reduce the mortality rate in various infections and can be taken ahead in case of COVID-19. Nevertheless, the use of corticosteroids in COVID-19 warrants the critical evaluation of their risk vs benefit ratio for a safe and efficacious therapeutic regimen.
6.4. Peroxisome proliferator–activated receptor-γ (PPAR-γ) agonist
PPAR-γ is a nuclear receptor which endogenously regulates metabolism, inflammation and immunity. It is expressed in airway epithelium, bronchial mucosa, smooth muscle cells and endothelial cells. T cells, B cells, dendritic cells, neutrophils, macrophages also express PPAR-γ during differentiation or upon activation. It plays a major role by modulating inflammation by interaction with NF-κB which is one of the main regulator of immune response and inflammatory cascade [89]. The activation and functioning of PPAR-γ takes place by heterodimerization of PPAR-γ with nuclear receptor Retinoid X Receptor (RXR) [90]. Upon ligand binding PPAR-RXR complex migrates into the nucleus and binds to peroxisome proliferator hormone response element (PPRE) causing target gene expression [91,92]. PPAR-γ has been shown to downregulate the production of IL-17 and IL-2 by CD4+ cells contributing to decreased proliferation and differentiation of Th1 and Th17 [89]. Xiong et al. demonstrated decreased expression of IL-6 and TNFα in Human Mesangial cells (HMCLs) upon treatment with PPAR-γ agonist [92]. Studies have suggested ovalbumin-induced murine model of asthma presented decreased eosinophilic inflammation and airway hyper-responsiveness upon PPAR-γ agonist treatment. Anti-inflammatory cytokine IL-10 expression increased in the lung tissues upon treatment with PPAR-γ agonist [93]. Reddy et al. demonstrated that PPAR-γ suppressed the expression of endothelial pro-inflammatory mediators such as cytokines and adhesion molecules facilitating migration of neutrophils into alveolar spaces in LPS mediated sepsis model in mice. PPAR-γ agonists not only down regulate the inflammatory response induced by cytokine storm but also promote their survival in mouse model of H1N1 influenza virus [94]. Sirtuin 1 activated PPAR-γ in gouty arthritis mouse model contributes to increased mRNA expression of PPAR-γ, inhibition of infiltration of inflammatory cells, decreased production of pro-inflammatory cytokines such as IL-1, IL-6 and chemokines [95]. The significant inhibition of wide variety of proinflammatory cytokines by PPAR agonists make them a better choice for the control of inflammatory conditions. Hence, repositioning PPAR-γ agonist such as Rosiglitazone, Pioglitazone can be one of the potential strategies to counteract cytokine storm and multi-organ failure involved in observed in SARS-CoV-2 patients.
6.5. JAK-STAT inhibitors
JAK-STAT pathway plays a major role in the progression and pathogenesis of Psoriatic Arthritis (PsA). A dysfunctional response from the innate and adaptive immune systems in arthritis results in overexpression of multiple inflammatory cytokines. Tofacitinib is a small molecule having a short half-life of 3 h which acts either as a kinase inhibitor of JAK or by competitive binding to the docking site with STAT. It has been reported that Tofacitinib significantly inhibited the secretion of IL-6, IL-8 and MCP-1 from PsA explant cultures. The long term safety and efficacy profile in patients receiving Tofacitinib as monotherapy or with non-biologic disease modifying anti-rheumatic drug is well demonstrated [96]. It is also used in the treatment of Inflammatory Bowel Disease (IBD) where JAK-STAT mediated release of cytokines such as IL-2, IL-4, IL-7, IL-9 is of significance [97]. Baricitinib, a selective inhibitor of JAK1/2, showed significant decrease in the expression of CD80/86 on LPS stimulated human monocyte-derived dendritic cells which inhibits the differentiation of naïve CD4+ T cells into Th1 and Th17 cells. It shows significant reduction in the production of chemokine MCP-1 as well as reduced expression of pSTAT3 which further inhibits the production of IL-17 and IL-22 in collagen induced arthritis in BALB/c mice [98]. On the other hand, Methotrexate (MTX) is a JAK/STAT inhibitor and a disease modifying anti-rheumatic drug (DMARD) licenced for treatment of Rheumatoid Arthritis (RA) in 1988 by Food and Drug Administration [99]. It has been established that MTX directly interferes with the binding of IL-1β to its receptor and inhibits the inflammatory activity of IL-1β. Moreover, there is an inverse relationship between long intergenic (noncoding) RNA–p21 (lincRNA-p21) and NF-κB activity in RA. Depressed levels of lincRNA-p21 leads to increased activity of NF-κB which then contributes to the release of cytokines. MTX increases the levels of lincRNA-p21 leading to decreased expression of NF-κB [100]. Additionally, it has also been demonstrated that MTX inhibits the expression of TNFα induced NF-κB activation by degradation of IκBα. The results have been found to be consistent in-vivo when mice were treated with MTX upon collagen induced arthritis [101]. As the JAK-STAT pathway is one of the important pathways for the cytokine storm observed in COVID-19, it can be deduced that the targeting of JAK-STAT pathway could be a successful approach to control inflammatory response in COVID-19.
6.6. PDE-4 inhibitors
PDE-4 inhibitors have attracted a considerable attention because of their potential use in pulmonary inflammatory diseases, contact dermatitis and psoriatic arthritis [35]. PDEs act by converting intracellular secondary messenger cyclic adenosine 3′,5′-monophosphate (cAMP) to Adenosine Monophosphate (AMP) thereby reducing Protein Kinase A activity which contributes to the production of pro-inflammatory mediators like TNFα, IL-22 and IL-17 in endothelial cells, inflammatory cells and keratinocytes [35,36]. Apremilast is a selective PDE-4 inhibitor approved by the US Food and Drug Administration for the treatment of moderate to severe plaque psoriasis as well as for psoriatic arthritis, which is a chronic, systemic inflammatory disease characterised by release of IL-23 and IL-12 [102]. Apremilast exhibits a significant reduction of cytokines mainly IL-17, IL-22 and TNFα involved in the pathogenesis of psoriasis [103]. In vivo studies in immune-deficient mice xenotransplanted with normal human skin and triggered with psoriatic Natural Killer (NK) cells when treated with Apremilast led to absence of lymphocyte infiltration and decreased proliferation index. Immunohistochemistry presented decreased TNFα, intracellular adhesion molecule-1 and human leukocyte antigen expression [104]. Analysis of clinical trial data of patients suffering from moderate to severe psoriasis upon treatment with 20 mg of Apremilast for 29 days presented a reduction in the number of T cells and dendritic cells in dermis and epidermis [105]. Similarly, another drug, Roflumilast belonging to same class is used in the treatment of Acute Lung Injury (ALI). ALI is characterised by recruitment of neutrophils, their activation followed by increased levels of TNFα, IL-6 and IL-8 in the early phase of ALI [103]. The overabundant release of cytokines destroys the endothelial layer of the lungs causing increased vascular permeability. In vivo data suggest that Roflumilast when administered to saline lavage-induced ALI demonstrated inhibition of endothelial barrier function, marked reduction in the concentration of IL-6, IL-8 and TNFα [106]. Interestingly, in another study, Roflumilast has been shown to act by inhibiting inflammatory inhibitors via NF-κB, p38 mitogen activated protein kinase and c-Jun NH2-terminal kinase inhibition in murine macrophage cell line RAW264.7 cells [107]. These studies suggesting a potential inhibition of induced cytokine signalling by PDE-4 inhibitors opens an opportunity to explore their effects in the SARS-CoV-2.
