Abstract
SARS-CoV-2 has a high transmission rate and shows frequent mutations, thus making vaccine development an arduous task. However, researchers around the globe are working hard to find a solution e.g. synthetic vaccine. Here, we have performed genome-wide analysis of 566 Indian SARS-CoV-2 genomes to extract the potential conserved regions for identifying peptide based synthetic vaccines, viz. epitopes with high immunogenicity and antigenicity. In this regard, different multiple sequence alignment techniques are used to align the SARS-CoV-2 genomes separately. Subsequently, consensus conserved regions are identified after finding the conserved regions from each aligned result of alignment techniques. Further, the consensus conserved regions are refined considering that their lengths are greater than or equal to 60nt and their corresponding proteins are devoid of any stop codons. Subsequently, their specificity as query coverage are verified using Nucleotide BLAST. Finally, with these consensus conserved regions, T-cell and B-cell epitopes are identified based on their immunogenic and antigenic scores which are then used to rank the conserved regions. As a result, we have ranked 23 consensus conserved regions that are associated with different proteins. This ranking also resulted in 34 MHC-I and 37 MHC-II restricted T-cell epitopes with 16 and 19 unique HLA alleles and 29 B-cell epitopes. After ranking, the consensus conserved region from NSP3 gene is obtained that is highly immunogenic and antigenic. In order to judge the relevance of the identified epitopes, the physico-chemical properties and binding conformation of the MHC-I and MHC-II restricted T-cell epitopes are shown with respect to HLA alleles.
Keywords: B-cell epitopes, Conserved regions, SARS-CoV-2, Peptide based vaccine, Physico-chemical properties, T-cell epitopes
1. Introduction
In December 2019, China reported a sudden outbreak of pneumonia due to an unknown source in Hubei province, Wuhan city [1] which later got attributed to a virus named SARS-CoV-2. SARS-CoV-2 belongs to the family of Coronaviridae which also houses SARS-CoV-1 [2], [3] and MERS-CoV [4] virus. Genomic sequence analysis of the newly reported virus was found to be highly similar to that of SARS-CoV (95%–100%), thus showing the evolutionary similarity between SARS-CoV and SARS-CoV-2 [5]. By October 2020, India has registered over 7.65 million cases [6], making it one of the most affected countries in the world. Symptoms of the COVID-19 vary from fever, cough, myalgia, dyspnoea and diarrhoea to severe respiratory distress which may require life support systems. In severe cases, it may even lead to death [7]. Considering these consequences, World Health Organisation (WHO) suggested to interrupt human–human contact in the form of total lock downs along with precautionary measures such as face masks and hand sanitizers to control the spread of COVID-19. Hence, it is the need of the hour to find a cure for COVID-19 in the form of vaccine.
Classical methods of vaccine design like attenuation of the virus through external sources such as micro-organisms to mitigate its harm or virulence usually depends on the response of the virus itself. Sometimes mutations in the virus genome can result in autoimmune response eventually making the virus even more virulent. Hence, such classic vaccine design approaches are time consuming, expensive and may not provide an effective response. With the evolution in bioinformatics and genome analysis, it is now possible to study the DNA, RNA and molecular evolution of a virus which can aid in development of vaccine through approaches such as reverse vaccinology. Reverse vaccinology involves pinpointing the protein sites that results into synthetic peptide based vaccines [8], [9]. The preparation of epitope based vaccine is carried out in sequential form, starting from scanning the genome of the pathogen to locating the surface proteins, followed by extracting the best epitopes situated on the surface and also testing these synthetic designs against any autoimmune response [9]. The antigens provided by the epitopes are the sites to which antibodies bind, hence selection of the best epitopes is one of the crucial and foremost steps in vaccine design. In regard to this, Skwarczynski et al. [8] have suggested several factors which influence the selection of epitopes, such as immune response to the pathogen, hypersensitivity responses and coverage of different peptide against different pathogen subtypes. Further, these epitopes can be classified into two classes i.e. MHC-I, MHC-II associated T-cell epitopes [10] and B-cell epitopes [11] based on their responses against recognized foreign pathogens. The antigens provided by MHC-I interact directly with the CD8 cells evoking the cellular response [8]. MHC-II antigens bind to the surface of the pathogens to initiate the T-helper cells (CD4) which are responsible for activating the Th1 and Th2 type helper cells in the form of cytotoxic T-lymphocyte (CTL) and humoral response through antigens loaded in MHC-I and B-cell epitopes. Hence, the selection of T-cell and B-cell epitopes is a crucial process in order to provide a reliable vaccine.
By considering the several advantages presented in form of peptide-based vaccine, many studies have been carried out to design a vaccine in order to provide a stable solution against the threat as presented by SARS-CoV-2 virus. Earlier, it was found that spike (S) glycoprotein of SARS-CoV-2 can act as an intermediary to bind to the host cells with a very strong affinity, thus eventually attracting various experiments towards targeting this protein site as the potential target for vaccine design and diagnostics [12]. Following this, many types of vaccine designs have been proposed based on RNA, vectored, recombinant protein sequence and cell-cultures while focusing on the spike protein or whole virion [13]. Additionally, in Lin et al. [14] heptad repeats 1 and 2 (HR1 and HR2) in the spike protein have been predicted followed by the peptides with the help of molecular dynamics simulation between the fusion of the viral membrane and the host cell membrane, eventually limiting the spread of the virus within the host cells. Another study carried out by Vashi et al. [15] predicted 24 potential epitope fragments of which 20 were on the surface of spike protein. This information can be helpful for designing potential immunogenic peptide-based vaccines. Similar study has been conducted by Rakib et al. [16] in which spike protein region has been analysed through multiple sequence analysis in different SARS-CoV-2 genomes to predict the most immunogenic peptide fragments. In this study, a multi-epitope based vaccine has been proposed through analysing the S1 and S2 domains of spike proteins of the SARS-CoV-2 genomes in order to provide the best epitopes [17] for designing a vaccine. However, it is important to note that other protein sites can also be targeted for vaccine design as well [18]. This depends on how the T-cell interacts inside the different protein region of SARS-CoV-2. Grifoni et al. [18] have identified that 70–100% of epitope pools detect CD8 and CD4 T-cells for SARS-CoV-2. CD4+ cells interact with the other proteins like membrane (M), nucleocapsid (N) and ORF1ab proteins like NSP3, NSP4 and NSP12, but the dominance of CD4+ cells is very high within the spike region. On the other hand, no such dominant reactivity was identified in case of CD8+ cells in spike protein region. Hence, MHC-I restricted epitopes derived from M, NSP6, ORF3a or N proteins can also be considered for vaccine design. Noorimotlagh et al. [19] have conducted a review on several papers and have inferred a set of T-cell and B-cell epitopes from the Spike and Nucleocapsid proteins with high antigenicity. Genomic analysis conducted by Yadav et al. [20] on the first two cases reported in India resulted in the introduction of two non-identical strains of SARS-CoV-2. With time, more mutation points have been discovered [21] as well. This alteration in the protein region of the genome can lead to vaccine failures as was noticed in the case of Influenza virus in 2013–14 [22]. Hence, stable vaccine design is the need of the hour. Moreover, for such RNA viruses which undergo rapid mutations, Nandy et al. [9] have suggested the extraction of genomic regions which are either not influenced or very less influenced by the process of mutation. This can be carried out by analysing large set of virus genomes with the help of sequence alignment techniques. Such similar regions inside different viral genomes can be then considered for synthetic peptide vaccine designs. In [23], Gupta et al. have developed a web resource “CoronaVR” and have identified a set of T-cell and B-cell epitopes that can be incorporated in vaccine design. On the other hand, Crooke et al. [24] have used available algorithms and webtools to identify 41 T-cell epitopes (5 HLA class I, 36 HLA class II) and 6 B-cell epitopes as probable targets for epitope-based vaccine design. Ong et al. [25] have used Vaxign and the recently developed Vaxign-ML reverse vaccinology tools to predict potential vaccine candidates for COVID-19. Apart from Spike, they have identified epitopes derived from NSP3, 3CL-pro, NSP8, NSP9 and NSP10 proteins to be highly likely candidates for vaccine design. There are other works like [26], [27], [28], [29], [30], [31], [32], [33] as well pertaining to epitope identification in SARS-CoV-2 for vaccine design.
In the above discussed literature, prediction of epitopes has been performed by analysing the virus proteins whereas genetical mutations are the primary reason for change in structure of the virus proteins. This fact motivated us to analyse the 566 available Indian SARS-CoV-2 genomes to identify the conserved regions to predict the immunogenic and antigenic epitopes. For this purpose, we have used four different multiple sequence alignment techniques viz. ClustalW [34], MUSCLE [35], ClustalO [36], [37] and MAFFT [38] to align the sequences. Consensus conserved regions (CCnR) are then identified after finding the conserved regions from each aligned results of the alignment techniques. Further, these conserved regions are filtered on the basis of (a) length should be greater than or equal to 60nt and (b) corresponding protein sequence should not have any stop codons. This is followed by the validation of specificity of the conserved regions as query coverage with the help of Nucleotide BLAST [39]. These filtered conserved regions are then used to identify the T-cell and B-cell epitopes based on their immunogenic and antigenic scores. Thereafter, these scores are used to rank the conserved regions. As a result, we have obtained 23 conserved regions encompassing NSP1, NSP2, NSP3, NSP4, 3CL-Proteinase, NSP10, RNA-directed RNA polymerase, Helicase, Spike glycoprotein and Nucleocapsid protein. Subsequently, the consensus conserved region in NSP3 gene has been found to be highly immunogenic and antigenic. It provides MHC-I and MHC-II restricted T-cell epitopes and B-cell epitopes, FLKKDAPYI, ITFLKKDAPYIVGDV, TLVSDIDITFLKKDAP as immunogenic and TAVVIPTKK, IDITFLKKDAPYIVG, LHPDSATLVSDIDITF as antigenic respectively. Also, different immunogenic and antigenic epitopes associated to other conserved regions are provided as well. Finally, to validate the identified epitopes, the conformational 2D non-covalent structure of the chosen epitopes is studied. Moreover, the physico-chemical properties of the epitopes along with Ramachandran plot and Z-scores are also reported in the paper.
2. Materials and methods
In this section, at first the data preparation is elaborated followed by the discussion on the pipeline of the proposed work. For the benefit of the readers, brief discussions on epitope based vaccine, T-cell and B-cell epitopes and their prediction tools, physico-chemical properties of epitopes and docking of T-cell epitopes are given in the supplementary file. Moreover, prediction tools for T-cell and B-cell epitopes are reported in Supplementary Tables S1 and S2.
