Abstract
Enteric coronaviruses (CoVs) are major pathogens that cause diarrhea in piglets. To date, four porcine enteric CoVs have been identified: transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), porcine deltacoronavirus (PDCoV), and HKU2-like porcine enteric alphacoronavirus (PEAV). In this study, we investigated the replicative capacity of these four enteric CoVs in LLC-PK1 cells, a porcine kidney cell line. The results showed that LLC-PK1 cells are susceptible to all four enteric CoVs, particularly to TGEV and PDCoV infections, indicating that LLC-PK1 cells can be applied to porcine enteric CoV research in vitro, particularly for coinfection studies.
Introduction
Coronaviruses (CoVs) are single-stranded, positive-sense enveloped RNA viruses. Coronavirus outbreaks, including severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and novel human severe acute respiratory syndrome CoV 2 (SARS-CoV-2), have posed enormous threats to human health and the economy [1–3]. In the pig-rearing industry, CoVs are also significant pathogens causing diarrhea in piglets, with fever, loss of appetite, vomiting, severe diarrhea, dehydration, and even death. Four porcine enteric CoVs have been identified, including transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), porcine deltacoronavirus (PDCoV), and the newly discovered HKU2-like porcine enteric alphacoronavirus (PEAV) [4–7].
The prevalence of coinfections by two or potentially more kinds of enteric CoVs is increasing in dozens of countries, including China, the USA, and South Korea [8–10]. For example, PEDV and PDCoV coinfections are frequently detected in piglets, and half of PDCoV-positive samples can also be positive for PEDV according to an investigation in our lab [7]. Liu et al. reported that the morbidity rate of mixed infections with PEDV, TGEV, and porcine rotavirus (PoRV) was 27.85% [11]. Zhou et al. performed a retrospective study and found that coinfections with PEAV and PEDV occurred most frequently with a rate of 17.65% [12]. As various experimental data have shown, coinfections are becoming more common. Therefore, experimental materials, especially cell lines, are urgently needed to facilitate research into coinfections.
A few cell lines have been used to model coinfections with different porcine enteric CoVs. IPI-FX cells, a subcloned line of homogeneous cells derived from IPI-2I cells (porcine intestinal epithelial cells), are susceptible to the four enteric CoVs mentioned above [13]. Additionally, the IPEC-J2 cell line and its subcloned cell line IPEC-DQ have been confirmed to be susceptible to multiple porcine enteric CoVs [14, 15]. It is known that ST (swine testicle) cells are permissive for TGEV and PDCoV infection, Vero (African green monkey kidney) cells are susceptible to PEDV and PEAV [16–18], and LLC-PK1 (porcine kidney epithelial) cells are extensively used in the proliferation and isolation of PDCoV [19]. However, it is unclear whether these widely used cell lines have the potential to become infected with different enteric CoVs.
In this study, we investigated the susceptibility of LLC-PK1 cells to infection by four porcine enteric CoVs and found that LLC-PK1 cells could be infected by TGEV, PEDV, PDCoV, and PEAV. Moreover, LLC-PK1 cells were more sensitive to the four porcine enteric CoVs than IPI-FX cells, particularly for TGEV and PDCoV infections, providing another ideal cell model for studying coinfection with different porcine enteric CoVs.
Materials and methods
Cells, viruses and reagents
LLC-PK1 and Vero cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA). ST and IPI-2I cells were acquired from the China Center for Type Culture Collection (CCTCC; Wuhan, China). IPI-FX cells were subcloned previously from IPI-2I by our group. TGEV strain WH1 (GenBank accession number HQ462571), PEDV strain AJ1102 (GenBank accession number JX188454.1), PDCoV strain CHN-HN-2014 (GenBank accession number KT336560), and PEAV strain CHN-GD-2017 (GenBank accession number MH539766) were isolated from neonatal piglets with acute diarrhea. Mouse monoclonal antibodies (mAbs) against TGEV nucleocapsid (N) protein, PEDV nucleocapsid (N) protein, PDCoV spike (S) protein, and PEAV spike (S) protein were produced previously by our group.
Virus inoculation and cytopathic effect (CPE)
LLC-PK1 cells were grown in 24-well plates to confluence and then inoculated with TGEV and PDCoV at a multiplicity of infection (MOI) of 0.01 and with PEDV and PEAV at an MOI of 0.25. The optional infection doses used for different CoVs were based on our preliminary experiments. After incubation for 1 h at 37 °C to allow virus attachment, the inocula were discarded and replaced with DMEM. CPE was observed at 6, 12, 18, 24, and 30 h postinfection (hpi).
