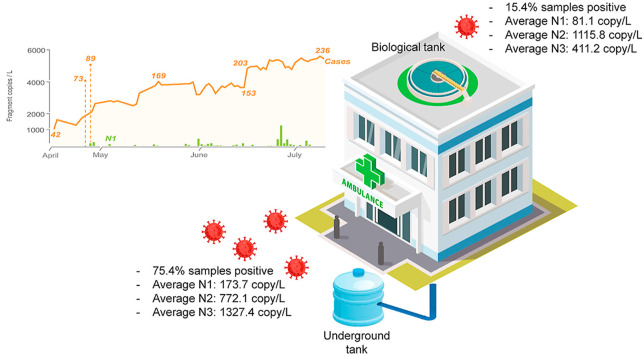
Abstract
There is increasing interest in wastewater-based epidemiology (WBE) of SARS-CoV-2 RNA to serve as an early warning system for a community. Despite successful detection of SARS-CoV-2 RNA in wastewaters sampled from multiple locations, there is still no clear idea on the minimal number of cases in a community that are associated with a positive detection of the virus in wastewater. To address this knowledge gap, we sampled wastewaters from a septic tank (n = 57) and biological activated sludge tank (n = 52) located on-site of a hospital. The hospital is providing treatment for SARS-CoV-2 infected patients, with the number of hospitalized patients per day known. It was observed that depending on which nucleocapsid gene is targeted by means of RT-qPCR, a range of 253–409 positive cases out of 10,000 persons are required prior to detecting RNA SARS-CoV-2 in wastewater. There was a weak correlation between N1 and N2 gene abundances in wastewater with the number of hospitalized cases. This correlation was however not observed for N3 gene. The frequency of detecting N1 and N2 gene in wastewater was also higher than that for N3 gene. Furthermore, nucleocapsid genes of SARS-CoV-2 were detected at lower frequency in the partially treated wastewater than in the septic tank. In particular, N1 gene abundance was associated with water quality parameters such as total organic carbon and pH. In instances of positive detection, the average abundance of N1 and N3 genes in the activated sludge tank were reduced by 50 and 70% of the levels detected in septic tank, suggesting degradation of the SARS-CoV-2 gene fragments already occurring in the early stages of the wastewater treatment process.
Keywords: Wastewater-based epidemiology, COVID-19, Reverse transcription quantitative PCR, Detection sensitivity, Log reduction
Graphical abstract
1. Introduction
Data from the meta-analysis of published medical literature suggests a wide variability and uncertainty in the percentage of COVID-19 infected individuals who are asymptomatic. This percentage can range from 16% as reported in one meta-analysis (He et al., 2020) to approximately 40–45% in another (Oran and Topol, 2020), and up to 80% as first suggested by World Health Organization during the early days of the pandemic (WHO, 2020). Asymptomatic cases are concerning as these individuals are not prioritized for swab testing protocols in some countries and can unknowingly transmit the virus to others. Although self-isolation, household quarantine and contact tracing are effective strategies in slowing the spread of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) (Kucharski et al., 2020), such intervention measures in the long term are unsustainable. The other solution lies in increasing our clinical and surveillance testing capacity. However, to swab everyone in a country for clinical surveillance over a long term period is also impractical, particularly for low resource countries, considering the amount of resources and labor hours needed. In contrast, monitoring for SARS-CoV-2 RNA directly in wastewaters over a longer duration would be complementary to the current clinical surveillance since about 39–65% of infected hosts, including asymptomatic carriers, shed the virus through their feces, while about 6% of patients shed it through urine (Chan et al., 2020; Cheung et al., 2020).
By sampling for the waste stream generated from a community, we therefore have a composite sample that would be suitable for wastewater-based epidemiology (WBE). WBE is an approach that includes the qualitative and quantitative determination of biomarkers in raw wastewater to provide information on the general trend of infection among inhabitants within that wastewater catchment area. Monitoring for SARS-CoV-2 RNA in wastewater has been demonstrated in many countries, including Australia, India, the Netherlands, Italy, Spain and US (Ahmed et al., 2020a; Haramoto et al., 2020; Kumar et al., 2020; La Rosa et al., 2020a; Medema et al., 2020; Randazzo et al., 2020; Sherchan et al., 2020). In these studies, SARS-CoV-2 RNA was sporadically detected from the sampled wastewaters, and the reported abundance of SARS-CoV-2 in wastewater ranged from 19 to 2.2 × 108 copies/L. In some instances, the WBE approach was able to detect SARS-CoV-2 RNA in wastewater before clinically diagnosed cases were made known (La Rosa et al., 2020b). WBE hence demonstrates potential to serve as an early warning of re (emergence) of COVID-19 in communities (Medema et al., 2020). However, without knowing the minimal number of positive cases needed in each community to achieve consistent detection of SARS-CoV-2 RNA from the sewage networks, the definition of how early into the outbreak WBE can inform us remains rather unclear (Cao et al., 2020).
Ideally, the detection sensitivity of WBE can be defined by the probability of a positive sample (or copy number above a certain threshold) associated with a specific total number of symptomatic and asymptomatic individuals present in the community served by the wastewater treatment system. To estimate the community served by the wastewater treatment plant (WWTP), one can obtain information on either the design capacity of the WWTP or estimate based on the volume of untreated sewage entering into the WWTP divided by the average municipal wastewater volume generated per capita. In contrast, the number of infected individuals within the community is difficult to obtain unless an active surveillance is conducted to swab test all individuals, including asymptomatic ones. To circumvent this limitation, a controlled community, for example, hospitals providing treatment to COVID-19 patients can be used as a model. In this manner, the number of patients contributing SARS-CoV-2 RNA to the hospital wastewaters are known through daily hospitalization and discharge records, and would provide inference to the minimal number of SARS-CoV-2 infected cases needed to generate quantitative PCR (qPCR) signals associated with the virus in a diluted wastewater stream.
Besides determining the detection sensitivity of WBE using hospital wastewaters as a study model, we further evaluate the correlation between detected abundance of individual SARS-CoV-2 gene fragments (i.e., N1, N2 and N3 nucleocapsid genes) in wastewater with the number of patients. This would provide insight to which nucleocapsid gene associates positively with infection numbers and suitable for use in future WBE. In addition, the abundance of nucleocapsid gene fragments as it moves from (i) the underground septic tank (i.e., equalization tank) to (ii) the first stage of the wastewater treatment plant (i.e., aerobic activated sludge tank) situated on-site was also evaluated. In this manner, the resistance of the nucleocapsid genes to biological wastewater treatment and their correlation with measurable water quality parameters can be inferred by determining the changes in their abundance along both stages of the wastewater treatment plant.
