Abstract
Objective
To study messenger ribonucleic acid (mRNA) and protein expressions of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) entry receptors (angiotensin 1-converting enzyme 2 [ACE2] and CD147) and proteases (transmembrane serine protease 2 [TMPRSS2] and cathepsin L [CTSL]) in human oocytes, embryos, and cumulus (CCs) and granulosa cells (GCs).
Design
Research study.
Setting
Clinical in vitro fertilization (IVF) treatment center.
Patients
Patients undergoing IVF were treated at the Colorado Center for Reproductive Medicine.
Interventions
Oocytes (germinal vesicle and metaphase II [MII]) and embryos (1-cell [1C] and blastocyst [BL]) were donated for research at the disposition by the patients undergoing IVF. Follicular cells (CC and GC) were collected from women undergoing egg retrieval after ovarian stimulation without an ovulatory trigger for in vitro maturation/IVF treatment cycles.
Main Outcome Measures
Presence or absence of ACE2, CD147, TMPRSS2, and CTSL mRNAs detected using quantitative reverse transcription polymerase chain reaction and proteins detected using capillary Western blotting in human oocytes, embryos, and ovarian follicular cells.
Results
The quantitative reverse transcription polymerase chain reaction analysis revealed high abundance of ACE2 gene transcripts in germinal vesicle and MII oocytes than in CC, GC, and BL. ACE2 protein was present only in the MII oocytes, and 1C and BL embryos, but other ACE2 protein variants were observed in all the samples. TMPRSS2 protein was present in all the samples, whereas mRNA was observed only in the BL stage. All the samples were positive for CD147 and CTSL mRNA expressions. However, CCs and GCs were the only samples that showed coexpression of both CD147 and CTSL proteins in low abundance.
Conclusions
CCs and GCs are the least susceptible to SARS-CoV-2 infection because of lack of the required combination of receptors and proteases (ACE2/TMPRSS2 or CD147/CTSL) in high abundance. The coexpression of ACE2 and TMPRSS2 proteins in the MII oocytes, zygotes, and BLs demonstrated that these gametes and embryos have the cellular machinery required and, thus, are potentially susceptible to SARS-CoV-2 infection if exposed to the virus. However, we do not know whether the infection occurs in vivo or in vitro in an assisted reproductive technology setting yet.
Key Words: SARS-CoV-2, human IVF, oocytes and embryos, ovarian cells
Discuss: You can discuss this article with its authors and other readers at https://www.fertstertdialog.com/posts/xfss-d-20-00072
Since the description of a novel coronavirus in Wuhan, China, in late 2019, the virus, now called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread worldwide, affecting 188 countries. More than 30 million people have been infected worldwide, and nearly 1 million people have died because of coronavirus disease 2019 (COVID-19), the disease caused by SARS-CoV-2 (1). Initially, assisted reproduction was also curtailed, being classified as nonessential in most instances, because countries rapidly shut down to control the spread of the novel coronavirus and avoid overburdening healthcare systems. Very little was known about potential of infection of the sperm, eggs, or embryos; potential of sexual transmission between partners; vertical transmission between a mother and conceptus; or possible effects of maternal infection on an early embryo and fetus. This lack of information raised an important concern in the field about how the virus might potentially affect laboratory and pregnancy outcomes in in vitro fertilization (IVF), in addition to speculation about potential changes in standard operating procedures in an IVF laboratory during the pandemic (2, 3).
As the pandemic continued, impact on couples desiring treatment increased. If their treatment continued to be delayed, specific groups of infertile patients faced significantly reduced prognoses (4). However, the unknown effects of SARS-CoV-2 on reproduction made it difficult to determine the best IVF laboratory practices necessary to mitigate the risks of contamination and infection to both patients and staff. For example, the current IVF practice often involves breaching the zona pellucida for biopsy and genetic testing, presenting a potential opportunity for the virus to gain access to embryonic cells both during culture before implantation and cryostorage. Therefore, it is of great importance that we advance our understanding of the potential of infection by SARS-CoV-2 in gametes and embryos.
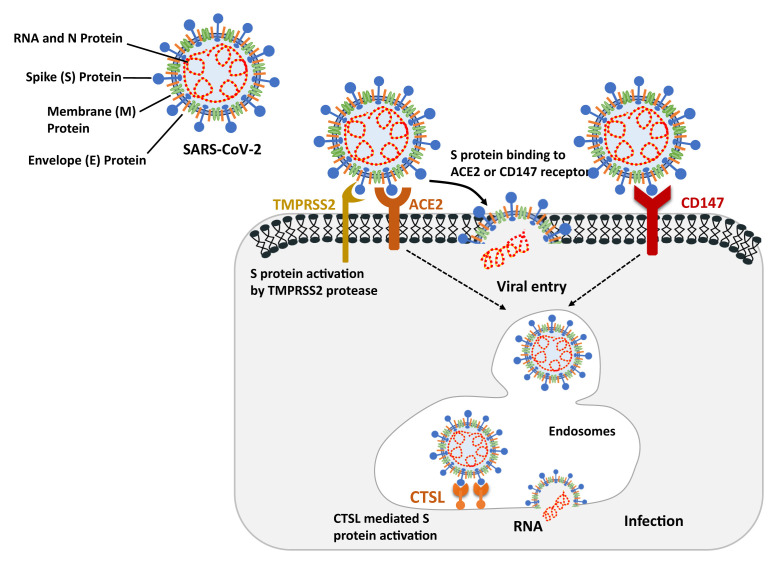
The role of proteins associated with SARS-CoV-2 host cell entry is outlined in Figure 1 . Although the understanding of the molecular mechanism of viral entry continues to get refined, the described mechanism involves viral spike (S) protein-mediated recognition and binding to the host cell’s receptor angiotensin 1-converting enzyme 2 (ACE2), after which the protease transmembrane serine protease 2 (TMPRSS2) cleaves S protein to facilitate cell entry (5). ACE2, a negative regulator of the renin-angiotensin system, has multiple physiologic functions and is widely expressed throughout the body (6). The S protein cleavage site in SARS-CoV-2 is identical to that in SARS-CoV, which is also known to take advantage of the endosomal cysteine protease cathepsin L (CTSL) to cleave specific peptide bonds and facilitate fusion of viral and host cell membranes and release of the viral genome into the host cell’s cytoplasm (7, 8). An alternative ACE2-independent mechanism, with basigin (BSG or CD147) as the cellular receptor and CTSL as the protease, may also mediate SARS-CoV-2 cell entry. Theoretically, tissues and cells that contain these receptors and proteases, particularly high coexpression of ACE2 and TMPRSS2, are more vulnerable to infection by SARS-CoV-2.
Figure 1.
Role of ACE2 and CD147 receptors and TMPRSS2 and CTSL proteases in SARS-CoV-2 entry into host cells. The primary pathway of SARS-CoV-2 to enter host cells is by S protein binding to the ACE2 receptors, which leads to either cell membrane fusion and release of the viral genome if activated by TMPRSS2 or endocytic uptake of the virus. CD147 is another host cell receptor that can be recognized by the SARS-S protein, which facilitates viral entry through endocytosis. After viral entry into the host cells via an endosome, viral RNA is released into the cytoplasm because of pH-dependent cleavage by CTSL and start encoding viral protein. Figure adapted from Heurich et al (2014) and Aguiar et al (2020). ACE2 = angiotensin 1-converting enzyme 2; CTSL = cathepsin L; S = spike; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; TMPRSS2 = transmembrane serine protease 2.