Indirect immunofluorescence assay (IFA)
Monolayer LLC-PK1 cells in 24-well plates were infected or mock infected with PDCoV (MOI = 0.01), TGEV (MOI = 0.01), PEAV (MOI = 0.25), or PEDV (MOI = 0.25). At 6, 12, 18, 24 and 30 hpi, the cells were washed three times with phosphate-buffered saline (PBS) and then fixed with 4% paraformaldehyde for 15 min and permeabilized with cold methyl alcohol for 10 min. The samples were then blocked with PBS containing 5% bovine serum albumin for 1 h and incubated with the corresponding mAbs for 1 h at 37 ℃. Afterwards, the cells were treated with Alexa Fluor 488–conjugated anti-mouse secondary antibody for 1 h at 37 ℃ and stained with 0.01% 4′,6′-diamidino-2-phenylindole (DAPI) for 15 min at room temperature. After three washes with PBS, fluorescent images were visualized using an inverted fluorescence microscope (Olympus IX73).
Median tissue culture infectious dose (TCID50) assay and proliferation curve
To compare the titers of TGEV, PDCoV, PEDV, and PEAV in different cells, LLC-PK1, IPI-FX, or ST cells were inoculated with TGEV or PDCoV at an MOI of 0.01, and LLC-PK1, IPI-FX, or Vero cells were inoculated with PEDV or PEAV at an MOI of 0.25. Samples were collected at 6, 12, 18, 24 and 30 hpi, frozen and thawed three times, and centrifuged at 4 ℃ 12000 rpm for 10 min, and the supernatants were collected. TCID50 assays were performed as described previously [14]. Briefly, cell monolayers seeded in 96-well plates were washed twice with DMEM containing 7.5 μg of trypsin per ml or 2% fetal bovine serum. Virus-containing supernatants were serially diluted tenfold, and appropriate dilutions of virus suspension were chosen to inoculate Vero or LLC-PK1 cells, with eight replicates at each dilution. After the cells were cultured at 37 ℃ with 5% CO2 for 3-4 days, the TCID50 was calculated by the Reed-Muench method, and the virus proliferation curves were plotted from three independent experiments.
Statistical analysis
Data are shown as the mean ± standard deviation of three independent experiments. Student’s t-test was used to evaluate differences in results, and P < 0.05 was considered statistically significant.