6.7. Anti-TNF-α therapy
TNF-α is one of the most important mediators of inflammatory process which is produced by a variety of cells including macrophages and T-cells [108]. Stimulation of TNF receptor leads to activation of NF-κB which then translocate into the nucleus and upregulates various transcriptional genes. Dysregulated TNF-α signalling has been shown to trigger cytokine storm which leads to cell death whereas optimal levels of TNF-α is required for tissue repairing upon acute injury [109]. Hence TNF-α act as an agent in inducing inflammatory cytokine storm at one hand and providing protection during tissue injury on the other side. Exaggerated TNF-α release occurs in various diseases including Rheumatoid Arthritis (RA), Crohn's Disease, PsA, spondyloarthritis and in our context COVID-19 has been reported [110]. Anti-TNFα therapy using Etanercept, Adalimumab, Infliximab, Golimumab, Certolizumab has been found to be successful in the treatment of various inflammatory diseases. TNF-α-targeting aptamer and its PEG-derivate with antagonistic functions have been shown to improve oxygen saturation, decrease fluid leakage and neutrophil infiltration in the alveolar spaces. Pro-inflammatory cytokines and chemokines in the tissue were reported to be suppressed [111]. Blockade of TNF-α in B27 transgenic rats, an experimental model of colitis prevented the progression of the disease [112]. In vivo findings suggest that treatment with a combination of TNF-α and IFN-γ neutralizing antibodies has provided protection against SARS-CoV-2 infection associated complications in mouse model [113]. Hence, it is not overstating to suggest the TNF-α therapy in management of cytokine storm conditions prevalent in COVID-19 patients. However, this remains entirely speculative and their use demands a better understanding of SARS-CoV-2 which could be achieved in future.
6.8. Oxidised-1-palmitoyl-2-arachidonoyl-phosphaticylcholic (OxPAPC) inhibitors
In lungs, phospholipids are found in lung surfactants which form a layer at the alveolar air-water interface and reduce surface tension. Surfactant is composed of phospholipids including unsaturated phosphatidylcholine. Administration of oxidised phospholipids induced increased levels of IL-6 in the lungs and triggered acute lung injury [114]. The inherent property to sense pathogen-associated molecular patterns (PAMPs) is found in humans which plays a major role in the early identification and host defence against pathogenic microorganisms [115]. Toll like receptors (TLRs) are one of the family of PAMP receptors. OxPAPC which stimulates TLR leads to intracellular signalling and production of TNF-α, IL-1 and other pro-inflammatory pathway causing capillary break and endothelial damage [116]. Eritoran, structurally analogous to lipid A portion of LPS, antagonizes LPS-mediated TLR signalling. Eritoran when administered to mice infected with influenza virus demonstrated decreased ALT, AST levels, blunted pro- and anti-inflammatory gene expression, decreased expression of IL-6 and IL-10 and normal lung architecture. In contrast, another study reported no improvement in lung histology, lung congestion, edema and haemorrhage after Eritoran treatment when compared to vehicle treated group. In the same study, plasma ALT, AST levels, gut permeability, liver inflammation were reduced and liver presented fewer and smaller areas of necrosis with decreased expression of NF-κB [117]. Another study demonstrated the prevention of uncontrolled inflammatory response along with inhibition of pro-inflammatory cytokine production and sepsis by Eritoran in endotoxin induced retinochoroidal damage [118]. Hence, we suggest that Eritoran can be a potential regimen in SARS-CoV-2 infected patients which can prevent multi-organ damage and decrease the mortality rate and benefit the patients.
6.9. Xanthine oxidase inhibitors
Xanthine oxidase (XO) is one of the major enzymes involved in the production of free radicals and ROS during inflammation and hypoxia like condition [119]. Under normal physiological conditions it is found in the form of Xanthine dehydrogenase, while under hypoxia like conditions, it gets converted to XO. XO oxidises purine substrates such as xanthine and hypoxanthine, to produce uric acid and ROS [120]. The expression of XO has been suggested to be upregulated in the lung tissues by various inflammatory stimuli such as endotoxin, hypoxia, and cytokines. Allopurinol, structural analogue of purines and pyrimidines, inhibits XO leading to the inhibition of uric acid production in gout. Febuxostat (FBX) is a non-purine inhibitor of XO and is advantageous over Allopurinol as it can suppress both oxidised as well as reduced forms of XO [120,121]. FBX has been reported to inhibit number of pathways such as MAPK, NF-κB, receptors for Advanced Glycation end product/Phosphatidyl inositol-1,4,5-bisphosphate-3-kinase (RAGE/PI3K/Akt) in various disease models including acute lung injury, Ulcerative Colitis, myocardial ischaemia [122]. FBX has been shown to reduce elevated lactate dehydrogenase (LDH), nitrogen oxides (NOx) levels, serum C-reactive protein (CRP), TNF-α levels and edema and alveolar thickening in LPS induced lung injury in rats [123]. Treatment with FBX resulted in improvement in the cardiac functions, improved haemodynamic and ventricular functions with characteristic decrease in the expression of inflammatory biomarkers have been reported in myocardial ischaemic reperfusion (IR) injured rats [124]. Similarly, FBX attenuated acetic acid induced Ulcerative Colitis in mice at a dose level of 20 mg/kg by inhibiting NF-κB [125]. A comparison of chest scans of patients upon 14 days treatment demonstrated 47% reduction in lung involvement when treated with FBX, whereas Hydroxychloroquine (HCQ) treated patients reported 58.3% reduction in lung involvement [126]. However, FBX could be a superior choice for COVID-19 patients as it has lower side-effects in comparison to HCQ where QT prolongation is one of the major concerns.
