Abstract
Purpose
Anti-programmed cell death protein 1 (PD1)±anti-cytotoxic T-lymphocyte associated protein 4 (CTLA4) immune checkpoint inhibitors (ICIs) are standard therapeutic options for metastatic melanoma. We assessed whether biologic subtype according to primary tumor type or genomic subtype can function as predictive biomarkers for anti-PD1±anti-CTLA4 ICI in patients with advanced melanoma.
Methods
We performed a single-center retrospective cohort analysis of patients who received anti-PD1±anti-CTLA4 ICI for advanced melanoma between 2012 and 2019. Primary tumor type, BRAF and NRAS mutation status, and other covariates were abstracted from chart review. Log-rank tests and multivariable Cox regression models were used to assess differences in clinical progression-free (cPFS) and overall survival (OS).
Results
We identified 230 patients who received 249 lines of anti-PD1±anti-CTLA4 ICI for unresectable or metastatic disease. Of these patients, 74% were cutaneous, 11% mucosal, 8% unknown primary and 7% acral. BRAF and NRAS mutations were identified in 35% and 28% of patients, respectively. In multivariable analyses of the entire cohort, acral or mucosal primary tumor type, >3 metastatic sites, elevated LDH were predictive of shorter cPFS and OS. Combination ICI therapy was associated with longer cPFS (HR 0.57, 95% CI 0.38 to 0.86, p=0.007) and OS (HR 0.42, 95% CI 0.28 to 0.65, p<0.001). Combination ICI was significantly associated with longer OS in unknown primary and mucosal melanoma. There was a non-significant trend toward longer OS with anti-PD1+anti-CTLA4 in cutaneous melanoma, but not in acral melanoma. In multivariable analyses, combination ICI was associated with longer OS in NRAS (HR 0.24, 95% CI 0.10 to 0.62, p=0.003, n=69) and BRAF V600E/K (HR 0.47, 95% CI 0.24 to 0.90, p=0.024, n=86) mutant melanoma but not BRAF/NRAS wild-type (n=94) melanoma.
Conclusions
In our cohort, primary melanoma tumor type and genomic subtype were independent predictive markers of cPFS and OS for patients with metastatic melanoma receiving anti-PD1 ICI. Primary tumor type and genomic subtype—including NRAS—should be further evaluated in prospective clinical trials to determine their value as predictive markers. Biologic subtypes may facilitate clinical decision-making when recommending combination ICI treatment (anti-PD1±anti-CTLA4) versus anti-PD1 alone for patients with metastatic melanoma.
Keywords: melanoma, genetic markers, immunotherapy, programmed cell death 1 receptor, tumor biomarkers
Introduction
Metastatic melanoma is a collection of diseases with distinct underlying tumor biology. Non-ocular melanomas can be classified into four groups based on the tissue type of the primary tumor: cutaneous, mucosal, acral and unknown primary. These are biologically distinct diseases with different etiologies, genomic alterations and natural histories.1 Comprehensive genomic analyses of melanoma indicate that cutaneous melanomas can also be divided into four molecular subtypes according to the presence or absence of mutations in oncogenic driver genes: BRAF, NRAS and NF1.2 BRAF V600E/K mutations are almost always mutually exclusive with NRAS mutations in melanoma. Patients with BRAF V600E/K mutant cutaneous melanoma are typically younger than average, whereas patients with NF1 mutations are typically older than average.2 The majority of BRAF, NRAS or NF1 mutant cutaneous melanomas are associated with ultraviolet (UV) light-induced DNA damage genomic profiles. BRAF mutant melanomas are typically associated with intermittent sun exposure, whereas NRAS or NF1 mutant melanomas are associated with chronic sun exposure. The etiology of acral and mucosal melanomas, however, is distinct from typical cutaneous melanomas, as these melanoma subtypes do not arise as a consequence of sun exposure and UV light-induced DNA damage.1 3 Acral melanomas arise on the glabrous skin of the volar aspects of the hands, feet and nail beds. Mucosal melanomas arise in the mucus membranes in the genital, anal, nasal or oral cavities. Neither acral nor mucosal melanoma are associated with UV exposure. Acral and mucosal melanomas have a lower burden of point mutations relative to cutaneous melanomas.1 4 Acral melanomas have a higher incidence of copy number variations, including multiple genomic amplifications.1 Genomic structural variations are more common in mucosal melanoma subtypes.4 Melanomas of unknown primary (MUP) are thought to arise from a primary tumor that has spontaneously regressed due to immune surveillance.5 Indeed, partially regressed melanomas were found to have higher numbers of infiltrating immune cells,6 which is a feature associated with improved melanoma survival.7 Some data suggest that the distribution of driver mutations in MUPs8 is similar to the distribution of driver genes in cutaneous melanomas that are associated with intermittent sun exposure,1 wherein BRAF V600 mutations predominate. However, this observation may not be broadly applicable to all MUPs, and some may be of acral or mucosal origin.
Despite these significant differences in tumor biology, all of the aforementioned subtypes of metastatic melanoma are amenable to first-line treatment with immune checkpoint inhibitors (ICIs).9 10 These ICI include the anti-PD1 inhibitors, pembrolizumab9 and nivolumab,11 and the anti-CTLA4 inhibitor, ipilimumab,12 which are standard-of-care treatment options for all patients with advanced melanomas. Anti-PD1 monotherapies9 10 and anti-PD1+anti-CTLA4 combination therapy10 have both been proven to be more effective than anti-CTLA4 monotherapies. In Checkmate 067, the 5-year overall survival (OS) of previously untreated patients randomized to receive first-line ipilimumab+nivolumab, single agent nivolumab or single agent ipilimumab were 52% and 44%, and 26% respectively.13 This study was not powered to definitively assess differences in survival between ipilimumab+nivolumab versus nivolumab alone. While not statistically significant, patients who received ipilimumab+nivolumab in Checkmate-067 had longer median OS (not reached, more than 60 months) than patients who received nivolumab alone (36.9 months). The 5-year follow-up report of Checkmate-067 confirmed that there were more grade 3/4 adverse events in the combination arm (59%) than in the nivolumab single agent arm (22%).13 Given the increased toxicity associated with combination immunotherapy, it is of great interest to clinicians to be able to identify those patients who could derive sufficient prolonged benefit from anti-PD1 monotherapy, and be spared the increased toxic side effects of combination immunotherapy.14
Subset analyses of Checkmate-067 suggest that tumor biology may function as predictive markers for therapeutic benefit from ipilimumab+nivolumab. For example, the 5-year survival advantage of ipilimumab +nivolumab over nivolumab was more pronounced in BRAF mutant melanoma (60% vs 46%) than it was in BRAF wild-type melanoma (35% vs 32%).13 Moreover, patients with mucosal melanomas were more likely to survive 5 years when treated with ipilimumab +nivolumab (36%) compared with nivolumab alone (17%), although the number of patients with mucosal melanoma enrolled on this study was small, so these data should be interpreted with caution.15 To date, there are no available published data from Checkmate-067 on the long-term outcomes of patients with NRAS mutations or other primary tumor types such as acral or unknown primary melanoma. Therefore, to address whether these distinct biologic subtypes of melanoma might represent predictive markers of therapeutic benefit of anti-PD1+anti-CTLA4 combination therapy versus anti-PD1 monotherapy, we performed a real-world retrospective analysis of survival outcomes of patients with metastatic melanoma treated with either anti-PD1+anti-CTLA4 or anti-PD1 ICI alone.