Results
Observation of typical CPE after enteric CoV infection
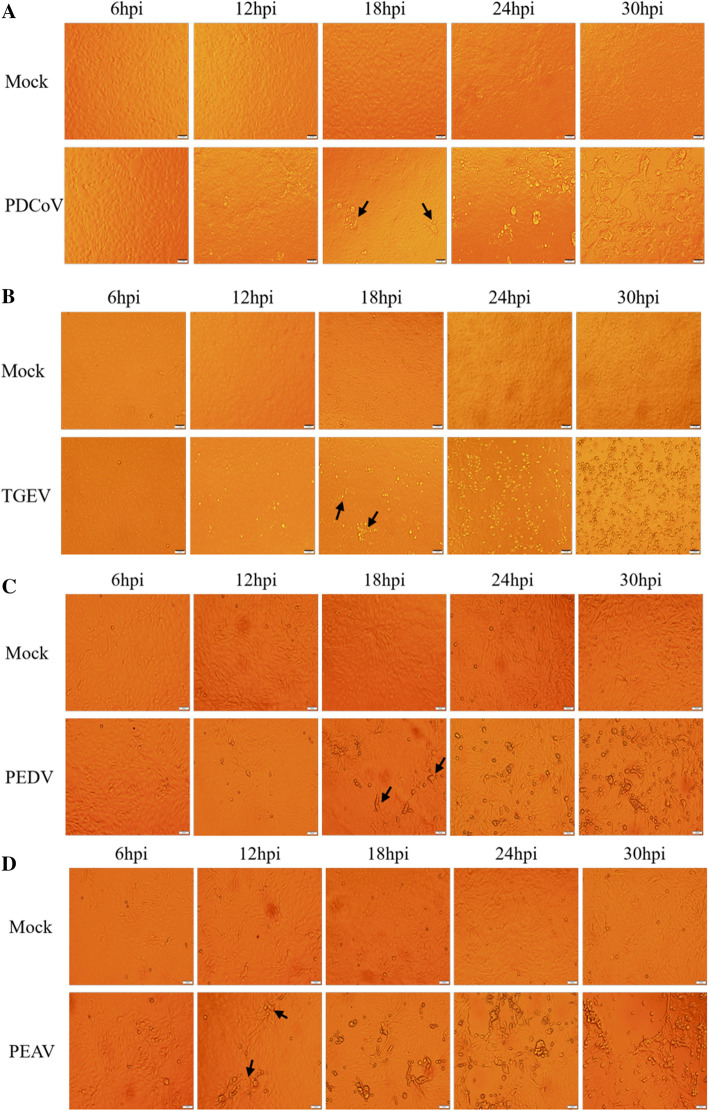
To determine the proliferation characteristics of four porcine enteric CoVs in LLC-PK1 cells, cells were infected with PDCoV or TGEV at an MOI of 0.01 or with PEDV or PEAV at an MOI of 0.25. CPE were examined at 6, 12, 18, 24 and 30 hpi. As shown in Fig. 1A, typical CPE characterized by enlarged, rounded, clustered cells became visible at 18 hpi and was increasingly apparent as PDCoV infection progressed. Similarly, CPE became observable at 12-30 hpi after TGEV infection, characterized by rounding, shrinkage, rupture, and shedding of cells (Fig. 1B). When LLC-PK1 cells were infected with PEDV or PEAV, fused syncytia were detected at 24 hpi, and cell detachment was observed at 30 hpi (Fig. 1C and D). These results indicate that LLC-PK1 cells are susceptible to all four porcine enteric CoVs.
Fig. 1.
Typical CPE after enteric CoV infection. (A) PDCoV CPE was detected at 6, 12, 18, 24, and 30 hpi. (B) TGEV CPE was detected at 6, 12, 18, 24, and 30 hpi. (C) PEDV CPE was detected at 6, 12, 18, 24, and 30 hpi. (D) PEAV CPE was detected at 6, 12, 18, 24, and 30 hpi. Typical CPE at the early stage of infection is indicated by black arrows.
Detection of specific fluorescence after enteric CoV infection
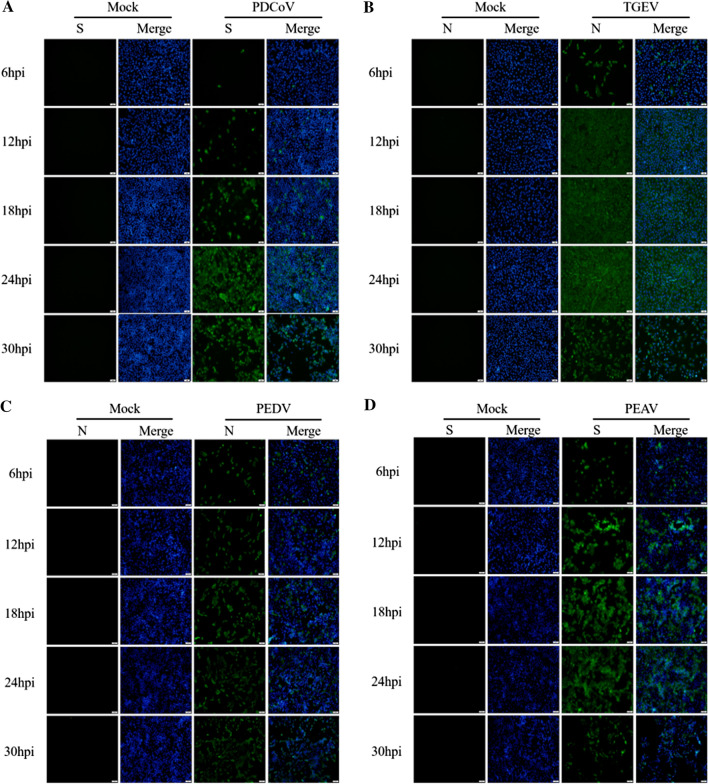
To further ascertain the replicative capacity of four porcine enteric CoVs in LLC-PK1 cells, mock-infected and infected cells were monitored by IFA with mAbs against the PDCoV S, TGEV N, PEDV N, and PEAV S protein, respectively. The results showed that only a small fraction of cells exhibited green fluorescence at 6, 12, and 18 hpi, but almost all of the cells were PDCoV positive at 24 hpi (Fig. 2A). Nearly 100% of the cells were infected by TGEV at 12 hpi (Fig. 2B), and PEDV-N- and PEAV-S-protein-specific immunofluorescence could be also observed throughout the infection (Fig. 2C and D). These results confirm the high susceptibility of LLC-PK1 cells to all four porcine enteric CoVs.