6.10. Histone Deacetylase (HDAC) inhibitors
Histones play a vital role in the regulation of gene expression by acetylation or deacetylation. Acetylation of histones leads to relaxation of chromatin following binding of transcription factors promoting transcription. In contrast, deacetylation of histone condenses chromatin and suppress gene transcription [127]. In mouse model of cecal ligation and puncture (CLP) induced lethal sepsis, Tubastin A, a selective HDAC class IIb and a specific HDAC 6 inhibitor has been reported to improve survival by decreasing the expression of pro-inflammatory cytokines and biomarkers such as TNF-α and IL-6 [128]. Suberoylanilide hydroxamic acid (SAHA) also known as Vorinostat is an HDAC 6 inhibitor approved by the U.S. Food and Drug Administration (FDA) for the treatment of Cutaneous T-Cell Lymphoma (CTCL) [129]. SAHA binds to the zinc containing pocket in the HDAC 6 and cause their reversible inhibition. Studies suggest that micromolar concentration of SAHA have anti-tumour effects, whereas nanomolar concentration decreases the secretion of cytokines [130]. It has been proposed that SAHA may suppress TLR4-MyD88-dependent pathway by acetylating STAT-1, which further reduces the nuclear translocation of NF-κB [131,132]. Since limited and elusive information exists regarding the actual mechanisms that are altered by different HDAC inhibitors but in the current scenario it appears attractive agent to be explored which may surmount cytokine storm in COVID-19.
6.11. Pyruvate dehydrogenase kinase inhibitors
Relating cytokine cycle with metabolism, pyruvate dehydrogenase is a key enzyme that regulates the glucose, lipid, lactate and ATP levels. Pyruvate dehydrogenase complex plays a major role in oxidative decarboxylation of pyruvate and links glycolysis, fatty acid synthesis and citric acid cycle. Pyruvate dehydrogenase kinase phosphorylates the α subunit of pyruvate dehydrogenase leading to its inactivation causing decreased metabolism and lack of energy [133]. The use of pyruvate dehydrogenase kinase inhibitors can be one of the strategies in order to prevent multi-organ failure due to cytokine storm. It has been reported through in-vivo studies that mice when infected with H1N1 virus at sub-lethal dose presents restoration of pyruvate dehydrogenase activity upon treatment with Diisopropylamine dichloroacetate (DADA). Results showed suppressed induction of pro-inflammatory cytokines, increased survival rate of mice and histopathology shows reversal of pathological treatment at 50 mg/kg twice daily for 14 days [134]. Therefore, these inhibitors could be explored in suitable preclinical models and also plan to evaluate in suitable clinical conditions to evaluate their efficacy against cytokine storm.
6.12. KV 1.3 channel blockers
Immune cells express a variety of ion channels and transporters that allow movement of ions across the plasma membrane and the membrane of intracellular organelles. In T cells, the interaction of the T cell receptors with its antigens induce an increase in the extracellular Ca2+ concentration, regulating numerous downstream signalling pathways that control clonal expansion, differentiation and cytokine production [135]. Upon activation, the number of Kv1.3 channels of effector memory (TEM) cells increase while that of KCa3.1 channels remain constant, emphasizing the role of Kv1.3 channels in TEM lymphocytes [136]. TEM cells rapidly produce and release inflammatory and cytotoxic mediators such as IFN-γ, IL-4 and perforin [137]. It has been reported that blocking of Kv1.3 channels in TEM cells reduced the influx of Ca2+, resulting in TEM cells specific immunomodulatory effects without compromising naïve and central memory lymphocytes effector functions such as protection against pathogens [138]. Similarly, in COVID-19, activation of T cells leads to generation of effector and memory T cells, which further directs the release the cytokines and chemokines [28,139]. Hence, selectively targeting TEM by blocking Kv1.3 channels to control the release of cytokines in COVID-19 can be a novel approach which requires further exploration. There are few selective Kv1.3 inhibitors which were proven to be safe upon clinical use, therefore, these inhibitors can be repurposed for the management of cytokine storm.
6.13. IL-12/23 axis
IL-12 and IL-23 are major players in activating adaptive immunity. Activation of dendritic cells and macrophages generates different cytokines such as IL-12, IL-23, IL-1β, transforming growth factor-β (TGF-β), and interferon γ (IFN-γ) [140]. IL-12 helps in differentiating a naive T cells into the Th1 subtype which further secretes TNF-α, IL-2, and IFN-γ [141]. On the other hand, IL-23 can activate the secretion of IL-17A, IL-17F, and IL-22. Moreover, IL-23 stabilises Th17 and suppresses T regulatory (Treg) cell differentiation from a naive T cell [142]. IL-17A and IL-17F enhance the expression of various inflammatory cytokines, chemokines and adhesion molecules. IL-17A induces stromal cells to produce extracellular matrix-degrading proteases which can cleave components of the extracellular matrix and promote tissue degradation. Inhibition of Treg differentiation further potentiates the inflammatory cascades [143]. Ustekinumab is a human IgG1 kappa (κ) monoclonal antibody that blocks the IL-12/23 p40 subunit and prevents the p40 subunit of IL-12 and IL-23 from interacting with their receptors. Prevention from binding subsequently neutralizes IL-12 and IL-23 mediated cell activation and cytokines generation [141]. Targeting this pro-inflammatory cytokine pathway can be an area of intense therapeutic exploration in case of novel coronavirus disease where cytokine storm syndrome has been identified as a major culprit to promote the damage. Fig. 5 summarises different potential targets for the management of cytokine storm in COVID-19.
Fig. 5.
Various potential targets and agents to control cytokine storm in COVID-19.