2.1. Data preparation
In order to map the SARS-CoV-2 proteins, we have used the reference SARS-CoV-2 genome (NC_045512.2)2 and 44583 available protein sequences from the National Center for Biotechnology (NCBI). To generate the protein sequence, we have taken the reference sequence of SARS-CoV-2 genome and considered the reading frame concepts. A reading frame divides the sequence of nucleotides of the reference sequence into a set of successive, non-overlapping triplets. There are three possible reading frames: Frame 1 which starts from the first nucleotide of a reference sequence and creates the triplets, Frame 2 which starts from the second nucleotide and creates the triplets and Frame 3 which starts from the third nucleotide and creates the triplets. For each frame, these triplets are then translated into the corresponding proteins based on the codon table3 . Finally, we have obtained 25 such unique proteins which were best matched to Frame 2. Also, the recent genomic sequences of Indian SARS-CoV-2 virus have been collected from Global Initiative on Sharing All Influenza Data (GISAID)4 in fasta format. It contains 566 complete and near complete genomes with sequence ID. The average length of the 566 genomes is 29,831 bp. These 566 SARS-CoV-2 sequences are aligned using multiple sequencing alignment (MSA) techniques to extract the conserved regions. Also, the coded protein associated to each conserved region are extracted. For the alignment of sequences, High Performance Computing (HPC) facility of NITTTR, Kolkata is used. The HPC cluster has a master node with dual Intel Xeon Gold 6130 Processor having 32 Cores, 2.10 GHz, 22 MB L3 Cache and 128 GB DDR4 RAM and 2 GPU and 4 CPU computing nodes with dual Intel Xeon Gold 6152 Processor having 44 Cores, 2.1 GHz, 30 MB L3 Cache and 192 GB DDR4 RAM each, while GPU nodes have NVIDIA Tesla V100 GPU with 16 GB memory each. MSA was performed using the 2 GPU and 4 CPU computing nodes.
2.2. Pipeline of the workflow
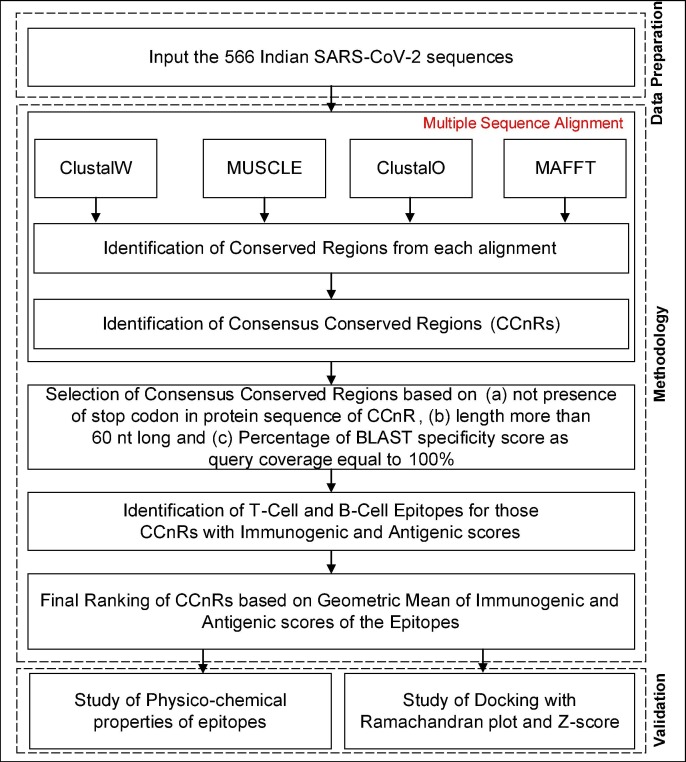
The pipeline of the workflow is shown in Fig. 1 . To start with, we have focused on finding the conserved regions in the 566 Indian SARS-CoV-2 genome sequence which are not affected by genetic mutations. For the same, initially we have constructed a Consensus Multiple Sequence Alignment (CMSA) approach in which we have used four different alignment techniques: ClustalW, MUSCLE, ClustalO and MAFFT in order to align the 566 SARS-CoV-2 sequences. Subsequently, consensus conserved regions (CCnR) are identified after finding the conserved regions from each aligned result of alignment techniques. ClustalW initially performs pairwise alignment of all sequences by using the k-tuple method. Thereafter, MSA is created by progressively aligning the most closely related sequences based on Neighbor-Joining guide tree method. In MUSCLE technique, two distance measures are used: k-mer for unaligned pairs and Kimura method for aligned pairs of sequences. Initially, a draft MSA is produced in MUSCLE using the k-mer method. Then, a progressive alignment is constructed based on the guide tree as produced by the UPGMA method. This initial tree is then re-estimated using the Kimura distance method after which UPGMA method is once again used to produce a new guide tree, thereby creating a second MSA. New MSAs are finally created by realigning the two sequences created previously. ClustalO uses the k-tuple method to produce pairwise alignment. Then mBed is used to cluster the sequences followed by k-means clustering algorithm. Next, the guide tree is built using Unweighted Pair Group Method with Arithmetic Mean (UPGMA) method. Finally, MSA is constructed using the HHalign package. MAFFT uses two different heuristic methods, progressive (FFT-NS-2) and iterative refinement (FFT-NS-i). The main aim of MAFFT is to merge local and global algorithms for MSA. Initially, FFT-NS-2 is used to calculate all-pairwise distances to create a provisional MSA from which refined distances are calculated. Then, FFT-NS-i is performed to get the final MSA. Thereafter, to identify the conserved regions, these aligned sequences are used to compute the entropy(E).
| (1) |
where Sx y indicates the frequency of each residue x occurring at position y and 5 represents the four possible residues as nucleotide plus gap. To identify the conserved regions (CnRs) for each alignment technique, a minimum segment length of 15 is considered with maximum average entropy as 0.2. Further, maximum entropy per position is taken as 0.2 with no gaps after finding the consensus sequence for the 566 genomic sequences. All these values are taken after following the literature. Thereafter, the CCnRs are identified considering the CnRs of all the alignment techniques. Next, a refinement process is carried out for the CCnRs based on the criteria that their length is greater then or equal to 60nt and no stop codon is present in the associated protein sequence. Moreover, Nucleotide BLAST is used to verify the specificity of the CCnRs as query coverage as well. Subsequently, T-cell and B-cell epitopes are identified from these CCnRs. To predict the T-cell and B-cell epitopes and to find their corresponding immunogenic scores, each CCnR is subjected to IEDB5 and ABCPred6 respectively. As recommended by IEDB, for the prediction of MHC-I and MHC-II T-cell epitopes, NetMHCpan7 and Consensus Approach8 [40] are selected respectively whereas for B-cell epitopes, prediction is carried out by ABCPred which uses Recurrent Neural Network. Then, by using the predicted epitopes, antigenic scores are calculated with the help of VaxiJen2.09 . For each CCnR, multiple T-cell and B-cell epitopes are identified along with their corresponding immunogenic and antigenic scores. Subsequently, for each CCnR the highest immunogenic and antigenic scores are considered to select the corresponding epitopes. Furthermore, these scores are used to rank the CCnRs based on geometric mean as given in Eq. (2). The use of geometric mean is to avoid the skewness of immunogenic and antigenic scores obtained for T-cell and B-cell epitopes so that proper ranking of the consensus conserved regions can be performed. Moreover, to validate the identified epitopes, the conformational 2D non-covalent structures of the identified epitopes are studied using LigPlot+ [41]. Furthermore, BepiPred2.0 server10 [42] is used for the verification of the predicted B-cell epitopes.Also, the physico-chemical properties of the epitopes along with Ramachandran plot are reported through PyMOL [43] and its extensive libraries Autodock Vina (for docking) [44] and PyMOD 3 [45] while for the Z-score calculation ProSA11 [46] online server is used.
| (2) |
where, RCCnR represents rank of consensus conserved region (CCnR) based on geometric mean of immunogenic and antigenic scores of T-cell and B-cell epitopes, ISi and ASi are the scaled immunogenic and antigenic scores for MHC-I, MHC-II and B-cell epitopes respectively.
Fig. 1.
Pipeline of the Workflow.
3. Results and discussions
3.1. Ranking of the CCnRs
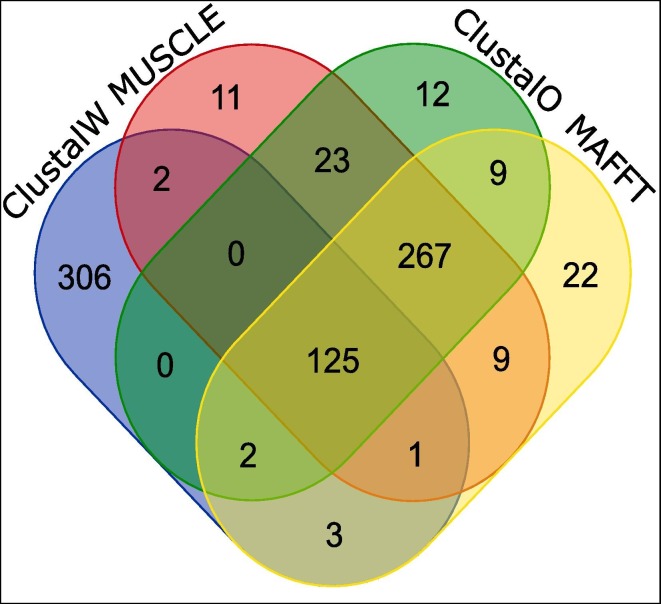
Experiments in this study are carried out according to the flowchart as mentioned in Fig. 1. Initially, 566 Indian SARS-CoV-2 genomes are aligned by using Consensus Multiple Sequence Alignment (CMSA) techniques, ClustalW, MUSCLE, ClustalO and MAFFT. Subsequently, we have obtained 125 CCnRs by considering all the alignment techniques. This is shown in Fig. 2 where 438, 439, 438 and 438 conserved regions (CnRs) from ClustalW, MUSCLE, ClustalO and MAFFT respectively are provided resulting in 125 CCnRs. This is followed by mapping of the CCnRs to 11 coding regions i.e. ORF1ab, Spike, ORF3a, Envelope, Membrane, ORF6, ORF7a, ORF7b, ORF8, Nucleocapsid and ORF10. The corresponding protein sequence for each CCnR has been taken from Frame 2. Now, the 125 CCnRs are filtered based on the criteria that (a) their length should be greater than or equal to 60nt and (b) no stop codons should be present in the corresponding proteins. A BLAST specificity score as query coverage equal to 100% is also considered during the filtering process. As a result, 23 CCnRs have been identified. Subsequently, these CCnRs are ranked on the basis of geometric mean of highly immunogenic and antigenic scores of the corresponding MHC-I, MHC-II T-cell and B-cell epitopes. It is worth mentioning that the immunogenic and antigenic scores are scaled within the range of 0–1 to bring the scores of all the epitopes for different CCnRs to a uniform scale and mentioned throughout the paper while the actual scores are given as Supplementary in excel file. After ranking, top 5 CCnRs along with their corresponding protein sequences, lengths, blast specificity scores, percentage of BLAST specificity scores as query coverage, coding regions with their starting and ending coordinates, lengths and coded proteins are also mentioned in Table 1 . Moreover, the ranking with the scores of these top 5 CCnRs is reported in Table 2 . It is found from Table 1, that the top 5 CCnRs belong to the coding region which codes NSP3, 3CL-Proteinase, NSP10 and NSP4 proteins respectively. Please note that all the 23 CCnRs are reported in Supplementary Table S3 while their ranking details are given in Supplementary Table S4.
Fig. 2.
125 Consensus Conserved Regions (CCnRs) from the four alignment techniques.
Table 1.