Scientifically, information on the effects of clinical application of assisted reproductive technologies (ARTs) continues to emerge. A popular method that quickly provides the needed information involves querying existing transcriptomic databases for the presence of the receptors and proteases necessary for SARS-CoV-2 infection of host cells. Gene expression studies have shown that the SARS-CoV-2 receptor ACE2 gene is transcribed in the ovary, where it is thought to regulate follicle development, hormone secretion, and oocyte maturation (9, 10). Another study has found that ACE2 is expressed in the ovary but only at low levels and that TMPRSS2 is not expressed, although CTSL is widely expressed; however, no cells coexpressed ACE2 and CTSL (11). Thus, the existing transcriptomic information does not provide much clarity on the potential of infection of the ovary and oocyte. The possibility remains that SARS-CoV-2 infection might affect female fertility by directly infecting ovarian granulosa cells (GCs) and/or oocytes, thereby negatively affecting ovarian function and reducing oocyte quality (12). Analysis of existing single-cell ribonucleic acid (RNA) sequencing datasets has also revealed the presence of transcripts of SARS-CoV-2-related genes, including the receptors ACE2 and BSG and the proteases TMPRSS2 and CTSL, in human embryos (13), suggesting that they may also have the potential to be infected by SARS-CoV-2. The fallopian tube expresses ACE2 in a small number of cells, but the protease TMPRSS2 is widely expressed in different cell types, although there was no detectable coexpression of ACE2 and TMPRSS2 (11), suggesting that the site of fertilization and early embryonic development is unlikely to be infected. In the uterus, ACE2 is expressed at a low level, but TMPRSS2 expression is absent, again suggesting that infection is unlikely (11). In periimplantation- and postimplantation-stage human embryos, the transcripts of ACE2 and TMPRSS2 are coexpressed in 6- and 7-day trophoblasts, 12- and 14-day syncytiotrophoblasts, and some hypoblast cells; TMPRSS2 alone is expressed in the epiblast (14). BSG and CTSL transcripts are expressed in all cell types at this stage (14). This again suggests the susceptibility of the human embryo to SARS-CoV-2 infection and its potential implications on implantation and pregnancy and the potential transmission of the virus from a mother to the fetus through the placenta. However, the transcript levels often do not accurately reflect protein abundance, and this has also been demonstrated for ACE2 (15). In fact, an analysis comparing publically available proteomic and transcriptomic databases has shown that despite its low messenger RNA (mRNA) level, ACE2 protein is abundant in the ovary (15). This is not terribly surprising because processes such as posttranscriptional, translational, and protein degradation processes occur after the mRNA is transcribed, leading to differences of as much as 15%–70% between mRNA levels and protein abundance (16, 17). Because of scarcity of human sample availability, protein expression analyses in oocytes, embryos, and follicular cells rely on immunohistochemistry (IHC). In addition to potential false-positive results due to antibody cross reactivity with similar proteins, IHC has been recognized for its false-negative results since its inception (18, 19, 20, 21). A recent study (22) has reported that IHC fails to detect ACE2 protein expression in either follicular or stromal ovarian cells, but high expression of ACE2 protein is detected by mass spectrometry. Western blot is a gold-standard technique to study protein expression because it reveals molecular mass of proteins along with their expression levels. However, the application of Western blot to the analysis of oocyte and embryo protein expressions has been limited because it requires a large number of samples (23, 24). In this study, we used a high-sensitivity capillary Western blot approach (ProteinSimple; Jess, ProteinSimple, San Jose, CA), which was further optimized in our laboratory to detect up to 10 proteins from a pool of 10 oocytes or embryos.
The objective of this work was to characterize the presence or absence of 4 proteins, ACE2, BSG (CD147), TMPRSS2, and CTSL, important for SARS-CoV-2 infection, in human follicular cells, mature oocytes, zygotes, and blastocysts (BLs). Our data demonstrated that these proteins are present in these tissues. Of interest, ACE2 and TMPRSS2 proteins are both present in high abundance in mature oocytes, zygotes, and BLs. This work demonstrated that human oocytes and embryos have the necessary machinery to be infected by SARS-CoV-2 if they are exposed, either in vivo or in vitro. This information is important for IVF clinics because they are working to provide safe infertility treatment during the global pandemic.
Materials and methods
Sample Collection
Oocyte, cumulus cell (CC), and GC samples were collected from fertility patients undergoing treatment at the Colorado Center for Reproductive Medicine who signed consent forms for biological materials discarded from their treatment cycles to be donated for research (WIRB #20142468). Surplus mature (metaphase II [MII]) eggs (n = 69 eggs from 6 patients, average age = 36.4 years), zygotes (n = 20 zygotes from 4 patients, average age = 35.0 years), and embryos (n = 80 embryos from 14 patients, average age = 28.6 years) were donated for research at the disposition by the patients. GCs were collected from fertility patients (n = 5 patients, average age = 34 years) undergoing egg retrieval after ovarian stimulation without an ovulatory trigger for in vitro maturation/IVF treatment. CCs were collected from standard IVF cycles after transvaginal ultrasound-guided oocyte retrieval and trimming of expanded CC-oocyte complexes before fertilization (n = 5 patients, average age = 38 years). Oocytes that were recovered from the standard IVF cycles, denuded, found to be in an immature state, and subsequently discarded from the treatment cycle were collected immediately at the germinal vesicle (GV) stage (n = 57 oocytes from 49 patients, average age = 37 years). The immature and mature oocytes completely devoid of GCs were washed 3 times with 1X phosphate-buffered saline (PBS) and stored at −80°C. Mural GCs and CCs were centrifuged at 1000 × g for 5 minutes to form a pellet, washed 3 times with 1X PBS, and stored at −80°C with a minimal amount of PBS. Mature eggs, zygotes, and BLs were washed 3 times in 1X PBS and stored at −80°C.
RNA and Protein Isolation
GCs and CCs from individual patients (n = 3 replicates per cell type) were used for RNA and protein isolations. Pools of 6 immature and 8 mature eggs or BLs (2 patients per pool) were used for RNA isolation (n = 3 replicates per sample type). Pools of 14–16 mature eggs (2–4 patients per pool), 10 zygotes (2 patients per pool), and 18 BLs (5 patients per pool) were used for protein isolation (n = 3 replicates per sample type). Not enough zygotes were available for both RNA and protein analyses; therefore, protein expression data for zygotes were derived from 2 biologic and 1 technical replicates. Total RNA from each sample was isolated using the RNeasy Micro Kit (QIAGEN #74004, Valencia, CA) according to the manufacturer’s protocol and eluted in 15 μL of nuclease-free water. All the RNA isolated from the oocytes and embryos was directly used for cDNA synthesis without quantification. The RNA isolated from CCs and GCs was first quantified using NanoDrop 2000 (Thermo Fisher, Waltham, MA), and then, 200 ng of the total RNA was used for cDNA synthesis. To isolate protein, a radioimmunoprecipitation assay buffer (Sigma #R0278) containing a mixture of protease (Sigma #11836170001, St. Louis, MO) and phosphatase (Thermo Fisher #78440) inhibitors was used to lyse the samples. The oocyte and embryo lysates were directly used for Western blot analysis by Jess (product number 004-650; ProteinSimple, San Jose, CA). GC and CC lysates were subjected to ultrasonication, followed by centrifugation at 14,000 × g for 10 minutes at 4°C to remove cellular debris. Supernatants collected from the cell lysates were then quantified using a Bio-Rad DC kit assay, and 900 ng of the protein was used for the Jess analysis.
