Abstract
Nucleic acid-binding proteins are traditionally divided into two categories: With the ability to bind DNA or RNA. In the light of new knowledge, such categorizing should be overcome because a large proportion of proteins can bind both DNA and RNA. Another even more important features of nucleic acid-binding proteins are so-called sequence or structure specificities. Proteins able to bind nucleic acids in a sequence-specific manner usually contain one or more of the well-defined structural motifs (zinc-fingers, leucine zipper, helix-turn-helix, or helix-loop-helix). In contrast, many proteins do not recognize nucleic acid sequence but rather local DNA or RNA structures (G-quadruplexes, i-motifs, triplexes, cruciforms, left-handed DNA/RNA form, and others). Finally, there are also proteins recognizing both sequence and local structural properties of nucleic acids (e.g., famous tumor suppressor p53). In this mini-review, we aim to summarize current knowledge about the amino acid composition of various types of nucleic acid-binding proteins with a special focus on significant enrichment and/or depletion in each category.
Keywords: DNA, RNA, protein binding, G-quadruplex, triplex, i-motif, Z-DNA, Z-RNA, cruciform, amino acid composition
1. Introduction
Interactions between proteins and nucleic acids (DNA and RNA) are central to all aspects of maintaining and accessing genetic information. Nucleic acid-binding proteins are mostly composed of at least one DNA or RNA-binding domain where the interfacing with amino acids takes place in a specific or nonspecific manner [1]. Identification of nucleic acid-binding proteins is one of the most important tasks in molecular biology. Currently, nucleic acid-binding proteins can be identified and further characterized by several experimental techniques, including pull-down assays [2,3], yeast one-hybrid system [4,5], electrophoretic mobility shift assays [6,7], chromatin immunoprecipitation [8,9], and by other specialized techniques [10,11]. However, it is time-consuming and expensive to identify nucleic acid-binding proteins by experimental approaches [12]. With the easy availability of a large amount of protein sequence data, there is a rapid development of computational approaches and prediction tools that can rapidly and reliably identify nucleic acid-binding proteins [13,14]. Several such tools model nucleic acid-binding abilities based on protein amino acid composition [15,16]. There is a growing interest in so-called noncanonical nucleic acid structures and proteins that preferentially bind them [17,18,19,20,21,22,23,24]. Noncanonical nucleic acid structures are DNA and RNA structures different from their basic form, i.e., double-stranded right-handed DNA or single-stranded RNA, and are often formed by simple nucleotide repeats [25,26,27,28]. Physiologically, they are represented mainly by G-quadruplexes [29], i-motifs [30], triplexes [31], R-loops [32], slipped hairpins [33], DNA cruciforms [34], RNA hairpins [35], and Z-DNA [36]. These DNA/RNA structures have important biological functions [37,38,39,40,41,42] and contribute to many human diseases [43,44,45,46]. It became more and more evident, that proteins preferentially interacting with these structures share distinct amino acid features/fingerprints [47,48]. This mini-review aims to focus on the amino acid composition of various types of DNA and RNA-binding proteins and to compare the amino acid composition of proteins that prefer binding to different noncanonical forms of nucleic acids.
2. Amino Acid Composition of Nucleic Acid-Binding Proteins
According to the Gene Ontology (GO) knowledgebase, there are 5037 nucleic acid-binding proteins (filtering GO:0003676 term by “protein”) with experimental evidence in Homo sapiens [49,50,51]. Of this number, 2572 are annotated as RNA-binding and 2439 as DNA-binding proteins (some proteins have both functions). 1768 human proteins are known to bind DNA in a sequence-specific manner. It would be interesting to quantify the overall amount of proteins binding nucleic acids in a structure-specific manner. Unfortunately, there is no such category yet. We strongly suggest revisions in this manner. Inspiration can be found in the following review papers/databases focused on specific properties of proteins binding to G-quadruplexes [19,52,53,54], cruciforms [55], and Z-DNA/Z-RNA [56].
2.1. History
Amino acid composition of some nucleic acid-binding proteins was intensively studied at the beginning of the 70s, when Koichi Iwai et al. determined that “calf-thymus histones comprise five main types which differ in amino acid composition and electrophoretic mobility: A glycine-rich, arginine-rich histone (also known as f2al or IV); a glutamic-acid-rich, arginine-rich histone (fe or III); a leucine-rich, intermediate type histone (f2a2 or IIb1); a serine-rich, slightly lysine-rich histone (f2b or IIb2); and an alanine-rich, very lysine-rich histone (f1 or I)” [57], by using specialized chromatographic technique followed by polyacrylamide gel electrophoresis. In 1975, from the comparison of 68 representative proteins and frequencies of 61 codons of the genetic code, it was found that the average amounts of lysine, aspartic acid, glutamic acid, and alanine are above the levels anticipated from the genetic code, and arginine, serine, leucine, cysteine, proline, and histidine are below such levels [58]. There are a couple of examples from the 90s and 2000s when amino acid substitution in nucleic acid-binding protein abolished its function, e.g., an arginine to lysine substitution in the bZIP (Basic Leucine Zipper) domain of an opaque-2 mutant in maize abolished specific DNA-binding [59], missense mutations (Met175Arg and Ser191Asn) abolishing DNA-binding of the osteoblast-specific transcription factor OSF2/CBFA1 in human patients with cleidocranial dysplasia [60], or impaired RNA-binding of fragile X mental retardation protein upon missense mutation IIe-304→Asn in one of its KH domain [61]. Recent advantages in sequencing and bioinformatic methods allow us to directly compare the amino acid composition of thousands of (not only) human nucleic acid-binding proteins [62,63]. One of the most popular programs for this purpose is, e.g., composition profiler [64], which is a web-based tool for semi-automatic discovery of enrichment or depletion of amino acids, either individually or grouped by their physicochemical or structural properties [64]. Scientists often find themselves in the situation when they only have a sequence of new “hypothetical” protein, derived mainly from transcriptome sequencing, and want to deduce its function [65]. In case that no meaningful alignment to protein with known function is available, there is still a way to get some useful information using only primary amino acid sequence and its composition. In 2003, Cai and Lin used a protein’s amino acid composition and support vector machine (SVM) prediction to decide if protein belongs to one of three classes—rRNA-, RNA-, or DNA-binding [66]. Currently, there are also user-friendly web-based prediction tools called DNAbinder and PseDNA-Pro, which can predict if the submitted protein sequence has DNA-binding ability [12,67].