Patients and methods
Patient population, treatment and response evaluation
We performed a single-center retrospective analysis of patients with advanced melanoma who received anti-PD1 immunotherapy at Princess Margaret Cancer Center, University Health Network (PM-UHN) between 2012 and 2019. The PM-UHN Tumor Immunotherapy Program (TIP) and melanoma referral databases were used to identify patients with melanoma who had received anti-PD1-based ICI (either alone or in combination with anti-CTLA4) as palliative treatment for advanced disease. Patients with uveal melanoma were excluded. Patients who did not have BRAF molecular testing or who were known to be BRAF wild type but did not have any subsequent molecular testing for NRAS were excluded from this analysis. KIT mutation data were collected when available, but was not assessed in all patients. Patients who received anti-PD1 ICI in combination with any non-anti-CTLA4 investigational therapy were also excluded (figure 1). We collected the following clinical variables: type of therapy received (ie, anti-PD1+anti-CTLA4 vs anti-PD1 alone), number of previous lines of systemic therapy, age at the time of starting anti-PD1 ICI, gender, primary tumor type (cutaneous, acral, mucosal or unknown primary), number and type of involved metastatic sites, M-stage according to AJCC V.8, neutrophil to lymphocyte ratio (NLR), lactate dehydrogenase (LDH) at time of starting anti-PD1 ICI were also collected. We collected the following efficacy data: clinical progression-free survival (cPFS) and overall survival (OS). Clinical progression was based on physician documentation of disease progression or death in the electronic medical record, according to either clinical or radiologic parameters of progression. cPFS was calculated from the date of initiation of anti-PD1 ICI to documented clinical progression, death or last follow-up. Patients who were alive without clinical progression at last follow-up were censored. OS was calculated from the date of initiation of anti-PD1 ICI to death or last follow-up. Patients who were alive at the time of last follow-up were censored.
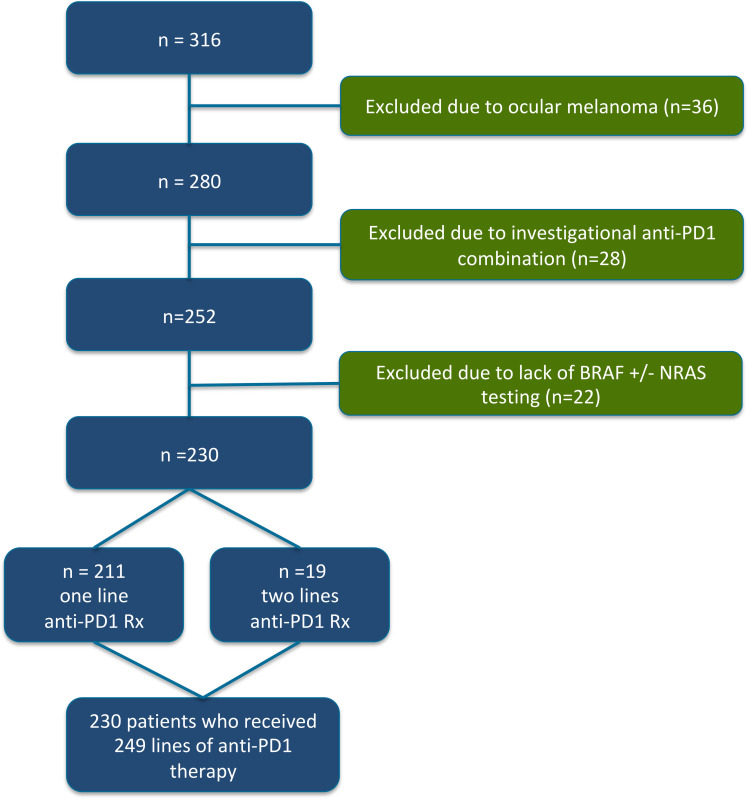
Figure 1.
CONSORT (Consolidated Standards of Reporting Trials) diagram. We identified 316 patients who had received anti-programmed cell death protein 1 (anti-PD1) containing therapy for advanced melanoma. Due to the lack of BRAF or NRAS mutations in uveal melanoma, we excluded 36 patients with uveal melanoma from our analysis. We also excluded 28 patients who had received anti-PD1 therapy combined with investigational agents (ie, not anti-CTLA4). Finally, we excluded 22 patients who did not have BRAF testing, or who were known to be BRAF wild type but did not have subsequent testing for NRAS mutations. After exclusion of these patients, we analyzed data from 230 individual patients who had received 249 lines of anti-PD1 monotherapy or anti-PD1+anti-CTLA4 combination immunotherapy.
Genomic driver mutation analysis
The presence of BRAF, NRAS and KIT oncogenic mutations and clinical covariates were abstracted from chart review by two investigators (AR and SA); the entire dataset was cross-checked for accuracy of data collection by AR. Between 2012 and 2019 at PM-UHN, advanced melanomas were initially assessed for the presence or absence of a BRAF V600E/K mutations using a PCR-based assay. If no BRAF V600E/K mutation was identified, subsequent testing for additional mutations was performed. The methodology for detecting for NRAS and other oncogenic mutations evolved between 2012 and 2019. Between 2012 and 2015, standard practice was to assess BRAF wild-type melanomas for the presence of NRAS (exons 1 and 2) and KIT mutations (exons 11, 13, 17 and 18) by PCR. In 2015, BRAF wild-type melanomas were assessed for NRAS mutations by next-generation sequencing (NGS).
Statistical analysis
Univariable and multivariable Cox models were used to assess differences in OS and cPFS. We performed multivariable analyses that include all variables in the initial multivariable model. We performed backward selection to identify potentially significant variables. The final multivariable models for PFS or OS included only those variables that were associated with p<0.10 in multivariable analysis. Analysis of variance was used to assess statistically significant differences in patient characteristics between groups. All statistical analyses were performed with Stata V.12.