Top 5 Consensus Conserved Regions (CCnRs) as derived from SARS-CoV-2 with associated details.
| Consensus Conserved Region (CCnR) | Protein Sequence of CCnR | Length of | BLAST Specificity | % of BLAST Specificity | Coding | Starting | Ending | Length of CR | Coded |
|---|---|---|---|---|---|---|---|---|---|
| CCnR | Score of CCnR | Score as Query Coverage | Region (CR) | Coordinate of CR | Coordinate of CR | Protein from CR | |||
| 4012-CACAGAAAACTTGTTACTTTATATTGACATTAATGGCAATCTTCATCCAGATTCTGCCACTCTTGTTAGTGACATTGACATCACTTTCTTAAAGAAAGATGCTCCATATATAGTGGGTGATGTTGTTCAAGAGGGTGTTTTAACTGCTGTGGTTATACCTACTAAAAAGGCTGGTGGCACTACTGAAATGCTAGCGAAAGCTTT-4215 | TENLLLYIDINGNLHPDSATLVSDIDITFLKKDAPYIVGDVVQEGVLTAVVIPTKKAGGTTEMLAKA | 204 | 377 | 100 | ORF1ab | 266 | 21555 | 21290 | NSP3 |
| 10463-TTAAGGGTTCATTCCTTAATGGTTCATGTGGTAGTGTTGGTTTTAACATAGATTATGACTGTGTCTCTTTTTGTTAC-10539 | KGSFLNGSCGSVGFNIDYDCVSFCY | 77 | 143 | 100 | ORF1ab | 266 | 21555 | 21290 | 3CL-Proteinase |
| 13291-TTTTGTGACTTAAAAGGTAAGTATGTACAAATACCTACAACTTGTGCTAATGACCCTGTGGGTTTTACACTTAAAAACACAGTCTGTACCGTCTGCGGTAT-13391 | FCDLKGKYVQIPTTCANDPVGFTLKNTVCTVCG | 101 | 187 | 100 | ORF1ab | 266 | 21555 | 21290 | NSP10 |
| 5307-TAACACTCCAACAAATAGAGTTGAAGTTTAATCCACCTGCTCTACAAGATGCTTATTACAG-5367 | TLQQIELKFNPPALQDAYY | 61 | 113 | 100 | ORF1ab | 266 | 21555 | 21290 | NSP3 |
| 9564-ATTCTTACCTGGTGTTTATTCTGTTATTTACTTGTACTTGACATTTTATCTTACTAATGATGTTTCTTTTTTAGCACATATTCAGTGGATGGTT-9657 | FLPGVYSVIYLYLTFYLTNDVSFLAHIQWMV | 94 | 174 | 100 | ORF1ab | 266 | 21555 | 21290 | NSP4 |
Table 2.
Ranking procedure done on the basis of Geometric Mean of Binding and Antigenic Scores of T-cell and B-cell epitopes from each CCnR.
| Consensus Conserved Region (CCnR) | Protein | Coded | MHC-I restricted T-cell |
MHC-II restricted T-cell |
B-cell Epitopes |
Final | |||
|---|---|---|---|---|---|---|---|---|---|
| Sequence | Protein | Immunogenic Score | Antigenic score | Immunogenic score | Antigenic Score | Immunogenic Score | Antigenic Score | Score | |
| 10463-CACAGAAAACTTGTTACTTTATATTGACATTAATGGCAATCTTCATCCAGATTCTGCCACTCTTGTTAGTGACATTGACATCACTTTCTTAAAGAAAGATGCTCCATATATAGTGGGTGATGTTGTTCAAGAGGGTGTTTTAACTGCTGTGGTTATACCTACTAAAAAGGCTGGTGGCACTACTGAAATGCTAGCGAAAGCTTT-10539 | TENLLLYIDINGNLHPDSATLVSDIDITFLKKDAPYIVGDVVQEGVLTAVVIPTKKAGGTTEMLAKA | NSP3 | 0.8640 | 0.7361 | 0.9804 | 0.6382 | 0.8810 | 1 | 0.84 |
| 9104-TTAAGGGTTCATTCCTTAATGGTTCATGTGGTAGTGTTGGTTTTAACATAGATTATGACTGTGTCTCTTTTTGTTAC-9211 | KGSFLNGSCGSVGFNIDYDCVSFCY | 3CL-Proteinase | 0.6552 | 0.9049 | 0.9114 | 0.7499 | 0.7143 | 0.7401 | 0.77 |
| 21661-TTTTGTGACTTAAAAGGTAAGTATGTACAAATACCTACAACTTGTGCTAATGACCCTGTGGGTTTTACACTTAAAAACACAGTCTGTACCGTCTGCGGTAT-21728 | FCDLKGKYVQIPTTCANDPVGFTLKNTVCTVCG | NSP10 | 0.9136 | 0.7542 | 0.9818 | 0.3852 | 0.9048 | 0.6813 | 0.74 |
| 5220-TAACACTCCAACAAATAGAGTTGAAGTTTAATCCACCTGCTCTACAAGATGCTTATTACAG-5288 | TLQQIELKFNPPALQDAYY | NSP3 | 0.8106 | 1 | 0.9485 | 0.6714 | 0.3333 | 0.8433 | 0.72 |
| 6706-ATTCTTACCTGGTGTTTATTCTGTTATTTACTTGTACTTGACATTTTATCTTACTAATGATGTTTCTTTTTTAGCACATATTCAGTGGATGGTT-6839 | FLPGVYSVIYLYLTFYLTNDVSFLAHIQWMV | NSP4 | 0.9980 | 0.7866 | 0.9933 | 0.3326 | 0.9762 | 0.4726 | 0.70 |
It is important to note that although structural proteins are the popular candidates for vaccine, vaccine protection can be correlated to non-structural proteins. In this regard, [47] showed that NS1 which is a non-structural protein can bring about protective immunity against flaviviruses. Though, no neutralizing effect was shown by antibodies against NS1, some exuded complement-fixing activity and even passive transfer of anti-NS1 antibody or immunization with NS1 can lead to protection against viruses [48]. Furthermore, anti-NS1 antibody could be responsible to block NS1-induced pathogenic effects, reduce viral replication by complement-dependent cytotoxicity of infected cells and even attenuate NS1-induced disease development. This has led to NS1 being a prospective vaccine candidate against Dengue virus [49], [50]. Another core advantage of NS1 is that being a non-structural protein, the anti-NS1 antibody will not instigate antibody-dependent enhancement (ADE), which is a virulence factor causing serious repercussions. Additionally, non-structural virus proteins can generate cytotoxic T lymphocytes which are important to control infection. In [51], the authors have shown that the non-structural proteins of the hepatitis-C virus could generate HCV-specific broad-spectrum T-cell responses. Non-structural proteins have been used by [52] for vaccine design against Usutu Virus. Also, as targets for prophylactic or therapeutic vaccines, the non-structural proteins of HIV-1 were shown to be quite important [53]. Moreover, Ong et al. [25] have predicted NSP3 in SARS-CoV-2 to produce high protective antigenicity. Thus, we can hypothesize that apart from structural proteins non-structural proteins of SARS-CoV-2 can be possible targets as well for vaccine design which may induce cell-mediated or humoral immunity that is necessary to prevent viral invasion and/or replication.
3.2. Identification of MHC-I restricted T-cell epitopes
For epitope prediction from the 23 CCnRs, the associated protein sequences are used as inputs to the prediction tools. In this regard, MHC-I binding predictions are performed using IEDB [54] recommended NetMHCpan EL 4.1 (published recently in September 2020) targeting 27 unique HLA alleles. As a result, for each CCnR good binders in the form of immunogenic score, 4 best HLA epitopes are selected, in total 92 epitopes of length 9–11 mer each are obtained. Their antigenic scores are evaluated using VaxiJen2.0 [55]. In order to rank the CCnRs, only the best immunogenic and antigenic MHC-I restricted T-cell epitopes are considered. As a consequence, 34 such epitopes are identified and reported in Supplementary Table S5 for all the CCnRs while for the top 5 CCnRs, 8 epitopes are provided in Table 3 . It is found that FLKKDAPYI and TAVVIPTKK are the highly immunogenic and antigenic MHC-I restricted T-cell epitopes from the NSP3 coded protein binded to HLA-A*31:01 and HLA-A*68:01 HLA alleles respectively. All the 92 MHC-I restricted T-cell epitopes along with their HLA alleles are provided in the supplementary as an excel file.
Table 3.
List of Immunogenic and Antigenic Epitopes for MHC-I, MHC-II restricted T-cell and B-cell Epitopes.
| Protein | Coded | Type | MHC-I restricted T-cell |
MHC-II restricted T-cell |
B-cell |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | Proteins | Epitope | Alleles | Scaled Score of |
Epitope | Alleles | Scaled Score of |
Epitope | Scaled Score of |
||||
| Immunogenicity | Antigenicity | Immunogenicity | Antigenicity | Immunogenicity | Antigenicity | ||||||||
| TENLLLYIDINGNLHPDSATLVSDIDITFLKKDAPYIVGDVVQEGVLTAVVIPTKKAGGTTEMLAKA | NSP3 | Immunogenic | FLKKDAPYI | HLA-A*31:01 | 0.8640 | 0.3890 | ITFLKKDAPYIVGDV | HLA-DRB3*01:01 | 0.9804 | 0.3036 | TLVSDIDITFLKKDAP | 0.8810 | 0.7314 |
| KGSFLNGSCGSVGFNIDYDCVSFCY | 3CL-Proteinase | Immunogenic | FLNGSCGSV | HLA-A*02:03 | 0.6552 | 0.3342 | CGSVGFNIDYDCVSF | HLA-DQA1*01:01/DQB1*05:01 | 0.9114 | 0.7499 | CGSVGFNIDYDCVSFC | 0.7143 | 0.7401 |
| Antigenic | |||||||||||||
| FCDLKGKYVQIPTTCANDPVGFTLKNTVCTVCG | NSP10 | Immunogenic | DLKGKYVQI | HLA-B*08:01 | 0.9136 | 0.7542 | KGKYVQIPTTCANDP | HLA-DRB1*04:01 | 0.9818 | 0.1892 | TTCANDPVGFTLKNTV | 0.9048 | 0.6813 |
| TLQQIELKFNPPALQDAYY | NSP3 | Immunogenic | NPPALQDAY | HLA-B*35:01 | 0.8106 | 0.4557 | QIELKFNPPALQDAY | HLA-DRB3*02:02 | 0.9485 | 0.6409 | LQQIELKFNPPALQDA | 0.3333 | 0.8433 |
| FLPGVYSVIYLYLTFYLTNDVSFLAHIQWMV | NSP4 | Immunogenic | VSFLAHIQW | HLA-B*57:01 | 0.9980 | 0.7866 | GVYSVIYLYLTFYLT | HLA-DPA1*01:03/DPB1*02:01 | 0.9933 | 0.3326 | YSVIYLYLTFYLTNDV | 0.9762 | 0.4726 |
| Antigenic | |||||||||||||
3.3. Identification of MHC-II restricted T-cell epitopes
Similar procedures are carried out for MHC-II restricted T-cell epitopes as well using MHC-II binding prediction tool provided by IEDB with consensus prediction targeting a different set of 27 unique HLA alleles. Subsequently, we obtained 92 epitopes of length 15–17 mer each which are bounded to their alleles along with their corresponding immunogenic and antigenic scores. In order to rank the CCnRs, the best immunogenic and antigenic MHC-II restricted T-cell epitopes are considered, resulting in 37 epitopes which are reported in Supplementary Table S5 for all the CCnRs. The 8 epitopes for the top 5 CCnRs are reported in Table 3. From this table, it is seen that ITFLKKDAPYIVGDV and IDITFLKKDAPYIVG are the most immunogenic and antigenic MHC-II restricted T-cell epitopes corresponding to HLA-DRB3*01:01 allele. All the 92 MHC-II restricted T-cell epitopes along with their HLA alleles are provided in the supplementary as an excel file.