7. Contradictions and unanswered questions
The involvement of cytokine storm in the pathogenesis of SARS-CoV-2 is well evident from clinical studies. However, the concept of cytokine storm is quite a complex phenomenon and a critical analysis is warranted before reaching any consensus. The first and foremost question is how relevant is the cytokine storm in SARS-CoV-2? Although, the clinical studies have shown upregulated levels of cytokines in SARS-CoV-2 patients; the levels do not show a strong, well-established correlation with severity of the infection. For exampleIL-6 is one of the most important mediators involved in pathogenesis of SARS-CoV-2 and is also considered as a potential biomarker of COVID-19 progression [144]. However, the induced expression of IL-6 in SARS-CoV-2 has been found to be erratic and heterogenous [145]. Also evidences from studies support the notion that timing is of the utmost importance in targeting inflammatory cytokines in COVID-19 [73]. A relevant question is that out of more than 20 cytokines/chemokines elevated in COVID-19, which one should be the prime focus at the earliest to prevent the complications in clinical conditions. Further, there are many limiting factors involved in analysis of cytokine expression such as time of sampling, methodology used, calibration of instrument, lab to lab variation, stage of infection and comorbid conditions etc. which are needed to be considered before evaluation. Another point of concern is that these mediators are mostly involved in pleiotropic effects covering a wide range of physiological functions. Hence, it is quite difficult to establish a direct correlation between their high levels and the severity of pathogenesis. In fact, the induced expression of these cytokines may not always translate to the pathological conditions, further limiting our inference. Furthermore, most of the cytokines have target cell specific functions and the effects of cytokines may vary with cell to cells. In addition, the effects of cytokines are defined by their cross talk with other cytokines/signalling molecules, hence the direct inhibition of a particular cytokine may not be a feasible approach apparently. Adding the complexity to the subject and limiting the efficacy of direct inactivation strategies.
In addition, the use of different immunosuppressants such as corticosteroids also brings their own complications with their use. As the hyperimmune response observed in patients is a consequence of activation/hyperactivation of body's protective mechanism with an aim to neutralise the infections; the suppression of immunity might also result in negative outcomes such as secondary infections and weakened ability of the body to eliminate SARS-CoV-2 infection.
As our knowledge of pathogenesis of SARS-CoV-2 infection is still limited; most of the agents tried in combating the infection are majorly speculative based on the similarities with other disease conditions such as influenza, rheumatoid arthritis etc. In this case, the likelihood of the success entirely depends on the similarity with other disease conditions. For example, Tocilizumab, which has shown effectiveness in rheumatoid arthritis is under clinical trials for SARS-CoV-2 due to resemblance on inflammatory basis. However, with increase in our knowledge about mechanisms involved and pathogenesis of virus, it is possible that these strategies could be fruitful approaches to control cytokine storm.
8. Concluding remarks
The SARS-CoV-2 has been a great burden for the global health, public and economy sectors which has affected almost every aspect of our life. Our limited knowledge about this novel virus is one of the major challenges for the discovery of safe and effective vaccine. Hence, there is an urgent need to explore the different pathways used by virus. One of the major events observed in the SARS-CoV-2 infection is the cytokine storm. The incidences of multiple complications associated with cytokine storm highlight its importance in SARS-CoV-2 infection and further present it as a possible major target. The different targeting strategies as discussed here are needed to be explored by keeping in mind the principles of viral pathogenesis and the physiological roles of inflammatory mediators. Further, as most of the studies are preliminary in nature; there is a need to replicate and reproduce the results of potential studies before declaring a valid target. Nevertheless, cytokine storm is a complex phenomenon associated with SARS-CoV-2 and its exploration may endow us with potential lead molecules for tackling one of the deadliest viruses, SARS-CoV-2. Moreover, the cytokine storm may also show its involvement with other future viral infections, hence, targeting cytokine storm may become a lifesaving strategy in different conditions.
CRediT authorship contribution statement
RH and MAS wrote the manuscript.
CG corrected and revised the manuscript.
Declaration of competing interest
None.
Acknowledgment
The authors would like to thank Department of Pharmaceuticals, Ministry of Chemicals and Fertilizers, Government of India and Director, NIPER Hyderabad for the support.
References
- 1.Guo Y-R, Cao Q-D, Hong Z-S, Tan Y-Y, Chen S-D, Jin H-J, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Military Medical Research. 2020;7(1):1–10. [DOI] [PMC free article] [PubMed]
- 2.Hui D.S., Zumla A. Severe acute respiratory syndrome: historical, epidemiologic, and clinical features. Infect. Dis. Clin. 2019;33(4):869–889. doi: 10.1016/j.idc.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Osail A.M., Al-Wazzah M.J. The history and epidemiology of Middle East respiratory syndrome corona virus. Multidisciplinary respiratory medicine. 2017;12(1) doi: 10.1186/s40248-017-0101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou G., Chen S., Chen Z. Advances in COVID-19: the virus, the pathogenesis, and evidence-based control and therapeutic strategies. Frontiers of Medicine. 2020:1–9. doi: 10.1007/s11684-020-0773-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong N, Yang X, Ye L, Chen K, Chan EW-C, Yang M, et al. Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China. BioRxiv. 2020.