Results
Patient characteristics
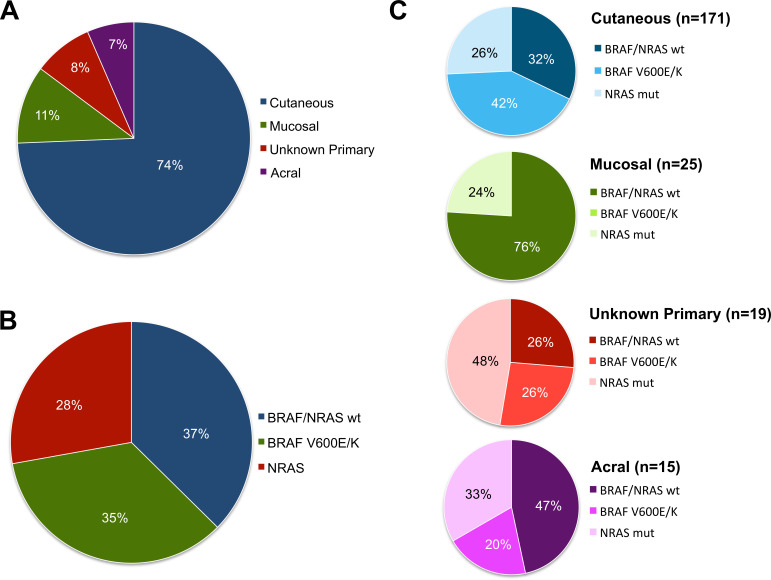
We reviewed a total of 739 medical records and identified 230 individual patients who received anti-PD1±anti-CTLA4 ICI and had molecular testing for BRAF and NRAS mutations. Of these 230 patients, 19 (8%) received two lines of anti-PD1 ICI; therefore, our entire cohort included 230 patients who cumulatively received 249 lines of anti-PD1±anti-CTLA4 ICI for advanced melanoma (figure 1). The median follow-up time was 20.5 and 36.0 months in the entire cohort and among survivors, respectively. Patient characteristics are detailed in table 1. Our cohort was comprised of 183 (74%) cutaneous, 28 (11%) mucosal, 20 (8%) unknown primary and 18 (7%) acral melanoma patients, respectively (figure 2A). The most common genomic subtype was BRAF/NRAS wild type, followed by BRAF V600E/K mutant and NRAS mutant (figure 2B). As expected, there were significant differences in the frequency of BRAF and NRAS mutations according to primary tumor type (table 1, figure 2). BRAF V600E/K mutations were most common in cutaneous melanomas (42%) and were absent in mucosal melanomas. NRAS mutations were most common in unknown primary melanomas (50%) and least common in mucosal melanomas (24%). BRAF/NRAS wild-type melanomas were most common among mucosal melanoma (76%) and least common among unknown primary melanomas (26%) (figure 2C). M-stage, LDH level and PD1 regimen received were also significantly different across primary tumor types (table 1). Patients with mucosal melanoma were most likely to be stage M1c or M1d (86%), those with acral and mucosal melanomas were most likely to have elevated LDH (50%), and patients with unknown primary melanoma were most likely to have received combination ICI with anti-PD1+antiCTLA4. There were no significant differences in the age or gender distribution of patients according to primary tumor type, nor were there are differences in the number of metastatic site, the NLR, or in whether patients were receiving anti-PD1 ICI as a first or later line systemic therapy.
Table 1.
Characteristics of the entire melanoma cohort and according to primary tumor type
| Entire cohort (n=249) |
Cutaneous (n=183) |
Mucosal (n=28) |
Unknown primary (n=20) |
Acral (n=18) | P value | |
| Genomic subtype | ||||||
| BRAF V600E/K | 86 (35%) | 78 (43%) | 0 (0%) | 5 (25%) | 3 (17%) | 0.0106 |
| NRAS | 69 (28%) | 46 (25%) | 7 (25%) | 10 (50%) | 6 (33%) | |
| BRAF/NRAS WT | 94 (38%) | 59 (32%) | 21 (75%) | 5 (25%) | 9 (50%) | |
| Age | ||||||
| <60 | 122 (49%) | 90 (49%) | 16 (57%) | 6 (30%) | 10 (56%) | 0.2703 |
| ≥60 | 127 (51%) | 93 (51%) | 12 (43%) | 14 (70%) | 8 (44%) | |
| Gender | ||||||
| Male | 145 (58%) | 115 (63%) | 12 (43%) | 11 (55%) | 7 (39%) | 0.2377 |
| Female | 104 (42%) | 68 (37%) | 16 (57%) | 9 (45%) | 11 (61%) | |
| No of metastatic sites | ||||||
| >3 | 184 (74%) | 134 (54%) | 21 (75%) | 17 (75%) | 12 (67%) | 0.6133 |
| ≤3 | 65 (26%) | 49 (46%) | 7 (25%) | 3 (25%) | 6 (33%) | |
| M-stage | ||||||
| M0/M1a/M1b | 93 (37%) | 78 (43%) | 4 (14%) | 3 (15%) | 8 (44%) | 0.0039 |
| M1c/M1d | 156 (63%) | 105 (57%) | 24 (86%) | 17 (85%) | 10 (56%) | |
| LDH | ||||||
| <1.5 x ULN | 183 (74%) | 146 (80%) | 14 (50%) | 14 (70%) | 9 (50%) | 0.0050 |
| ≥1.5 x ULN | 65 (26%) | 36 (20%) | 14 (50%) | 6 (30%) | 9 (50%) | |
| NLR | ||||||
| <5 | 188 (76%) | 139 (76%) | 18 (64%) | 15 (75%) | 16 (89%) | 0.2933 |
| ≥5 | 60 (24%) | 43 (24%) | 10 (36%) | 5 (25%) | 2 (11%) | |
| Anti-PD1 regimen | ||||||
| Anti-PD1 | 172 (69%) | 136 (74%) | 15 (54%) | 10 (50%) | 11 (61%) | 0.0234 |
| Anti-PD1+anti-CTLA4 | 77 (31%) | 47 (26%) | 13 (46%) | 10 (50%) | 7 (39%) | |
| Line of therapy | ||||||
| First | 135 (54%) | 94 (51%) | 18 (64%) | 14 (70%) | 9 (50%) | 0.2776 |
| Second or later | 114 (46%) | 89 (49%) | 10 (36%) | 6 (30%) | 9 (50%) |
WT, wild type.
Figure 2.
Characterization of primary tumor and genomic subtypes of patients with melanoma. (A) Primary tumor type of the entire cohort of 230 individual patients was analyzed. Our cohort was comprised of 171 (74%) cutaneous, 25 (11%) mucosal, 19 (8%) unknown primary and 15 (7%) acral melanoma patients, respectively. (B) Within the entire cohort, 86 (37%) of patients were BRAF/NRAS wild-type (WT), 80 (35%) had BRAF V600E/K mutations and 64 (28%) had NRAS mutations. (C) The incidence of genomic subtypes according to primary tumor type is indicated.