Reverse Transcription and Quantitative Polymerase Chain Reaction
RNA obtained from each sample was converted into cDNA using iScriptReverse Transcription Supermix (Bio-Rad #1708840) based on the manufacturer’s protocol. The resulting cDNA was diluted in nuclease-free water in 1:1 for the oocytes and embryos and 1:3 for CCs and GCs and stored at −20°C. For quantitative polymerase chain reaction (qPCR), primers for each target gene were designed for the conserved sequence of their protein-coding mRNA isoforms published in Ensemble genome browser using the PerlPrimer software, version 1.1.21. All the oligonucleotides used in this study were synthesized using integrated DNA technology and are listed in Supplementary Table 1 (available online). For qPCR, a total of 2 μL of the diluted cDNA was used in a reaction mixture (25 μL) containing 2X Sso Advanced SYBR green (Bio-Rad) master mix, 5 pM each of the gene-specific forward and reverse primers, and nuclease-free water. The reaction mixtures were divided in half (12.5 μL each) to prepare 2 technical replicates per sample and subjected to qPCR amplification at 95°C for 10 minutes for initial denaturation, followed by 45 cycles of 95°C denaturation for 15 seconds, and an annealing or extension step at 60°C for 1 minute. For each reaction product, a melt-curve analysis after the amplification was performed to ensure a single and specific amplicon. Relative abundance of endogenous GAPDH was used as an internal control for each sample, and qPCR data were analyzed using the 2−ΔΔCt method.
Jess Simple Western Blotting
Protein expression analysis was performed by capillary Western blotting using the Jess system. All antibodies used in this study were produced against human peptides in rabbit and predicated (in silico) to cross react with mouse and rat homologous proteins. A list of the antibodies used is presented in Supplementary Table 2 (available online). All the antibodies were first validated using protein lysates from various human tissues, including the liver, lung, colon, ovary, small intestine (OriGene, Rockville, MD), and cell lines, including white blood cells, HeLa, and HEK293T (Santa Cruz, CA). Based on molecular weight (MW) of the target proteins and source of primary antibodies, the 12–230 kDa separation module (ProteinSimple, SM-W004) and anti-rabbit detection module (ProteinSimple DM-001) were used according to the manufacturer’s protocol. Briefly, the protein lysate from each sample was mixed with a fluorescent master mix in a 4:1 ratio and heated at 95°C for 10 minutes, followed by 5-minute incubation at room temperature. After a brief period of vortexing and spin down, a total of 3 μL of a mixture of the sample, protein-normalization reagent, blocking reagent (antibody diluent 2), wash buffer, primary antibodies diluted at 1:20, horseraddish peroxidase-conjugated ready-to-use secondary antibodies (ProteinSimple DM-001), and chemiluminescent substrate were pipetted into designated wells in the manufacturer-provided microplate. After centrifugation at 1,000 × g for 5 minutes, the plates were loaded onto the Jess instrument, and the default setting was used to run the chemiluminescent and protein normalization-based Jess program for immunodetection of the target proteins. After completion of each run, electropherograms generated by Simple Western Compass software for each capillary were analyzed to determine whether automatic peak detection required manual correction. The target and total protein quantities were determined based on area-under-the-peak analysis by the Compass software. Expression of each target protein was normalized by total protein signal in the same capillary. To discriminate the target protein signal from background, a peak signal-to-noise ratio of ≥10 for any target protein was considered significant and included in the data analysis. All 4 proteins (ACE2, TMPRSS2, CTSL, and CD147) were analyzed in each individual sample lysate using 3 biologic replicates. The blot image in the figures (Figure 2, Figure 3, Figure 4, Figure 5) shows the chemiluminescence signal for each target protein generated by the Compass software, and the bar graphs depict the relative expression of the target protein normalized by total protein detected in the same sample.
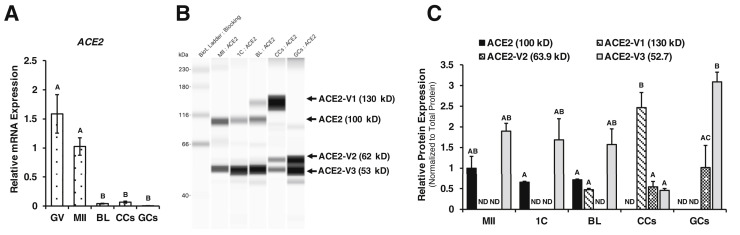
Figure 2.
ACE2 mRNA and protein expressions in human oocytes, embryos, and follicular cells. (A) RT-qPCR analysis of human ACE2 mRNA expression in GV- and MII-stage oocytes, BL (n = 3 pools of oocytes or embryos), CC, and GC (n = 3). Relative expression of ACE2 mRNA was normalized to GAPDH expression and presented as a bar graph. (B) Capillary Western blot of total ACE2 protein expression and (C) fold change in the abundance of ACE2 protein (100 kDa) and putative variants of 130 kDa (ACE2-V1), 62 kDa (ACE2-V2), and 53 kDa (ACE2-V3) in MII-stage oocytes, 1C- and BL-stage embryos, CC, and GC (n = 3 replicates). Arrow indicates the target protein. ACE2 protein expression was normalized to the total protein present in each sample, and data were presented as average protein fold change from 3 experiments. All bar graphs show the mean ± SEM. Different letters above the bars in graph represent the statistical difference (P < .05) determined by 1-way ANOVA followed by Tukey’s test. 1C = 1-cell; ACE2 = angiotensin 1-converting enzyme 2; ANOVA = analysis of variance; BL = blastocyst; CC = cumulus cells; GC = granulosa cells; GV = germinal vesicle; MII = metaphase II; mRNA = messenger ribonucleic acid; ND = nondetectable; RT-qPCR = quantitative reverse transcription polymerase chain reaction; SEM = standard error of the mean.
Figure 3.