Fig. 2.
Specific immunofluorescence after enteric CoV infection. (A) PDCoV S. (B) TGEV N. (C) PEDV N. (D) PEAV S
Proliferation of enteric CoVs in LLC-PK1 cells
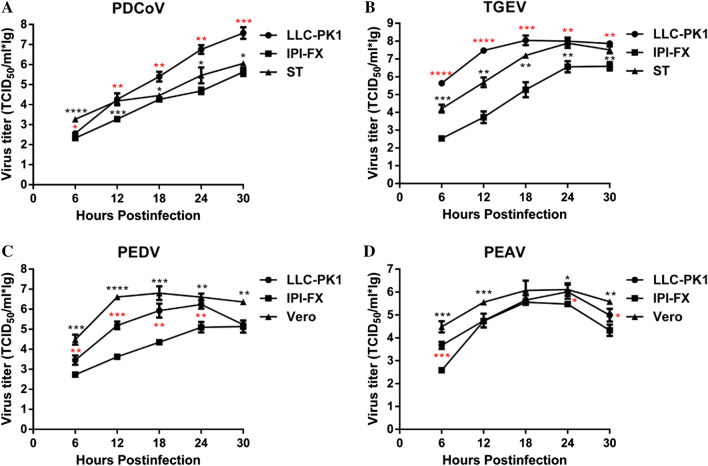
To study the growth kinetics of the four porcine enteric CoVs in LLC-PK1 cells in more detail, the growth curves of PDCoV, TGEV, PEDV and PEAV were compared in different susceptive cell lines, including LLC-PK1, IPI-FX, and ST/Vero cells. As shown in Fig. 3A, the PDCoV titer in LLC-PK1 cells reached 107.8 TCID50/mL, while in IPI-FX and ST cells, it only reached 105.8 TCID50/mL and 106.2 TCID50/mL, respectively. Similar growth curves were observed in TGEV-infected LLC-PK1 and ST cells, while TGEV infection in IPI-FX cells was roughly 1.4 log lower (Fig. 3B). Comparing the PEDV and PEAV proliferation curves in different cell lines, the viral titers in LLC-PK1 cells reached 106.4 TCID50/ml and 106.2 TCID50/mL at 24 hpi, respectively (Fig. 3C and D). Although the PEDV and PEAV titers propagated in LLC-PK1 cells were slightly lower than those in Vero cells, they were significantly higher than those in IPI-FX cells. These results show that the four porcine enteric CoVs reached a higher titer in LLC-PK1 cells than in ST or IPI-FX cells, indicating that LLC-PK1 is an ideal cell line for studying CoV infections or coinfections in vitro.
Fig. 3.
Growth curves of four porcine enteric CoVs measured by virus titration. (A) PDCoV growth curves in LLC-PK1, ST and IPI-FX cells. (B) TGEV growth curves in LLC-PK1, ST and IPI-FX cells. (C) PEDV growth curves in LLC-PK1, Vero and IPI-FX cells. (D) PEAV growth curves in LLC-PK1, Vero and IPI-FX cells. The mean titer and standard deviation were calculated from three independent experiments. Red marks represent differences between LLC-PK1 cells and IPI-FX cells, and black marks represent differences between ST and Vero cells. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05
Discussion
In recent years, emerging and re-emerging porcine enteric CoVs have presented new challenges for the pig industry. In addition to the first discovered TGEV, re-emerging PEDV variants possess greater virulence and lead to more serious economic losses. Furthermore, the emerging PDCoV and the bat-HUK2-like CoV PEAV are also more common in Asia and America. Mixed infections with two or more CoVs are also increasingly common on farms in various countries. Therefore, we should pay more attention to studying the mechanisms of porcine enteric CoV infection and host immune responses. Moreover, the development of suitable experimental materials, including cell lines, for coinfection studies should also be continually enriched.