2. Materials and methods
2.1. Quality control of sample processing and RNA extraction protocols
To determine recovery and viral nucleic acid extraction efficiency, a known concentration of murine norovirus (MNV), ranging from 106-107 genome copies/L, was spiked into untreated wastewater. RAW 264.7 cells (ATCC TIB-71) were used as MNV host cells and cultured in modified Dulbecco's Eagle's medium (DMEM) containing 10% (vol/vol) fetal bovine serum (FBS), 0.075% NaHCO3, 2 mM L-glutamine, 1000 U/mL penicillin-streptomycin. Cells were cultured at 37 °C and in atmospheric headspace containing 5% CO2. MNV propagation in RAW 264.7 cells was conducted by inoculating the initial MNV stock into the cells and then incubated for 5–7 d until cytopathic effect (CPE) was observed. To harvest the virus and to separate the virus from the host cell debris, the medium containing host cells and virus were subjected to freeze (−80 °C) and thaw (37 °C) cycles for 3 times. The medium is then centrifuged at 10,481 g for at least 15 min. The supernatant containing MNV was later filtered through 0.2 μm filter before being kept at −80 °C prior to use.
MNV was spiked as surrogate because it is a positive-sense single-stranded RNA virus like SARS-CoV-2, and will likely be similar in terms of RNA extraction efficiency. Quantitative PCR (qPCR) for MNV was performed using forward primer (5′- CCGCAGGAACGCTCAGCAG-3′), reverse primer (5′- GGYTGAATGGGGACGGCCTG-3′) and Taqman probe (5′- FAM-ATGAGTGATGGCGCA-ZEN/IBFQ-3′) (Integrated DNA Technologies, Leuven, Belgium (Kitajima et al., 2010). A six-points standard curve was generated using a synthetic oligonucleotide (gblocks@ gene fragment, Integrated DNA technologies, IA, USA) containing 300–350 bp DNA sequence that encompasses a complementary region for which the primers and probes would anneal to (Table S1). An amplification efficiency of 102.7% was achieved. LOD is determined by taking multiple 10-fold dilution of MNV gblocks oligonucleotide fragment and determining when the Cq value is still detectable without deviating from a linear trend. In our case, a serial dilution from 108 to 101 genome copy/reaction of solution was conducted, and 10 copy/well was determined as our LOD for MNV. MNV is not anticipated to be present in municipal wastewaters and hence abundances detected by means of qPCR would be due to the spiking event.
2.2. Sampling of hospital wastewaters from jeddah, Saudi Arabia
The hospital monitored in this study is located in Jeddah, Saudi Arabia and is one of the three main hospitals designated to provide treatment for COVID-19 infected patients in the city. During the sampling period, no visitors are allowed at the hospital and would not contribute to any waste streams collected from the on-site wastewater treatment plant (WWTP). In addition to the daily number of patients hospitalized (Table S2), workers and hospital staff only underwent clinical swab testing when symptoms appear and hence we cannot discount potential asymptomatic cases from this cohort that may be contributing to SARS-CoV-2 into the wastewater. Wastewater discharged from the hospital was collected at two sampling points of the WWTP located on-site. The first sampling point was the underground septic tank of 1150 m3 collection volume. This tank also serves as an equalization tank that averaged out daily flow and volume fluctuations. The second sampling point was from the aerobic biological tank located on the roof of the hospital. The daily wastewater intake for the on-site WWTP is 750 m3. The volume of wastewater generated by this hospital approximates the size of a community with ca. 2884 persons based on an average capita water usage of 260 L/person/d (GaStat, 2018). The hospital wastewater is treated first by biological activated sludge system, operated with hydraulic retention time of 9 h. The secondary-treated wastewater is then disinfected with 100 mg/L chlorine at a contact time of 2 h. Approximately 1 L of grab sample was individually collected from the underground tank (i.e., representing untreated wastewater) and from the top aqueous layer of the biological activated sludge tank (i.e., partially treated wastewater) in the morning (approximately at 9 a.m.) of all sampling dates. The entire sampling duration was from 15 April to July 9, 2020, with frequencies ranging from 3 to 5 daily samples per week (Table S2 and Table S3). Grab samples instead of 24 h composite samples were collected during this period as automated samplers were not available on site for both sampling points, and these samplers were not commercially available for immediate deployment during the study period. Samples were stored at 4 °C for not more than 1 week before they were transported to our laboratory and immediately processed. Daily transferal of samples from hospital to laboratory during the sampling period was not possible as Saudi Arabia imposed a strict movement restriction order at that time. In addition, the storage condition used on-site was limited by the resources available at the hospital. Although the storage temperature was not ideal, recent studies suggest high stability of SARS-CoV-2 virus at 4 °C (Chin et al., 2020) and prolonged persistence of SARS-CoV-2 RNA at 4 °C without significant degradation for up to 20 d in untreated wastewater (Ahmed et al., 2020b).