Expression of TMPRSS2 mRNA and protein in human oocytes, embryos, and follicular cells. (A) TMPRSS2 mRNA level measured by RT-qPCR analysis in GV- and MII-stage oocytes, BL, CC, and GC. The expression data were normalized to GAPDH mRNA abundance and presented as a bar graph (n = 3 replicates). (B) Capillary Western blot of total TMPRSS2 protein (54 kDa) expression and (C) fold change in the TMPRSS2 protein abundance in MII-stage oocytes, 1C- and BL-stage embryos, CC, and GC (n = 3). Arrow indicates the target protein. TMPRSS2 protein expression was normalized to the total protein present in each sample, and data were presented as average protein fold change from 3 experiments. All bar graphs show the mean ± SEM. Different letters above the bars in graph indicate statistical difference (P < .05) determined by 1-way ANOVA followed by Tukey’s test. 1C = 1-cell; ANOVA = analysis of variance; BL = blastocyst; CC = cumulus cells; GC = granulosa cells; GV = germinal vesicle; MII = metaphase II; mRNA = messenger ribonucleic acid; ND = nondetectable; RT-qPCR = quantitative reverse transcription polymerase chain reaction; SEM = standard error of the mean; TMPRSS2 = transmembrane serine protease 2.
Figure 4.
Expression of CD147 mRNA and protein in human oocytes, embryos, and follicular cells. (A) RT-qPCR analysis of CD147 mRNA in GV- and MII-stage oocytes, BL (n = 3 pools of 6 oocytes or embryos), CC, and GC (n = 3). Expression data were normalized to GAPDH and presented as a bar graph. (B) Capillary Western blot of total CD147 protein (60 kDa) expression and (C) fold change in the CD147 protein abundance in MII-stage oocytes, 1C- and BL-stage embryo, CC, and GC (n = 3 replicates). Arrow indicates the target protein. All bar graphs show the mean ±SEM. Different letters indicate statistical difference (P < .05), determined by 1-way ANOVA followed by Tukey’s test. 1C = 1-cell; ANOVA = analysis of variance; BL = blastocyst; CC = cumulus cells; GC = granulosa cells; GV = germinal vesicle; MII = metaphase II; mRNA = messenger ribonucleic acid; ND = nondetectable; RT-qPCR = quantitative reverse transcription polymerase chain reaction; SEM = standard error of the mean; TMPRSS2 = transmembrane serine protease 2.
Figure 5.
CTSL mRNA and protein expressions in human oocytes, embryos, and follicular cells. (A) CTSL mRNA expression analyzed by RT-qPCR in GV- and MII-stage oocytes, BL (n = 3 pools of 6 oocytes or embryos), CC, and GC (n = 3). Expression data were normalized to GAPDH and presented as a bar graph. (B) Capillary Western blot of total CTSL protein expression and (C) fold change in the abundance of CTSL protein (55 kDa) and possible variants (60 kDa) in MII-stage oocytes, 1C- and BL-stage embryos, CC, and GC (n = 3 replicates). Arrow indicates the target protein. All bar graphs show the mean ± SEM. Significant differences in expression between samples were determined using 1-way ANOVA followed by Tukey’s test. No significant difference was observed in CTSL mRNA and protein expressions present in the samples. 1C = 1-cell; ANOVA = analysis of variance; BL = blastocyst; CC = cumulus cells; CTSL = cathepsin L; GC = granulosa cells; GV = germinal vesicle; MII = metaphase II; mRNA = messenger ribonucleic acid; ND = nondetectable; RT-qPCR = quantitative reverse transcription polymerase chain reaction; SEM = standard error of the mean; TMPRSS2 = transmembrane serine protease 2.
Statistical Analysis
All data were analyzed with 1-way analysis of variance (ANOVA) using Prism software, version 8.2.1. Ct values obtained from quantitative reverse transcription polymerase chain reaction (RT-qPCR) were used to perform statistical analysis. The Western blot data were first checked for assumptions of normality and log-transformed before the analysis if required. Differences between means were determined using 1-way ANOVA followed by Tukey’s test. Data are presented as untransformed mean ± standard error of the mean.
Results
Antibody Validation
The protein lysates from the human lung, liver, ovary, colon, small intestine, white blood cells, HeLa cells, and HEK293T cells were used as positive controls to determine the specificity of all the antibodies and their compatibility with capillary electrophoresis-based immunodetection on the Jess system. We observed a specific signal for anti-ACE2 with lung and liver (100 kDa) (Supplemental Fig. 1A, available online), anti-TMPRSS2 with HeLa cells, liver, and small intestine (55 kDa) (Supplemental Fig. 1B), and anti-CD147 with colon (43, 60 kDa), ovary (60 kDa), and lung (60 kDa) (Supplemental Fig. 1C). Antibody for procathepsin L (38–42 kDa) and CTSL (25–35 kDa) was tested with HEK293T and HeLa cell lysates because it is a well-characterized positive control (25). We observed a higher-than-expected (54 kDa) MW (38 kDa) band with the positive controls in the capillary Western blot (Supplemental Fig. 1D), and therefore, a 54-kDa band observed in any target sample (oocyte, embryos, and follicular cells) was considered CTSL-positive and included in the analysis. In addition to the specific band, some of the cell and tissue lysates showed additional bands when probed with the antibody against ACE2 (53, 62, and130 kDa) and CD147 (43 kDa) (Supplemental Fig. 1). ACE2 protein consists of 805 amino acids and a predicted MW of 92.5 kDa. However, a glycosylated form (105–130 kD) and 2 truncated forms (63.9 kDa and 52.7 kDa) of ACE2 have been previously reported to be expressed in human tissues (26, 27, 28, 29) (Supplemental Fig. 2, available online). Similarly, CD147 (30, 31) and CTSL (32) are also known to have protein variants of different MWs resulting from alternative mRNA splicing or posttranslational modification; we identified these bands as probable protein variants (V) based on their respective MWs compared with those of reported variants of these proteins (Supplemental Table 3, available online).
To confirm that the origin of these bands was not a result of secondary antibody cross reactivity with target protein lysates, we used only a secondary antibody (without primary antibody) with each sample type as a negative control in the Jess analysis. The results demonstrated that the secondary antibody had no significant direct reactivity with any of the tissue, cell, oocyte, or embryo lysates used in this study (Supplemental Figs. 1 and 3, available online). With each run on the Jess system, the radioimmunoprecipitation assay buffer was used as a negative control for all the antibodies to avoid any false-positive chemiluminescent signal due to cross contamination (Supplemental Fig. 1).
ACE2 Receptors are Present in Human Oocytes and Preimplantation Embryos
The RT-qPCR analysis showed that the ACE2 transcript was present in both GV- and MII-stage human oocytes and was significantly reduced (P < .05) in the BL stage. ACE2 mRNA was also preferentially (P < .05) expressed in the oocytes than in CCs and GCs (Fig. 2 A). Analysis of ACE2 protein abundance in the oocytes, embryos, and follicular cells revealed the presence of ACE2 protein and/or varying variants in all the samples (Fig. 2B and C).We observed a 100-kDa MW band specific to the ACE2 protein in the MII oocytes, 1-cell (1C)-stage embryos, and BL-stage embryos, with no expression in CCs and GCs. A possible glycosylated form of approximately 130-kDa ACE2 (ACE2-V1) was barely detected in BLs and was present in significantly higher (P < .05) abundance in CCs but absent in the MII oocytes, zygotes, and GCs. Another 62-kDa variant (ACE2-V2) was present in both GCs and CCs but was completely absent in the MII-stage oocytes and embryos (1C and BL). Expression of an approximately 53-kDa variant (ACE2-V3) was observed in all the samples, but CC showed significantly lower abundance of ACE2-V3 compared with GC.