Although PEDV is highly pathogenic to pigs, some porcine-derived cell lines appear not to be susceptible to PEDV infection [11]. Thus, Vero cells, a cell line derived from African green monkey kidney, are extensively used for PEDV and PEAV isolation and proliferation. However, it has been reported that Vero cells cannot express interferon because of their inherent genetic defects in the beta and alpha genes [20]. Some scholars believed that the attenuated vaccines produced by successive passage of highly virulent PEDV strains in non-pig-derived Vero cells often could not induce sufficient protective antibodies to counter a challenge with virulent PEDV [21]. Thus, Vero cells may not be an ideal candidate cell line for immunology studies of PEDV and PEAV. In contrast, multiple viral infections have been reported to inhibit the production of interferon in LLC-PK1 cells. For example, it has been reported that PDCoV infection suppresses the RIG-I-mediated IFN signaling pathway in LLC-PK1 cells [19]. In this study, we found that PEDV and PEAV could strongly infect LLC-PK1 cells despite attaining lower titers than those in Vero cells. This cumulative evidence indicates that pig-derived LLC-PK1 cells are applicable for PEDV and PEAV research and have an advantage over Vero cells in immunological studies.
TGEV, PEDV, PDCoV and PEAV are the four currently prevalent porcine enteric CoVs, and mixed infections are relatively common clinically under the current domestic intensive pig farming circumstances. Coinfections with two or more enteric CoVs have also been increasingly reported, but how CoVs interplay with each other remains unknown. A recent study indicated that mixing of viral components during coinfection altered pathogenic outcomes or viral replication. For example, overexpression of TGEV N increased production of PEDV RNA and virions [22]; therefore, we inferred that one coinfecting virus might augment the proliferation of the other, leading to more-severe diarrheal disease. Notably, new strains are likely to emerge in cases of coinfection, such as the emergence of recombinant S-INDEL-variant PEDV [23]. Hence, it is necessary to understand the mechanisms of coinfection. However, there are currently few cell lines that can be used for studies of multiple intestinal CoVs. IPI-FX, IPEC-J2, and subcloned IPEC-DQ cells can be infected by multiple enteric CoVs, as reported previously. However, all of these cell lines have the disadvantage of low viral titers. The results of this study showed that all four enteric CoVs could infect LLC-PK1 cells, and their ability to proliferate in LLC-PK1 cells was stronger than in IPI-FX cells. From this perspective, the LLC-PK1 cell line is a new ideal cell model for coinfection studies of enteric CoVs. In contrast, pathological changes occur mainly in the ileum and jejunum after CoV infection in vivo, and IPI-2I and IPEC-J2 cells are derived from swine ileal epithelium and jejunum epithelium, respectively [14, 15]. Lin et al. confirmed that the proteomic data from in vitro PDCoV-infected IPEC-J2 cells were more consistent with those obtained in vivo [24]. Furthermore, Jung et al. showed that PDCoV did not induce apoptosis in infected intestinal enterocytes in vivo but did in two infected cell lines of swine origin: LLC-PK and ST cells [25]. This highlights some limitations of LLC-PK1 cells as an in vitro model. However, LLC-PK1 cells can be transfected more efficiently than IPEC-J2 and IPI-FX cells. Based on the advantages and disadvantages of LLC-PK1 and enteric cells, suitable cell lines can be selected for different experimental purposes.
In conclusion, we have investigated the susceptibility of LLC-PK1 cells to infection with four different porcine enteric CoVs and found that they are highly susceptible to TGEV, PEDV, PDCoV, and PEAV and thus could be applied to enteric CoV research and coinfection studies.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 31730095; U1704231; 31672569).
Compliance with ethical standards
Conflict of interest
The authors of this paper have no conflict of interest. All authors have seen and approved the manuscript.
Ethical approval
All applicable international, national, and/or institutional guidelines were followed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenwen Xiao and Xunlei Wang contributed equally to this work.