2.3. Processing of wastewater samples
250–500 mL of samples were individually concentrated for viral particles using electronegative membrane (Merck Millipore HAWP09000, Cork, Ireland) method with slight modifications (Haramoto et al., 2004). Briefly, 2 mL of 2.5 M MgCl2 was added to every 100 mL of the sample, agitated for 3 min and left to stand for 3 min. The samples were then filtered through the electronegative membrane. The electronegative membrane was then filtered through with 200 mL of 0.5 mM H2SO4 to adjust the isoelectric point of viruses. The viral particles retained on the electronegative membrane were eluted with 10 mL of 1 mM NaOH into a sterile collection tube that contained 100 μL of 100X tris-EDTA buffer and 50 μL of 100 mM H2SO4. The eluate was concentrated with Centripep YM-50 (Millipore) to approximately 680 μL based on manufacturer's protocol. Briefly, the 10 mL eluate is dispensed into the Centripep YM-50 column and centrifuged at 1000 g for 10 min at 20 °C. The filtrate was discarded, and the column was centrifuged again at the same centrifugal speed and temperature for 5 min. The remaining retentate serves as the final concentrate. 140 μL of the final concentrate was extracted for its RNA using QIAmp Viral RNA kit following manufacturer's protocol (Thermo Fisher Scientific, Carlsbad, CA). QIAmp Viral RNA kit is suggested by the US CDC as one of the extraction protocols to generate highly purified RNA for clinical specimens (CDC, 2020b) but was also used for WBE (Haramoto et al., 2020; Rimoldi et al., 2020). Extracted RNA was converted to complementary DNA (cDNA) using Superscript III First Strand Synthesis System (Thermo Fisher Scientific, Carlsbad, CA). Briefly, 4–5 μL of viral RNA was first mixed with 50 ng/μL random hexamers and 1 μL of annealing buffer, and subsequently incubated in a thermal cycler at 65οC for annealing before 5 min cooling on ice. The annealed product is then mixed with 10 μL of 2X first-strand reaction mix and 2 μL of superscript III/RNAase out enzyme mix, prior to incubation at 25οC for 10 min, 50οC for 50 min, and 85οC for 5 min to terminate the reaction. The whole process generates the cDNA, which is kept at −20οC before qPCR.
2.4. Quantitative(q) PCR inhibition assay and performance characteristic of SARS-CoV-2 qPCR
To determine whether there is any sample inhibitor affecting detection by RT-qPCR assay, we performed two separate experiments. First, pepper mild mottle viruses (PMMoV) are the most abundant RNA virus in human feces and proposed as a potential viral indicator for human fecal contamination due to its global ubiquitous distribution without substantial seasonal fluctuations (Kitajima et al., 2018). A negative detection of PMMoV in any of the wastewater samples collected in this study and an abundance that is below the abundance range of PMMoV reported by earlier studies would be indicative of inhibitors that detrimentally impacted RT and qPCR. qPCR for PMMoV was performed using forward primer (5′- GAGTGGTTTGACCTTAACGTTTGA-3′), reverse primer (5′-TTGTCGGTTGCAATGCAAGT-3′), and Taqman probe (5′- FAM-CCTACCGAAGCAAATG-ZEN/IBFQ-3′) (Integrated DNA Technologies, Leuven, Belgium). Thermal cycler profile of 95 °C for 15 min, followed by 40 cycles of 95 °C and 55 °C for 15 s and 30 s was used (Haramoto et al., 2013). For PMMoV qPCR, a LOD of 10 copy/well and an amplification efficiency 0f 101.5% was achieved. The synthetic oligonucleotide of PMMoV which was used to generate standard curve is provided in Table S1.
Second, we conducted an experiment by spiking a known copy number (105 copy per reaction) of synthetic oligonucleotides containing 300–350 bp of MNV's DNA sequence into our sample's cDNA. Separately, we also prepared a serial dilution of MNV oligonucleotides with a factor of 10 ranging from 107 to 102 copy/reaction to generate Cq (quantification cycle) value references in a standard curve. Wastewater samples with a difference ≥ 2-Cq values were previously considered to have qPCR inhibition (Staley et al., 2012).
Genes associated with nucleocapsid (N) proteins N1, N2 and N3 of SARS-CoV-2 were targeted for quantitative PCR because these genes are listed to be used for the purposes of respiratory virus surveillance and research by US Centers for Disease Control and Prevention (CDC, 2020c), and detection of these genes were also demonstrated in earlier studies performing WBE (Guerrero-Latorre et al., 2020; Medema et al., 2020; Peccia et al., 2020; Randazzo et al., 2020; Sherchan et al., 2020; Wu et al., 2020b). G-blocks that include the annealing regions of N1, N2 and N3 primers-probes were synthesized based on the published sequences of SARS-CoV-2 (Forster et al., 2020) and sequences were listed in Table S1. Six-point standard curves were generated for each of the primer-probe pair to determine their respective amplification efficiencies. Limit of detection is estimated based on the lowest copy number of G-block template with detectable threshold Cq value. Based on the standard curves, the average amplification efficiency were 92.5%, 91.5%, 90.8% and the R2 value ranged from 0.98 to 0.99, 0.97–0.99, and 0.97–0.99 for N1, N2, and N3, respectively. The LOD was 2 copy/well for all N1, N2, and N3 annealed region target. A non-template control serves as negative control. A positive control made up from cDNA derived from RNA of SARS-CoV-2-positive clinical swab specimens was also included for qPCR. Positive controls have average Cq value of 29 for N1, 30 to 31 for N2 and N3 genes. qPCR reaction was conducted by mixing 2.5 μL of sample cDNA with 12.5 μL TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific, Carlsbad, CA), 6.5 μL of nuclease free water, 400 nm of each forward and reverse primer, and 300 nm of probe for all N1, N2, and N3 for total of 25 μL solution mix. All qPCR reaction was run using QuantStudio™ 3 real time PCR system, 96 wells plate of 0.1 mL, and analyzed with QuantStudio design and analysis software v1.5.1. Thermal cycling conditions include 50 cycles of 95 °C for denaturation (3 s) and 55 °C for annealing and amplification (30 s). All samples, standards and controls were performed in technical duplicates.
2.5. Measurement of water quality
Wastewater samples were first filtered through 0.45 μm syringe filter and then measured for total dissolved organic carbon (DOC) using Fusion TOC UV/Persulfate Analyzer (Teledyne Tekmar, Mason, OH). pH and conductivity were measured with HI-98136 portable meter (Hanna Instruments, Woonsocket, RI). Turbidity was measured with 2100Q portable turbidimeter (Hach, Loveland, CO). To explore the association of water quality parameters (i.e., DOC, pH, turbidity and conductivity) with reported abundance of nucleocapsid genes, a regression analysis was conducted. A multivariable model of the log abundance of nucleocapsid genes against the water quality parameters and reported number COVID-19 patients was carried out, and the strength of association reported.