TMPRSS2 Protease is Also Present in Human Oocytes and Preimplantation Embryos
The results demonstrated no mRNA expression of TMPRRSS2 in CCs, GCs, or oocytes (GV and MII), but TMPRSS2 transcripts were present in human BLs (Fig. 3 A). In contrast, a specific band of TMPRSS2 protein (54 kDa) was observed in the MII oocytes, 1C- and BL-stage embryos, CC, and GC. The expression of TMPRSS2 was significantly (P < .05) higher in GC compared with that in CC (Fig. 3B and C).
Mature Oocytes and Embryos Lack Basigin (CD147) Protein
We observed significantly higher CD147 mRNA expression in the immature oocytes than in the MII oocytes, BL embryos, CC, and GC (Fig. 4 A). CD147 is an N-linked glycosylated protein and contains multiple glycosylation sites, which result in its migration between 43-kDa and 66-kDa MW in Western blot analysis (30). Our results demonstrated the presence of glycosylated CD147 (60 kDa) protein, the expression of which was significantly higher (P < .05) in GCs than in CCs and was completely undetected in the oocytes or embryos (Fig. 4B and C).
Procathepsin L and Cathepsin L Proteins are not Present in Human Oocytes and 1-cell Embryos
The RT-qPCR analysis revealed the expression of the CTSL transcript in the oocytes, embryos, and follicular cells (Fig. 5 A). The CTSL antibody used to analyze protein expression in all the target samples was designed to recognize both procathepsin L and CTSL proteins. Procathepsin L is an inactive zymogen of 38–42 kDa that undergoes autolysis to give mature forms (CTSL) of a 25–35-kDa enzyme. CTSL is a lysosomal endopeptidase expressed in most eukaryotic cells but has also been reported in the nucleus, with 34-, 55-, and 60-kDa molecular mass variants (32). Since the CTSL antibody used in this study resulted in a 55-kDa band with the HEK293T and HeLa cells used as positive controls in the capillary Western blot, we considered this molecular mass specific to the CTSL protein. Our results demonstrated that CTSL (55 kDa) was completely absent in the MII-stage oocytes and 1C embryos, but its expression was observed in the BL-stage embryos, CC, and GC. In addition, CC and GC also showed a 60-kDa variant (CTSL-V1) (Fig. 5B and C).
Discussion
Our results demonstrated that the ACE2 receptor protein and TMPRSS2 protease protein, both of which are crucial for SARS-CoV-2 infection of host cells, are present and coexpressed in mature human oocytes, zygotes, and BLs. This work demonstrated that human oocytes and embryos have the machinery necessary for infection in other tissues. However, we do not know whether the infection happens in vivo or in vitro; we only showed that the machinery to support the infection is present. An ACE2-independent receptor system, composed of the BSG (CD147) receptor protein and CTSL, was not expressed in oocytes, zygotes, or BLs, although CTSL alone was found in the BL-stage embryos. GCs and CCs of the ovary expressed all 4 of these proteins or their variants. However, both the receptor and protease were not abundantly coexpressed in CCs and GCs, suggesting that the likelihood of infection of the oocyte via CCs in the ovarian follicle is low.
We found that the ACE2 mRNA was mainly expressed in the oocytes, but the protein was present in the oocytes and embryos, suggesting the typical pattern of oocyte-expressed transcripts that are translated into protein to support early embryonic development and then degraded (33). The ACE2 mRNA has multiple transcript variants that have been predicted to encode a full-length protein of 92.4 kDa (Q9BYF1.1, NP_001358344.1), with an observed molecular mass of 100–130 kDa because of multiple sites of glycosylation (34) and truncated proteins of 90.3 kDa (XP_011543851.1), 88.8 kDa (XP_011543853.1), 79.4 kDa (XP_011543854.1), and 63.9 kDa (Q9BYF1.2) (35). Recently (36), a short 11-exon ACE2 novel isoform encoding a 52.7-kDa protein has also been reported to be exclusively expressed in the human airways, liver, and kidney. When the ACE2 antibody was tested with the different human tissues, we also observed this isoform in the human liver, lung, and small intestine. Our results demonstrated an approximately 53-kDa (ACE2-V3) protein in the oocytes, embryos, and CCs and approximately 62-kDa (ACE2-V2) protein in CCs and GCs. However, ACE2-V1 lacks the signal peptide required for SARS-CoV2 spike binding (36), and ACE2-V2 is a soluble isoform that lacks the transmembrane domain necessary for membrane attachment and SARS-CoV-2 entry (29, 37). Because both ACE2-V1 and ACE2-V2 are likely to be truncated isoforms lacking the key residues required for SARS-CoV-2 entry, we hypothesized that the presence of these proteins is not a probable viral entry point in oocytes, embryos, and ovarian somatic cells. CCs also had a 130-kDa ACE2 isoform, which might be the result of hyperglycosylation of ACE2 (100 kDa) (28). However, further studies using PNGase-mediated deglycosylation are required to confirm whether the higher-MW protein observed in CCs was because of glycosylation or cross reactivity of the antibody with a similar protein. The oocytes, zygotes, and BLs had a specific 100-kDa molecular mass of ACE2 protein along with TMPRSS2, demonstrating the presence of an optimum environment for SARS-CoV-2 infection. The presence of the TMPRSS2 protein without any detectable mRNA level in oocytes, zygotes, CCs, and GCs suggests that the protein is translated from mRNA present in the oocytes and ovarian somatic cells during early stages of follicular development (22).
Transcript and protein expressions of these genes in various reproductive tissues have been an area of interest since the COVID-19 pandemic began. A published RNA sequence database of a nonhuman primate ovarian tissue has described the expression of CD147 and CTSL in ovarian somatic cells, whereas ACE2 and TMPRSS2 expressions were restricted to germ cells (22). This is in agreement with our protein findings. Protein expression from 2 public databases, Human Protein Atlas and Human Proteome Map, found the presence of ACE2 protein in ovarian cells (22). Because we did not detect a specific band of ACE2 protein (100 kDa) in either CCs or GCs in this study, the protein expression reported in the ovarian somatic cells in the Human Protein Atlas and Proteome Map databases might have been derived from truncated protein variants, such as ACE2-V2 (62 kDa), ACE2-V3 (53 kDa), or glycosylated ACE2 (ACE2-130I) observed in this study. The higher abundance of CD147 mRNA in the immature oocytes than in the matured oocytes, embryos, and ovarian cell suggests that the CD147 transcript was degraded during the end of oocyte maturation because no protein was detected in the matured oocytes and early stage of the embryos. Despite the low mRNA abundance, the protein expressions of CD147 in CCs and GCs suggest that CD147 protein was translated during early stages of follicular development. The coexpression of CD147 receptor and CTSL proteases, analyzed as an alternative pathway for SARS-CoV-2 host entry, was absent in the oocytes and embryos but was found in CCs and GCs, with low abundance of CTSL. These results were in accordance with previously reported CD147 (22) and CTSL (38) protein expressions in human ovarian cells.