- 9.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindson J. COVID-19: faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020;17(5):259. doi: 10.1038/s41575-020-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vardavas C.I., Nikitara K. COVID-19 and smoking: a systematic review of the evidence. Tob. Induc. Dis. 2020;18 doi: 10.18332/tid/119324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. bioRxiv. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 15.Tang X., Wu C., Li X., Song Y., Yao X., Wu X., et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walls A.C., Tortorici M.A., Bosch B.-J., Frenz B., Rottier P.J., DiMaio F., et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531(7592):114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye Y., Hogue B.G. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 2007;81(7):3597–3607. doi: 10.1128/JVI.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., et al. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87(5):e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 22.Wang W., He J., Wu S. The definition and risks of cytokine release syndrome-like in 11 COVID-19-infected pneumonia critically ill patients: disease characteristics and retrospective analysis. Medrxiv. 2020 doi: 10.1093/infdis/jiaa387. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduction and Targeted Therapy. 2020;5(1):1–10. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133(1):13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swain S.L., McKinstry K.K., Strutt T.M. Expanding roles for CD4+ T cells in immunity to viruses. Nat. Rev. Immunol. 2012;12(2):136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong C., Lam C., Wu A., Ip W., Lee N., Chan I., et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clinical & Experimental Immunology. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salti S.M., Hammelev E.M., Grewal J.L., Reddy S.T., Zemple S.J., Grossman W.J., et al. Granzyme B regulates antiviral CD8+ T cell responses. J. Immunol. 2011;187(12):6301–6309. doi: 10.4049/jimmunol.1100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012;76(1):16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen S., Thomsen A.R. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 2012;86(6):2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García-Sastre A., Biron C.A. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312(5775):879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 33.Yu H., Pardoll D., Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ara T., DeClerck Y.A. Interleukin-6 in bone metastasis and cancer progression. Eur. J. Cancer. 2010;46(7):1223–1231. doi: 10.1016/j.ejca.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumer W., Hoppmann J., Rundfeldt C., Kietzmann M. Highly selective phosphodiesterase 4 inhibitors for the treatment of allergic skin diseases and psoriasis. Inflammation & Allergy-Drug Targets (Formerly Current Drug Targets-Inflammation & Allergy) 2007;6(1):17–26. doi: 10.2174/187152807780077318. [DOI] [PubMed] [Google Scholar]
- 36.Lefkimmiatis K., Zaccolo M. cAMP signaling in subcellular compartments. Pharmacol. Ther. 2014;143(3):295–304. doi: 10.1016/j.pharmthera.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81(16):8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner M.D., Nedjai B., Hurst T., Pennington D.J. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2014;1843(11):2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Zhang W., Zhao Y., Zhang F., Wang Q., Li T., Liu Z., et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): the experience of clinical immunologists from China. Clin. Immunol. 2020;214 doi: 10.1016/j.clim.2020.108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg S.A. IL-2: the first effective immunotherapy for human cancer. J. Immunol. 2014;192(12):5451–5458. doi: 10.4049/jimmunol.1490019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu K, Fang Y-Y, Deng Y, Liu W, Wang M-F, Ma J-P, et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin. Med. J.. 2020. [DOI] [PMC free article] [PubMed]
- 42.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin C., Zhou L., Hu Z., Yang S., Zhang S., Chen M., et al. Clinical characteristics and outcomes of COVID-19 patients with a history of stroke in Wuhan, China. Stroke. 2020;51(7):2219–2223. doi: 10.1161/STROKEAHA.120.030365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C., Zhang X., Ju Z., He W. Advances in the research of mechanism and related immunotherapy on the cytokine storm induced by coronavirus disease 2019. Zhonghua shao shang za zhi= Zhonghua shaoshang zazhi= Chinese journal of burns. 2020;36(6):471–475. doi: 10.3760/cma.j.cn501120-20200224-00088. [DOI] [PubMed] [Google Scholar]
- 45.Saghazadeh A., Rezaei N. Immune-epidemiological parameters of the novel coronavirus–a perspective. Expert. Rev. Clin. Immunol. 2020:1–6. doi: 10.1080/1744666X.2020.1750954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet (London, England) 2020;395(10229):1033. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mal X., Trinchieri G. 2001. Regulation of Interleukin-12 Production in Antigen-presenting Cells. [DOI] [PubMed] [Google Scholar]
- 48.Rengarajan J., Szabo S.J., Glimcher L.H. Transcriptional regulation of Th1/Th2 polarization. Immunol. Today. 2000;21(10):479–483. doi: 10.1016/s0167-5699(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 49.McGeachy M.J., Cua D.J., Gaffen S.L. The IL-17 family of cytokines in health and disease. Immunity. 2019;50(4):892–906. doi: 10.1016/j.immuni.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan S., Yi Q., Fan S., Lv J., Zhang X., Guo L., et al. MedRxiv. 2020. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP) [Google Scholar]
- 51.Chockalingam S., Ghosh S.S. Macrophage colony-stimulating factor and cancer: a review. Tumor Biol. 2014;35(11):10635–10644. doi: 10.1007/s13277-014-2627-0. [DOI] [PubMed] [Google Scholar]
- 52.Hartung T., Sv Aulock, Wendel A. Role of granulocyte colony-stimulating factor in infection and inflammation. Med. Microbiol. Immunol. 1998;187(2):61–69. doi: 10.1007/s004300050075. [DOI] [PubMed] [Google Scholar]
- 53.Shiomi A., Usui T. Pivotal roles of GM-CSF in autoimmunity and inflammation. Mediat. Inflamm. 2015;2015 doi: 10.1155/2015/568543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruffilli I., Ferrari S.M., Colaci M., Ferri C., Fallahi P., Antonelli A. IP-10 in autoimmune thyroiditis. Horm. Metab. Res. 2014;46(09):597–602. doi: 10.1055/s-0034-1382053. [DOI] [PubMed] [Google Scholar]
- 55.Deshmane S.L., Kremlev S., Amini S., Sawaya B.E. Monocyte chemoattractant protein-1 (MCP-1): an overview. J. Interf. Cytokine Res. 2009;29(6):313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chu H, Chan JF-W, Wang Y, Yuen TT-T, Chai Y, Hou Y, et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis.. 2020. [DOI] [PMC free article] [PubMed]
- 57.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lees J.R. Interferon gamma in autoimmunity: a complicated player on a complex stage. Cytokine. 2015;74(1):18–26. doi: 10.1016/j.cyto.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 59.W-j Guan, Ni Z.-y., Hu Y., Liang W.-h., C-q Ou, He J.-x., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paul N.S., Roberts H., Butany J., Chung T., Gold W., Mehta S., et al. Radiologic pattern of disease in patients with severe acute respiratory syndrome: the Toronto experience. Radiographics. 2004;24(2):553–563. doi: 10.1148/rg.242035193. [DOI] [PubMed] [Google Scholar]