Survival outcomes of the entire cohort according to anti-PD1 regimen
The survival of the entire cohort of patients with metastatic melanoma (n=249) was evaluated. The median cPFS and OS for the entire cohort were 8.3 and 32.9 months respectively. Patients who received anti-PD1+anti-CTLA4 ICI experienced longer median cPFS (15.6 months) compared with those who received anti-PD1 alone (6.3 months), p=0.0098 (online supplemental figure S1a). Similarly, patients who received anti-PD1 anti-CTLA4 ICI also experienced longer median OS (42.6 months) compared with those who received anti-PD1 alone (21.3 months), p=0.0132 (online supplemental figure S1b). Some characteristics were significantly different between the group of patients who received combination ICI compared with those who received anti-PD1 monotherapy (online supplemental table S1). Specifically, patients who received combination ICI were more likely to have mucosal, acral or unknown primary melanoma (p=0.022), they were younger (p=0.006), more likely to be male (p=0.035), and more likely to have received it as their first line of systemic therapy (p<0.001). In multivariable analyses, accounting for these variables, anti-PD1+anti-CTLA4 remained significantly associated with longer cPFS (HR 0.57, 95% CI 0.38 to 0.86, p=0.007) and OS (HR 0.42, 95% CI 0.28 to 0.65, p<0.001) (table 2).
Table 2.
Cox regression analysis for cPFS and OS for entire cohort of patients with advanced melanoma
| cPFS univariable | cPFS multivariable | OS univariable | OS multivariable | |||||||||
| HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | |
| Primary tumor type | ||||||||||||
| Acral or mucosal vs cutaneous or unknown | 1.57 | 1.09 to 2.26 | 0.016 | 1.85 | 1.23 to 2.79 | 0.003 | 1.84 | 1.23 to 2.76 | 0.003 | 2.13 | 1.36 to 3.34 | 0.001 |
| Genomic subtype | ||||||||||||
| BRAF or NRAS mt vs WT | 1.28 | 0.94 to 1.77 | 0.120 | 1.40 | 0.99 to 1.96 | 0.056 | 1.16 | 0.81 to 1.65 | 0.423 | 1.46 | 1.00 to 2.13 | 0.052 |
| Anti-PD1 regimen | ||||||||||||
| PD1+CTLA4 vs PD1 | 0.63 | 0.45 to 0.90 | 0.011 | 0.57 | 0.38 to 0.86 | 0.007 | 0.60 | 0.40 to 0.90 | 0.014 | 0.42 | 0.28 to 0.65 | <0.001 |
| Line of therapy | ||||||||||||
| First vs second or later | 0.56 | 0.42 to 0.76 | <0.001 | 0.72 | 0.51 to 1.01 | 0.062 | 0.53 | 0.38 to 0.75 | <0.001 | – | ||
| No of metastatic sites | ||||||||||||
| >3 vs ≤3 | 2.15 | 1.56 to 2.98 | <0.001 | 2.06 | 1.47 to 2.88 | <0.001 | 2.23 | 1.56 to 3.18 | <0.001 | 2.16 | 1.50 to 3.11 | <0.001 |
| M-stage | ||||||||||||
| M1c/1d vs M0/1a/1b | 1.45 | 1.06 to 2.00 | 0.022 | – | 1.79 | 1.24 to 2.57 | 0.002 | – | ||||
| LDH | ||||||||||||
| ≥1.5X ULN vs <1.5 x | 1.83 | 1.32 to 2.54 | <0.001 | 1.49 | 1.06 to 2.11 | 0.023 | 2.41 | 1.68 to 3.46 | <0.001 | 1.88 | 1.28 to 2.76 | <0.001 |
| NLR | ||||||||||||
| ≥5 vs <5 | 1.39 | 0.98 to 1.96 | 0.062 | – | 1.72 | 1.19 to 2.51 | 0.004 | 1.59 | 1.06 to 2.36 | 0.023 | ||
| Age | ||||||||||||
| ≥60 vs <60 | 0.83 | 0.61 to 1.11 | 0.215 | – | 1.21 | 0.86 to 1.71 | 0.267 | – | ||||
| Gender | ||||||||||||
| Male vs female | 0.78 | 0.58 to 1.06 | 0.114 | – | 0.82 | 0.58 to 1.15 | 0.254 | – | ||||
cPFS, clinical progression-free survival; LDH, lactate dehydrogenase; NLR, neutrophil to lymphocyte ratio; OS, overall survival.
jitc-2020-001642supp001.pdf (1.6MB, pdf)
Survival outcomes of the entire cohort according to primary tumor type and anti-PD1 regimen
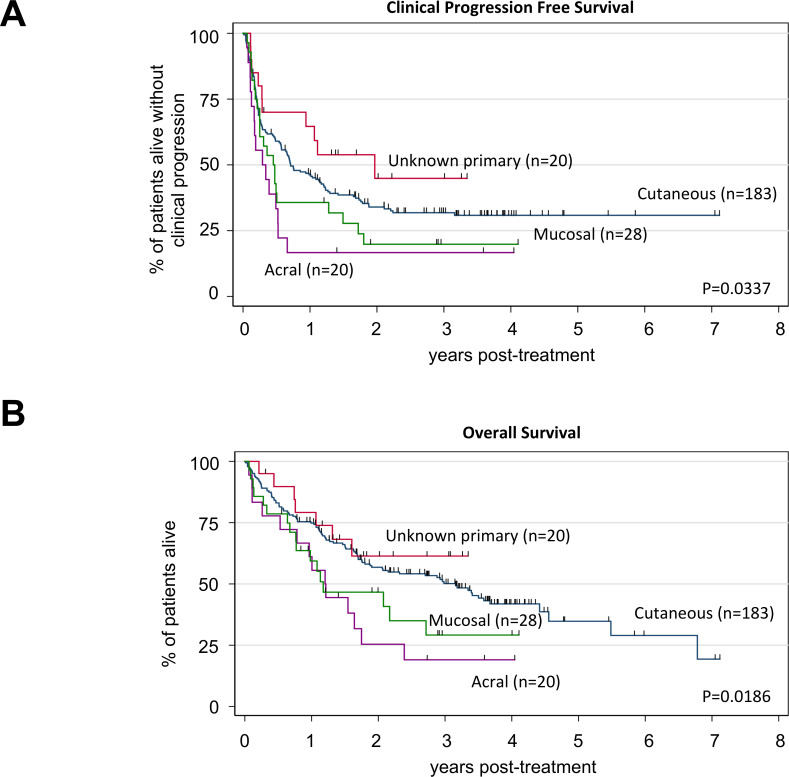
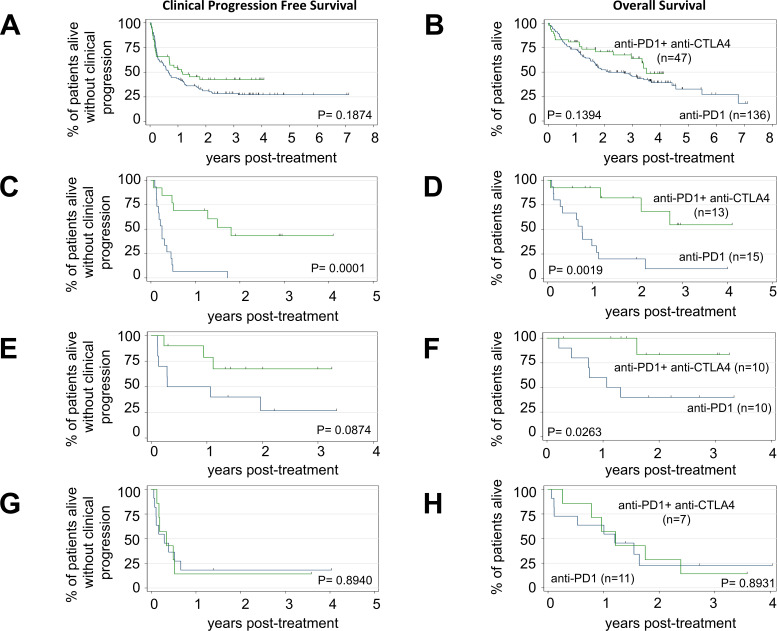