3.4. Identification of B-cell epitopes
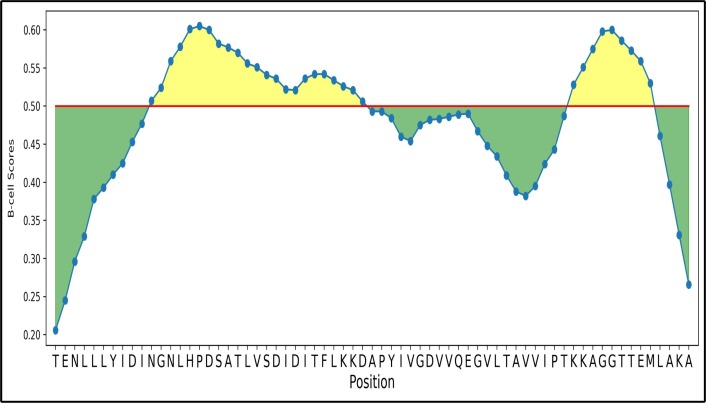
After obtaining MHC-I and MHC-II T-cell epitopes, B-cell epitopes which are responsible for antigen productions are predicted using ABCPred [56] with the length of 15–18 mer and their antigenic scores are evaluated from the VaxiJen server. As a result, 61 epitopes are found. In order to rank the CCnRs, the best immunogenic and antigenic B-cell epitopes are considered which resulted in 29 epitopes. These epitopes are reported in Supplementary Table S5 for all the CCnRs while for the top 5 CCnRs, 6 B-cell epitopes are reported in Table 3. In this table, it is found that TLVSDIDITFLKKDAP and LHPDSATLVSDIDITF are the most immunogenic and antigenic B-cell epitopes. Here, it should be noted that for antigenicity evaluation, a threshold of 0.4 is maintained throughout the experiment by following the literature [20]. The graphical representation of TLVSDIDITFLKKDAP and LHPDSATLVSDIDITF is shown in Fig. 3 using BepiPred 2.0 where the total green and yellow regions represent the protein sequence TENLLLYIDINGNLHPDSATLVSDIDITFLKKDAPYIVGDVVQEGVLTAVVIPTKKAGGTTEMLAKA while the two yellow regions denote the B-cell epitopes TLVSDIDITFLKKDAP and LHPDSATLVSDIDITF respectively. The red line in the figure represents the threshold which is set to 0.5. For all the 23 CCnRs the results are shown in Supplementary Fig. S1 while the 61 B-cell epitopes are provided in the supplementary as an excel file.
Fig. 3.
Graphical representation of B-cell epitopes for TLVSDIDITFLKKDAP and LHPDSATLVSDIDITF with the threshold marked by red line.
3.5. Final panel of epitopes
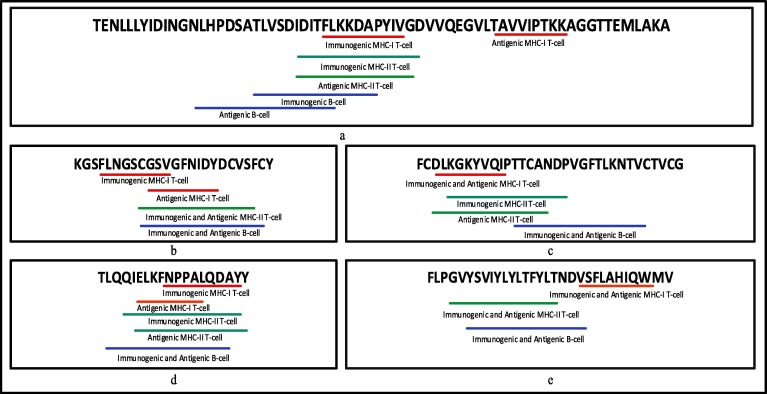
Table 4 summarises the final panel of the 34 MHC-I, 37 MHC-II restricted T-cell epitopes and 29 B-cell epitopes for 23 CCnRs based on their highest immunogenic and antigenic scores. There are 16 unique HLA alleles for MHC-I and 19 unique HLA alleles for MHC-II restricted T-cell epitopes. The associated coded proteins for the 23 CCnRs are NSP1, NSP2, NSP3, NSP4, 3CL-Proteinase, NSP10, RNA-directed RNA polymerase, Helicase, Spike glycoprotein and Nucleocapsid protein. For better readability, the epitopes associated with the top 5 CCnRs are underlined in Fig. 4 whereas the epitopes for 23 CCnRs are underlined in Supplementary Fig. S2. The red lines, green lines and the blue lines respectively denote the MHC-I, MHC-II T-cells and B-cells respectively. Moreover, for the ease of the readers, all the details related to the 125 CCnRs, 92 MHC-I and MHC-II restricted T-cell epitopes and 61 B-cell epitopes are provided in the supplementary as an excel file, the link of which is given in Table S6. Additionally, a list of MHC-I and MHC-II restricted T-cell and B-cell epitopes for SARS-CoV-2 as collected from different sources in the literature like [26], [27], [17], [28], [16], [15], [20], [24], [23], [29], [30], [31], [32], [33], [25] are reported in Table 5. For space constraint, 3 of each MHC-I and MHC-II restricted T-cell and B-cell epitopes from each paper are mentioned in this table while the list of all the MHC-I and MHC-II restricted T-cell and B-cell epitopes are given in the supplementary as an excel file as given in Table S6. Thus, Table 4, Table 5 can provide the readers a better insight into the epitopes identified so far.
Table 4.
Overview of MHC-I, MHC-II restricted T-cell and B-cell epitopes for the 23 CCnRs.
| Coded | Type | MHC-I restricted T-cell |
MHC-II restricted T-cell |
B-cell Epitopes | ||
|---|---|---|---|---|---|---|
| Proteins | Epitopes | HLA Alleles | Epitopes | HLA Alleles | ||
| NSP3 | Immunogenic | FLKKDAPYI | HLA-A*31:01 | ITFLKKDAPYIVGDV | HLA-DRB3*01:01 | TLVSDIDITFLKKDAP |
| Antigenic | TAVVIPTKK | HLA-A*68:01 | IDITFLKKDAPYIVG | HLA-DRB3*01:01 | LHPDSATLVSDIDITF | |
| 3CL-Proteinase | Immunogenic | FLNGSCGSV | HLA-A*02:03 | CGSVGFNIDYDCVSF | HLA-DQA1*01:01/DQB1*05:01 | CGSVGFNIDYDCVSFC |
| Antigenic | GSVGFNIDY | HLA-A*30:02 | ||||
| NSP10 | Immunogenic | DLKGKYVQI | HLA-B*08:01 | KGKYVQIPTTCANDP | HLA-DRB1*04:01 | TTCANDPVGFTLKNTV |
| Antigenic | DLKGKYVQIPTTCAN | HLA-DRB1*04:01 | ||||
| NSP3 | Immunogenic | NPPALQDAY | HLA-B*35:01 | QIELKFNPPALQDAY | HLA-DRB3*02:02 | LQQIELKFNPPALQDA |
| Antigenic | IELKFNPPAL | HLA-B*40:01 | IELKFNPPALQDAYY | HLA-DRB3*02:02 | ||
| NSP4 | Immunogenic | VSFLAHIQW | HLA-B*57:01 | GVYSVIYLYLTFYLT | HLA-DPA1*01:03/DPB1*02:01 | YSVIYLYLTFYLTNDV |
| Antigenic | ||||||
| NSP3 | Immunogenic | QVNGLTSIKW | HLA-B*57:01 | PQVNGLTSIKWADNN | HLA-DQA1*01:02/DQB1*06:02 | KYPQVNGLTSIKWADN |
| Antigenic | KYPQVNGLTSIKWAD | HLA-DQA1*01:02/DQB1*06:02 | ||||
| Helicase | Immunogenic | RAQNMTMSY | HLA-A*30:02 | YQLKLLIHHRAQNMT | HLA-DRB4*01:01 | FWDYQLKLLIHHRAQN |
| Antigenic | DYQLKLLIHHRAQNM | HLA-DRB4*01:02 | IHHRAQNMTMSYSLKP | |||
| Spike glycoprotein | Immunogenic | HADQLTPTW | HLA-B*58:01 | DIPIGAGICASYQTQ | HLA-DQA1*05:01/DQB1*03:01 | GCLIGAEHVNNSYECD |
| Antigenic | ||||||
| NSP4 | Immunogenic | ICISTKHFYW | HLA-B*57:01 | KHFYWFFSNYLKRRV | HLA-DPA1*01:03/DPB1*04:01 | ISTKHFYWFFSNYLKR |
| Antigenic | TKHFYWFFSNYLKRR | HLA-DPA1*01:03/DPB1*04:01 | ||||
| Nucleocapsid protein | Immunogenic | AQFAPSASAF | HLA-B*15:01 | ATKAYNVTQAFGRR | HLA-DRB5*01:01 | KSAAEASKKPRQKRTA |
| Antigenic | KAYNVTQAFGRRGP | HLA-DRB5*01:01 | GRRGPEQTQGNFGDQE | |||
| Spike glycoprotein | Immunogenic | FERDISTEI | HLA-B*40:01 | VEGFNCYFPLQSYGF | HLA-DQA1*01:01/DQB1*05:01 | GSTPCNGVEGFNCYFP |
| Antigenic | YFPLQSYGF | HLA-A*24:02 | NGVEGFNCYFPLQSY | HLA-DRB3*01:01 | EGFNCYFPLQSYGFQP | |
| NSP4 | Immunogenic | NVLEGSVAY | HLA-B*35:01 | PVPYCYDTNVLEGSV | HLA-DRB1*04:01 | SGKPVPYCYDTNVLEG |
| Antigenic | SGKPVPYCY | HLA-A*30:02 | GKPVPYCYDTNVLEG | HLA-DRB1*04:01 | ||
| Helicase | Immunogenic | VLAYVDHSY | HLA-B*15:01 | VDHSYVVNAVTTMSY | HLA-DRB3*02:02 | LAYVDHSYVVNAVTTM |
| Antigenic | ||||||
| NSP3 | Immunogenic | NYMPYFFTL | HLA-A*24:02 | CTNYMPYFFTLLLQL | HLA-DPA1*03:01/DPB1*04:02 | VCTNYMPYFFTLLLQL |
| Antigenic | ||||||
| NSP10 | Immunogenic | FAVDAAKAY | HLA-B*35:01 | LSFCAFAVDAAKAYK | HLA-DRB3*01:01 | GTGQAITVTPEANMDQ |
| Antigenic | VPANSTVLSF | HLA-B*35:01 | KMLCTHTGTGQAITVT | |||
| 3CL-Proteinase | Immunogenic | GTTTLNGLW | HLA-B*57:01 | TTTLNGLWLDDVVYC | HLA-DQA1*01:01/DQB1*05:01 | QVTCGTTTLNGLWLDD |
| Antigenic | TLNGLWLDDVVYCPR | HLA-DQA1*01:01/DQB1*05:01 | ||||
| NSP1 | Immunogenic | HVGEIPVAY | HLA-B*15:01 | VAYRKVLLRKNGNKG | HLA-DRB1*11:01 | PHVGEIPVAYRKVLLR |
| Antigenic | HVGEIPVAYR | HLA-A*68:01 | IPVAYRKVLLRKNGN | HLA-DRB1*11:01 | ||
| NSP4 | Immunogenic | RPDTRYVLM | HLA-B*07:02 | LMDGSIIQFPNTYLE | HLA-DRB1*15:01 | GSIIQFPNTYLEGSVR |
| Antigenic | LRPDTRYVLMDGSIIQ | |||||
| NSP4 | Immunogenic | VCVSTSGRW | HLA-B*57:01 | TSGRWVLNNDYYRSL | HLA-DRB3*02:02 | YCRHGTCERSEAGVCV |
| Antigenic | STSGRWVLNNDYYRS | HLA-DRB3*02:02 | ||||
| RNA-directed | Immunogenic | DTLSLTTNMK | HLA-A*68:01 | TTNMKKQFIIYLRIV | HLA-DPA1*02:01/DPB1*05:01 | LRDTLSLTTNMKKQFI |
| RNA polymerase | Antigenic | LSLTTNMKK | HLA-A*11:01 | |||
| NSP2 | Immunogenic | VTHSKGLYR | HLA-A*31:01 | ETFVTHSKGLYRKCV | HLA-DRB5*01:01 | LNLGETFVTHSKGLYR |
| Antigenic | VTHSKGLYRK | HLA-A*03:01 | LGETFVTHSKGLYRK | HLA-DRB5*01:01 | ||
| Spike glycoprotein | Immunogenic | VYYPDKVFR | HLA-A*31:01 | TRGVYYPDKVFRSSV | HLA-DRB1*03:01 | RGVYYPDKVFRSSVLH |
| Antigenic | GVYYPDKVFR | HLA-A*31:01 | ||||
| NSP2 | Immunogenic | LEQPTSEAV | HLA-B*40:01 | GDLQPLEQPTSEAVE | HLA-DQA1*03:01/DQB1*03:02 | TGDLQPLEQPTSEAVE |
| Antigenic | EVVLKTGDL | HLA-A*26:01 | EVVLKTGDLQPLEQP | HLA-DRB1*08:02 | ||
Fig. 4.