- 61.Das K.M., Lee E.Y., Langer R.D., Larsson S.G. Middle East respiratory syndrome coronavirus: what does a radiologist need to know? Am. J. Roentgenol. 2016;206(6):1193–1201. doi: 10.2214/AJR.15.15363. [DOI] [PubMed] [Google Scholar]
- 62.Li Y., Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am. J. Roentgenol. 2020;214(6):1280–1286. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 63.Esser S., Lampugnani M.G., Corada M., Dejana E., Risau W. Vascular endothelial growth factor induces VE-cadherin tyrosine phosphorylation in endothelial cells. J. Cell Sci. 1998;111(13):1853–1865. doi: 10.1242/jcs.111.13.1853. [DOI] [PubMed] [Google Scholar]
- 64.Mittal M., Siddiqui M.R., Tran K., Reddy S.P., Malik A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014;20(7):1126–1167. doi: 10.1089/ars.2012.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen L., Li X., Chen M., Feng Y., Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020;116(6):1097–1100. doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li L., Zhou Q., Xu J. Changes of laboratory cardiac markers and mechanisms of cardiac injury in coronavirus disease 2019. Biomed. Res. Int. 2020;2020 doi: 10.1155/2020/7413673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Öztürk F., Karaduman M., Çoldur R., İncecik Ş., Güneş Y., Tuncer M. Interpretation of arrhythmogenic effects of COVID-19 disease through ECG. The Aging Male. 2020:1–4. doi: 10.1080/13685538.2020.1769058. [DOI] [PubMed] [Google Scholar]
- 68.Pirola C.J., Sookoian S. SARS-CoV-2 virus and liver expression of host receptors: putative mechanisms of liver involvement in COVID-19. Liver Int. 2020;40(8):2038–2040. doi: 10.1111/liv.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su S., Shen J., Zhu L., Qiu Y., He J.-S., Tan J.-Y., et al. covid19 involvement of digestive system in COVID-19: manifestations, pathology, management and challenges. Ther. Adv. Gastroenterol. 2020;13 doi: 10.1177/1756284820934626. (1756284820934626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tian D., Ye Q. Hepatic complications of COVID-19 and its treatment. J. Med. Virol. 2020;92(10):1818–1824. doi: 10.1002/jmv.26036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ronco C., Reis T. Kidney involvement in COVID-19 and rationale for extracorporeal therapies. Nat. Rev. Nephrol. 2020:1–3. doi: 10.1038/s41581-020-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Del Valle D.M., Kim-Schulze S., Hsin-hui H., Beckmann N.D., Nirenberg S., Wang B., et al. medRxiv. 2020. An inflammatory cytokine signature helps predict COVID-19 severity and death. [Google Scholar]
- 73.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klemm C., Bruchhagen C., Van Krüchten A., Niemann S., Löffler B., Peters G., et al. Mitogen-activated protein kinases (MAPKs) regulate IL-6 over-production during concomitant influenza virus and Staphylococcus aureus infection. Sci. Rep. 2017;7 doi: 10.1038/srep42473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murray P.J. The JAK-STAT signaling pathway: input and output integration. J. Immunol. 2007;178(5):2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 76.Xu X., Han M., Li T., Sun W., Wang D., Fu B., et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alattar R., Ibrahim T.B., Shaar S.H., Abdalla S., Shukri K., Daghfal J.N., et al. Tocilizumab for the treatment of severe COVID-19. J. Med. Virol. 2020;92:2042–2049. doi: 10.1002/jmv.25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dinarello C.A., Simon A., Van Der Meer J.W. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat. Rev. Drug Discov. 2012;11(8):633–652. doi: 10.1038/nrd3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burrage P.S., Mix K.S., Brinckerhoff C.E. Matrix metalloproteinases: role in arthritis. Front. Biosci. 2006;11(1):529–543. doi: 10.2741/1817. [DOI] [PubMed] [Google Scholar]
- 80.Bresnihan B, Alvaro-Gracia JM, Cobby M, Doherty M, Domljan Z, Emery P, et al. Treatment of rheumatoid arthritis with recombinant human interleukin-1 receptor antagonist. Arthritis & Rheumatism. 1998;41(12):2196–204. [DOI] [PubMed]
- 81.Latourte A., Bardin T., Richette P. Prophylaxis for acute gout flares after initiation of urate-lowering therapy. Rheumatology. 2014;53(11):1920–1926. doi: 10.1093/rheumatology/keu157. [DOI] [PubMed] [Google Scholar]
- 82.Yam L.Y.-C., Lau A.C.-W., Lai F.Y.-L., Shung E., Chan J., Wong V., et al. Corticosteroid treatment of severe acute respiratory syndrome in Hong Kong. J. Infect. 2007;54(1):28–39. doi: 10.1016/j.jinf.2006.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen R.-c., X-p Tang, S-y Tan, B-l Liang, Z-y Wan, J-q Fang, et al. Treatment of severe acute respiratory syndrome with glucosteroids: the Guangzhou experience. Chest. 2006;129(6):1441–1452. doi: 10.1378/chest.129.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Z., Zhang F., Xu M., Huang K., Zhong W., Cai W., et al. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J. Med. Microbiol. 2003;52(8):715–720. doi: 10.1099/jmm.0.05320-0. [DOI] [PubMed] [Google Scholar]
- 85.Ziesché E., Scheiermann P., Bachmann M., Sadik C., Hofstetter C., Zwissler B., et al. Dexamethasone suppresses interleukin-22 associated with bacterial infection in vitro and in vivo. Clinical & Experimental Immunology. 2009;157(3):370–376. doi: 10.1111/j.1365-2249.2009.03969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arabi Y.M., Mandourah Y., Al-Hameed F., Sindi A.A., Almekhlafi G.A., Hussein M.A., et al. Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am. J. Respir. Crit. Care Med. 2018;197(6):757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- 87.Auyeung T.W., Lee J.S., Lai W.K., Choi C.H., Lee H.K., Lee J.S., et al. The use of corticosteroid as treatment in SARS was associated with adverse outcomes: a retrospective cohort study. J. Infect. 2005;51(2):98–102. doi: 10.1016/j.jinf.2004.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bassaganya-Riera J., Song R., Roberts P.C., Hontecillas R. PPAR-γ activation as an anti-inflammatory therapy for respiratory virus infections. Viral Immunol. 2010;23(4):343–352. doi: 10.1089/vim.2010.0016. [DOI] [PubMed] [Google Scholar]
- 89.Miyata K.S., McCaw S.E., Marcus S.L., Rachubinski R.A., Capone J.P. The peroxisome proliferator-activated receptor interacts with the retinoid X receptor in vivo. Gene. 1994;148(2):327–330. doi: 10.1016/0378-1119(94)90707-2. [DOI] [PubMed] [Google Scholar]