2.2. Methods to Inspect the Amino Acid Composition of Proteins
Several approaches are used to inspect the amino acid composition of nucleic acid-binding proteins. Basically, we can divide the methods into in vitro and in silico. In vitro approaches are necessary to obtain a sequence of the protein of interest. Although the development of large-scale genomic sequencing has greatly simplified the procedure of determining the primary structures of proteins, the genomic sequences of many organisms are still unknown, and also modifications such as post-translational events (citrullination, deamidation, polyglutamylation,…) may prevent proper determination of the protein sequence [68]. Then, the complete characterization of the primary protein structure often requires a mass spectrometry method with minimal assistance from genomic data, i.e., de novo protein sequencing [68,69]. In silico approaches are based mostly on previous knowledge about primary protein sequence. There is currently a plentitude of bioinformatics tools designed for that purpose, see, e.g., [64,70,71,72,73].
2.3. Amino Acid Composition of Nucleic Acid-Binding Proteins
Nucleic acid-binding proteins are traditionally divided into two categories. The first category comprises proteins with the ability to bind DNA, and the second category comprises proteins that bind to RNA. This division is quite outdated, mainly because, from the historical perspective, proteins that bind RNA were typically considered as functionally distinct from proteins that bind DNA and studied independently. Interestingly, current gene ontology analyses reveal that DNA-binding is potentially a major function of the mRNA-binding proteins [74]. Nonetheless, several studies inspecting amino acid composition of DNA and/or RNA-binding proteins were published [75,76] and find that particular amino acid residues are generally enriched or depleted within these protein categories (see Table 1).
Table 1.
Types of nucleic acid-binding proteins. This table summarizes the main categories of nucleic acid-binding proteins. There are two points of view. At first, we can simply divide these proteins into DNA and RNA-binding ones (and a relatively small category of proteins that are able to bind both DNA and RNA). Secondly (and more importantly), we can distinguish proteins that specifically bind known sequence motifs (sequence-specific DNA/RNA-binding) and proteins, which specifically bind local DNA/RNA structures. Besides, keep in mind that this table is very simplified, and categories are divided to be reader-friendly. In fact, many of the DNA/RNA-binding proteins combine sequence and structure-specific binding mechanisms.
| Important Notes | References | |
|---|---|---|
| DNA-binding | Arginine, tryptophan, tyrosine, histidine, phenylalanine, and lysine residues enrichment. Glutamate, aspartate, and proline depletion in the protein-DNA interface. | [76,79] |
| RNA-binding | Arginine, methionine, histidine, and lysine residues enrichment. Glutamate, aspartate residues depletion in protein-RNA interface. | [75,76] |
| DNA and RNA-binding | Proteins that are able to bind both DNA and RNA. | [74,80] |
| Sequence-specific | ||
| Zinc finger proteins | Cysteine and histidine amino acid residues are crucially important to coordinate Zn2+ binding in the Cys2His2 subgroup of zinc-finger proteins | [77,78] |
| Helix-turn-helix (HTH) | Conserved “shs” and “phs” patterns, where ‘s’ is a small residue, most frequently glycine in the first position, ‘h’ is a hydrophobic residue, and ‘p’ is a charged residue, most frequently glutamate. “shs” pattern lies in the turn between helix-2 and helix-3 of the core HTH structure, and “phs” is present in helix-2. | [81] |
| Basic Helix-loop-helix (bHLH) | Mostly arginine, lysine or histidine amino acid residues are present within conserved positions of this motif | [82,83] |
| Leucine zipper proteins | Leucine amino acid residues are crucial for leucine zipper motifs | [84,85] |
| Structure specific | ||
| G-quadruplex binding proteins | Global enrichment for glycine, arginine, aspartic acid, asparagine, valine, and depletion for cysteine, histidine, leucine, proline, glutamine, and tryptophan residues | [47,86,87,88] |
| Cruciform binding proteins | Global enrichment for lysine and serine, and depletion for alanine, glycine, glutamine, arginine, tyrosine, and tryptophan residues | [48,55] |
| Triplex binding proteins | Global enrichment for asparagine, aspartic acid, isoleucine, tyrosine, and depletion for cysteine, histidine, and proline residues | [89] |
| Z-DNA/RNA-binding proteins | Global enrichment for isoleucine, aspartic acid, lysine, and depletion for cysteine residues | [89] |
Another, even more important division of nucleic acid-binding proteins is based on a so-called sequence or structure-specific type of binding. Proteins able to bind nucleic acids in a sequence-specific manner usually contain one or more of the well-defined structural motifs. One of such motifs, zinc-finger, binds DNA (or RNA) through specific interaction with nucleotides and sugar-phosphate backbone. Tandem repeating of slightly different zinc-finger motifs in protein then allows to recognizing its consensus nucleic acid-binding sequence specifically. Cysteine and histidine amino acid residues are crucially important to coordinate Zn2+ binding in the largest and best-characterized subgroup of zinc-finger binding proteins named the Cys2His2 fold subgroup [77,78]. Other well-defined sequence-specific motifs—leucine zipper, helix-turn-helix, or helix-loop-helix—are listed in Table 1, together with their common signatures of amino acid residues.
In contrast, many proteins do not recognize nucleic acid sequence but rather local DNA or RNA structures (G-quadruplexes, i-motifs, triplexes, cruciforms, left-handed DNA/RNA form, and others) [19,30,41,55,90]. Finally, there are also proteins recognizing both sequence and local structural properties of nucleic acids (e.g., famous tumor suppressor p53 [91], Myc-associated zinc finger protein (MAZ) [92,93], and many RNA-binding proteins [94])—these proteins usually contain sequence-specific binding domain(s) together with domain(s)/region(s) with preference to noncanonical nucleic acid structures [95,96,97]. In 2016, Wang et al. analyzed the abundance of intrinsic disorder in the DNA- and RNA-binding proteins in over 1000 species from Eukaryota, Bacteria, and Archaea domains of life [98]. They have revealed a very interesting phenomenon that DNA-binding proteins had significantly increased disorder content and were significantly enriched in disordered domains in Eukaryotes but not in Archaea and Bacteria. The RNA-binding proteins were significantly enriched in the disordered domains in Bacteria, Archaea, and Eukaryota, while the overall abundance of disorder in these proteins was significantly increased in Bacteria, Archaea, animals, and fungi [98]. Disordered domains or regions are also extensively present in chromatin-binding proteins [99,100]. Interestingly, some disordered proteins or regions show very high structural specificity to the different types of noncanonical nucleic acids. For instance, human protein SRSF1 (Serine/arginine-rich splicing factor 1) contains several intrinsically disordered regions [101], which are compositionally enriched in glycine (14.11% of overall amino acid residues) and arginine (17.39% of overall amino acid residues) content. It was previously shown that SRSF1 has a high affinity to RNA G-quadruplex structure [102]. Subsequent analyses have shown that the dataset of 77 G-quadruplex binding proteins is significantly globally enriched in arginine, glycine, aspartic acid, asparagine, and valine, and depleted in cysteine and other amino acid residues [47] (Figure 1). Finally, the common amino acid motif in the form of RGRGRGRGGGSGGSGGRGRG was derived, and most of the currently known G-quadruplex binding proteins contain at least some modification of it [47]. Using this motif, a new dataset of G-quadruplex binding proteins was predicted from the set of all human DNA/RNA-binding proteins [47], and some of them were independently experimentally validated (e.g., CIRBP, which is a cold-inducible RNA-binding protein in the study by Huang and colleagues [103]). A similar study focused on an amino acid composition of cruciform binding proteins was also published, and the significant enrichment for lysine and serine amino acid residues has been revealed [48] (Figure 1). Unpublished results also indicate distinct amino acid profiles in Z-DNA/RNA and triplex binding proteins, both significantly enriched in aspartic acid and isoleucine and depleted in cysteine residues [89] (Figure 1). In future studies, it would be interesting to specifically analyze local amino acid composition (only in the nucleic acid interaction sites and their close neighborhood) in these proteins. Unfortunately for the vast majority of them, the knowledge about exact DNA/RNA binding site(s) is still missing.