MHC-I, MHC-II restricted T-cell and B-cell epitopes underlined in the protein sequences of top 5 CCnRs for (a) NSP3 (b) 3CL-Proteinase (c) NSP10 (d) NSP3 and (e) NSP4.
Table 5.
List of proposed epitopes for SARS-CoV-2 as given in the literature.
| Source | Coded Proteins | MHC-I restricted T-cell Epitopes | MHC-II restricted T-cell Epitopes | B-cell Epitopes |
|---|---|---|---|---|
| Bhattacharya et al. [26] | Spike glycoprotein | SQCVNLTTR | IHVSGTNGT | SQCVNLTTRTQLPPAYTNSFTRGVY |
| YTNSFTRGV | VYYHKNNKS | FSNVTWFHAIHVSGTNGTKRFDN | ||
| GVYYHKNNK | LVRDLPQGF | DPFLGVYYHKNNKSWME | ||
| Chen et al. [27] | Spike glycoprotein | LSPRWYFYY | IKLDDKDPN | EVRQIAPGQTGKIADY |
| RSRNSSRNS | RSGARSKQR | GCLIGAEHVNNSYECD | ||
| IGYYRRATR | RIGMEVTPS | FAMQMAYRFNGIGVTQ | ||
| Naz et al. [17] | Spike glycoprotein | GVYFASTEK | EFVFKNIDGYFKIYS | YNSASFSTFKCYGVSPTKLNDLCFT |
| STQDLFLPF | QPYRVVVLSFELLHA | |||
| KTSVDCTMY | MTKTSVDCTMYICGD | |||
| Kar et al. [28] | Spike glycoprotein | QIITTDNTF | INITRFQTLLALHRS | FSYTESLAGKREMAII |
| YQPYRVVVL | GINITRFQTLLALHR | HAGPGPGPY | ||
| FTISVTTEI | GWTFGAGAALQIPFA | KMGPGPGTRFA | ||
| Rakib et al. [16] | Spike glycoprotein | WTAGAAAYY | LIVNNATNV | RTQLPPAYTNS |
| CNDPFLGVY | IVNNATNVV | SGTNGTKRFDN | ||
| GAAAYYVGY | SKTQSLLIV | LTPGDSSSGWTAG | ||
| Vashi et al. [15] | Spike glycoprotein | RTQLPPAY | MFVFLVLLPLVSSQC | PPAYTNSFTRGVYY |
| RTQLPPA | MFVFLVLLPLVSSQCVN | HVSGTNGTKRFDN | ||
| LPPAYTNSF | QGNFKNLREFVFKNI | YYHKNNKSWMES | ||
| Yadav et al. [20] | Spike glycoprotein | GVYFASTEK | NA | HRSYLTPGDSSSGWTA |
| FEYVSQPFL | NA | FPNITNLCPFGEVFNA | ||
| WTAGAAAYY | NA | EVIQIAPGQTGKIADY | ||
| Crooke et al. [24] | Membrane glycoprotein | ATSRTLSYY | TLSYYKLGASQRVAG | EVTPSGTWL |
| RLFARTRSM | RTLSYYKLGASQRVA | KLDDKDPNFK | ||
| YANRNRFLY | ASFRLFARTRSMWSF | KTFPPTEPKKDKKKKADETQALPQ | ||
| Gupta et al. [23] | Spike glycoprotein | VRFPNITNL | NVTWFHAIHV | GDEVRQIAPGQTGKIADYNYKLP |
| YQPYRVVVL | ||||
| PYRVVVLSF | ||||
| Bhatnager et al. [29] | Spike glycoprotein | LTDEMIAQY | VASQSIIAYTMSLGA | KEEQIGKCSTR |
| LLTDEMIAQY | LTDEMIAQYTSALLA | ELGKYEQYGPGPGKWP | ||
| IPFAMQMAY | VLNDILSRLDKVEAE | IRAGPGPGGNC | ||
| Kwarteng et al. [30] | Nucleocapsid protein | KTFPPTEPK | AQFAPSASAFFGMSR | AGLPYGANK |
| SSPDDQIGY | IAQFAPSASAFFGMS | SKQLQQSMSSADS | ||
| SSPDDQIGYY | PQIAQFAPSASAFFG | RRIRGGDGKMKDL | ||
| Baruah et al. [31] | Spike glycoprotein | YLQPRTFLL | NA | CVNLTTRTQLPPAYTN |
| GVYFASTEK | NVTWFHAIHVSGTNG | |||
| EPVLKGVKL | SFSTFKCYGVSPTKLND | |||
| Bency et al. [32] | Spike glycoprotein | KIADYNYKL | VVFLHVTYV | MDLEGKQGNFKNL |
| CYGVSPTKL | IGINITRFQ | YYVGYLQPR | ||
| VVVLSFELL | FNCYFPLQS | NITNLCPFGE | ||
| Singh et al. [33] | Nucleocapsid protein | AQFAPSASA | AQFAPSASAFFGMSR | KEDLKFP |
| GDAALALLL | GDAALALLLLDRLNQ | IKLDDKDPNFKDQ | ||
| GMSRIGMEV | ASAFFGMSRIGMEVT | PPTEPKKDKKKKADETQALPQRQKKQQTVT | ||
| Ong et al. [25] | NSP3 | STNVTIATY | ISNSWLMWLIINLVQ | EDEEEGDCEEEEFEPSTQYEYGTEDDYQGKPLEFGATS |
| RMYIFFASF | LAYILFTRFFYVLGL | EEEQEEDWLDDD | ||
| AEWFLAYIL | AAIMQLFFSYFAVHF | VGQQDGSEDNQ | ||
3.6. Study of physico-chemical properties of epitopes
To judge the relevance of the epitopes as found in this work, we have evaluated the physico-chemical properties for each selected epitope. The values of each physico-chemical property lie between 0 and 1. Table 6, Table 7, Table 8 show the physico-chemical properties for MHC-I, MHC-II restricted T-cell and B-cell epitopes respectively for the top 5 CCnRs whereas for all the 23 CCnRs, the results are reported in Supplementary Tables S7-S9 respectively. For example, in Table 6 MHC-I restricted T-cell epitope FLKKDAPYI has a positively charged value of 0.222, a negatively charged value of 0.111, polarity of 0.111, non-polarity of 0.556, alphaticity of 0.444, aromaticity of 0.222, acidicity of 0.111, Basicity of 0.222, hydrophobicity of 0.556, hydrophilicity of 0.333, a neutral value of 0.111, hydroxylic value of 0 and sulphur content is 0 as well. Similarly, for other epitopes their physico-chemical properties can be found in the tables.
Table 6.
List of physico-chemical properties of MHC-I restricted T-cell epitopes.
| MHC-I restricted T-cell epitopes | Positively charged | Negatively charged | Polarity | Non Polarity | Aliphaticity | Aromaticity | Acidicity | Basicity | Hydrophobicity | Hydrophilicity | Neutral | Hydroxylic | Sulphur Content |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FLKKDAPYI | 0.222 | 0.111 | 0.111 | 0.556 | 0.444 | 0.222 | 0.111 | 0.222 | 0.556 | 0.333 | 0.111 | 0 | 0 |
| TAVVIPTKK | 0.222 | 0 | 0.222 | 0.556 | 0.556 | 0 | 0 | 0.222 | 0.778 | 0.333 | 0.222 | 0.222 | 0 |
| FLNGSCGSV | 0 | 0 | 0.333 | 0.556 | 0.444 | 0.111 | 0 | 0 | 0.444 | 0.111 | 0.444 | 0.222 | 0.111 |
| GSVGFNIDY | 0 | 0.111 | 0.222 | 0.556 | 0.444 | 0.222 | 0.111 | 0 | 0.333 | 0.111 | 0.444 | 0.111 | 0 |
| DLKGKYVQI | 0.222 | 0.111 | 0.222 | 0.444 | 0.444 | 0.111 | 0.111 | 0.222 | 0.333 | 0.222 | 0.333 | 0 | 0 |
| NPPALQDAY | 0 | 0.111 | 0.222 | 0.556 | 0.556 | 0.111 | 0.111 | 0 | 0.556 | 0.333 | 0.222 | 0 | 0 |
| IELKFNPPAL | 0.1 | 0.1 | 0 | 0.7 | 0.6 | 0.1 | 0.1 | 0.1 | 0.7 | 0.4 | 0.1 | 0 | 0 |
| VSFLAHIQW | 0.111 | 0 | 0.222 | 0.667 | 0.444 | 0.222 | 0 | 0.111 | 0.667 | 0.111 | 0.222 | 0.111 | 0 |
Table 7.
List of physico-chemical properties of MHC-II restricted T-cell epitopes.
| MHC-II restricted T-cell epitopes | Positively charged | Negatively charged | Polarity | Non Polarity | Aliphaticity | Aromaticity | Acidicity | Basicity | Hydrophobicity | Hydrophilicity | Neutral | Hydroxylic | Sulphur Content |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITFLKKDAPYIVGDV | 0.133 | 0.133 | 0.133 | 0.6 | 0.533 | 0.133 | 0.133 | 0.133 | 0.6 | 0.2 | 0.267 | 0.067 | 0 |
| IDITFLKKDAPYIVG | 0.133 | 0.133 | 0.133 | 0.6 | 0.533 | 0.133 | 0.133 | 0.133 | 0.6 | 0.2 | 0.267 | 0.067 | 0 |
| CGSVGFNIDYDCVSF | 0 | 0.133 | 0.333 | 0.467 | 0.333 | 0.2 | 0.133 | 0 | 0.467 | 0.067 | 0.4 | 0.133 | 0.133 |
| KGKYVQIPTTCANDP | 0.133 | 0.067 | 0.333 | 0.4 | 0.4 | 0.067 | 0.067 | 0.133 | 0.533 | 0.333 | 0.333 | 0.133 | 0.067 |
| DLKGKYVQIPTTCAN | 0.133 | 0.067 | 0.333 | 0.4 | 0.4 | 0.067 | 0.067 | 0.133 | 0.533 | 0.267 | 0.333 | 0.133 | 0.067 |
| QIELKFNPPALQDAY | 0.067 | 0.133 | 0.2 | 0.533 | 0.467 | 0.133 | 0.133 | 0.067 | 0.533 | 0.267 | 0.267 | 0 | 0 |
| IELKFNPPALQDAYY | 0.067 | 0.133 | 0.2 | 0.533 | 0.467 | 0.2 | 0.133 | 0.067 | 0.533 | 0.267 | 0.2 | 0 | 0 |
| GVYSVIYLYLTFYLT | 0 | 0 | 0.467 | 0.533 | 0.467 | 0.333 | 0 | 0 | 0.6 | 0 | 0.267 | 0.2 | 0 |
Table 8.