3. Results and discussion
3.1. Recovery and extraction efficiency
A total of 6.89-log copies/L MNV was spiked into the untreated wastewater. The overall recovery and RNA extraction efficiency of MNV was ca. 45%, and falls within the range reported by an earlier study that utilized similar protocol but with different surrogate virus. To illustrate, Ahmed et al. reported an average recovery efficiency of 65.7% ± 23.8% for murine hepatitis virus (MHV) spiked to untreated wastewater (Ahmed et al., 2020c). Our reported recovery and RNA extraction efficiency was also within the range of 10%–73% reported by other studies using different concentration procedures. For example, direct ultracentrifugation of 20 mL of wastewater through a 50% sucrose phase layer recovered 12% of the deactivated SARS-CoV-2 spiked into the sample (Green et al., 2020). Aluminium hydroxide adsorption-precipitation (adjusted to pH 6) method recovered on average 11% of spiked porcine epidemic diarrhea virus and mengovirus, respectively, from 200 mL wastewater (Randazzo et al., 2020). Ultrafiltration of 100–200 mL using Centricon Plus-70 (molecular weight cut-off of 10 kDa) recovered an average 73% of the spiked F-specific RNA phages but with high standard deviation of 50% (Medema et al., 2020). Ahmed et al. further evaluated a wide range of concentration methods, including ultrafiltration using Centricon Plus-70, to recover MHV at 28.0% ± 9.1% (Ahmed et al., 2020c).
The reported differences in recovery efficiencies can be due to the varying type of viral surrogates spiked. Different viruses have different isoelectric points (pI), which denotes the pH values at which the net surface charge switches. The determination of pI is particularly relevant for concentration protocols that utilize electrocharged membranes or coagulant adsorption-precipitation. Norovirus was previously reported to have a pI ranging from 5.5 to 6 (Goodridge et al., 2004), while the spike protein of porcine epidemic diarrhea virus has a pI of 5.0 (Pickett et al., 2012). In contrast, there is no experimental data currently available that reports on the pI of SARS-CoV-2 but predictive calculations for some of its key proteins put the range of pI between 4.2 and 10.1 (Scheller et al., 2020). However, it is noted that MNV is a nonenveloped virus while SARS-CoV-2 is an enveloped virus. Attachment and adsorption factors are hence likely to differ between both viruses. These differences incur uncertainty in which particular surrogate used thus far among studies would best approximate the recovery of SARS-CoV-2.
Regardless of the viral surrogates, recovery of viral particles from untreated wastewater has been challenging as evidenced from the wide range of reported efficiencies. High turbidity and organic carbon content in raw wastewater matrix can foul ultrafiltration membranes and decrease flux. In this study, the raw wastewater samples had an average dissolved organic carbon (DOC) concentration of 6.4 mg/L and average turbidity of 10.6 NTU (Table 1 ). DOC can facilitate unintended adsorption of virus onto the filtering glassware (Gerba and Betancourt, 2017), and hence lower the probability of recovering viral particles that are already present in low abundance and/or not homogenously distributed in the wastewater matrix. Furthermore, suspended particles that accounted for turbidity can clog the membranes rapidly and permit only a limited volume of untreated wastewater (<500 mL) to be filtered through without significantly lengthening the processing time.
Table 1.
Summary of the percentage of samples detected positive for nucleocapsid genes of SARS-CoV-2 and the average abundance among positive detection. Average pH, dissolved organic carbon (DOC), turbidity (NTU) and conductivity ± standard error were also measured for wastewaters collected at both sampling points. * denotes no standard error applicable since only one occurrence of positive detection.
| % occurrence | Average copy numbers/L ± std error | Average pH ± std error | Average DOC (mg/L) ± std error | Average NTU ± std error | Average conductivity (mS/cm) ± std error | |
|---|---|---|---|---|---|---|
| Underground septic tank | 75.4 | N1: 173.7 ± 32.2 | 7.03 ± 0.04 | 6.40 ± 0.28 | 10.57 ± 0.99 | 0.89 ± 0.04 |
| N2: 772.1 ± 172.5 | ||||||
| N3: 1327.4 ± 176.6 | ||||||
| Biological activated sludge tank | 15.4 | N1: 81.1 ± 11.0 | 7.48 ± 0.04 | 4.51 ± 0.27 | 6.72 ± 0.93 | 1.44 ± 0.05 |
| N2: 1115.8 ± 173.1 | ||||||
| N3: 411.2 * |
3.2. Assessment of RT-qPCR inhibitors in wastewater samples
All wastewater samples collected in this study (i.e., n = 57 for untreated wastewater from underground tank, n = 52 for partially treated wastewater from activated sludge tank) were detected positive of PMMoV. The average abundance of PMMoV was 7.98 ± 0.58-log copy/L and 6.96 ± 0.75-log copy/L in the untreated and treated wastewater, respectively (Tables S2 and S3). Earlier literature reported the abundance of PMMoV to range from 6 to 9 log copies/L in untreated wastewater (Hamza et al., 2011; Hughes et al., 2017; Kitajima et al., 2014; Schmitz et al., 2016; Symonds et al., 2014, 2017) and 5 to 7 copies/L in treated wastewater (Betancourt et al., 2014; Hughes et al., 2017; Kitajima et al., 2014; Kuroda et al., 2015). The ubiquitous detection of PMMoV at abundance within the range reported by earlier studies suggest that RT was not inhibited. In addition, all samples that were spiked with synthetic DNA of MNV generate less than 2 Cq value difference compared to the 105 copy reference at the standard curve (Figs. S1 and S2). There is hence no significant amount of inhibitors present in our samples that would inhibit subsequent qPCR reactions.
3.3. Detected abundance of SARS-CoV-2 in untreated hospital wastewaters
Overall, 43 out of the 57 (75.4%) collected samples of untreated hospital wastewaters tested positive for either one of the 3 nucleocapsid genes of SARS-CoV-2 (Table S2). Specifically, N1 has a positive detection frequency of 54.4% (31 out of 57 samples), N2 has a positive detection frequency of 45.6% (26 out of 57 samples) while N3 has a positive detection frequency of 40.4% (23 out of 57 samples). Compared to earlier studies which reported occurrence frequency to range from 15.6 to 50% of the collected municipal wastewaters (Ahmed et al., 2020a; Döhla et al., 2020; La Rosa et al., 2020a; Sharif et al., 2020; Sherchan et al., 2020), the number of samples that were positive for SARS-CoV-2 RNA occurred at higher frequency in this study. This is not surprising since the wastewaters originated from a hospital that provides treatment for COVID-19 infected patients.