In men, a previously published single-cell transcriptome analysis has demonstrated low levels of ACE2 and TMPRSS2 expression in the testes, with little coexpression, suggesting that SARS-CoV-2 infection is unlikely to occur in testicular cells (39). Other transcriptomic studies have also found expression of ACE2 and TMPRSS2 in testicular cells, including elongated spermatids and spermatogonial stem cells, but again, coexpression was rare (22, 40, 41). BSG was widely expressed across testicular cell types and was coexpressed with CTSL in early and late primary spermatocytes and to a lesser degree in Leydig cells, myoid cells, endothelial cells, and differentiating spermatogonia (22). An analysis of protein expression in the testicular tissue using the 2 existing databases showed the presence of ACE2 and BSG in both the databases, whereas TMPRSS2 was not detected, and CTSL was detected in 1 of the 2 databases (22). This information suggests that SARS-CoV-2 is unlikely to infect testicular cells using the traditional ACE2/TMPRSS2 mechanism but that the testes may be susceptible to the ACE2-independent mechanism of infection. It remains unclear whether the virus is present in the semen and whether it can potentially be sexually transmitted. In patients in the recovery phase after COVID-19 and in a man who died of the disease, no SARS-CoV-2 viral RNA was detected in the semen (39, 42). In a separate study, 6 of 38 men (15.8%) in both acute and recovery phases tested positive for SARS-CoV-2 in their semen (43).
There are several specific concerns while operating in an ART laboratory during the SARS-CoV-2 pandemic, namely, whether oocytes can be infected at retrieval, whether embryos can be infected while in the laboratory, and whether embryos can infect other embryos during cryostorage. To minimize the risk of viral infection while retrieving oocytes, most practices have implemented testing patients before egg retrieval. However, viral RNA amplification tests can be influenced by viral load, sample site, collection method, and specimen shipment, resulting in potential false-negative results. The current antibody-based test lacks sensitivity and specificity until many days after symptom onset. These limitations create confusion about the utility of the patient test and increase the possibility of treating infected patients even if the test is in place (44). Coupled with the existing delays in obtaining test results, it is difficult for clinics to guarantee that no infected patients undergo IVF. Given these limitations and based on the prevalence of asymptomatic people and our findings that the mechanism necessary for SARS-CoV-2 infection is present, oocytes and embryos handled in laboratories during the pandemic should be treated with special precautions in the same manner as those for oocytes and embryos from patients infected with other viruses. Currently, no guidelines exist for doing so for patients infected with SARS-CoV-2 undergoing infertility treatment or with oocyte or sperm donors (45), although this may be a relevant safety precaution to implement. To avoid the infection of embryos during their time in the laboratory, a staff-testing program may be implemented, although it carries the same limitations as that for patients. Certainly, a rigorous protocol over and above normal safety measures for cleaning should be in place to prevent SARS-CoV-2 from entering IVF laboratories. However, the SARS-CoV-2 genome was not detected in oocytes analyzed from infected women (38) despite the cellular machinery required for SARS-CoV2 infection being present in the oocytes. This suggests that the zona pellucida can serve as a protective barrier for oocytes in vivo. This is particularly important in IVF procedures because the zona pellucida, which may serve as a viral barrier, is breached during treatment if intracytoplasmic sperm injection and/or biopsy are performed. Laboratories are already equipped to safely store cryopreserved samples in a way that prevents cross contamination by previously existing pathogens (46). The risk of cross contamination with SARS-CoV-2 between embryos during cryostorage in modern, closed cryodevices and liquid nitrogen-vapor storage containers may be virtually negligible (47) despite the presence of the receptor and protease required for the viral infection, as we have demonstrated. However, the possibility to reinfect a mother and respread the virus at a later date after warming and transfer of an infected embryo cannot be ignored. Thus, robust patient- and staff-testing programs in addition to rigorous laboratory protocols to prevent the infection are critical pieces of practicing IVF during the SARS-CoV-2 pandemic. In the current scenario, IVF laboratory directors must make all of these decisions without much information because we are still in the process of learning about this virus and how it may affect reproduction. In fact, it is this lack of information that has prevented professional societies from establishing their own SARS-CoV-2-specific recommendations (48).
Given the presence of the mechanism of SARS-CoV-2 infection in oocytes and embryos that we have described, some patients might wish to postpone infertility treatment until the pandemic is better controlled. However, it has now been widely recognized that the postponement of treatment is detrimental for some patients, although a recent study has found no decrease in live birth rate among patients with diminished ovarian reserve who initiated treatment immediately or delayed treatment for up to 6 months (49). Assisted reproduction, in the form of IVF and frozen embryo transfer, offers an opportunity to postpone pregnancy to avoid the disease during gestation while respecting the importance of time in successful treatment (46). This approach does not completely avoid all potential risks, such as active infection in the patient during egg retrieval or infection of the embryo in vitro or during cryostorage. However, this strategy may be a viable alternative because it postpones pregnancy until we have more information about the effects of the disease during gestation on the fetus, until the pandemic is less of a threat to pregnant women, or until there is a safe and effective vaccine for both mothers and babies.
Acknowledgments
The authors thank Rachel West, Ph.D., for assistance with protein cell lysates to test the antibodies and helpful discussion.
Footnotes
S.K.R. has nothing to disclose. D.M.L. has nothing to disclose. B.K. has nothing to disclose. H.J.E. has nothing to disclose. B.G. has nothing to disclose. S.K. has nothing to disclose. J.S. has nothing to disclose. S.M. has nothing to disclose. W.B.S. has nothing to disclose. Y.Y. has nothing to disclose. R.L.K. has nothing to disclose.
Supported by the Colorado Center for Reproductive Medicine.
Supplementary data
Antibody validation for SARS-CoV-2 virus host entry receptors (ACE2 and CD147) and proteases (TMPRSS2 and CTSL) using human tissue and cell lysates. Liver, lung, small intestine, colon, and ovary tissues and HEK293T and HeLa cells were used as positive controls to test all the target antibodies. (A) ACE2 antibody showed a specific band of 100 kDa in the lung and liver, a 130-kDa variant in the liver, and 53- and 62-kDa variants in the lung and small intestine. (B) HeLa cell, liver, and small intestine protein lysates showed a specific band of 54 kDa with TMPRSS2 antibody. (C) Glycosylated forms of CD147 protein were observed, with a molecular mass of 43 and 60 kDa in the colon and 60 kDa in the ovary and lung. (D) HEK293T and HeLa cell protein lysates showed a 54-kDa band with CTSL antibody. None of the protein lysates used as a positive control showed significant reactivity with only secondary antibody. RIPA buffer showed no chemiluminescent signal with any of the target antibodies. Red boxes indicate the target proteins. ACE2 = angiotensin 1-converting enzyme 2; CTSL = cathepsin L; RIPA = radioimmunoprecipitation assay; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; TMPRSS2 = transmembrane serine protease 2.