Acral or mucosal melanomas were independently associated with shorter cPFS (HR 1.85, 95% CI 1.23 to 2.79, p=0.003) and OS (HR 2.13, 95% CI 1.36 to 3.34, p=0.001) compared cutaneous or unknown primary melanomas in multivariable analyses of the entire cohort (table 2). Patients with unknown primary melanoma experienced the longest median cPFS (23.9 months) followed by cutaneous (8.6 months), mucosal (5.5 months) and acral melanoma (3.5 months), p=0.0337 (figure 3A). Patients with unknown primary melanoma also experienced the longest median OS (not reached) followed by cutaneous (38.4 months), acral (14.6 months) and mucosal melanoma (14.3 months), p=0.0186 (figure 3B). Among the four primary tumor types described, we observed a significant trend toward improved cPFS with anti-PD1+anti-CTLA4 ICI versus anti-PD1 alone in mucosal melanoma (p=0.0001, figure 4C). There were non-significant trends toward prolonged cPFS with combination ICI in patients with cutaneous (figure 4A) or unknown primary melanoma (figure 4E), but not in patients with acral melanoma (figure 4G). Combination ICI was associated with prolonged OS compared with anti-PD1 alone in patients with mucosal (p=0.0019, figure 4D) or unknown primary melanoma (p=0.0263, figure 4F). Again, we observed a non-significant trend toward prolonged OS with combination ICI in patients with cutaneous melanoma (figure 4B) but not in acral melanoma (figure 4H). In multivariable analyses of cutaneous melanomas, adjusting for the number of involved metastatic sites, LDH level, NLR, and the presence of BRAF or NRAS mutations, combination ICI was significantly associated with longer OS compared with anti-PD1 monotherapy (HR 0.57, 95% CI 0.33 to 0.96, p=0.035 (online supplemental table S2).
Figure 3.
Association between primary tumor type and survival in patients with melanoma. Kaplan-Meier curves of clinical progression-free survival (cPFS) (A) and overall survival (OS) (B) among patients with melanoma stratified according to primary tumor type: cutaneous (n=183, blue line), mucosal (n=28, green line), unknown primary (n=20, red line), acral (n=18, purple line). Log-rank tests were p=0.0337 (cPFS) and p=0.0186 (OS). Tick marks indicate censored patients.
Figure 4.
Survival according to primary melanoma tumor type and anti-programmed cell death protein 1 (anti-PD1) regimen. (A) Clinical progression-free survival (cPFS) and (B) overall survival (OS) for cutaneous melanoma; anti-PD1 (n=136) anti-PD1+anti-CTLA4 (n=47), log-rank test p=0.1874 (cPFS); p=0.1394 (OS). (C) cPFS and (D) OS for mucosal melanoma; anti-PD1 (n=15) anti-PD1+anti-CTLA4 (n=13), log-rank test p=0.0001 (cPFS); p=0.0019 (OS). (E) cPFS and (F) OS for unknown primary melanoma; anti-PD1 (n=10) anti-PD1+anti-CTLA4 (n=10), log-rank test p=0.0874 (cPFS); p=0.0263 (OS). (G) cPFS and (H) OS for acral melanoma; anti-PD1 (n=11) anti-PD1+anti-CTLA4 (n=7), log-rank test p=0.8940 (cPFS); p=0.8931 (OS). Blue line=anti-PD1 monotherapy and green line=anti-PD1+anti-CTLA4. Tick marks indicate censored patients.
Survival outcomes of the entire cohort according to genomic subtype and anti-PD1 regimen
In multivariable analyses of the entire cohort, there was a non-significant trend toward shorter cPFS among patients who had either a BRAF V600E/K or NRAS mutation (HR 1.40, 95% CI 0.99 to 1.96, p=0.056) and OS (HR 1.46, 95% CI 1.00 to 2.13, p=0.052) compared with those who were BRAF/NRAS wild type (table 2). In multivariable analyses of survival outcomes of patients with cutaneous melanoma (n=183), the presence of either a BRAF or NRAS mutation was significantly associated with shorter cPFS (HR 1.63, 95% CI 1.07 to 2.49, p=0.022) and OS (HR 1.65, 95% CI 1.02 to 2.69, p=0.043) (online supplemental table S2).