List of physico-chemical properties of B-cell epitopes.
| B-cell epitopes | Positively charged | Negatively charged | Polarity | Non Polarity | Aliphaticity | Aromaticity | Acidicity | Basicity | Hydrophobicity | Hydrophilicity | Neutral | Hydroxylic | Sulphur Content |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TLVSDIDITFLKKDAP | 0.125 | 0.188 | 0.188 | 0.500 | 0.438 | 0.062 | 0.188 | 0.125 | 0.625 | 0.188 | 0.375 | 0.188 | 0 |
| LHPDSATLVSDIDITF | 0.062 | 0.188 | 0.250 | 0.500 | 0.438 | 0.062 | 0.188 | 0.062 | 0.625 | 0.125 | 0.438 | 0.250 | 0 |
| CGSVGFNIDYDCVSFC | 0 | 0.125 | 0.375 | 0.438 | 0.312 | 0.188 | 0.125 | 0 | 0.500 | 0.062 | 0.375 | 0.125 | 0.188 |
| TTCANDPVGFTLKNTV | 0.062 | 0.062 | 0.312 | 0.438 | 0.375 | 0.062 | 0.062 | 0.062 | 0.688 | 0.250 | 0.375 | 0.250 | 0.062 |
| LQQIELKFNPPALQDA | 0.062 | 0.125 | 0.188 | 0.562 | 0.500 | 0.062 | 0.125 | 0.062 | 0.562 | 0.250 | 0.312 | 0 | 0 |
| YSVIYLYLTFYLTNDV | 0 | 0.062 | 0.438 | 0.438 | 0.375 | 0.312 | 0.062 | 0 | 0.562 | 0.062 | 0.25 | 0.188 | 0 |
3.7. Study of docking with Ramachandran plot and Z-score
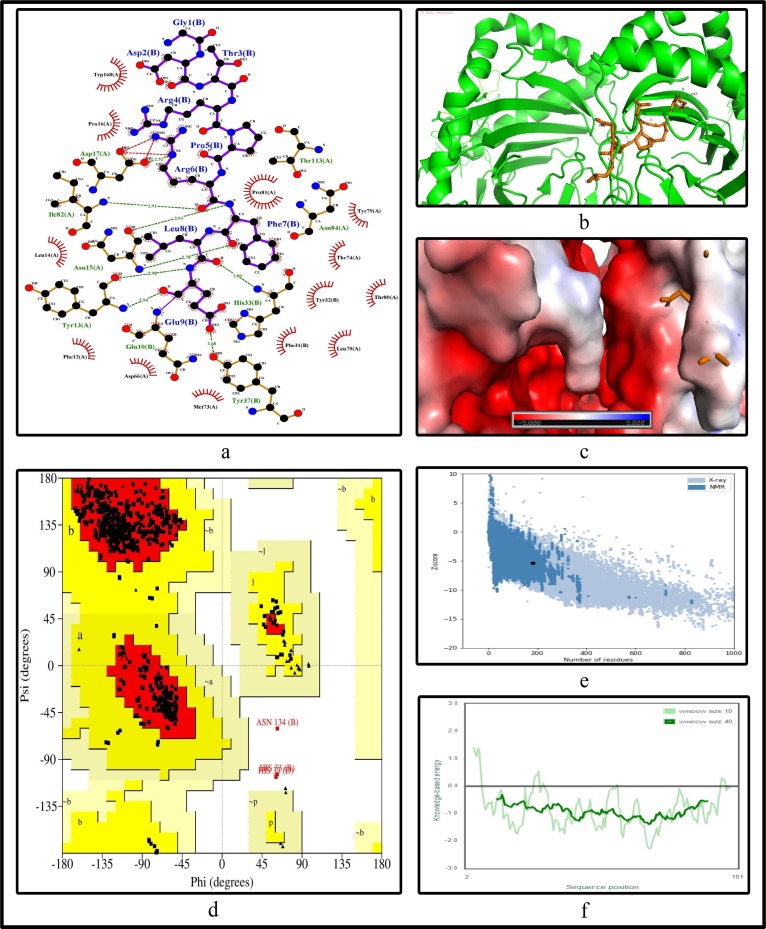
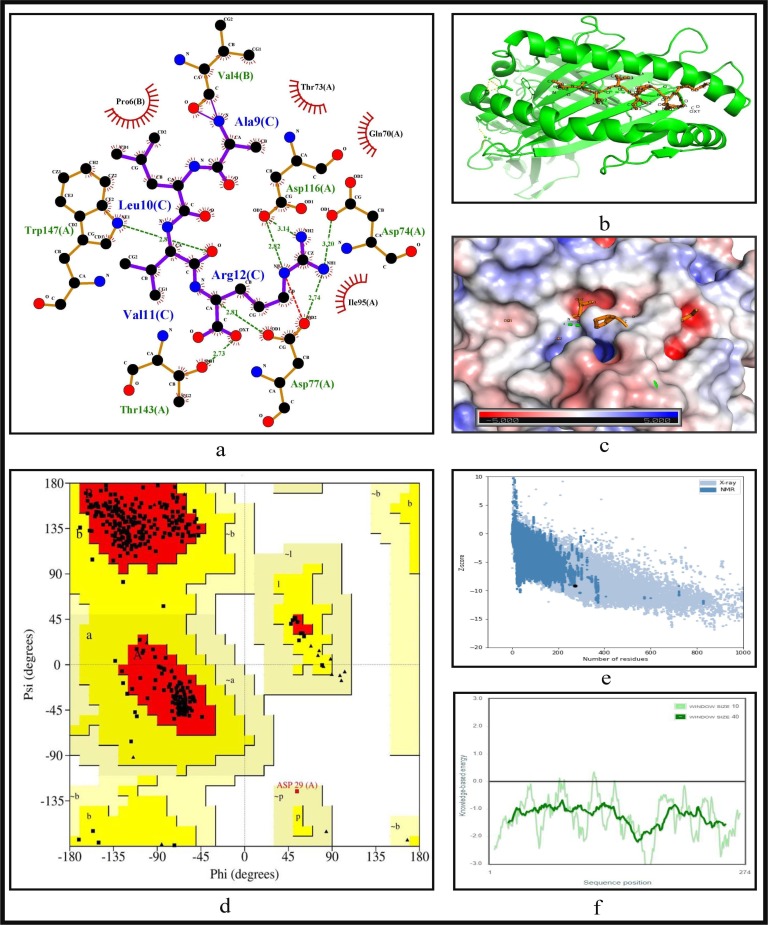
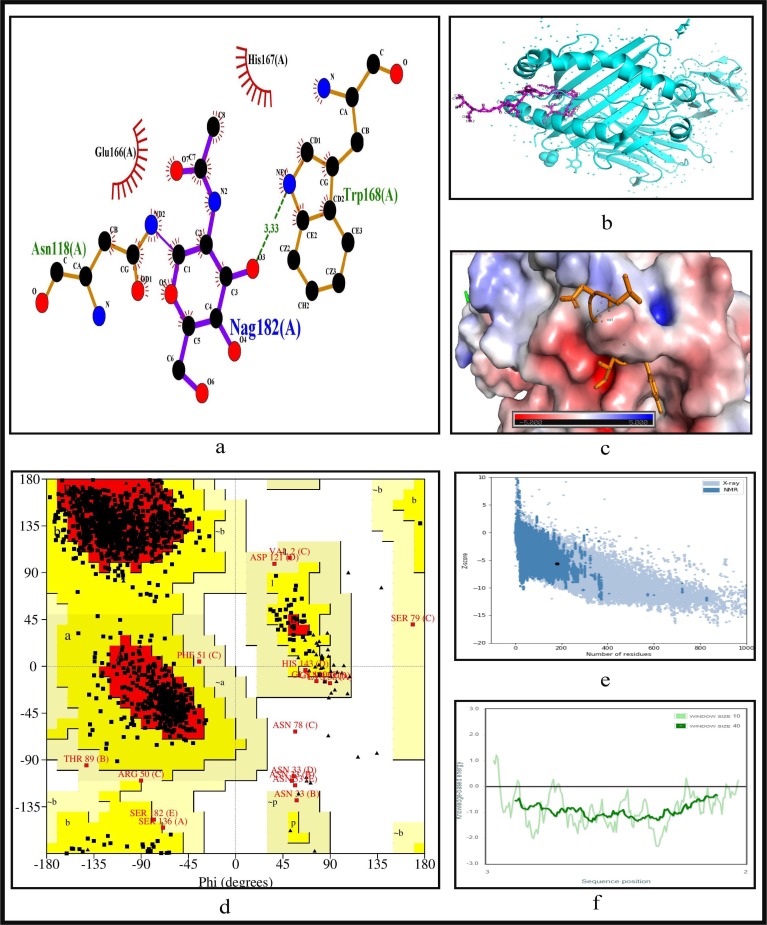
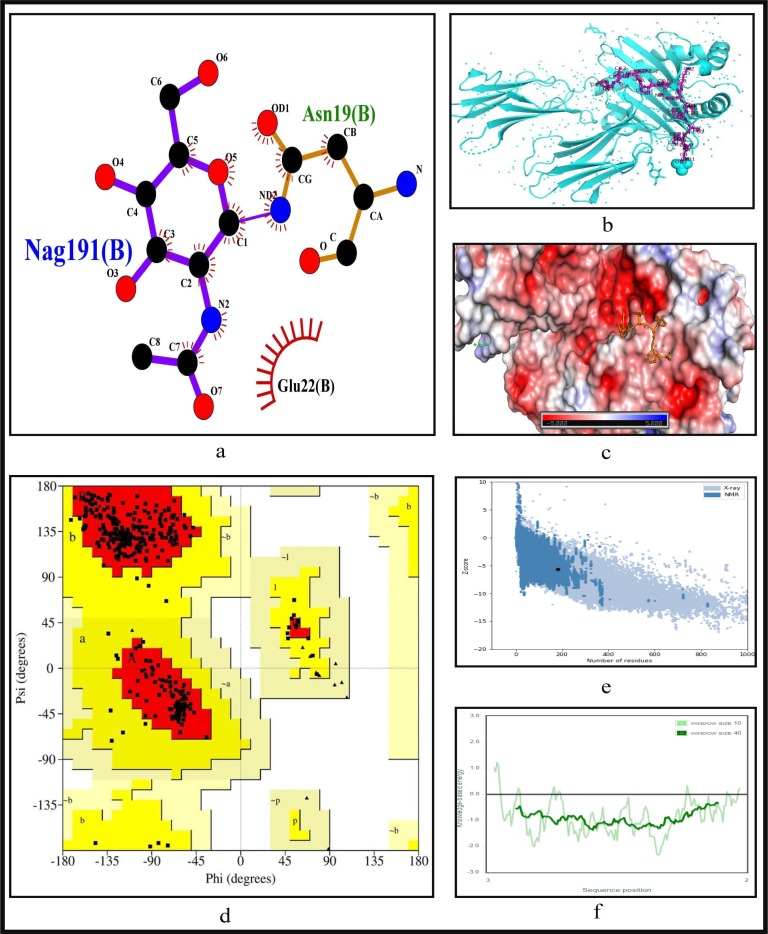
To further validate the identified epitopes, the conformational 2D non-covalent structures of the identified MHC-I and MHC-II restricted T-cell epitopes are studied using LigPlot+. For the highly immunogenic and antigenic epitopes of each CCnR, molecular docking is computed using Autodock Vina in order to extract the stable binding conformation of each predicted epitope allele pair. For MHC-I restricted T-cell epitopes, 12 binding scores are generated from Autodock Vina while for MHC-II 9 binding scores are generated. For some epitopes, the docking structures are unable to generate due to the unavailability of the corresponding structure of the HLA alleles. Furthermore, Ramachandran plot and Z-score are also evaluated for further validation using PyMod 3 and ProSA server respectively. The results of docking along with Z-scores are reported in Table 9 . The results for FLKKDAPYI and TAVVIPTKK which are the most highly immunogenic and antigenic MHC-I restricted T-cell epitopes are shown in Fig. 5, Fig. 6 while ITFLKKDAPYIVGDV and IDITFLKKDAPYIVG which are the most highly immunogenic and antigenic MHC-II restricted T-cell epitopes are shown in Fig. 7, Fig. 8 respectively. In these four figures, (a) shows the binding pose of the molecules of the two epitopes, (b) shows the exact binding position of the epitopes in the binding grooves of the alleles obtained from Autodock Vina with docking scores of −8.2 and −8.1 for MHC-I and −9 and −8.8 for MHC-II for both immunogenic and antigenic epitopes respectively and (c) depicts the surface interaction between the alleles and the identified epitopes showing the fitting sites in binding grooves. Further, quality of the residues inside the epitopes are evaluated on the basis of rotational spin of the atoms around bonds. This is depicted in (d) of Fig. 5, Fig. 6 for MHC-I and Fig. 7, Fig. 8 for MHC-II through Ramachandran plot in which points lying in the red region represents much more stable state of their bond orientations inside a molecule. This is followed by the Z-Score evaluation in (e) where the negative values of Z-score which are −9.81 and −5.9 for MHC-I and −5.53 and −5.59 for MHC-II as shown in Table 9 and Fig. 5, Fig. 6, Fig. 7, Fig. 8 verify the stability of the structures and (f) shows the overall negative energy values of the entire residues inside the whole structures which confirm the molecular stability of the identified epitopes. The results for docking along with Z-scores for all the 23 CCnRs are reported in Supplementary Table S10 while the corresponding structural analysis are given in Supplementary Figs. S3 and S4.