Supplemental Figure 2. Block diagram of ACE2 gene and mRNA and protein variants. The ACE2 gene contains 18 exons (E) (yellow), 3 mRNA splice variants (E1–18, E1–12, and E10–18), and downstream protein variants. Full-length ACE2 protein comprises an N-terminal signal peptide (gray), an ectodomain (green), a transmembrane domain (orange), and a C-terminal cytoplasmic domain (blue), with a predicted molecular weight of 92.5 kDa. ACE2 protein has a total of 7 N-glycosylation sites (N53, 90, 103, 332, 432, and 546) and 3 O-glycosylation sites (O155, 496, and 730), which can result in 105–130-kDa molecular weight depending upon the glycosylation level. The protein variant resulting from E1–12 lacks the transmembrane and cytoplasmic domains and is, therefore, referred to as soluble ACE2 (63.9 kDa). Short ACE2 (52.7 kDa) is another protein variant resulting from E10–18 that lacks the signal peptide and ectodomain parts, which are critical for interaction with SARS-CoV-2. ACE2 = angiotensin 1-converting enzyme 2; mRNA = messenger ribonucleic acid.
Supplemental Figure 3. Validation of oocyte, embryo, and ovarian follicular cell lysates for false-positive signals. (A) MII oocyte, 1C- and BL-stage embryos, and CC and GC protein lysates were analyzed on Jess in the absence of a primary antibody. No false-positive chemiluminescent signal was observed as a result of cross reactivity of total protein lysates with a secondary antibody. (B) Autogenerated electropherogram by Compass software showing area under the blue line as total protein abundance of MII, 1C, BL, CC, and GC samples. 1C = 1-cell; BL = blastocyst; CC = cumulus cell; GC = granulosa cell; MII = metaphase II.
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anifandis G., Messini C.I., Daponte A., Messinis I.E. COVID-19 and fertility: a virtual reality. Reprod BioMed Online. 2020;41:157–159. doi: 10.1016/j.rbmo.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arav A. A recommendation for IVF lab practice in light of the current COVID-19 pandemic. J Assist Reprod Genet. 2020;37:1543. doi: 10.1007/s10815-020-01841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alviggi C., Esteves S.C., Orvieto R., Conforti A., La Marca A., Fischer R. COVID-19 and assisted reproductive technology services: repercussions for patients and proposal for individualized clinical management. Reprod Biol Endocrinol. 2020;18:45. doi: 10.1186/s12958-020-00605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elshabrawy H.A., Fan J., Haddad C.S., Ratia K., Broder C.C., Caffrey M. Identification of a broad-spectrum antiviral small molecule against severe acute respiratory syndrome coronavirus and Ebola, Hendra, and Nipah viruses by using a novel high-throughput screening assay. J Virol. 2014;88:4353–4365. doi: 10.1128/JVI.03050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jing Y., Run-Qian L., Hao-Ran W., Hao-Ran C., Ya-Bin L., Yang G. Potential influence of COVID-19/ACE2 on the female reproductive system. Mol Hum Reprod. 2020;26:367–373. doi: 10.1093/molehr/gaaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan P.P., Zhan Q.T., Le F., Zheng Y.M., Jin F. Angiotensin-converting enzymes play a dominant role in fertility. Int J Mol Sci. 2013;14:21071–21086. doi: 10.3390/ijms141021071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goad J., Rudolph J., Rajkovic A. Female reproductive tract has low concentration of SARS-CoV2 receptors. bioRxiv. 2020;15 doi: 10.1371/journal.pone.0243959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R., Yin T., Fang F., Li Q., Chen J., Wang Y. Potential risks of SARS-CoV-2 infection on reproductive health. Reprod Biomed Online. 2020;41:89–95. doi: 10.1016/j.rbmo.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colaco S., Chhabria K., Singh D., Bhide A., Singh N., Singh A. A single-cell RNA expression map of coronavirus receptors and associated factors in developing human embryos. Cell Rep. 2020;32:108175. doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weatherbee B.A.T., Glover D.M., Zernicka-Goetz M. Expression of SARS-CoV-2 receptor ACE2 and the protease TMPRSS2 suggests susceptibility of the human embryo in the first trimester. Open Biol. 2020;10 doi: 10.1098/rsob.200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Wang Y., Luo W., Huang L., Xiao J., Li F. A comprehensive investigation of the mRNA and protein level of ACE2, the putative receptor of SARS-CoV-2, in human tissues and blood cells. Int J Med Sci. 2020;17:1522–1531. doi: 10.7150/ijms.46695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Sousa Abreu R., Penalva L.O., Marcotte E.M., Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan J.K., Wong C.S., Ku W.T., Kwan M.Y. Reflections on the use of controls in immunohistochemistry and proposal for application of a multitissue spring-roll control block. Ann Diagn Pathol. 2000;4:329–336. doi: 10.1053/adpa.2000.17892. [DOI] [PubMed] [Google Scholar]
- 19.Leong A.S. Pitfalls in diagnostic immunohistology. Adv Anat Pathol. 2004;11:86–93. doi: 10.1097/00125480-200403000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Hammond M.E., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuovo G.J. Elsevier; Amsterdam; Boston: 2013. In situ molecular pathology and co-expression analyses. [Google Scholar]
- 22.Stanley K.E., Thomas E., Leaver M., Wells D. Coronavirus disease-19 and fertility: viral host entry protein expression in male and female reproductive tissues. Fertility and Sterility. 2020;114:33–43. doi: 10.1016/j.fertnstert.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajput S.K., Yang C., Ashry M., Folger J.K., Knott J.G., Smith G.W. Role of bone morphogenetic protein signaling in bovine early embryonic development and stage specific embryotropic actions of follistatindagger. Biol Reprod. 2020;102:795–805. doi: 10.1093/biolre/ioz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhenhua G., Rajput S.K., Folger J.K., Di L., Knott J.G., Smith G.W. Pre- and peri-/post-compaction follistatin treatment increases in vitro production of cattle embryos. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavcic N., Butinar M., Sobotic B., Hafner Cesen M., Petelin A., Bojic L. Intracellular cathepsin C levels determine sensitivity of cells to leucyl-leucine methyl ester-triggered apoptosis. FEBS J. 2020;287:5148–5166. doi: 10.1111/febs.15326. [DOI] [PubMed] [Google Scholar]
- 26.Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wysocki J., Schulze A., Batlle D. Novel variants of angiotensin converting enzyme-2 of shorter molecular size to target the kidney renin angiotensin system. Biomolecules. 2019;9:886. doi: 10.3390/biom9120886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lew R.A., Warner F.J., Hanchapola I., Smith A.I. Characterization of angiotensin converting enzyme-2 (ACE2) in human urine. Int J Pept Res Ther. 2006;12:283–289. doi: 10.1007/s10989-006-9031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rehman S.U., Tabish M. Alternative splicing of ACE2 possibly generates variants that may limit the entry of SARS-CoV-2: a potential therapeutic approach using SSOs. Clin Sci (Lond) 2020;134:1143–1150. doi: 10.1042/CS20200419. [DOI] [PubMed] [Google Scholar]