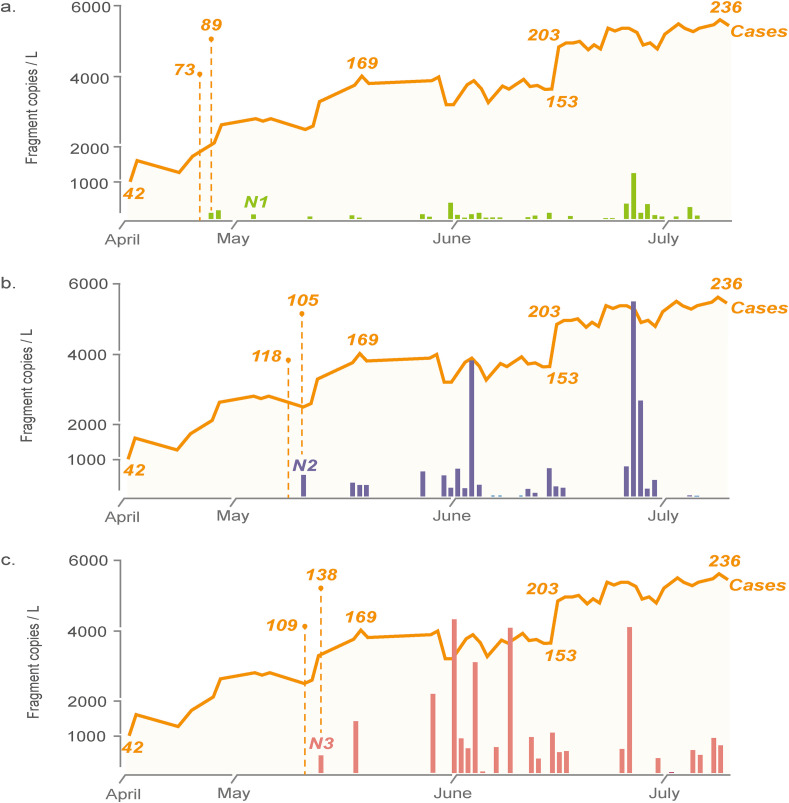
However, despite the confirmed presence of infected patients in this hospital since our first sampling date, we did not detect SARS-CoV-2 in the untreated wastewaters sampled on 15, 16, 22 and April 24, 2020. Instead, a relatively consistent detection of the SARS-CoV-2 N1 gene was only observed after April 27, 2020. Wu et al. determined correlation between wastewater viral titers and daily reports of new clinically confirmed COVID-19 cases in Boston, US, and found that the best correlation was seen with 4 to 10 days time-lag (Wu et al., 2020a). Hence, it is likely that a consistent detection of SARS-CoV-2 in the hospital wastewaters can be achieved when the cumulative number of COVID-19 patients in the hospital was >73 if detection is based on N1 gene (Fig. 1 a). Likewise, > 118 and > 109 hospitalized patients are required for consistent detection of N2 and N3 gene, respectively, in the hospital wastewaters (Fig. 1b and c).
Fig. 1.
Abundance of nucleocapsid genes associated with SARS-CoV-2 and the number of hospitalized patients throughout the sampling period. (a) N1 gene, (b) N2 gene, (c) N3 gene. The right orange dashed line indicates the number of hospitalized patients needed to obtain qPCR signals for that N gene without time-lag while the left dashed line indicates the number of hospitalized patients needed to obtain qPCR signals for that N gene with time-lag. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Considering that the daily wastewater intake for the on-site hospital WWTP is 750 m3, the volume of wastewater generated by this hospital approximates the size of a community with ca. 2884 persons based on an average capita water usage of 260 L/person/d (GaStat, 2018). This therefore suggests that the procedure described in this study has a detection sensitivity ranging from 253 to 409 infected persons per 10,000 inhabitants, depending on which nucleocapsid gene is used as the monitoring marker. However, it is noted that the range estimated in this study is derived based on our described procedures of grab sampling, HA filtration, RT-qPCR efficiency and gene markers, and may differ from other scenarios in which other concentration protocols and/or more sensitive detection methods are used. To exemplify, 24 h composite samples increase analytical sensitivity by lowering the coefficient of variation in detected abundance of enteric pathogens compared to hourly grab samples (Ahmed et al., 2020d). The use of E (envelope protein) or RdRp (RNA-dependent RNA polymerase) gene assays was also shown to have a higher sensitivity than the N assays, albeit tested only against purified SARS-CoV strain Frankfurt-1 (Corman et al., 2020). Therefore, more effective protocols would likely further lower the minimum number of infected cases required for reliable detection of SARS-CoV-2 RNA in wastewater to levels lower than that estimated in this study.
Similar to that reported by Medema et al. (2020), N1 and N2 genes were observed in the hospital wastewater first before N3 gene. The simultaneous detection of N1, N2 and N3 gene was observed after a cumulative 138 patients or more were hospitalized (i.e., >464 infected persons per 10,000 inhabitants). With the exception of N1 and N2 gene exhibiting a weak positive correlation (r = 0.21 and 0.24, respectively), there was no apparent correlation between abundance of N3 genes and the number of hospitalized patients. A recent study also evaluated the analytical sensitivity and efficiency of the CDC primer-probe sets and determined that the N3 forward primer could be problematic to detect viruses with T→ C substitution at the 8th position of the primer binding region (Vogels et al., 2020). US CDC has since removed their 2019-nCoV N3 primer-probe set for diagnostic testing (CDC, 2020a). Collectively, our observations reiterate that N3 gene detected by the CDC N3 primer-probe pair is also not an ideal marker for WBE.
The weak correlation and sporadic detection of SARS-CoV-2 N1 and N2 genes in hospital wastewaters may be dependent on the viral shedding load per patient and the shedding duration. In this study, information on whether patients are exhibiting gastrointestinal distress was not provided to us due to concerns of breaching patients’ confidentiality. Hence it is uncertain how many percentage of the patients may be shedding the virus into the wastewater to result in the sporadic detection of SARS-CoV-2. The weak correlation between N gene abundance and the patient numbers can also be potentially affected by the grab sampling strategy adopted in this study. However, based on our observed detection frequencies of each genes, coupled with an earlier study that reported N1 primer-probe set is more sensitive than the N2 primer-probe set for human clinical specimens (Vogels et al., 2020), N1 gene may be a more appropriate nucleocapsid gene marker for use in WBE.