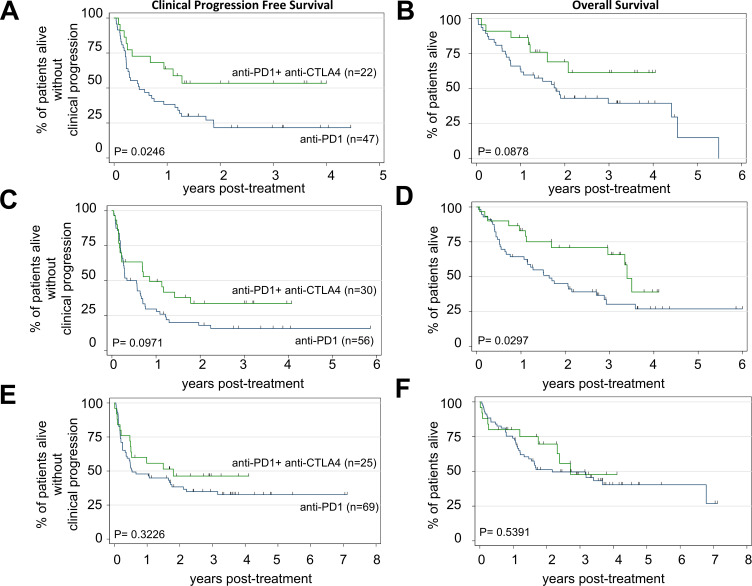
NRAS mutant cohort
Patients with NRAS mutant melanoma (n=69) experienced significantly longer cPFS when treated with anti-PD1+anti-CTLA4 ICI (median cPFS not reached) versus anti-PD1 (median cPFS=7.0 months) (figure 5A). Similarly, there was a non-significant trend toward prolonged OS with anti-PD1+anti-CTLA4 ICI (median OS not reached) versus anti-PD1 (median OS=21.9 months) (figure 5B). In multivariable analyses of NRAS mutant melanoma, treatment with anti-PD1 +anti-CTLA4 ICI was associated with significantly improved cPFS (HR 0.34, 95% CI 0.16 to 0.71, p=0.004) and OS (HR 0.24, 95% CI 0.10 to 0.62, p=0.003) (online supplemental table S3). Clinical variables that were independently associated with poor cPFS and OS in NRAS mutant melanoma included >3 metastatic sites and elevated LDH. Mucosal or acral primary tumor type was significantly associated with shorter OS but not cPFS (online supplemental table S3).
Figure 5.
Survival according to genomic subtype and anti-programmed cell death protein 1 (anti-PD1) regimen. (A) Clinical progression-free survival (cPFS) and (B) overall survival (OS) for NRAS mutant melanoma; anti-PD1 (n=47) anti-PD1+anti-CTLA4 (n=22), log-rank test p=0.0246 (cPFS); p=0.0878 (OS). (C) cPFS and (D) OS for BRAF V600E/K mutant melanoma; anti-PD1 (n=56) anti-PD1+anti-CTLA4 (n=30), log-rank test p=0.0971 (cPFS); p=0.0297 (OS). (E) cPFS and (F) OS for BRAF/NRAS wild-type melanoma; anti-PD1 (n=69) anti-PD1+anti-CTLA4 (n=25), log-rank test p=0.3226 (cPFS); p=0.5391 (OS). Blue line=anti-PD1 monotherapy and green line=anti-PD1+anti-CTLA4. Tick marks indicate censored patients.
BRAF V600E/K mutant cohort
In BRAF V600E/K mutant melanoma (n=86), there was a significant trend toward longer OS and non-significant trend toward longer cPFS with combination immunotherapy versus anti-PD1 alone (figure 5C, D). In multivariable analyses of survival outcomes among patients with BRAF V600E/K mutant melanoma, anti-PD1+anti-CTLA4 ICI was associated with longer OS (HR 0.47, 95% CI 0.24 to 0.90, p=0.024), as was >3 metastatic sites and NLR >5. Anti-PD1 regimen type was not significantly associated with cPFS in a multivariable model of BRAF V600E/K mutant melanoma (online supplemental table S4).
BRAF/NRAS wild-type cohort
In our cohort of BRAF/NRAS wild-type melanoma (n=94), we did not observe any significant trend toward improved cPFS or OS with combination immunotherapy versus anti-PD1 alone (figure 5E, F). In this genomic subtype of melanoma, only primary tumor type and line of systemic therapy were significantly associated with cPFS and OS in multivariable analyses (online supplemental table S5). Indeed, there was no association with cPFS (HR 1.01, 95% CI 0.47 to 2.18, p=0.982) or OS (HR 1.36, 95% CI 0.62 to 3.02, p=0.436) with anti-PD1+anti-CTLA4 versus anti-PD1 alone in non-mucosal BRAF/NRAS wild-type melanoma (n=73) (online supplemental figure S2a, b). However, anti-PD1+anti-CTLA4 ICI treatment was associated with improved cPFS (HR 0.15, 95% CI 0.04 to 0.50, p=0.002) and OS (HR 0.13, 95% CI 0.03 to 0.63, p=0.011) in BRAF/NRAS wild-type mucosal melanoma (n=21) (online supplemental figure S2c, d). A subset of patients with BRAF wild-type melanoma were also assessed for the presence of a KIT mutation. We identified seven patients with KIT mutant melanoma, including one who had two lines of anti-PD1 containing ICI therapy. Compared with patients BRAF/NRAS/KIT wild-type melanoma, those patients with a KIT mutation did not experience any significant differences in cPFS or OS (online supplemental figure S3).
Discussion
In this single-center retrospective cohort study, we found that distinct biologic subtypes of melanoma can serve as important predictors of ICI response in metastatic melanoma, and that the relationship between oncogenic mutations and therapeutic outcomes with anti-PD1±anti-CTLA4 varied across melanomas derived from different primary tumor types.
In our study, acral or mucosal melanoma subtypes predicted poor response to anti-PD1 monotherapy, in line with previous studies.16–18 These tumor types are rare, and the reported numbers of patients with acral or mucosal melanoma that were treated with anti-PD1 ICI were small; our data provide further validation of these observations. The unique genomic make-up of acral and mucosal melanomas, as described above, may explain their relative insensitivity to anti-PD1 ICI compared with cutaneous melanomas. We observed a clear overall survival benefit with combination ICI in mucosal and unknown primary melanoma, but there was no survival benefit for combination ICI in acral melanoma. These data warrant further validation in additional cohorts, such as the Checkmate-067 study to determine if patients with acral melanoma could be spared the toxicity associated with combination ICI. Together, these data suggest that the primary tumor type of melanoma should be considered when selecting ICI regimens for metastatic disease, and that further validation of these observations is warranted.19
The dismal prognosis of patients with acral melanoma highlights the need to develop more effective precision immunogenomic therapies for this subtype. KIT mutations, while more common in acral melanoma than other subtypes, are still only present in a minority of these tumors.3 As such, targeting KIT alone will not benefit the majority of patients with metastatic acral melanoma. Inhibitors of angiogenesis, such as multitargeted receptor tyrosine kinase inhibitors (ie, VEGFR, PDGFR, KIT and so on) have been shown to reprogram immunosuppressive tumor microenvironments to facilitate antitumor immune infiltration.20 Combined inhibition of the PD1/PD-L1 axis along with multitargeted receptor tyrosine kinase inhibitors has demonstrated efficacy in renal cell carcinoma21 22 and it is emerging as a promising combination therapy in many other cancer types, including melanoma.23 This may represent a promising treatment option for acral melanoma, and there are a number of ongoing clinical trials actively investigating this strategy (NCT03955354, NCT03991975).