Figure 1.
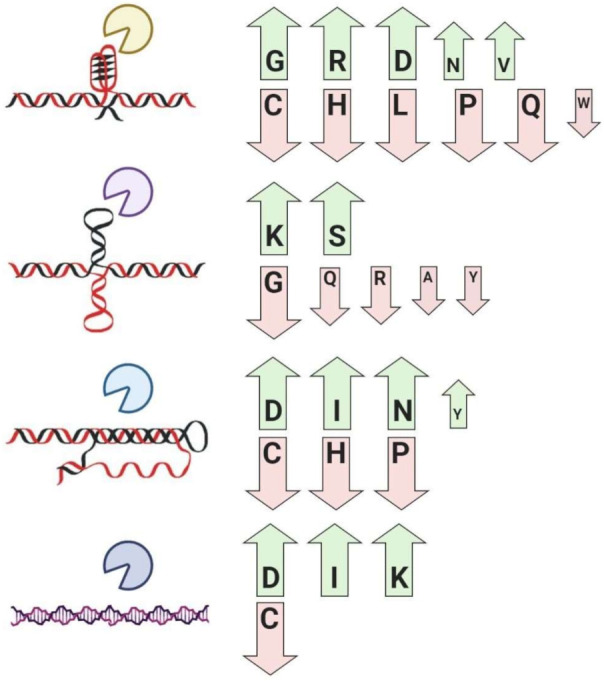
Significantly enriched and depleted amino acid residues (one letter aa code) in the dataset of G-quadruplex binding proteins (top), cruciform binding proteins, triplex binding proteins, and Z-DNA-binding proteins. Using Bonferroni correction, only values lower than 0.0025 were taken as significant (p <0.0025; p <0.0010; p <0.0001). The size of arrows indicates the significance of enrichment/depletion on scale (highest, moderate, lowest). Figure compiled using data from [47,48,89]. Created with BioRender.com.
As was shown above, proteins that preferentially recognize noncanonical nucleic acid structures often have a distinct amino acid composition with particular significant global enrichment and/or depletion of different amino acid residues. Noncanonical structures and proteins preferentially binding them often play a critical role in physiological molecular processes [32,104,105], but also in the progression of human diseases, such as various cancer types and neurodegenerative diseases, reviewed in [55,106,107]. Knowledge about the amino acid composition of various proteins binding noncanonical nucleic acids can be utilized as an additional clue/fingerprint in discovering novel noncanonical nucleic acid-binding protein candidates and therapeutically utilized [108,109,110].
The scheme below depicts sequence and structure-specific nucleic acid-binding phenomena in a nutshell (Figure 2).
Figure 2.
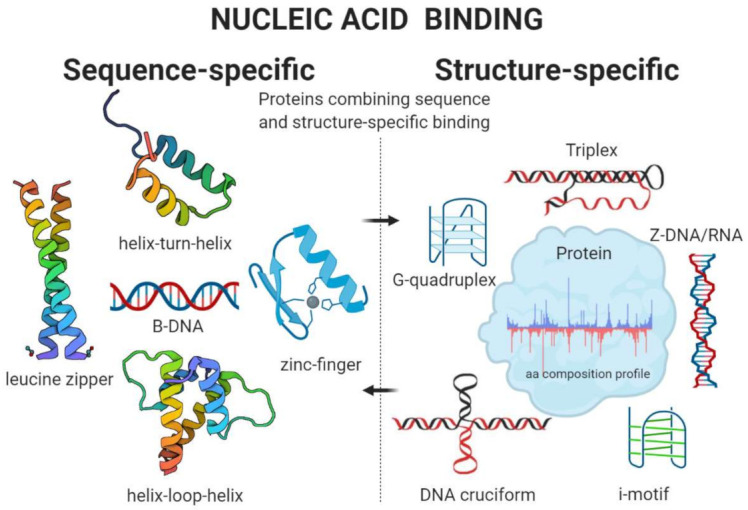
Types of nucleic acid-binding. The nucleic acid-binding mechanism can be basically divided into two main categories—sequence and structure-specific binding. (Left) Sequence-specific binding proteins recognize the variety of known DNA/RNA sequences via specific interaction with well-characterized protein motifs (zinc-fingers, helix-loop-helix, leucine zipper, helix-turn-helix, etc.). (Right) Structure-specific binding proteins recognize specific local structure(s) of nucleic acids, e.g., G-quadruplexes, i-motifs, cruciforms, triplexes, Z-DNA, and many others. In fact, it is a very common phenomenon that protein with the sequence-specific binding also prefers local DNA/RNA structure in its binding site or within the near neighborhood (e.g., p53), which is indicated by vertical black dashed line and arrows. Created with BioRender.com.
Almost every year, multiple novel noncanonical nucleic acid-binding proteins are identified. This year was, for instance, found that Guanine Nucleotide-Binding Protein-Like 1 (GNL1) binds RNA G-quadruplex structures in genes associated with Parkinson’s disease [111], or that Small Nuclear Ribonucleoprotein Polypeptide A (SNRPA) directly binds to the BAG-1 mRNA through the G-quadruplex which can modulate BAG-1 expression level [112] (anti-apoptotic BAG-1 protein is known to be overexpressed in colorectal cancers [113]). Prediction of proteins that preferentially bind noncanonical DNA/RNA structures, therefore, should be a logical first step towards rapid identification of novel therapeutic targets for future treatment of severe human diseases.
3. Closing Remarks
The global or local amino acid composition of nucleic acid-binding proteins is often overlooked and an unjustly underestimated parameter. Mainly statistically significant enrichment or depletion of particular amino acid residues may serve as a promising tool to predict novel proteins with a similar function, as it was confirmed e.g., for G-quadruplex binding proteins.