By utilizing a semi-controlled confined environment (i.e., a hospital with known number of COVID-19 cases), this study determined the detection sensitivity of wastewater-based epidemiology for SARS-CoV-2 RNA at this site to be about >253 persons per 10,000 inhabitants based on the more sensitive N1 gene. The correlation between nucleocapsid gene abundances of SARS-CoV-2 and the number of infected hospitalized patients was weak, suggesting the difficulty in correlating the number of cases based on the gene abundances detected in wastewaters. It is not known if infected cases in the hospitals would excrete a higher viral load than those who are residing in the community, possibly because the latter individuals typically do not exhibit severe symptoms that would warrant hospitalization. This might suggest that an even higher number of infected individuals would be needed for detecting SARS-CoV-2 N1 gene in the municipal wastewaters. Regardless, it is likely that when the nucleocapsid genes were detected, there is already a substantial number of cases circulating in the community that would warrant immediate intervention measures to be taken.
3.4. Log reduction of SARS-CoV-2 gene fragments at different stage of treatment process
A recent systematic review and meta-analysis of previous literature suggests that the persistence of viral surrogates like human/animal coronaviruses and bacteriophage Phi6 dec in wastewater are dependent on temperature, the nature of wastewater (i.e., sterilized vs. non-sterilized wastewater), and the disinfectant used (Silverman and Boehm, 2020). Likewise, these factors also affect the persistence of SARS-CoV-2 and its RNA (Table 2 a), for example, untreated wastewater and increasing temperatures detrimentally affect the persistence of both virus particles and RNA (Ahmed et al., 2020b; Bivins et al., 2020). We therefore further examine how N genes abundance would associate with some of these parameters, particularly focusing on certain aspects of the untreated wastewater that can be measured in this study.
Table 2.
A collation of peer-reviewed data related to the decay kinetics of SARS-CoV-2 virus and RNA, evaluated based on (a) factors that can affect persistence, and (b) treatment process.
| a) | Reference | Factor 1 | Factor 2 | Target | Time for 1-log reduction (d) |
|---|---|---|---|---|---|
| Ahmed et al. (2020b) | Untreated wastewater | 4 | N1 gene, quantified by RT-qPCR | 11.9 | |
| 15 | 8.8 | ||||
| 25 | 5.5 | ||||
| 37 | 3.5 | ||||
| Autoclaved wastewater | 4 | 18.5 | |||
| 15 | 13.0 | ||||
| 25 | 5.8 | ||||
| 37 | 2.5 | ||||
| Tap water | 4 | 25.6 | |||
| 15 | 22.2 | ||||
| 25 | 6.6 | ||||
| 37 | 4.1 | ||||
| Bivins et al. (2020) | Untreated wastewater | 20 | SARS-CoV-2 virus, quantified based on 50% tissue culture infective dose (TCID-50) | 1.5 | |
| 50 | 0.01 | ||||
| Tap water | 20 | 14.3 | |||
| b) | Reference | Treatment | Target | Log reduction | |
| Kumar et al. (2021) | Upflow anaerobic sludge blanket system | N genes | 1.53 | ||
| S protein | 1.57 | ||||
| ORF1ab | 1.64 | ||||
| This study | Open air biological activated sludge tank with hydraulic retention time of 9 h | N1 | 0.3 | ||
| N2 | Increase by 44% | ||||
| N3 | 0.5 | ||||
The biological activated sludge tank is an open-air tank placed on the roof of the hospital, and wastewater inside the tank was subjected to an increasing ambient temperature and solar irradiance during the sampling period as KSA enters into summer. To exemplify, maximum diurnal temperature increased from 41.5 °C in April to 45.3 °C in May and 46.3 °C in June 2020, while minimum diurnal temperature also increased from 14.2 °C in April to 21 °C in June (personal communication, Matthew McCabe). The maximum net solar irradiation measured at 1 p.m. also increased from 1184.9 W/m2 in April to 1333.0 W/m2 in June (personal communication, Matthew McCabe). However, the sporadic detection of SARS-CoV-2 RNA in the wastewater samples collected from biological activated sludge tank and their abundance (Table S3) exhibit no apparent association with the ambient temperatures and solar irradiance.
Parameters related to wastewater quality, namely DOC and turbidity were lower in values for wastewater sampled from the biological activated sludge tank compared to the underground septic tank (Table 2). This indicates a certain degree of partial treatment was achieved at the sampling point of the activated sludge tank. Furthermore, the nature of the wastewater also changed from one stage to the other of the treatment process, as seen from the increase in pH and conductivity in wastewater from the septic tank to the rooftop biological activated sludge tank (Table 1). Through correlation analysis, the N1 gene copy number was determined to associate with TOC and pH but not with turbidity and conductivity (Tables S4–S6), suggesting that TOC and pH may influence the copy number within samples and can potentially be monitored along with the primary abundance data. However, the sample size was relatively small, and these parameters were not associated with the copy number of the N2 and N3 genes, so the relevance of these findings should be treated with caution.
Nevertheless, the decrease in DOC level within the activated sludge tank and certain extent of partial treatment resulted in a lower detection frequency of SARS-CoV-2 in wastewaters sampled from the biological activated sludge tank. Only 8 out of 52 samples (15.4%) collected from the biological activated sludge tank were tested positive for N genes associated with SARS-CoV-2. Specifically, N1 genes were detected in 5 samples, N2 genes were detected in 4 samples, and 1 positive occurrence of N3 genes among the collected samples. N1 genes were again detected first before N2 and N3. The average abundance of N1 and N3 genes decreased by 0.3-log (i.e., 50%) and 0.5-log (i.e., 70%), respectively, from the underground septic tanks to the biological activated sludge tank. The use of upflow anaerobic sludge blanket reactor for municipal wastewater treatment resulted in a higher reduction of N genes (1.53-log), with a similar log reduction of >1.5-log also observed for other genes associated with SARS-CoV-2 (Kumar et al., 2021) (Table 2b).
Besides the lower log reduction of N1 and N3 genes compared to that reported by Kumar et al. (2021), there was also an increase by 44% in the average abundance of N2 genes sampled from the activated sludge tank compared to that in the underground septic tank. This suggests a potential accumulation of N2 genes in the activated sludge tank, possibly due to better persistence or sorption of N2 compared to N1 and N3 genes in the sludge tank. Peccia et al. detected up to 105 copies of SARS-CoV-2 gene copies per mL of sludge, which is at a higher concentration than that detected in our untreated wastewaters (Peccia et al., 2020), suggesting that these gene fragments accumulate within the sludge tanks. Most of the positive detection in the biological activated sludge tank occurred in late June, and coincides after a period of high cumulative numbers of hospitalized patients.