In addition to primary tumor type, we found that genomic subtype could function as a predictive biomarker for ICI regimens in metastatic melanoma. We found that combination ICI was associated with improved OS over anti-PD1 monotherapy in multivariable analyses of BRAF V600E/K mutant and in NRAS mutant melanoma, but not in BRAF/NRAS wild-type melanoma. Our retrospective analysis provides real-world data supporting the results of the BRAF mutation subset analyses of the Checkmate-067 study,13 and validates the use of anti-CTLA4+anti-PD1 combination as the preferred ICI regimen for BRAF V600E/K mutant melanoma. Previous retrospective studies have examined the relationship between NRAS mutations and ICI outcomes in melanoma, but have not established a relationship between NRAS and poor survival. However, these studies were limited because they include a very small number of NRAS mutant patients who received anti-PD1 ICI24 or analyzed only those patients who had already progressed on anti-PD1 ICI.25 To our knowledge, the observation that NRAS mutations status can function as a predictive marker for anti-PD1 ICI outcomes is a novel finding.
NRAS is a GTPase that acts upstream of BRAF in the MAPK pathway. NRAS mutations are mutually exclusive with BRAF V600E/K mutations. To date, no targeted therapies have demonstrated OS benefit in NRAS-mutated melanoma26; as such, NRAS mutations status is not generally assessed in prospective clinical trials with ICIs and testing for NRAS it is not mandatory for routine clinical practice. However, NRAS mutations are generally associated with poor prognosis.27 The impact of an activating NRAS mutation on the immune composition of the melanoma tumor microenvironment is not well described. Activating BRAF mutations are well known to orchestrate an immune-suppressed tumor microenvironment via various mechanisms, including downregulation of MHC class 1 molecules on the surface of tumor cells, enhanced production of chemokines, which recruit myeloid-derived suppressor cells (MDSCs),28 and increased expression of vascular endothelial growth factor (VEGF), which promotes myeloid cell maturation.29 Inhibition of the BRAF V600E/K oncogene in melanoma with BRAF and/or MEK inhibitors also leads to an increase in tumor-infiltrating lymphocytes30 and other molecular alterations that facilitate immune activation within the tumor immune microenvironment.29 31 32 Therefore, it is conceivable that NRAS mutations, via activation of the downstream MAPK pathway could function similarly to BRAF mutations in this respect. Indeed, it has been reported that activating KRAS mutations has been shown to facilitate immune suppression via metabolic reprogramming, which leads to increased glycolysis and MDSC recruitment in triple negative breast cancer33 and colon cancer.34 Our data are hypothesis-generating and identify NRAS as a potential novel biomarker for specific ICI regimens. These data also highlight the importance of testing NRAS in melanoma, and reporting NRAS mutation status in prospective clinical trials investigating novel immunotherapeutic strategies.
Cutaneous melanomas that lack BRAF V600E/K or NRAS mutations belong to either the NF1 mutant or triple wild-type genomic subgroups.2 NF1 mutant melanoma in particular have a high tumor mutation burden (TMB),2 resulting in a high number of neoantigens, which can be readily detected by T-cells. Indeed, high TMB has been associated with increased responsiveness to anti-PD1 monotherapy.35 We did not assess NF1 mutation status or TMB in our cohort, but we hypothesize that NF1 mutations and/or high TMB would be overrepresented in our cohort of n=55 BRAF/NRAS wild-type cutaneous melanomas. This would explain why we observed longer relative survival in our cohort of BRAF/NRAS wild-type non-mucosal melanomas that were treated with anti-PD1 alone, and no added survival benefit with combination ICI. If validated in independent studies, these data provide rationale for examining the predictive value of prospectively testing for TMB and/or NF1 specifically within BRAF/NRAS wild-type cutaneous melanomas. Biomarkers that could identify which patients with cutaneous melanoma are most likely to derive substantial benefit from anti-PD1 alone, thereby potentially mitigating the added toxicities associated with anti-CTLA4 ICI, would be of important clinical value.
Our study was subject to a number of limitations. This was a retrospective cohort study, and as such, unknown confounders that we did not assess could explain the associations that we observed. This was a single center study, therefore, the patient population and treatment selection strategies may not be broadly generalizable. Moreover, the sample sizes of our non-cutaneous melanoma types and genomic subgroups were small and therefore subject to bias. Finally, subsequent lines of therapy could confound OS results. Due to these limitations, our results should be considered hypothesis-generating.
In conclusion, our study highlights that metastatic melanoma constitutes a diverse group of biologically and genomically distinct diseases. Response to immunotherapy differs across these unique subsets. Primary tumor type and genomic subtype are emerging as potential predictive biomarkers that can aid in selection of specific ICI regimens for patients with metastatic melanoma.
Acknowledgments
AANR is a recipient of a Hold’em For Life Clinician Scientist Award. TPM and AANR acknowledge fellowship funding from Alamos Gold Inc.
Footnotes
Contributors: Conception and design of study: AANR and AS. Acquisition of data: AANR, SMA, DH, MOB, SDS, DPA, TPM, DK, DG, IK, ZSK, KR and AS. Analysis and interpretation of data: AANR and AS. Writing and review of manuscript: AANR, SMA, DH, MOB, SS, DPA, TPM, DK, DG, IK, ZSK, KR and AS.
Funding: This research has been supported by funding from a University of Toronto Douglas Wright Melanoma Award to AANR, and research funds of AS.
Competing interests: AANR: Immediate family member is an employee of Merck. AS: Consultant/Advisory Board: Merck, Bristol-Myers Squibb, Novartis, Oncorus, Janssen. Grant/Research support from Clinical Trials: Novartis, Bristol-Myers Squibb, Symphogen AstraZeneca/Medimmune, Merck, Bayer, Surface Oncology, Northern Biologics, Janssen Oncology/Johnson & Johnson, Roche, Regeneron, Alkermes, Array Biopharma. SDS: Consultant/Advisory Board: Novartis, Janssen.
Patient consent for publication: Not required.