Table 9.
Docking and Z-scores of MHC-I and MHC-II restricted T-cell epitopes for the top 5 ranked CCnRs.
| MHC-I restricted | Score from | Z Score | MHC-II restricted | Score from | Z Score |
|---|---|---|---|---|---|
| T-cell epitopes | Autodock Vina | T-cell epitopes | Autodock Vina | ||
| FLKKDAPYI | −8.2 | −9.81 | ITFLKKDAPYIVGDV | −9 | −5.53 |
| TAVVIPTKK | −8.1 | −5.9 | IDITFLKKDAPYIVG | −8.8 | −5.59 |
| FLNGSCGSV | Not Generated | Not Generated | CGSVGFNIDYDCVSF | Not Generated | Not Generated |
| GSVGFNIDY | −7.1 | −5.4 | |||
| DLKGKYVQI | −8.1 | −8.81 | KGKYVQIPTTCANDP | Not Generated | Not Generated |
| DLKGKYVQIPTTCAN | Not Generated | Not Generated | |||
| NPPALQDAY | Not Generated | Not Generated | QIELKFNPPALQDAY | Not Generated | Not Generated |
| IELKFNPPAL | Not Generated | Not Generated | IELKFNPPALQDAYY | Not Generated | Not Generated |
| VSFLAHIQW | −8.8 | −9.26 | GVYSVIYLYLTFYLT | −8 | −5.02 |
Fig. 5.
Structural analysis for the highly immunogenic MHC-I restricted T-cell epitope “FLKKDAPYI” for NSP3 coded protein (a) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (b) Docking structure of MHC-I restricted T-cell epitope (c) The surface interaction between the allele and epitopes showing the fitting sites in binding grooves (d) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frame (e) Z-score plot and (f) all residue energy.
Fig. 6.
Structural analysis for the highly antigenic MHC-I restricted T-cell epitope “TAVVIPTKK” for NSP3 coded protein (a) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (b) Docking structure of MHC-I restricted T-cell epitope (c) The surface interaction between the allele and epitopes showing the fitting sites in binding grooves (d) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frame (e) Z-score plot and (f) all residue energy.
Fig. 7.
Structural analysis for the highly immunogenic MHC-II restricted T-cell epitope “ITFLKKDAPYIVGDV” for NSP3 coded protein (a) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (b) Docking structure of MHC-II restricted T-cell epitope (c) The surface interaction between the allele and epitopes showing the fitting sites in binding grooves (d) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frame (e) Z-score plot and (f) all residue energy.
Fig. 8.
Structural analysis for the highly antigenic MHC-II restricted T-cell epitope “IDITFLKKDAPYIVG” for NSP3 coded protein (a) 2D pose representation between the epitope and HLA allele showing the different non-covalent bonds (b) Docking structure of MHC-II restricted T-cell epitope (c) The surface interaction between the allele and epitopes showing the fitting sites in binding grooves (d) Ramachandran plot of the epitope allele structure showing lower energy sites of the residues in different frame (e) Z-score plot and (f) all residue energy.
Due to the worldwide pandemic caused by SARS-CoV-2, development of safe and effective vaccines is the need of the hour. This study has identified T-cell and B-cell epitopes using computational methods which can be used for probable vaccine design. The main advantages of this work can be summarised as (a) whole genome analysis of 566 Indian SARS-CoV-2 genomes in order to consider the genetic mutations to understand and target the virus proteins, (b) finding consensus conserved regions from four alignment techniques viz. ClustalW, MUSCLE, ClustalO and MAFFT and (c) using latest tools like NetMHCpan EL 4.1 (published in September 2020), PyMod 3 and BepiPred 2.0 for computational purposes. Furthermore, we have used our own developed tool ABCpred to predict the B-cell epitopes.
4. Conclusion
In this work, genome-wide analysis of 566 Indian SARS-CoV-2 genomes have been performed to extract the potential conserved regions for epitope-based synthetic vaccine design which show high immunogenicity and antigenicity. In this regard, 125 CCnRs have been identified after extracting the conserved regions from the aligned sequences of the four multiple sequence alignment techniques. These CCnRs are then filtered based on three major criteria of length greater than or equal to 60nt, no stop codons in the proteins and percentage of BLAST specificity score as query coverage equal to 100%. Such filtering resulted in 23 CCnRs covering NSP1, NSP2, NSP3, NSP4, 3CL-Proteinase, NSP10, RNA-directed RNA polymerase, Helicase, Spike glycoprotein and Nucleocapsid protein. This ranking also resulted in 34 MHC-I and 37 MHC-II restricted T-cell epitopes with 16 and 19 unique HLA alleles and 29 B-cell epitopes for the 23 CCnRs. These CCnRs are then ranked based on their immunogenic and antigenic scores to identify the MHC-I and MHC-II restricted T-cell and B-cell epitopes. This ranking identified CCnR from NSP3 coded protein to be highly immunogenic and antigenic, providing MHC-I and MHC-II restricted T-cell and B-cell epitopes, FLKKDAPYI, ITFLKKDAPYIVGDV, TLVSDIDITFLKKDAP as most immunogenic and TAVVIPTKK, IDITFLKKDAPYIVG, LHPDSATLVSDIDITF as most antigenic respectively. These epitopes can be considered for designing of synthetic vaccines. Furthermore, to validate the relevance of these epitopes, their binding confirmation and physico-chemical properties are also shown with respect to HLA alleles. This study thus provides the potential MHC-I and MHC-II restricted T-cell and B-cell epitopes to design epitope-based synthetic vaccines.
Ethics approval and consent to participate
The ethical approval or individual consent was not applicable.
Availability of data and materials
The aligned 566 Indian SARS-CoV-2 genomes with reference as well as consensus sequences and the final results of this work are available at “http://www.nitttrkol.ac.in/indrajit/projects/COVID-EpitopeVaccine-India/”. Moreover, Indian SARS-CoV-2 genomes used in this work are publicly available at GISAID database.
Consent for publication
Not applicable.
Funding
This work has been partially supported by CRG short term research grant on COVID-19 (CVD/2020/000991) from Science and Engineering Research Board (SERB), Department of Science and Technology, Govt. of India.
Author contributions
Nimisha Ghosh: Formal analysis; Methodology, Coding; Visualization; Writing - original draft & editing, Nikhil Sharma: Methodology; Coding; Visualization; Writing - review & editing, Indrajit Saha: Conceptualization; Data curation; Supervision; Funding acquisition; Formal analysis; Investigation; Methodology; Project administration; Resources; Validation; Visualization; Writing - review & editing, Sudipto Saha: Conceptualization; Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgement
We thank all those who have contributed sequences to GISAID database.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.intimp.2020.107276.
Supplementary material
The following are the Supplementary data to this article:
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C., Chen H., Chen J., Luo Y., Guo H., Jiang R., Liu M., Chen Y., Shen X., Wang X., Zheng X., Zhao K., Chen Q., Deng L.L.F., Yan B., Zhan F., Wang Y., Xiao G., Shi Z. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ksiazek T., Erdman D., Goldsmith C., Zaki S., Peret T., Emery S., Tong S., Urbani C., Comer J., Lim W., Rollin P., Dowell S., Ling A., Humphrey C., Shieh W.J., Guarner J., Paddock C., Rota P., Fields B., Anderson L. A novel coronavirus associated with severe acute respiratory syndrome. New Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Holmes K., Enjuanes L. The sars coronavirus: A postgenomic era. Science (New York, N.Y.) 2003;300:1377–1378. doi: 10.1126/science.1086418. [DOI] [PubMed] [Google Scholar]
- 4.Groot R.D., Baker S., Baric R., Brown C., Drosten C., Enjuanes L., Fouchier R., Galiano M., Gorbalenya A., Memish Z., Perlman S., Poon L., Snijder E., Stephens G., Woo P., Zaki A., Zambon M., Ziebuhr J. Middle east respiratory syndrome coronavirus (mers-cov): Announcement of the coronavirus study group. J. Virol. 2013;87 doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J., Zhao S., Teng T., Abdalla A., Zhu W., Xie L., Wang Y., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: Sars-cov-2 and sars-cov. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worldometer, Coronavirus disease 2019 (covid-19) cases in india, https://www.worldometers.info/coronavirus/country/india/, accessed: 2020-10-21 (2020).