Despite the slight increase of N2 genes in a small group of samples and the low log reduction of N1 and N3 genes achieved by the biological activated sludge tank, it is unlikely that the N genes will persist in the final treated wastewater. An earlier study determined that 6.5 mg/L of free chlorine and 1.5 h of contact time was already sufficient to remove 0.5 to 18.7 × 103 copies/L of SARS-CoV-2 viral RNA to levels below detection limits (Zhang et al., 2020). Considering that the last stage of the hospital WWTP includes a final disinfection step at a very high chlorine concentration (100 mg/L) and at long contact time (2 h), remnant SARS-CoV-2 RNA is likely to be fully inactivated or degraded prior to discharge of effluent.
4. Conclusion
This study utilized a semi-controlled hospital environment that has a known number of COVID-19 patients to estimate the detection sensitivity of wastewater monitoring for SARS-CoV-2 RNA. An estimated range of 253–409 positive cases out of 10,000 persons are required prior to detecting SARS-CoV-2 RNA in wastewater/\///. However, this detection range may be a system- or site-specific estimation and that the values would depend on factors such as sampling frequencies (e.g. grab vs. 24 h composite samples), recovery efficiencies, RT-qPCR detection limits, the volume of wastewater (and hence dilution factor) in the sampling tank, the geographical locations and the associated environmental factors that can influence persistence and abundance of SARS-CoV-2 RNA, as well as the type of marker gene used for WBE.
It was observed that N1 gene may be a more sensitive marker than N2 and N3 genes. Furthermore, N1 and N2 gene abundances exhibited weak positive correlation with the number of hospitalized patients. In contrast, N3 gene marker as detected by the CDC 2019-nCoV-N3 primer-probe pair do not exhibit any correlation and likely does not work well for both clinical diagnostic and environmental surveillance.
The N1 and N3 gene abundances decayed by 0.3-log and 0.5-log, respectively, from the underground septic tanks to the rooftop biological activated sludge tank. The persistence of SARS-CoV-2 RNA, including the N genes, were previously determined to be affected by the type of water matrix, temperature and treatment technology. This study further determined a correlation between the N1 gene abundance and DOC and pH of the wastewater matrix. In contrast to N1 and N3 gene, N2 gene abundance increased slightly by 44% from the underground septic tanks to the rooftop biological activated sludge tank, suggesting a better persistence or sorption of this gene to the sludge.
To further improve WBE, future studies would be needed to optimize the various aspects that would affect detection sensitivity. This includes for instance, but not limited to, the recovery efficiency of viral particles from wastewater matrix, and better molecular assays that enable specific and sensitive detection. There is also a need for more studies that look into the decay kinetics of the SARS-CoV-2 associated genes in various environmental conditions to facilitate better estimates of the actual abundance at the original points of dissemination and to devise intervention measures necessary to protect public health.
Authors’ contribution statement
ATR, DM and PYH conceived the experimental design, performed the experiments and analyzed the data; MA, YMB and HAQ performed sampling at the hospital, KO performed linear regression analysis and provided feedback on statistical model, JZ measured all water qualities of samples. PYH wrote the original draft, ATR and DM edited the draft and provided illustrations. All authors have reviewed and provided feedback to the draft.
Funding
This work was funded by KAUST baseline grant BAS/1/1033-01-01 awarded to PYH.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank the medical team of the studied hospital for providing access to wastewater samples and for saving lives during this pandemic. The authors would also like to thank Professor Arnab Pain for providing the RNA sample that serves as positive control, Professor Matthew McCabe and Mr Samir Almashharawi for providing the ambient temperature and solar irradiance measured at the Hada Al Sham monitoring station.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envres.2021.110748.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191:110092. doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739:139960. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Gyawali P., Sherchan S.P., et al. Intraday variability of indicator and pathogenic viruses in 1-h and 24-h composite wastewater samples: implications for wastewater-based epidemiology. Environ. Res. 2020:110531. doi: 10.1016/j.envres.2020.110531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Kitajima M., Wing A.D., Regnery J., Drewes J.E., Pepper I.L., et al. Assessment of virus removal by managed aquifer recharge at three full-scale operations. Journal of Environmental Science and Health, Part A. 2014;49:1685–1692. doi: 10.1080/10934529.2014.951233. [DOI] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., et al. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Gu A.Z., Hong P.-Y., Ivanek R., Li B., Wang A., et al. Editorial perspective: viruses in wastewater: wading into the knowns and unknowns. Environ. Res. 2020:110255. doi: 10.1016/j.envres.2020.110255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2020. CDC's Diagnostic Test for COVID-19 Only and Supplies.https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html 30 Sept 2020 [Google Scholar]
- CDC . 2020. Real-Time RT-PCR Panel for Detection 2019-Novel Coronavirus.https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-for-detection-instructions.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . 2020. Research Use Only 2019-novel Coronavirus (2019-nCoV) Real-Time RT-PCR Panel Primer and Probe Information.https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html [Google Scholar]
- Chan V.W.-S., Chiu P.K.-F., Yee C.-H., Yuan Y., Ng C.-F., Teoh J.Y.-C. A systematic review on COVID-19: urological manifestations, viral RNA detection and special considerations in urological conditions. World J. Urol. 2020:1–12. doi: 10.1007/s00345-020-03246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., et al. Gastroenterology; 2020. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples from a Hong Kong Cohort: Systematic Review and Meta-Analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., et al. Stability of SARS-CoV-2 in different environmental conditions. The Lancet Microbe. 2020;1:e10. doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhla M., Wilbring G., Schulte B., Kümmerer B.M., Diegmann C., Sib E., et al. medRxiv; 2020. SARS-CoV-2 in Environmental Samples of Quarantined Households. 2020.05.28.20114041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. Unit. States Am. 2020:202004999. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GaStat . 2018. Per Capita Water Consumption Drops for First Time in 5 Years.https://saudigazette.com.sa/article/535696 31 July 2020 [Google Scholar]