Ethics approval: This study was approved by the University Health Network Institutional Research Ethics Board (REB# 19-5186).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Primary data are available upon reasonable request. All aggregate data relevant to the study are included in the article or uploaded as supplementary information.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Elder DE, Bastian BC, Cree IA, et al. . The 2018 World Health organization classification of cutaneous, mucosal, and uveal melanoma: detailed analysis of 9 distinct subtypes defined by their evolutionary pathway. Arch Pathol Lab Med 2020;144:500–22. 10.5858/arpa.2019-0561-RA [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell 2015;161:1681–96. 10.1016/j.cell.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayward NK, Wilmott JS, Waddell N, et al. . Whole-Genome landscapes of major melanoma subtypes. Nature 2017;545:175–80. 10.1038/nature22071 [DOI] [PubMed] [Google Scholar]
- 4.Newell F, Kong Y, Wilmott JS, et al. . Whole-Genome landscape of mucosal melanoma reveals diverse drivers and therapeutic targets. Nat Commun 2019;10:3163. 10.1038/s41467-019-11107-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith JL, Stehlin JS. Spontaneous regression of primary malignant melanomas with regional metastases. Cancer 1965;18:1399–415. [DOI] [PubMed] [Google Scholar]
- 6.Saleh FH, Crotty KA, Hersey P, et al. . Primary melanoma tumour regression associated with an immune response to the tumour-associated antigen Melan-A/MART-1. Int J Cancer 2001;94:551–7. 10.1002/ijc.1491 [DOI] [PubMed] [Google Scholar]
- 7.Clemente CG, Mihm MC, Bufalino R, et al. . Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer 1996;77:1303–10. [DOI] [PubMed] [Google Scholar]
- 8.Gos A, Jurkowska M, van Akkooi A, et al. . Molecular characterization and patient outcome of melanoma nodal metastases and an unknown primary site. Ann Surg Oncol 2014;21:4317–23. 10.1245/s10434-014-3799-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robert C, Schachter J, Long GV, et al. . Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med 2015;372:2521–32. 10.1056/NEJMoa1503093 [DOI] [PubMed] [Google Scholar]
- 10.Wolchok JD, Rollin L, Larkin J. Nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2017;377:2503–4. 10.1056/NEJMc1714339 [DOI] [PubMed] [Google Scholar]
- 11.Robert C, Long GV, Brady B, et al. . Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med 2015;372:320–30. 10.1056/NEJMoa1412082 [DOI] [PubMed] [Google Scholar]
- 12.Hodi FS, O'Day SJ, McDermott DF, et al. . Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. . Five-Year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2019;381:1535–46. 10.1056/NEJMoa1910836 [DOI] [PubMed] [Google Scholar]
- 14.LoRusso PM, Schalper K, Sosman J. Targeted therapy and immunotherapy: emerging biomarkers in metastatic melanoma. Pigment Cell & Melanoma Research 2019. [DOI] [PubMed] [Google Scholar]
- 15.Shoushtari AN, Wagstaff J, Ascierto PA, et al. . CheckMate 067: long-term outcomes in patients with mucosal melanoma. Journal of Clinical Oncology 2020;38:10019. 10.1200/JCO.2020.38.15_suppl.10019 [DOI] [Google Scholar]
- 16.Klemen ND, Wang M, Rubinstein JC, et al. . Survival after checkpoint inhibitors for metastatic acral, mucosal and uveal melanoma. J Immunother Cancer 2020;8:e000341. 10.1136/jitc-2019-000341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoushtari AN, Munhoz RR, Kuk D, et al. . The efficacy of anti-PD-1 agents in acral and mucosal melanoma. Cancer 2016;122:3354–62. 10.1002/cncr.30259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamid O, Robert C, Ribas A, et al. . Antitumour activity of pembrolizumab in advanced mucosal melanoma: a post-hoc analysis of KEYNOTE-001, 002, 006. Br J Cancer 2018;119:670–4. 10.1038/s41416-018-0207-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eroglu Z, Zaretsky JM, Hu-Lieskovan S, et al. . High response rate to PD-1 blockade in desmoplastic melanomas. Nature 2018;553:347–50. 10.1038/nature25187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Yuan J, Righi E, et al. . Vascular normalizing doses of antiangiogenic treatment reprogram the immunosuppressive tumor microenvironment and enhance immunotherapy. Proc Natl Acad Sci U S A 2012;109:17561–6. 10.1073/pnas.1215397109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rini BI, Plimack ER, Stus V, et al. . Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1116–27. 10.1056/NEJMoa1816714 [DOI] [PubMed] [Google Scholar]
- 22.Motzer RJ, Penkov K, Haanen J, et al. . Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1103–15. 10.1056/NEJMoa1816047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor MH, Lee C-H, Makker V, et al. . Phase Ib/II trial of lenvatinib plus pembrolizumab in patients with advanced renal cell carcinoma, endometrial cancer, and other selected advanced solid tumors. J Clin Oncol 2020;38:1154–63. 10.1200/JCO.19.01598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson DB, Lovly CM, Flavin M, et al. . Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Cancer Immunol Res 2015;3:288–95. 10.1158/2326-6066.CIR-14-0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrinely JR, Baker LX, Davis EJ, et al. . Outcomes after progression of disease with anti-PD-1/PD-L1 therapy for patients with advanced melanoma. Cancer 2020;126:3448–55. 10.1002/cncr.32984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dummer R, Schadendorf D, Ascierto PA, et al. . Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 2017;18:435–45. 10.1016/S1470-2045(17)30180-8 [DOI] [PubMed] [Google Scholar]
- 27.Jakob JA, Bassett RL, Ng CS, et al. . Nras mutation status is an independent prognostic factor in metastatic melanoma. Cancer 2012;118:4014–23. 10.1002/cncr.26724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khalili JS, Liu S, Rodríguez-Cruz TG, et al. . Oncogenic BRAF(V600E) promotes stromal cell-mediated immunosuppression via induction of interleukin-1 in melanoma. Clin Cancer Res 2012;18:5329–40. 10.1158/1078-0432.CCR-12-1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sumimoto H, Imabayashi F, Iwata T, et al. . The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J Exp Med 2006;203:1651–6. 10.1084/jem.20051848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frederick DT, Piris A, Cogdill AP, et al. . Braf inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res 2013;19:1225–31. 10.1158/1078-0432.CCR-12-1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu-Lieskovan S, Mok S, Homet Moreno B, et al. . Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAF(V600E) melanoma. Sci Transl Med 2015;279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose AAN, Annis MG, Frederick DT, et al. . Mapk pathway inhibitors sensitize BRAF-mutant melanoma to an antibody-drug conjugate targeting GPNMB. Clin Cancer Res 2016;22:6088–98. 10.1158/1078-0432.CCR-16-1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franklin DA, Sharick JT, Ericsson-Gonzalez PI, et al. . Mek activation modulates glycolysis and supports suppressive myeloid cells in TNBC. JCI Insight 2020;5. 10.1172/jci.insight.134290. [Epub ahead of print: 06 Aug 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao W, Overman MJ, Boutin AT, et al. . KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell 2019;35:559–72. 10.1016/j.ccell.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500–1. 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001642supp001.pdf (1.6MB, pdf)