Abbreviations
| bZIP | Basic Leucine Zipper |
| CIRBP | Cold inducible RNA-binding protein |
| GNL1 | Guanine Nucleotide-Binding Protein-Like 1 |
| GO | Gene Ontology |
| bHLH | Helix-loop-helix |
| HTH | Helix-turn-helix |
| MAZ | Myc-associated zinc finger protein |
| SNRPA | Small Nuclear Ribonucleoprotein Polypeptide A |
| SRSF1 | Serine/arginine-rich splicing factor 1 |
| SVM | Support vector machine |
Author Contributions
Conceptualization, M.B. and P.P.; methodology, M.B.; resources, M.B., S.G., and K.S.; writing—original draft preparation, M.B., S.G, K.S., and J.Č.; writing—review and editing, J.Č and P.P.; visualization, M.B.; supervision, P.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by University of Ostrava, SGS01/PřF/2020, and by National Agency for Agricultural Research (NAZV) of Czech Republic grant no. QK1810391 “Utilization of genomic and transcriptomic approaches to create genetic resources and breeding materials of poppy with specific traits”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ghani N.S.A., Firdaus-Raih M., Ahmad S. Computational Prediction of Nucleic acid-binding Residues From Sequence. In: Ranganathan S., Gribskov M., Nakai K., Schönbach C., editors. Encyclopedia of Bioinformatics and Computational Biology. Academic Press; Oxford, UK: 2019. pp. 678–687. [Google Scholar]
- 2.Jutras B.L., Verma A., Stevenson B. Identification of Novel DNA-Binding Proteins Using DNA-Affinity Chromatography/Pull Down. Curr. Protoc. Microbiol. 2012;24:1F.1.1–1F.1.13. doi: 10.1002/9780471729259.mc01f01s24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang I.X., Grunseich C., Fox J., Burdick J., Zhu Z., Ravazian N., Hafner M., Cheung V.G. Human Proteins That Interact with RNA/DNA Hybrids. Genome Res. 2018;28:1405–1414. doi: 10.1101/gr.237362.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ouwerkerk P.B., Meijer A.H. Plant Reverse Genetics. Springer; Basel, Switzerland: 2011. Yeast one-hybrid screens for detection of transcription factor DNA interactions; pp. 211–227. [DOI] [PubMed] [Google Scholar]
- 5.Gaudinier A., Tang M., Bågman A.-M., Brady S.M. Identification of Protein–DNA Interactions Using Enhanced Yeast One-Hybrid Assays and a Semiautomated Approach. In: Busch W., editor. Plant Genomics: Methods and Protocols. Springer; New York, NY: 2017. pp. 187–215. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 6.Hellman L.M., Fried M.G. Electrophoretic Mobility Shift Assay (EMSA) for Detecting Protein–Nucleic Acid Interactions. Nat. Protoc. 2007;2:1849. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo M., Lei L., Egli M. Label-Free Electrophoretic Mobility Shift Assay (EMSA) for Measuring Dissociation Constants of Protein-RNA Complexes. Curr. Protoc. Nucleic Acid Chem. 2019;76:e70. doi: 10.1002/cpnc.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carey M.F., Peterson C.L., Smale S.T. Chromatin Immunoprecipitation (Chip) Cold Spring Harb. Protoc. 2009;2009:pdb-prot5279. doi: 10.1101/pdb.prot5279. [DOI] [PubMed] [Google Scholar]
- 9.de Barsy M., Herrgott L., Martin V., Pillonel T., Viollier P.H., Greub G. Identification of New DNA-Associated Proteins from Waddlia Chondrophila. Sci. Rep. 2019;9:4885. doi: 10.1038/s41598-019-40732-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunová N., Ondrovičová G., Bauer J.A., Bellová J., Ambro Ľ., Martináková L., Kotrasová V., Kutejová E., Pevala V. The Role of Lon-Mediated Proteolysis in the Dynamics of Mitochondrial Nucleic Acid-Protein Complexes. Sci. Rep. 2017;7:631. doi: 10.1038/s41598-017-00632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haronikova L., Coufal J., Kejnovska I., Jagelska E.B., Fojta M., Dvořáková P., Muller P., Vojtesek B., Brazda V. IFI16 Preferentially Binds to DNA with Quadruplex Structure and Enhances DNA Quadruplex Formation. PLoS ONE. 2016;11:e0157156. doi: 10.1371/journal.pone.0157156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B., Wang S., Wang X. DNA-binding Protein Identification by Combining Pseudo Amino Acid Composition and Profile-Based Protein Representation. Sci. Rep. 2015;5:15479. doi: 10.1038/srep15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y., Guo Y., Feng Y., Li M. Predicting DNA-Binding Proteins: Approached from Chou’s Pseudo Amino Acid Composition and Other Specific Sequence Features. Amino Acids. 2008;34:103–109. doi: 10.1007/s00726-007-0568-2. [DOI] [PubMed] [Google Scholar]
- 14.Wei L., Tang J., Zou Q. Local-DPP: An Improved DNA-Binding Protein Prediction Method by Exploring Local Evolutionary Information. Inf. Sci. 2017;384:135–144. doi: 10.1016/j.ins.2016.06.026. [DOI] [Google Scholar]
- 15.Liu B., Xu J., Lan X., Xu R., Zhou J., Wang X., Chou K.-C. IDNA-Prot|dis: Identifying DNA-Binding Proteins by Incorporating Amino Acid Distance-Pairs and Reduced Alphabet Profile into the General Pseudo Amino Acid Composition. PLoS ONE. 2014;9:e106691. doi: 10.1371/journal.pone.0106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi S., Han K. Prediction of RNA-Binding Amino Acids from Protein and RNA Sequences. BMC Bioinform. 2011;12:S7. doi: 10.1186/1471-2105-12-S13-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brázda V., Coufal J., Liao J.C.C., Arrowsmith C.H. Preferential Binding of IFI16 Protein to Cruciform Structure and Superhelical DNA. Biochem. Biophys. Res. Commun. 2012;422:716–720. doi: 10.1016/j.bbrc.2012.05.065. [DOI] [PubMed] [Google Scholar]