- Gerba C.P., Betancourt W.Q. Viral aggregation: impact on virus behavior in the environment. Environ. Sci. Technol. 2017;51:7318–7325. doi: 10.1021/acs.est.6b05835. [DOI] [PubMed] [Google Scholar]
- Goodridge L., Goodridge C., Wu J., Griffiths M., Pawliszyn J. Isoelectric point determination of norovirus virus-like particles by capillary isoelectric focusing with whole column imaging detection. Anal. Chem. 2004;76:48–52. doi: 10.1021/ac034848s. [DOI] [PubMed] [Google Scholar]
- Green H., Wilder M., Middleton F.A., Collins M., Fenty A., Gentile K., et al. medRxiv; 2020. Quantification of SARS-CoV-2 and Cross-Assembly Phage (crAssphage) from Wastewater to Monitor Coronavirus Transmission within Communities. 2020.05.21.20109181. [Google Scholar]
- Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Granda M.G., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Total Environ. 2020;743:140832. doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I.A., Jurzik L., Uberla K., Wilhelm M. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 2011;45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Katayama H., Ohgaki S. Detection of noroviruses in tap water in Japan by means of a new method for concentrating enteric viruses in large volumes of freshwater. Appl. Environ. Microbiol. 2004;70:2154–2160. doi: 10.1128/AEM.70.4.2154-2160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Kishida N., Konno Y., Katayama H., Asami M., et al. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79:7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737:140405. doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Guo Y., Mao R., Zhang J. Proportion of asymptomatic coronavirus disease 2019: a systematic review and meta-analysis. J. Med. Virol. 2020 doi: 10.1002/jmv.26326. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes B., Beale D.J., Dennis P.G., Cook S., Ahmed W. Cross-comparison of human wastewater-associated molecular markers in relation to fecal indicator bacteria and enteric viruses in recreational beach waters. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.00028-17. e00028-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Pepper I.L., Gerba C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes--identification of potential viral indicators. Sci. Total Environ. 2014;488–489:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Oka T., Takagi H., Tohya Y., Katayama H., Takeda N., et al. Development and application of a broadly reactive real-time reverse transcription-PCR assay for detection of murine noroviruses. J. Virol Methods. 2010;169:269–273. doi: 10.1016/j.jviromet.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. npj Clean Water. 2018;1:19. [Google Scholar]
- Kucharski A.J., Klepac P., Conlan A.J.K., Kissler S.M., Tang M.L., Fry H., et al. Effectiveness of isolation, testing, contact tracing, and physical distancing on reducing transmission of SARS-CoV-2 in different settings: a mathematical modelling study. Lancet Infect. Dis. 2020;20:1151–1160. doi: 10.1016/S1473-3099(20)30457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Kuroda K., Patel A.K., Patel N., Bhattacharya P., Joshi M., et al. Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with Upflow Anaerobic Sludge Blanket (UASB) system evaluated through two sample concentration techniques. Sci. Total Environ. 2021;754:142329. doi: 10.1016/j.scitotenv.2020.142329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., et al. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746:141326. doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda K., Nakada N., Hanamoto S., Inaba M., Katayama H., Do A.T., et al. Pepper mild mottle virus as an indicator and a tracer of fecal pollution in water environments: comparative evaluation with wastewater-tracer pharmaceuticals in Hanoi. Vietnam. Science of The Total Environment. 2015;506–507:287–298. doi: 10.1016/j.scitotenv.2014.11.021. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., et al. medRxiv; 2020. SARS-CoV-2 Has Been Circulating in Northern Italy since December 2019: Evidence from Environmental Monitoring. 2020.06.25.20140061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Oran D.P., Topol E.J. Prevalence of asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744:140911. doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller C., Krebs F., Minkner R., Astner I., Gil-Moles M., Wätzig H. Physicochemical properties of SARS-CoV-2 for drug targeting, virus inactivation and attenuation, vaccine formulation and quality control. Electrophoresis. 2020;41:1137–1151. doi: 10.1002/elps.202000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Kitajima M., Campillo M.E., Gerba C.P., Pepper I.L. Virus reduction during advanced bardenpho and conventional wastewater treatment processes. Environ. Sci. Technol. 2016;50:9524–9532. doi: 10.1021/acs.est.6b01384. [DOI] [PubMed] [Google Scholar]
- Sharif S., Ikram A., Khurshid A., Salman M., Mehmood N., Arshad Y., et al. medRxiv; 2020. Detection of SARS-Coronavirus-2 in Wastewater, Using the Existing Environmental Surveillance Network: an Epidemiological Gateway to an Early Warning for COVID-19 in Communities. 2020.06.03.20121426. [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., et al. Science of The Total Environment; USA: 2020. First Detection of SARS-CoV-2 RNA in Wastewater in North America: A Study in Louisiana; p. 140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman A.I., Boehm A.B. Systematic review and meta-analysis of the persistence and disinfection of human coronaviruses and their viral surrogates in water and wastewater. Environ. Sci. Technol. Lett. 2020;7:544–553. doi: 10.1021/acs.estlett.0c00313. [DOI] [PubMed] [Google Scholar]
- Staley C., Gordon K.V., Schoen M.E., Harwood V.J. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Appl. Environ. Microbiol. 2012;78:7317. doi: 10.1128/AEM.01430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Verbyla M.E., Lukasik J.O., Kafle R.C., Breitbart M., Mihelcic J.R. A case study of enteric virus removal and insights into the associated risk of water reuse for two wastewater treatment pond systems in Bolivia. Water Res. 2014;65:257–270. doi: 10.1016/j.watres.2014.07.032. [DOI] [PubMed] [Google Scholar]
- Symonds E.M., Young S., Verbyla M.E., McQuaig-Ulrich S.M., Ross E., Jiménez J.A., et al. Microbial source tracking in shellfish harvesting waters in the Gulf of Nicoya, Costa Rica. Water Res. 2017;111:177–184. doi: 10.1016/j.watres.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nature Microbiology. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Q&A: Similarities and Differences – COVID-19 and Influenza.https://www.who.int/news-room/q-a-detail/q-a-similarities-and-differences-covid-19-and-influenza [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., et al. medRxiv; 2020. SARS-CoV-2 Titers in Wastewater Foreshadow Dynamics and Clinical Presentation of New COVID-19 Cases. 2020.06.15.20117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5:e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., et al. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of Fangcang Hospital. Sci. Total Environ. 2020;741:140445. doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.