- 7.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Cao B. Clinical features of patients infected with 2019 novel coronavirus in wuhan, china. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skwarczynski M., Toth I. Peptide-based synthetic vaccines. Chem. Sci. 2015;7 doi: 10.1039/C5SC03892H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nandy A., Basak S. Bioinformatics in Design of Antiviral Vaccines. Encycl. Biomed. Eng. 2018 doi: 10.1016/B978-0-12-801238-3.10878-5. [DOI] [Google Scholar]
- 10.Patronov A., Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3:120139. doi: 10.1098/rsob.120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad T., Ewida A., Sheweita S. B-cell epitope mapping for the design of vaccines and effective diagnostics. Trials Vaccinol. 2016;5:71–83. doi: 10.1016/j.trivac.2016.04.003. [DOI] [Google Scholar]
- 12.D. Wrapp, N. Wang, K. Corbett, J. Goldsmith, C. Hsieh, O. Abiona, B. Graham, J. Mclellan, Cryo-em structure of the 2019-ncov spike in the prefusion conformation, bioRxiv: the preprint server for biology (2020). doi:10.1101/2020.02.11.944462. [DOI] [PMC free article] [PubMed]
- 13.Amanat F., Krammer F. Sars-cov-2 vaccines: Status report. Immunity. 2020;52 doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling R., Dai Y., Huang B., Huang W., Lu X., Jiang Y. In silico design of antiviral peptides targeting the spike protein of sars-cov-2. Peptides. 2020;130:170328. doi: 10.1016/j.peptides.2020.170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vashi Y., Jagrit V., Kumar S. Understanding the b and t cells epitopes of spike protein of severe respiratory syndrome coronavirus-2: A computational way to predict the immunogens. Infect. Genet. Evol. 2020;84:104382. doi: 10.1016/j.meegid.2020.104382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rakib A., Saad A., Sami S., Mimi N.J., Chowdhury M., Eva T., Nainu F., Paul A., Shahriar A., Tareq A., Laam N.U., Chakraborty S., Shil S., Mily D., Hadda T.B., Almalki F., Emran T. Immunoinformatics-guided design of an epitope-based vaccine against severe acute respiratory syndrome coronavirus 2 spike glycoprotein. Comput. Biol. Med. 2020;124:103967. doi: 10.1016/j.compbiomed.2020.103967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naz A., Shahid F., Butt T., Awan F., Ali A., Malik D. Designing multi-epitope vaccines to combat emerging coronavirus disease 2019 (covid-19) by employing immuno-informatics approach. Front. Immunol. 2020;11:1663. doi: 10.3389/fimmu.2020.01663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grifoni A., Weiskopf D., Ramirez S., Mateus J., Dan J., Moderbacher C., Rawlings S., Sutherland A., Premkumar L., Jadi R., Marrama D., Silva A., Frazier A., Carlin A., Greenbaum J., Peters B., Krammer F., Smith D., Crotty S., Sette A. Targets of t cell responses to sars-cov-2 coronavirus in humans with covid-19 disease and unexposed individuals. Cell. 2020;181 doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noorimotlagh Z., Karami C., Mirzaee S.A., Kaffashian M., Mami S., Azizi M. Immune and bioinformatics identification of t cell and b cell epitopes in the protein structure of sars-cov-2: A systematic review. Int. Immunopharmacol. 2020;86:106738. doi: 10.1016/j.intimp.2020.106738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadav P.D., Potdar V., Choudhary M.L., Nyayanit D.A., Agrawal M., Jadhav S.M., Majumdar T.D., Aich A.S., Basu A., Abraham P., Cherian S.S. Full-genome sequences of the first two sars-cov-2 viruses from india. Indian J Med Res. 2020;151 doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saha I., Ghosh N., Maity D., Sharma N., Sarkar J., Mitra K. Genome-wide analysis of indian sars-cov-2 genomes for the identification of genetic mutation and snp. Infect. Genet. Evol. 2020;85:104457. doi: 10.1016/j.meegid.2020.104457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu W., Wang C., Wang B.Z. From variation of influenza viral proteins to vaccine development. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18071554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A.K., Khan M.S., Choudhury S., Mukhopadhyay A., Sakshi, Rastogi A., Thakur A., Kumari P., Kaur M., Shalu, Saini C., Sapehia V., Barkha, Patel P.K., Bhamare K.T., Kumar M. Coronavr: A computational resource and analysis of epitopes and therapeutics for severe acute respiratory syndrome coronavirus-2. Frontiers in Microbiology. 2020;11:1858. doi: 10.3389/fmicb.2020.01858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crooke S.N., Ovsyannikova I.G., Kennedy R.B., Poland G.A. Immunoinformatic identification of b cell and t cell epitopes in the sars-cov-2 proteome. Sci. Rep. 2020;10:14179. doi: 10.1038/s41598-020-70864-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong E., Wong M.U., Huffman A., He Y. Covid-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front. Immunol. 2020;11:1581. doi: 10.3389/fimmu.2020.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharya M., Sharma A., Patra P., Ghosh P., Sharma G., Patra B., Lee S.S., Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (sars-cov-2): Immunoinformatics approach. J. Med. Virol. 2020;92 doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H.Z., Tang L.L., Yu X.L., Zhou J., Chang Y.F., Wu X. Bioinformatics analysis of epitope-based vaccine design against the novel sars-cov-2. Infect. Dis. Poverty. 2020;9 doi: 10.1186/s40249-020-00713-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kar T., Narsaria U., Basak S., Deb D., Castiglione F., Mueller D., Srivastava A. A candidate multi-epitope vaccine against sars-cov-2. Sci. Rep. 2020;10:10895. doi: 10.1038/s41598-020-67749-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhatnager R., Bhasin M., Arora J., Dang A. Epitope based peptide vaccine against sars-cov2: an immune-informatics approach. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1787227. [DOI] [PubMed] [Google Scholar]
- 30.Kwarteng A., Asiedu E., Sakyi S.A., Asiedu S.O. Targeting the sars-cov2 nucleocapsid protein for potential therapeutics using immuno-informatics and structure-based drug discovery techniques. Biomed. Pharmacotherapy. 2020:132. doi: 10.1016/j.biopha.2020.110914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baruah V., Bose S. Immunoinformatics-aided identification of t cell and b cell epitopes in the surface glycoprotein of 2019-ncov. J. Med. Virol. 2020;92 doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bency J., Helen M. Novel epitope based peptides for vaccine against sars-cov-2 virus: immunoinformatics with docking approach. Int. J. Res. Med. Sci. 2020;8:2385. doi: 10.18203/2320-6012.ijrms20202875. [DOI] [Google Scholar]
- 33.Singh A., Thakur M., Sharma L., Chandra K. Designing a multi-epitope peptide based vaccine against sars-cov-2. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-73371-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson J.D., Higgins D.G., Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sievers F., Wilm A., Dineen D., Gibson T., Karplus K., Li W., Lopez R., Mcwilliam H., Remmert M., Söding J., Thompson J., Higgins D. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sievers F., Higgins D. Clustal omega. Curr. Protocols Bioinformat. 2014;48 doi: 10.1002/0471250953.bi0313s48. 3.13.1–3.13.16. [DOI] [PubMed] [Google Scholar]
- 38.Katoh K., Kuma K.I., Miyata T., Toh H. Improvement in the accuracy of multiple sequence alignment program mafft, Genome informatics. Int. Conf. Genome Informat. 2005;16:22–33. [PubMed] [Google Scholar]
- 39.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T. Ncb blast: a better web interface. Nucleic Acids Res. 2008;36:W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sidney J., Dow C., Mothé B., Sette A., Peters B. A systematic assessment of mhc class ii peptide binding predictions and evaluation of a consensus approach. PLoS Comput. Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace A.C., Laskowski A.R., Thornton J.M. Ligplot: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Select. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 42.Jespersen M., Peters B., Nielsen M., Marcatili P. Bepipred-2.0: Improving sequence-based b-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan S., Chan H.S., Hu Z. Using pymol as a platform for computational drug design. WIREs Comput. Mol. Sci. 2017;7(2):e1298. doi: 10.1002/wcms.1298. [DOI] [Google Scholar]
- 44.Rauf M.A. Ligand docking and binding site analysis with pymol and autodock/vina. Int. J. Basic Appl. Sci. 2015;4:168–177. doi: 10.14419/ijbas.v4i2.4123. [DOI] [Google Scholar]
- 45.Janson G., Paiardini A. Pymod 3: a complete suite for structural bioinformatics in pymol. Bioinformatics. 2020:1367–4803. doi: 10.1093/bioinformatics/btaa849. [DOI] [PubMed] [Google Scholar]
- 46.Wiederstein M., Sippl M. Prosa-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–10. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salat J., Mikulasek K., Larralde O., Formanova P.P., Chrdle A., Haviernik J., Elsterova J., Teislerova D., Palus M., Eyer L., Zdrahal Z., Petrik J., Ruzek D. Tick-borne encephalitis virus vaccines contain non-structural protein 1 antigen and may elicit ns1-specific antibody responses in vaccinated individuals. Vaccines. 2020;8 doi: 10.3390/vaccines8010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibson C.A., Schlesinger J.J., Barrett A.D.T. Prospects for a virus non-structural protein as a subunit vaccine. Vaccine. 1988;6(1):7–9. doi: 10.1016/0264-410X(88)90004-7. [DOI] [PubMed] [Google Scholar]
- 49.Chen H.R., Lai Y.C., Yeh T.M. Dengue virus non-structural protein 1: a pathogenic factor, therapeutic target, and vaccine candidate. J. Biomed. Sci. 2018;25(58) doi: 10.1186/s12929-018-0462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lan J., Zhou J.M., Yin Y., Fang D.Y., Tang Y.X., Jiang L.F. Selection and identification of b-cell epitope on ns1 protein of dengue virus type 2. Virus Res. 2010;150:49–55. doi: 10.1016/j.virusres.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 51.Ip P.P., Boerma A., Regts J., Meijerhof T., Wilschut J., Nijman H.W., Daemen T. Alphavirus-based vaccines encoding nonstructural proteins of hepatitis c virus induce robust and protective t-cell responses. Mol. Therapy. 2014;22(4) doi: 10.1038/mt.2013.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Satyam R., Janahi E., Bhardwaj T., Somvanshi P., Haque S., Najm M. In silico identification of immunodominant b-cell and t-cell epitopes of non-structural proteins of usutu virus. Microbial Pathogen. 2018;125 doi: 10.1016/j.micpath.2018.09.019. [DOI] [PubMed] [Google Scholar]
- 53.Cafaro A., Tripiciano A., Picconi O., Sgadari C., Moretti S., Buttò S., Monini P., Ensoli B. Anti-tat immunity in hiv-1 infection: Effects of naturally occurring and vaccine-induced antibodies against tat on the course of the disease. Mol. Ther. 2019;7(3) doi: 10.3390/vaccines7030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vita R., Mahajan S., Overton J., Dhanda S., Martini S., Cantrell J., Wheeler D., Sette A., Peters B. The immune epitope database (iedb): 2018 update. Nucleic Acids Res. 2018:gky1006. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doytchinova I., Flower D. Vaxijen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. bmc bioinformatics 8:4. BMC Bioinformat. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saha S., Raghava G. Prediction methods for b-cell epitopes. Methods Mol. Biol. (Clifton, N.J.) 2007;409:387–394. doi: 10.1007/978-1-60327-118-9_29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The aligned 566 Indian SARS-CoV-2 genomes with reference as well as consensus sequences and the final results of this work are available at “http://www.nitttrkol.ac.in/indrajit/projects/COVID-EpitopeVaccine-India/”. Moreover, Indian SARS-CoV-2 genomes used in this work are publicly available at GISAID database.