- 18.Čechová J., Coufal J., Jagelská E.B., Fojta M., Brázda V. P73, like Its P53 Homolog, Shows Preference for Inverted Repeats Forming Cruciforms. PLoS ONE. 2018;13:e0195835. doi: 10.1371/journal.pone.0195835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brázda V., Hároníková L., Liao J.C., Fojta M. DNA and RNA Quadruplex-Binding Proteins. Int. J. Mol. Sci. 2014;15:17493–17517. doi: 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helma R., Bažantová P., Petr M., Adámik M., Renčiuk D., Tichỳ V., Pastuchová A., Soldánová Z., Pečinka P., Bowater R.P. P53 Binds Preferentially to Non-B DNA Structures Formed by the Pyrimidine-Rich Strands of GaA· TTC Trinucleotide Repeats Associated with Friedreich’s Ataxia. Molecules. 2019;24:2078. doi: 10.3390/molecules24112078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyons S.M., Kharel P., Akiyama Y., Ojha S., Dave D., Tsvetkov V., Merrick W., Ivanov P., Anderson P. EIF4G Has Intrinsic G-Quadruplex Binding Activity That Is Required for TiRNA Function. Nucleic Acids Res. 2020;48:6223–6233. doi: 10.1093/nar/gkaa336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porubiaková O., Bohálová N., Inga A., Vadovičová N., Coufal J., Fojta M., Brázda V. The Influence of Quadruplex Structure in Proximity to P53 Target Sequences on the Transactivation Potential of P53 Alpha Isoforms. Int. J. Mol. Sci. 2020;21:127. doi: 10.3390/ijms21010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oyoshi T., Masuzawa T. Modulation of Histone Modifications and G-Quadruplex Structures by G-Quadruplex-Binding Proteins. Biochem. Biophys. Res. Commun. 2020;531:39–44. doi: 10.1016/j.bbrc.2020.02.178. [DOI] [PubMed] [Google Scholar]
- 24.Bartas M., Brázda V., Bohálová N., Cantara A., Volná A., Stachurová T., Malachová K., Jagelská E.B., Porubiaková O., Červeň J. In-Depth Bioinformatic Analyses of Nidovirales Including Human SARS-CoV-2, SARS-CoV, MERS-CoV Viruses Suggest Important Roles of Non-Canonical Nucleic Acid Structures in Their Lifecycles. Front. Microbiol. 2020;11:1583. doi: 10.3389/fmicb.2020.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tateishi-Karimata H., Sugimoto N. Chemical Biology of Non-Canonical Structures of Nucleic Acids for Therapeutic Applications. Chem. Commun. 2020;56:2379–2390. doi: 10.1039/C9CC09771F. [DOI] [PubMed] [Google Scholar]
- 26.Cer R.Z., Donohue D.E., Mudunuri U.S., Temiz N.A., Loss M.A., Starner N.J., Halusa G.N., Volfovsky N., Yi M., Luke B.T. Non-B DB v2. 0: A Database of Predicted Non-B DNA-Forming Motifs and Its Associated Tools. Nucleic Acids Res. 2012;41:D94–D100. doi: 10.1093/nar/gks955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brazda V., Fojta M., Bowater R.P. Structures and Stability of Simple DNA Repeats from Bacteria. Biochem. J. 2020;477:325–339. doi: 10.1042/BCJ20190703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brázda V., Luo Y., Bartas M., Kaura P., Porubiaková O., Št’astnỳ J., Pečinka P., Verga D., Da Cunha V., Takahashi T.S. G-Quadruplexes in the Archaea Domain. Biomolecules. 2020;10:1349. doi: 10.3390/biom10091349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhodes D., Lipps H.J. G-Quadruplexes and Their Regulatory Roles in Biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeraati M., Langley D.B., Schofield P., Moye A.L., Rouet R., Hughes W.E., Bryan T.M., Dinger M.E., Christ D. I-Motif DNA Structures Are Formed in the Nuclei of Human Cells. Nat. Chem. 2018;10:631–637. doi: 10.1038/s41557-018-0046-3. [DOI] [PubMed] [Google Scholar]
- 31.Brázdová M., Tichý V., Helma R., Bažantová P., Polášková A., Krejčí A., Petr M., Navrátilová L., Tichá O., Nejedlý K., et al. P53 Specifically Binds Triplex DNA In Vitro and in Cells. PLoS ONE. 2016;11:e0167439. doi: 10.1371/journal.pone.0167439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chedin F., Benham C.J. Emerging Roles for R-Loop Structures in the Management of Topological Stress. J. Biol. Chem. 2020;295:4684–4695. doi: 10.1074/jbc.REV119.006364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu P., Pan F., Roland C., Sagui C., Weninger K. Dynamics of Strand Slippage in DNA Hairpins Formed by CAG Repeats: Roles of Sequence Parity and Trinucleotide Interrupts. Nucleic Acids Res. 2020;48:2232–2245. doi: 10.1093/nar/gkaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleming A.M., Zhu J., Jara-Espejo M., Burrows C.J. Cruciform DNA Sequences in Gene Promoters Can Impact Transcription upon Oxidative Modification of 2′-Deoxyguanosine. Biochemistry. 2020;59:2616–2626. doi: 10.1021/acs.biochem.0c00387. [DOI] [PubMed] [Google Scholar]
- 35.Bevilacqua P.C., Ritchey L.E., Su Z., Assmann S.M. Genome-Wide Analysis of RNA Secondary Structure. Annu. Rev. Genet. 2016;50:235–266. doi: 10.1146/annurev-genet-120215-035034. [DOI] [PubMed] [Google Scholar]
- 36.Shin S.-I., Ham S., Park J., Seo S.H., Lim C.H., Jeon H., Huh J., Roh T.-Y. Z-DNA-Forming Sites Identified by ChIP-Seq Are Associated with Actively Transcribed Regions in the Human Genome. DNA Res. 2016;23:477–486. doi: 10.1093/dnares/dsw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spiegel J., Adhikari S., Balasubramanian S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020;2:123–136. doi: 10.1016/j.trechm.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S. The Regulation and Functions of DNA and RNA G-Quadruplexes. Nat. Rev. Mol. Cell Biol. 2020;21:459–474. doi: 10.1038/s41580-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaushik M., Kaushik S., Roy K., Singh A., Mahendru S., Kumar M., Chaudhary S., Ahmed S., Kukreti S. A Bouquet of DNA Structures: Emerging Diversity. Biochem. Biophys. Rep. 2016;5:388–395. doi: 10.1016/j.bbrep.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masai H., Tanaka T. G-Quadruplex DNA and RNA: Their Roles in Regulation of DNA Replication and Other Biological Functions. Biochem. Biophys. Res. Commun. 2020;531:25–38. doi: 10.1016/j.bbrc.2020.05.132. [DOI] [PubMed] [Google Scholar]
- 41.Herbert A. Z-DNA and Z-RNA in Human Disease. Commun. Biol. 2019;2:1–10. doi: 10.1038/s42003-018-0237-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan W.-F., Wan L.-Y., Peng H., Zhong Y.-M., Cai W.-L., Zhang Y.-Q., Ai W.-B., Wu J.-F. The Influencing Factors and Functions of DNA G-Quadruplexes. Cell Biochem. Funct. 2020;38:524–532. doi: 10.1002/cbf.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bacolla A., Cooper D.N., Vasquez K.M., Tainer J.A. eLS. American Cancer Society; Atlanta, GA, USA: 2018. Non-B DNA Structure and Mutations Causing Human Genetic Disease; pp. 1–15. [Google Scholar]