- 30.Bai Y., Huang W., Ma L.T., Jiang J.L., Chen Z.N. Importance of N-glycosylation on CD147 for its biological functions. Int J Mol Sci. 2014;15:6356–6377. doi: 10.3390/ijms15046356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao C.G., Kong L.M., Song F., Xing J.L., Wang L.X., Sun Z.J. Characterization of basigin isoforms and the inhibitory function of basigin-3 in human hepatocellular carcinoma proliferation and invasion. Mol Cell Biol. 2011;31:2591–2604. doi: 10.1128/MCB.05160-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puchi M., Garcia-Huidobro J., Cordova C., Aguilar R., Dufey E., Imschenetzky M. A new nuclear protease with cathepsin L properties is present in HeLa and Caco-2 cells. J Cell Biochem. 2010;111:1099–1106. doi: 10.1002/jcb.22712. [DOI] [PubMed] [Google Scholar]
- 33.Evsikov A.V., Marin de Evsikova C. Gene expression during the oocyte-to-embryo transition in mammals. Mol Reprod Dev. 2009;76:805–818. doi: 10.1002/mrd.21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 35.Manna V.J., Caradonna S.J. The heterogeneous nature of the Coronavirus receptor, angiotensin-converting enzyme 2 (ACE2) in differentiating airway epithelia. EBioMedicine. 2020;60:102976. doi: 10.1016/j.bbadva.2022.100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blume C., Jackson C.L., Spalluto C.M., Legebeke J., Nazlamova L., Conforti F. A novel isoform of ACE2 is expressed in human nasal and bronchial respiratory epithelia and is upregulated in response to RNA respiratory virus infection. bioRxiv. 2020 doi: 10.1101/2020.07.31.230870. [DOI] [PubMed] [Google Scholar]
- 37.Batlle D., Wysocki J., Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci (Lond) 2020;134:543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 38.Garcia V., Kohen P., Maldonado C., Sierralta W., Munoz A., Villarroel C. Transient expression of progesterone receptor and cathepsin-l in human granulosa cells during the periovulatory period. Fertil Steril. 2012;97:707–713.e1. doi: 10.1016/j.fertnstert.2011.12.039. [DOI] [PubMed] [Google Scholar]
- 39.Pan F., Xiao X., Guo J., Song Y., Li H., Patel D.P. No evidence of severe acute respiratory syndrome-coronavirus 2 in semen of males recovering from coronavirus disease 2019. Fertil Steril. 2020;113:1135–1139. doi: 10.1016/j.fertnstert.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma S., Saksena S., Sadri-Ardekani H. ACE2 receptor expression in testes: implications in COVID-19 pathogenesis. Biol Reprod. 2020;103:449–451. doi: 10.1093/biolre/ioaa080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song C., Wang Y., Li W., Hu B., Chen G., Xia P. Absence of 2019 novel coronavirus in semen and testes of COVID-19 patients†. Biol Reprod. 2020;103:4–6. doi: 10.1093/biolre/ioaa050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li D., Jin M., Bao P., Zhao W., Zhang S. Clinical characteristics and results of semen tests among men with coronavirus disease 2019. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.La Marca A., Nelson S.M. SARS-CoV-2 testing in infertile patients: different recommendations in Europe and America. J Assist Reprod Genet. 2020:1–6. doi: 10.1007/s10815-020-01887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segars J., Katler Q., McQueen D.B., Kotlyar A., Glenn T., Knight Z. Prior and novel coronaviruses, Coronavirus Disease 2019 (COVID-19), and human reproduction: what is known? Fertil Steril. 2020;113:1140–1149. doi: 10.1016/j.fertnstert.2020.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alteri A., Pisaturo V., Somigliana E., Vigano P. Cryopreservation in reproductive medicine during the COVID-19 pandemic: rethinking policies and European safety regulations. Hum Reprod. 2020;35:2650–2657. doi: 10.1093/humrep/deaa210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pomeroy K.O., Schiewe M.C. Cryopreservation and IVF in the time of Covid-19: what is the best good tissue practice (GTP)? J Assist Reprod Genet. 2020:1–6. doi: 10.1007/s10815-020-01904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hickman C., Rogers S., Huang G., MacArthur S., Meseguer M., Nogueira D. Managing the IVF laboratory during a pandemic: international perspectives from laboratory managers. Reprod Biomed Online. 2020;41:141–150. doi: 10.1016/j.rbmo.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romanski P.A., Bortoletto P., Rosenwaks Z., Schattman G.L. Delay in IVF treatment up to 180 days does not affect pregnancy outcomes in women with diminished ovarian reserve. Hum Reprod. 2020;35:1630–1636. doi: 10.1093/humrep/deaa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Antibody validation for SARS-CoV-2 virus host entry receptors (ACE2 and CD147) and proteases (TMPRSS2 and CTSL) using human tissue and cell lysates. Liver, lung, small intestine, colon, and ovary tissues and HEK293T and HeLa cells were used as positive controls to test all the target antibodies. (A) ACE2 antibody showed a specific band of 100 kDa in the lung and liver, a 130-kDa variant in the liver, and 53- and 62-kDa variants in the lung and small intestine. (B) HeLa cell, liver, and small intestine protein lysates showed a specific band of 54 kDa with TMPRSS2 antibody. (C) Glycosylated forms of CD147 protein were observed, with a molecular mass of 43 and 60 kDa in the colon and 60 kDa in the ovary and lung. (D) HEK293T and HeLa cell protein lysates showed a 54-kDa band with CTSL antibody. None of the protein lysates used as a positive control showed significant reactivity with only secondary antibody. RIPA buffer showed no chemiluminescent signal with any of the target antibodies. Red boxes indicate the target proteins. ACE2 = angiotensin 1-converting enzyme 2; CTSL = cathepsin L; RIPA = radioimmunoprecipitation assay; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; TMPRSS2 = transmembrane serine protease 2.
Supplemental Figure 2. Block diagram of ACE2 gene and mRNA and protein variants. The ACE2 gene contains 18 exons (E) (yellow), 3 mRNA splice variants (E1–18, E1–12, and E10–18), and downstream protein variants. Full-length ACE2 protein comprises an N-terminal signal peptide (gray), an ectodomain (green), a transmembrane domain (orange), and a C-terminal cytoplasmic domain (blue), with a predicted molecular weight of 92.5 kDa. ACE2 protein has a total of 7 N-glycosylation sites (N53, 90, 103, 332, 432, and 546) and 3 O-glycosylation sites (O155, 496, and 730), which can result in 105–130-kDa molecular weight depending upon the glycosylation level. The protein variant resulting from E1–12 lacks the transmembrane and cytoplasmic domains and is, therefore, referred to as soluble ACE2 (63.9 kDa). Short ACE2 (52.7 kDa) is another protein variant resulting from E10–18 that lacks the signal peptide and ectodomain parts, which are critical for interaction with SARS-CoV-2. ACE2 = angiotensin 1-converting enzyme 2; mRNA = messenger ribonucleic acid.
Supplemental Figure 3. Validation of oocyte, embryo, and ovarian follicular cell lysates for false-positive signals. (A) MII oocyte, 1C- and BL-stage embryos, and CC and GC protein lysates were analyzed on Jess in the absence of a primary antibody. No false-positive chemiluminescent signal was observed as a result of cross reactivity of total protein lysates with a secondary antibody. (B) Autogenerated electropherogram by Compass software showing area under the blue line as total protein abundance of MII, 1C, BL, CC, and GC samples. 1C = 1-cell; BL = blastocyst; CC = cumulus cell; GC = granulosa cell; MII = metaphase II.