- 90.Viswakarma N., Jia Y., Bai L., Vluggens A., Borensztajn J., Xu J., et al. Coactivators in PPAR-regulated gene expression. PPAR Res. 2010;2010 doi: 10.1155/2010/250126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiong Z., Huang H., Li J., Guan Y., Wang H. Anti-inflammatory effect of PPARγ in cultured human mesangial cells. Ren. Fail. 2004;26(5):497–505. doi: 10.1081/jdi-200031747. [DOI] [PubMed] [Google Scholar]
- 92.Kim S.R., Lee K.S., Park H.S., Park S.J., Min K.H., Jin S.M., et al. Involvement of IL-10 in peroxisome proliferator-activated receptor γ-mediated anti-inflammatory response in asthma. Mol. Pharmacol. 2005;68(6):1568–1575. doi: 10.1124/mol.105.017160. [DOI] [PubMed] [Google Scholar]
- 93.Reddy A.T., Lakshmi S.P., Kleinhenz J.M., Sutliff R.L., Hart C.M., Reddy R.C. Endothelial cell peroxisome proliferator–activated receptor γ reduces endotoxemic pulmonary inflammation and injury. J. Immunol. 2012;189(11):5411–5420. doi: 10.4049/jimmunol.1201487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang J., Chen G., Lu L., Zou H. Sirt1 inhibits gouty arthritis via activating PPARγ. Clin. Rheumatol. 2019;38(11):3235–3242. doi: 10.1007/s10067-019-04697-w. [DOI] [PubMed] [Google Scholar]
- 95.Wollenhaupt J., Silverfield J., Lee E.B., Curtis J.R., Wood S.P., Soma K., et al. Safety and efficacy of tofacitinib, an oral Janus kinase inhibitor, for the treatment of rheumatoid arthritis in open-label, longterm extension studies. J. Rheumatol. 2014;41(5):837–852. doi: 10.3899/jrheum.130683. [DOI] [PubMed] [Google Scholar]
- 96.Danese S., Grisham M., Hodge J., Telliez J.-B. JAK inhibition using tofacitinib for inflammatory bowel disease treatment: a hub for multiple inflammatory cytokines. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2016;310(3):G62–G155. doi: 10.1152/ajpgi.00311.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kubo S., Nakayamada S., Sakata K., Kitanaga Y., Ma X., Lee S., et al. Janus kinase inhibitor baricitinib modulates human innate and adaptive immune system. Front. Immunol. 2018;9:1510. doi: 10.3389/fimmu.2018.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weinblatt M.E. Methotrexate in rheumatoid arthritis: a quarter century of development. Trans. Am. Clin. Climatol. Assoc. 2013;124:16. [PMC free article] [PubMed] [Google Scholar]
- 99.Spurlock C.F., III, Tossberg J.T., Matlock B.K., Olsen N.J., Aune T.M. Methotrexate inhibits NF-κB activity via long intergenic (noncoding) RNA–p21 induction. Arthritis & rheumatology. 2014;66(11):2947–2957. doi: 10.1002/art.38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Majumdar S., Aggarwal B.B. Methotrexate suppresses NF-κB activation through inhibition of IκBα phosphorylation and degradation. J. Immunol. 2001;167(5):2911–2920. doi: 10.4049/jimmunol.167.5.2911. [DOI] [PubMed] [Google Scholar]
- 101.Poole R.M., Ballantyne A.D. Apremilast: first global approval. Drugs. 2014;74(7):825–837. doi: 10.1007/s40265-014-0218-4. [DOI] [PubMed] [Google Scholar]
- 102.Matthay M.A., Ware L.B., Zimmerman G.A. The acute respiratory distress syndrome. J. Clin. Invest. 2012;122(8):2731–2740. doi: 10.1172/JCI60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schafer P., Parton A., Gandhi A., Capone L., Adams M., Wu L., et al. Apremilast, a cAMP phosphodiesterase-4 inhibitor, demonstrates anti-inflammatory activity in vitro and in a model of psoriasis. Br. J. Pharmacol. 2010;159(4):842–855. doi: 10.1111/j.1476-5381.2009.00559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gottlieb A., Strober B., Krueger J., Rohane P., Zeldis J., Hu C., et al. An open-label, single-arm pilot study in patients with severe plaque-type psoriasis treated with an oral anti-inflammatory agent, apremilast. Curr. Med. Res. Opin. 2008;24(5):1529–1538. doi: 10.1185/030079908x301866. [DOI] [PubMed] [Google Scholar]
- 105.Kosutova P., Mikolka P., Kolomaznik M., Balentova S., Adamkov M., Calkovska A., et al. Reduction of lung inflammation, oxidative stress and apoptosis by the PDE4 inhibitor roflumilast in experimental model of acute lung injury. Physiol. Res. 2018;67:S54–S645. doi: 10.33549/physiolres.934047. [DOI] [PubMed] [Google Scholar]
- 106.Kwak H.J., Song J.S., Heo J.Y., Yang S.D., Nam J.-Y., Cheon H.G. Roflumilast inhibits lipopolysaccharide-induced inflammatory mediators via suppression of nuclear factor-κB, p38 mitogen-activated protein kinase, and c-Jun NH2-terminal kinase activation. J. Pharmacol. Exp. Ther. 2005;315(3):1188–1195. doi: 10.1124/jpet.105.092056. [DOI] [PubMed] [Google Scholar]
- 107.Bradley J. TNF-mediated inflammatory disease. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 2008;214(2):149–160. doi: 10.1002/path.2287. [DOI] [PubMed] [Google Scholar]
- 108.Billmeier U., Dieterich W., Neurath M.F., Atreya R. Molecular mechanism of action of anti-tumor necrosis factor antibodies in inflammatory bowel diseases. World J. Gastroenterol. 2016;22(42):9300. doi: 10.3748/wjg.v22.i42.9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Menegatti S., Bianchi E., Rogge L. Anti-TNF therapy in spondyloarthritis and related diseases, impact on the immune system and prediction of treatment responses. Front. Immunol. 2019;10:382. doi: 10.3389/fimmu.2019.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lai W-Y, Wang J-W, Huang B-T, Lin EP-Y, Yang P-C. A novel TNF-α-targeting aptamer for TNF-α-mediated acute lung injury and acute liver failure. Theranostics. 2019;9(6):1741. [DOI] [PMC free article] [PubMed]
- 111.Milia A.F., Manetti M., Generini S., Polidori L., Benelli G., Cinelli M., et al. TNFα blockade prevents the development of inflammatory bowel disease in HLA-B27 transgenic rats. J. Cell. Mol. Med. 2009;13(1):164–176. doi: 10.1111/j.1582-4934.2008.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., et al. COVID-19 cytokines and the hyperactive immune response: synergism of TNF-α and IFN-γ in triggering inflammation, tissue damage, and death. bioRxiv. 2020 preprint. [Google Scholar]
- 113.Capote K.R., McCormack F.X., Possmayer F. Pulmonary surfactant protein-a (SP-A) restores the surface properties of surfactant after oxidation by a mechanism that requires the Cys6 interchain disulfide bond and the phospholipid binding domain. J. Biol. Chem. 2003;278(23):20461–20474. doi: 10.1074/jbc.M212697200. [DOI] [PubMed] [Google Scholar]
- 114.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 115.Imai Y., Kuba K., Neely G.G., Yaghubian-Malhami R., Perkmann T., van Loo G., et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133(2):235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Korff S., Loughran P., Cai C., Lee Y.S., Scott M., Billiar T.R. Eritoran attenuates tissue damage and inflammation in hemorrhagic shock/trauma. J. Surg. Res. 2013;184(2):e17–e25. doi: 10.1016/j.jss.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ekici F, Karaca EE, Korkmaz Ş, Yüksel O, Gülbahar Ö, Alper M, et al. Effect of the toll-like receptor 4 antagonist eritoran on retinochoroidal inflammatory damage in a rat model of endotoxin-induced inflammation. Mediat. Inflamm.. 2014;2014. [DOI] [PMC free article] [PubMed]