- 44.Bacolla A., Tainer J.A., Vasquez K.M., Cooper D.N. Translocation and Deletion Breakpoints in Cancer Genomes Are Associated with Potential Non-B DNA-Forming Sequences. Nucleic Acids Res. 2016;44:5673–5688. doi: 10.1093/nar/gkw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cammas A., Millevoi S. RNA G-Quadruplexes: Emerging Mechanisms in Disease. Nucleic Acids Res. 2017;45:1584–1595. doi: 10.1093/nar/gkw1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kharel P., Balaratnam S., Beals N., Basu S. The Role of RNA G-Quadruplexes in Human Diseases and Therapeutic Strategies. Wiley Interdiscip. Rev. RNA. 2020;11:e1568. doi: 10.1002/wrna.1568. [DOI] [PubMed] [Google Scholar]
- 47.Brázda V., Cerveň J., Bartas M., Mikysková N., Coufal J., Pečinka P. The Amino Acid Composition of Quadruplex Binding Proteins Reveals a Shared Motif and Predicts New Potential Quadruplex Interactors. Molecules. 2018;23 doi: 10.3390/molecules23092341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartas M., Bažantová P., Brázda V., Liao J., Červeň J., Pečinka P. Identification of Distinct Amino Acid Composition of Human Cruciform Binding Proteins. Mol. Biol. 2019;53:97–106. doi: 10.1134/S0026893319010023. [DOI] [PubMed] [Google Scholar]
- 49.Consortium G.O. Expansion of the Gene Ontology Knowledgebase and Resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Consortium G.O. Gene Ontology Consortium: Going Forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S., Hub A., Group W.P.W. AmiGO: Online Access to Ontology and Annotation Data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mishra S.K., Tawani A., Mishra A., Kumar A. G4IPDB: A Database for G-Quadruplex Structure Forming Nucleic Acid Interacting Proteins. Sci. Rep. 2016;6:38144. doi: 10.1038/srep38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moccia F., Platella C., Musumeci D., Batool S., Zumrut H., Bradshaw J., Mallikaratchy P., Montesarchio D. The Role of G-Quadruplex Structures of LIGS-Generated Aptamers R1.2 and R1.3 in IgM Specific Recognition. Int. J. Biol. Macromol. 2019;133:839–849. doi: 10.1016/j.ijbiomac.2019.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riccardi C., Napolitano E., Platella C., Musumeci D., Melone M.A.B., Montesarchio D. Anti-VEGF DNA-Based Aptamers in Cancer Therapeutics and Diagnostics. Med. Res. Rev. 2021;41:464–506. doi: 10.1002/med.21737. [DOI] [PubMed] [Google Scholar]
- 55.Brázda V., Laister R.C., Jagelská E.B., Arrowsmith C. Cruciform Structures Are a Common DNA Feature Important for Regulating Biological Processes. BMC Mol. Biol. 2011;12:33. doi: 10.1186/1471-2199-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim C. How Z-DNA/RNA-binding Proteins Shape Homeostasis, Inflammation, and Immunity. BMB Rep. 2020;53:453–457. doi: 10.5483/BMBRep.2020.53.9.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iwai K., Ishikawa K., Hayashi H. Amino-Acid Sequence of Slightly Lysine-Rich Histone. Nature. 1970;226:1056–1058. doi: 10.1038/2261056b0. [DOI] [PubMed] [Google Scholar]
- 58.Jukes T.H., Holmquist R., Moise H. Amino Acid Composition of Proteins: Selection against the Genetic Code. Science. 1975;189:50–51. doi: 10.1126/science.237322. [DOI] [PubMed] [Google Scholar]
- 59.Aukerman M.J., Schmidt R.J., Burr B., Burr F.A. An Arginine to Lysine Substitution in the BZIP Domain of an Opaque-2 Mutant in Maize Abolishes Specific DNA-binding. Genes Dev. 1991;5:310–320. doi: 10.1101/gad.5.2.310. [DOI] [PubMed] [Google Scholar]
- 60.Lee B., Thirunavukkarasu K., Zhou L., Pastore L., Baldini A., Hecht J., Geoffrey V., Ducy P., Karsenty G. Missense Mutations Abolishing DNA-binding of the Osteoblast-Specific Transcription Factor OSF2/CBFA1 in Cleidocranial Dysplasia. Nat. Genet. 1997;16:307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 61.Siomi H., Choi M., Siomi M.C., Nussbaum R.L., Dreyfuss G. Essential Role for KH Domains in RNA-binding: Impaired RNA-binding by a Mutation in the KH Domain of FMR1 That Causes Fragile X Syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 62.Cheng S., Melkonian M., Smith S.A., Brockington S., Archibald J.M., Delaux P.-M., Li F.-W., Melkonian B., Mavrodiev E.V., Sun W., et al. 10KP: A Phylodiverse Genome Sequencing Plan. GigaScience. 2018;7 doi: 10.1093/gigascience/giy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kriventseva E.V., Kuznetsov D., Tegenfeldt F., Manni M., Dias R., Simão F.A., Zdobnov E.M. OrthoDB V10: Sampling the Diversity of Animal, Plant, Fungal, Protist, Bacterial and Viral Genomes for Evolutionary and Functional Annotations of Orthologs. Nucleic Acids Res. 2019;47:D807–D811. doi: 10.1093/nar/gky1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vacic V., Uversky V.N., Dunker A.K., Lonardi S. Composition Profiler: A Tool for Discovery and Visualization of Amino Acid Composition Differences. BMC Bioinform. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sivashankari S., Shanmughavel P. Functional Annotation of Hypothetical Proteins – A Review. Bioinformation. 2006;1:335–338. doi: 10.6026/97320630001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai Y., Lin S.L. Support Vector Machines for Predicting RRNA-, RNA-, and DNA-Binding Proteins from Amino Acid Sequence. Biochim. Et Biophys. Acta (Bba) - Proteins Proteom. 2003;1648:127–133. doi: 10.1016/S1570-9639(03)00112-2. [DOI] [PubMed] [Google Scholar]