References
- 1.Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthews KL, Coleman CM, van der Meer Y, Snijder EJ, Frieman MB. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J Gen Virol. 2014;95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ouyang Y, Yin J, Wang W, Shi H, Shi Y, et al. Down-regulated gene expression spectrum and immune responses changed during the disease progression in COVID-19 patients. Clin Infect Dis. 2020;71:2052–2060. doi: 10.1093/cid/ciaa462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enjuanes L, Smerdou C, Castilla J, Anton IM, Torres JM, et al. Development of protection against coronavirus induced diseases. A review. Adv Exp Med Biol. 1995;380:197–211. doi: 10.1007/978-1-4615-1899-0_34. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Li H, Liu Y, Pan Y, Deng F, et al. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis. 2012;18:1350–1353. doi: 10.3201/eid1803.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong N, Fang L, Zeng S, Sun Q, Chen H, Xiao S. Porcine deltacoronavirus in Mainland China. Emerg Infect Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong L, Li J, Zhou Q, Xu Z, Chen L, et al. A New Bat-HKU2-like Coronavirus in Swine, China, 2017. Emerg Infect Dis. 2017;23:1607–1609. doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Wang L, Zheng Y, Zhang J, Guo B, et al. Metagenomic analysis of the RNA fraction of the fecal virome indicates high diversity in pigs infected by porcine endemic diarrhea virus in the United States. Virol J. 2018;15:95. doi: 10.1186/s12985-018-1001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhai SL, Wei WK, Li XP, Wen XH, Zhou X, et al. Occurrence and sequence analysis of porcine deltacoronaviruses in southern China. Virol J. 2016;13:136. doi: 10.1186/s12985-016-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang G, Lee KK, Kim SH, Lee C. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014–2016. Transbound Emerg Dis. 2017;64:1364–1370. doi: 10.1111/tbed.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, et al. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou L, Sun Y, Lan T, Wu R, Chen J, et al. Retrospective detection and phylogenetic analysis of swine acute diarrhoea syndrome coronavirus in pigs in southern China. Transbound Emerg Dis. 2019;66:687–695. doi: 10.1111/tbed.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Fang L, Liu S, Ke W, Wang D, et al. Susceptibility of porcine IPI-2I intestinal epithelial cells to infection with swine enteric coronaviruses. Vet Microbiol. 2019;233:21–27. doi: 10.1016/j.vetmic.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung K, Miyazaki A, Hu H, Saif LJ. Susceptibility of porcine IPEC-J2 intestinal epithelial cells to infection with porcine deltacoronavirus (PDCoV) and serum cytokine responses of gnotobiotic pigs to acute infection with IPEC-J2 cell culture-passaged PDCoV. Vet Microbiol. 2018;221:49–58. doi: 10.1016/j.vetmic.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q, Ke H, Blikslager A, Fujita T, Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J Virol. 2018;92:e01677-17. doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Shi HY, Chen JF, Shi D, Lang HW, et al. Identification of cellular proteome using two-dimensional difference gel electrophoresis in ST cells infected with transmissible gastroenteritis coronavirus. Proteome Sci. 2013;11:31. doi: 10.1186/1477-5956-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann M, Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J Clin Microbiol. 1988;26:2235–2239. doi: 10.1128/JCM.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan Y, Tian X, Qin P, Wang B, Zhao P, et al. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo J, Fang L, Dong N, Fang P, Ding Z, et al. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology. 2016;495:10–17. doi: 10.1016/j.virol.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett PN, Mundt W, Kistner O, Howard MK. Vero cell platform in vaccine production: moving towards cell culture-based viral vaccines. Expert Rev Vaccines. 2009;8:607–618. doi: 10.1586/erv.09.19. [DOI] [PubMed] [Google Scholar]
- 21.Hou Y, Wang Q. Emerging highly virulent porcine epidemic diarrhea virus: molecular mechanisms of attenuation and rational design of live attenuated vaccines. Int J Mol Sci. 2019;20:5478. doi: 10.3390/ijms20215478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sungsuwan S, Jongkaewwattana A, Jaru-Ampornpan P. Nucleocapsid proteins from other swine enteric coronaviruses differentially modulate PEDV replication. Virology. 2020;540:45–56. doi: 10.1016/j.virol.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Q, Thomas JT, Gimenez-Lirola LG, Hardham JM, Gao Q, et al. Evaluation of serological cross-reactivity and cross-neutralization between the United States porcine epidemic diarrhea virus prototype and S-INDEL-variant strains. BMC Vet Res. 2016;12:70. doi: 10.1186/s12917-016-0697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin H, Li B, Chen L, Ma Z, He K, et al. Differential protein analysis of IPEC-J2 cells infected with porcine epidemic diarrhea virus pandemic and classical strains elucidates the pathogenesis of infection. J Proteome Res. 2017;16:2113–2120. doi: 10.1021/acs.jproteome.6b00957. [DOI] [PubMed] [Google Scholar]
- 25.Jung K, Hu H, Saif LJ. Porcine deltacoronavirus induces apoptosis in swine testicular and LLC porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Vet Microbiol. 2016;182:57–63. doi: 10.1016/j.vetmic.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]