- 118.Nomura J., Busso N., Ives A., Matsui C., Tsujimoto S., Shirakura T., et al. Xanthine oxidase inhibition by febuxostat attenuates experimental atherosclerosis in mice. Sci. Rep. 2014;4(1):1–9. doi: 10.1038/srep04554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Granger D.N., Kvietys P.R. Reperfusion injury and reactive oxygen species: the evolution of a concept. Redox Biol. 2015;6:524–551. doi: 10.1016/j.redox.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sabán-Ruiz J., Alonso-Pacho A., Fabregate-Fuente M., Gonzalez-Quevedo CdlP. Xanthine oxidase inhibitor febuxostat as a novel agent postulated to act against vascular inflammation. Anti-Inflammatory & Anti-Allergy Agents in Medicinal Chemistry (Formerly Current Medicinal Chemistry-Anti-Inflammatory and Anti-Allergy Agents) 2013;12(1):94–99. doi: 10.2174/1871523011312010011. [DOI] [PubMed] [Google Scholar]
- 121.Ahmed M.A., El Morsy E.M., Ahmed A.A. Protective effects of febuxostat against paraquat-induced lung toxicity in rats: impact on RAGE/PI3K/Akt pathway and downstream inflammatory cascades. Life Sci. 2019;221:56–64. doi: 10.1016/j.lfs.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 122.Fahmi A.N., Shehatou G.S., Shebl A.M., Salem H.A. Febuxostat protects rats against lipopolysaccharide-induced lung inflammation in a dose-dependent manner. Naunyn Schmiedeberg’s Arch. Pharmacol. 2016;389(3):269–278. doi: 10.1007/s00210-015-1202-6. [DOI] [PubMed] [Google Scholar]
- 123.Khan S.I., Malhotra R.K., Rani N., Sahu A.K., Tomar A., Garg S., et al. Febuxostat modulates MAPK/NF-κBp65/TNF-α signaling in cardiac ischemia-reperfusion injury. Oxidative Med. Cell. Longev. 2017;2017 doi: 10.1155/2017/8095825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Miyata H., Takada T., Toyoda Y., Matsuo H., Ichida K., Suzuki H. Identification of febuxostat as a new strong ABCG2 inhibitor: potential applications and risks in clinical situations. Front. Pharmacol. 2016;7:518. doi: 10.3389/fphar.2016.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davoodi L., Abedi S.M., Salehifar E., Alizadeh-Navai R., Rouhanizadeh H., Khorasani G., et al. Febuxostat therapy in outpatients with suspected COVID-19: a clinical trial. Int. J. Clin. Pract. 2020:e13600. doi: 10.1111/ijcp.13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu W., Parmigiani R., Marks P. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26(37):5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 127.Bradner J.E., West N., Grachan M.L., Greenberg E.F., Haggarty S.J., Warnow T., et al. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 2010;6(3):238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Duvic M., Talpur R., Ni X., Zhang C., Hazarika P., Kelly C., et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109(1):31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Leoni F., Zaliani A., Bertolini G., Porro G., Pagani P., Pozzi P., et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc. Natl. Acad. Sci. 2002;99(5):2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kimura A., Naka T., Nakahama T., Chinen I., Masuda K., Nohara K., et al. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J. Exp. Med. 2009;206(9):2027–2035. doi: 10.1084/jem.20090560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Krämer O.H., Baus D., Knauer S.K., Stein S., Jäger E., Stauber R.H., et al. Acetylation of Stat1 modulates NF-κB activity. Genes Dev. 2006;20(4):473–485. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wu P. Regulation of the activity of the pyruvate dehydrogenase complex. Adv. Enzym. Regul. 2002;42:249. doi: 10.1016/s0065-2571(01)00061-9. [DOI] [PubMed] [Google Scholar]
- 133.Yamane K., Indalao I.L., Chida J., Yamamoto Y., Hanawa M., Kido H. Diisopropylamine dichloroacetate, a novel pyruvate dehydrogenase kinase 4 inhibitor, as a potential therapeutic agent for metabolic disorders and multiorgan failure in severe influenza. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0098032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Feske S., Wulff H., Skolnik E.Y. Ion channels in innate and adaptive immunity. Annu. Rev. Immunol. 2015;33:291–353. doi: 10.1146/annurev-immunol-032414-112212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wulff H., Calabresi P.A., Allie R., Yun S., Pennington M., Beeton C., et al. The voltage-gated Kv1. 3 K+ channel in effector memory T cells as new target for MS. J. Clin. Invest. 2003;111(11):1703–1713. doi: 10.1172/JCI16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol.. 2004;22:745–63. [DOI] [PubMed]
- 137.Tarcha E.J., Chi V., Muñoz-Elías E.J., Bailey D., Londono L.M., Upadhyay S.K., et al. Durable pharmacological responses from the peptide ShK-186, a specific Kv1. 3 channel inhibitor that suppresses T cell mediators of autoimmune disease. J. Pharmacol. Exp. Ther. 2012;342(3):642–653. doi: 10.1124/jpet.112.191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kazama I. Targeting lymphocyte Kv1. 3-channels to suppress cytokine storm in severe COVID-19: can it be a novel therapeutic strategy? Drug Discoveries & Therapeutics. 2020;14(3):143–144. doi: 10.5582/ddt.2020.03046. [DOI] [PubMed] [Google Scholar]
- 139.Blanco P., Palucka A.K., Pascual V., Banchereau J. Dendritic cells and cytokines in human inflammatory and autoimmune diseases. Cytokine Growth Factor Rev. 2008;19(1):41–52. doi: 10.1016/j.cytogfr.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Benson J.M., Heavner G.A., Giles-Komar J.M., et al. In: MAbs. Peritt D., Scallon B.J., Shealy D.J., editors. Taylor & Francis; 2011. Discovery and mechanism of ustekinumab: a human monoclonal antibody targeting interleukin-12 and interleukin-23 for treatment of immune-mediated disorders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shen W., Durum S.K. Synergy of IL-23 and Th17 cytokines: new light on inflammatory bowel disease. Neurochem. Res. 2010;35(6):940–946. doi: 10.1007/s11064-009-0091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.De Nitto D., Sarra M., Laura Cupi M., Pallone F., Monteleone G. Targeting IL-23 and Th17-cytokines in inflammatory bowel diseases. Curr. Pharm. Des. 2010;16(33):3656–3660. doi: 10.2174/138161210794079164. [DOI] [PubMed] [Google Scholar]
- 144.Ulhaq Z.S., Soraya G.V. Interleukin-6 as a potential biomarker of COVID-19 progression. Med. Mal. Infect. 2020;50(4):382–383. doi: 10.1016/j.medmal.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Coomes E.A., Haghbayan H. Interleukin-6 in COVID-19: a systematic review and meta-analysis. MedRxiv. 2020;30(6):1–9. doi: 10.1002/rmv.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]