- 67.Kumar M., Gromiha M.M., Raghava G.P. Identification of DNA-Binding Proteins Using Support Vector Machines and Evolutionary Profiles. BMC Bioinform. 2007;8:463. doi: 10.1186/1471-2105-8-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Standing K.G. Peptide and Protein de Novo Sequencing by Mass Spectrometry. Curr. Opin. Struct. Biol. 2003;13:595–601. doi: 10.1016/j.sbi.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 69.Vitorino R., Guedes S., Trindade F., Correia I., Moura G., Carvalho P., Santos M.A.S., Amado F. De Novo Sequencing of Proteins by Mass Spectrometry. Expert Rev. Proteom. 2020;17:595–607. doi: 10.1080/14789450.2020.1831387. [DOI] [PubMed] [Google Scholar]
- 70.Gasteiger E., Hoogland C., Gattiker A., Wilkins M.R., Appel R.D., Bairoch A. The proteomics protocols handbook. Springer; Basel, Switzerland: 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [Google Scholar]
- 71.Cao D.-S., Xu Q.-S., Liang Y.-Z. Propy: A Tool to Generate Various Modes of Chou’s PseAAC. Bioinformatics. 2013;29:960–962. doi: 10.1093/bioinformatics/btt072. [DOI] [PubMed] [Google Scholar]
- 72.Vishnoi S., Garg P., Arora P. Physicochemical N-Grams Tool: A Tool for Protein Physicochemical Descriptor Generation via Chou’s 5-Step Rule. Chem. Biol. Drug Des. 2020;95:79–86. doi: 10.1111/cbdd.13617. [DOI] [PubMed] [Google Scholar]
- 73.Zuo Y., Li Y., Chen Y., Li G., Yan Z., Yang L. PseKRAAC: A Flexible Web Server for Generating Pseudo K-Tuple Reduced Amino Acids Composition. Bioinformatics. 2017;33:122–124. doi: 10.1093/bioinformatics/btw564. [DOI] [PubMed] [Google Scholar]
- 74.Hudson W.H., Ortlund E.A. The Structure, Function and Evolution of Proteins That Bind DNA and RNA. Nat. Rev.. Mol. Cell Biol. 2014;15:749–760. doi: 10.1038/nrm3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Terribilini M., Lee J.-H., Yan C., Jernigan R.L., Honavar V., Dobbs D. Prediction of RNA-binding Sites in Proteins from Amino Acid Sequence. RNA. 2006;12:1450–1462. doi: 10.1261/rna.2197306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J., Ma Z., Kurgan L. Comprehensive Review and Empirical Analysis of Hallmarks of DNA-, RNA-and Protein-Binding Residues in Protein Chains. Brief. Bioinform. 2019;20:1250–1268. doi: 10.1093/bib/bbx168. [DOI] [PubMed] [Google Scholar]
- 77.Michalek J.L., Besold A.N., Michel S.L.J. Cysteine and Histidine Shuffling: Mixing and Matching Cysteine and Histidine Residues in Zinc Finger Proteins to Afford Different Folds and Function. Dalton Trans. 2011;40:12619–12632. doi: 10.1039/c1dt11071c. [DOI] [PubMed] [Google Scholar]
- 78.Laity J.H., Lee B.M., Wright P.E. Zinc Finger Proteins: New Insights into Structural and Functional Diversity. Curr. Opin. Struct. Biol. 2001;11:39–46. doi: 10.1016/S0959-440X(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 79.Yesudhas D., Batool M., Anwar M.A., Panneerselvam S., Choi S. Proteins Recognizing DNA: Structural Uniqueness and Versatility of DNA-Binding Domains in Stem Cell Transcription Factors. Genes. 2017;8:192. doi: 10.3390/genes8080192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahmad M., Xu D., Wang W. Type IA Topoisomerases Can Be “Magicians” for Both DNA and RNA in All Domains of Life. RNA Biol. 2017;14:854–864. doi: 10.1080/15476286.2017.1330741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aravind L., Anantharaman V., Balaji S., Babu M.M., Iyer L.M. The Many Faces of the Helix-Turn-Helix Domain: Transcription Regulation and Beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 82.Atchley W.R., Fitch W.M. A Natural Classification of the Basic Helix–Loop–Helix Class of Transcription Factors. Proc. Natl. Acad. Sci. USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Casey B.H., Kollipara R.K., Pozo K., Johnson J.E. Intrinsic DNA-binding Properties Demonstrated for Lineage-Specifying Basic Helix-Loop-Helix Transcription Factors. [(accessed on 2 January 2021)]; doi: 10.1101/gr.224360.117. Available online: http://genome.cshlp.org. [DOI] [PMC free article] [PubMed]
- 84.Hakoshima T. eLS. American Cancer Society; Atlanta, GA, USA: 2014. Leucine Zippers. [Google Scholar]
- 85.Miller M. The Importance of Being Flexible: The Case of Basic Region Leucine Zipper Transcriptional Regulators. Curr. Protein Pept. Sci. 2009;10:244–269. doi: 10.2174/138920309788452164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yagi R., Miyazaki T., Oyoshi T. G-Quadruplex Binding Ability of TLS/FUS Depends on the β-Spiral Structure of the RGG Domain. Nucleic Acids Res. 2018;46:5894–5901. doi: 10.1093/nar/gky391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ishiguro A., Kimura N., Noma T., Shimo-Kon R., Ishihama A., Kon T. Molecular Dissection of ALS-Linked TDP-43 – Involvement of the Gly-Rich Domain in Interaction with G-Quadruplex MRNA. FEBS Lett. 2020;594:2254–2265. doi: 10.1002/1873-3468.13800. [DOI] [PubMed] [Google Scholar]
- 88.Takahama K., Oyoshi T. Specific Binding of Modified RGG Domain in TLS/FUS to G-Quadruplex RNA: Tyrosines in RGG Domain Recognize 2′-OH of the Riboses of Loops in G-Quadruplex. J. Am. Chem. Soc. 2013;135:18016–18019. doi: 10.1021/ja4086929. [DOI] [PubMed] [Google Scholar]
- 89.Bartas M., Červeň J., Pečinka P. Identification of Distinct Amino Acid Composition of Z-DNA/RNA and Triplex-Binding Proteins. Mol. Bio. 53:97–106. doi: 10.1134/S0026898419010026. [DOI] [PubMed] [Google Scholar]
- 90.Ribeiro de Almeida C., Dhir S., Dhir A., Moghaddam A.E., Sattentau Q., Meinhart A., Proudfoot N.J. RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol. Cell. 2018;70:650–662.e8. doi: 10.1016/j.molcel.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cai B.-H., Chao C.-F., Huang H.-C., Lee H.-Y., Kannagi R., Chen J.-Y. Roles of P53 Family Structure and Function in Non-Canonical Response Element Binding and Activation. Int. J. Mol. Sci. 2019;20:3681. doi: 10.3390/ijms20153681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bossone S.A., Asselin C., Patel A.J., Marcu K.B. MAZ, a Zinc Finger Protein, Binds to c-MYC and C2 Gene Sequences Regulating Transcriptional Initiation and Termination. Proc. Natl. Acad. Sci. USA. 1992;89:7452–7456. doi: 10.1073/pnas.89.16.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cogoi S., Zorzet S., Rapozzi V., Géci I., Pedersen E.B., Xodo L.E. MAZ-Binding G4-Decoy with Locked Nucleic Acid and Twisted Intercalating Nucleic Acid Modifications Suppresses KRAS in Pancreatic Cancer Cells and Delays Tumor Growth in Mice. Nucleic Acids Res. 2013;41:4049–4064. doi: 10.1093/nar/gkt127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dominguez D., Freese P., Alexis M.S., Su A., Hochman M., Palden T., Bazile C., Lambert N.J., Van Nostrand E.L., Pratt G.A., et al. Sequence, Structure, and Context Preferences of Human RNA-binding Proteins. Mol. Cell. 2018;70:854–867. doi: 10.1016/j.molcel.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laptenko O., Tong D.R., Manfredi J., Prives C. The Tail That Wags the Dog: How the Disordered C-Terminal Domain Controls the Transcriptional Activities of the P53 Tumor-Suppressor Protein. Trends Biochem. Sci. 2016;41:1022–1034. doi: 10.1016/j.tibs.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Petr M., Helma R., Polášková A., Krejčí A., Dvořáková Z., Kejnovská I., Navrátilová L., Adámik M., Vorlíčková M., Brázdová M. Wild-Type P53 Binds to MYC Promoter G-Quadruplex. Biosci. Rep. 2016;36 doi: 10.1042/BSR20160232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Inukai S., Kock K.H., Bulyk M.L. Transcription Factor–DNA-binding: Beyond Binding Site Motifs. Curr. Opin. Genet. Dev. 2017;43:110–119. doi: 10.1016/j.gde.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang C., Uversky V.N., Kurgan L. Disordered Nucleiome: Abundance of Intrinsic Disorder in the DNA- and RNA-Binding Proteins in 1121 Species from Eukaryota, Bacteria and Archaea. Proteomics. 2016;16:1486–1498. doi: 10.1002/pmic.201500177. [DOI] [PubMed] [Google Scholar]
- 99.Watson M., Stott K. Disordered Domains in Chromatin-Binding Proteins. Essays Biochem. 2019;63:147–156. doi: 10.1042/EBC20180068. [DOI] [PubMed] [Google Scholar]
- 100.Turner A.L., Watson M., Wilkins O.G., Cato L., Travers A., Thomas J.O., Stott K. Highly Disordered Histone H1−DNA Model Complexes and Their Condensates. Proc. Natl. Acad. Sci. USA. 2018;115:11964–11969. doi: 10.1073/pnas.1805943115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Serrano P., Aubol B.E., Keshwani M.M., Forli S., Ma C.-T., Dutta S.K., Geralt M., Wüthrich K., Adams J.A. Directional Phosphorylation and Nuclear Transport of the Splicing Factor SRSF1 Is Regulated by an RNA Recognition Motif. J. Mol. Biol. 2016;428:2430–2445. doi: 10.1016/j.jmb.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Von Hacht A.V., Seifert O., Menger M., Schütze T., Arora A., Konthur Z., Neubauer P., Wagner A., Weise C., Kurreck J. Identification and Characterization of RNA Guanine-Quadruplex Binding Proteins. Nucleic Acids Res. 2014;42:6630–6644. doi: 10.1093/nar/gku290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang Z.-L., Dai J., Luo W.-H., Wang X.-G., Tan J.-H., Chen S.-B., Huang Z.-S. Identification of G-Quadruplex-Binding Protein from the Exploration of RGG Motif/G-Quadruplex Interactions. J. Am. Chem. Soc. 2018;140:17945–17955. doi: 10.1021/jacs.8b09329. [DOI] [PubMed] [Google Scholar]
- 104.Rigo R., Palumbo M., Sissi C. G-Quadruplexes in Human Promoters: A Challenge for Therapeutic Applications. Biochim. Et Biophys. Acta (Bba)-Gen. Subj. 2017;1861:1399–1413. doi: 10.1016/j.bbagen.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 105.Poggi L., Richard G.-F. Alternative DNA Structures In Vivo: Molecular Evidence and Remaining Questions. Microbiol. Mol. Biol. Rev. 2020;85 doi: 10.1128/MMBR.00110-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sissi C., Gatto B., Palumbo M. The Evolving World of Protein-G-Quadruplex Recognition: A Medicinal Chemist’s Perspective. Biochimie. 2011;93:1219–1230. doi: 10.1016/j.biochi.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brázda V., Coufal J. Recognition of Local DNA Structures by P53 Protein. Int. J. Mol. Sci. 2017;18:375. doi: 10.3390/ijms18020375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun Z.-Y., Wang X.-N., Cheng S.-Q., Su X.-X., Ou T.-M. Developing Novel G-Quadruplex Ligands: From Interaction with Nucleic Acids to Interfering with Nucleic Acid–Protein Interaction. Molecules. 2019;24:396. doi: 10.3390/molecules24030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kharel P., Becker G., Tsvetkov V., Ivanov P. Properties and Biological Impact of RNA G-Quadruplexes: From Order to Turmoil and Back. Nucleic Acids Res. 2020;48:12534–12555. doi: 10.1093/nar/gkaa1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee T., Pelletier J. The Biology of DHX9 and Its Potential as a Therapeutic Target. Oncotarget. 2016;7:42716–42739. doi: 10.18632/oncotarget.8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Turcotte M.-A., Garant J.-M., Cossette-Roberge H., Perreault J.-P. Guanine Nucleotide-Binding Protein-Like 1 (GNL1) Binds RNA G-Quadruplex Structures in Genes Associated with Parkinson’s Disease. RNA Biol. 2020:1–15. doi: 10.1080/15476286.2020.1847866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bolduc F., Turcotte M.-A., Perreault J.-P. The Small Nuclear Ribonucleoprotein Polypeptide A (SNRPA) Binds to the G-Quadruplex of the BAG-1 5′UTR. Biochimie. 2020;176:122–127. doi: 10.1016/j.biochi.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 113.Clemo N.K., Collard T.J., Southern S.L., Edwards K.D., Moorghen M., Packham G., Hague A., Paraskeva C., Williams A.C. BAG-1 Is up-Regulated in Colorectal Tumour Progression and Promotes Colorectal Tumour Cell Survival through Increased NF-ΚB Activity. Carcinogenesis. 2008;29:849–857. doi: 10.1093/carcin/bgn004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.


