Abstract
Members of the APOBEC family of cytidine deaminases show antiviral activities in mammalian cells through lethal editing in the genomes of small DNA viruses, herpesviruses and retroviruses, and potentially those of RNA viruses such as coronaviruses. Consistent with the latter, APOBEC-like directional C→U transitions of genomic plus-strand RNA are greatly overrepresented in SARS-CoV-2 genome sequences of variants emerging during the COVID-19 pandemic. A C→U mutational process may leave evolutionary imprints on coronavirus genomes, including extensive homoplasy from editing and reversion at targeted sites and the occurrence of driven amino acid sequence changes in viral proteins. If sustained over longer periods, this process may account for the previously reported marked global depletion of C and excess of U bases in human seasonal coronavirus genomes. This review synthesizes the current knowledge on APOBEC evolution and function and the evidence of their role in APOBEC-mediated genome editing of SARS-CoV-2 and other coronaviruses.
Keywords: SARS-CoV-2, Coronavirus, Virus evolution, APOBEC, Innate immunity
Highlights
-
-
SARS-CoV-2 sequence variants contain an overabundance of C- > U transitions
-
-
C- > U transitions are the hallmark of the activity of APOBEC cytosine deaminases
-
-
Further work is needed to determine APOBEC's role in coronavirus evolution
1. Introduction
The sustained spread of SARS coronavirus 2 (SARS-CoV-2), coupled with readily deployable and accessible sequencing technologies, has given researchers an unprecedented ability to track virus evolution from spillover through early adaptation to human hosts. Mining this data, several authors have identified an overabundance of homoplastic cytidine to uridine transitions in the SARS-CoV-2 genome, particularly in the context of 5′-UC-3′, resulting in a large proportion of nonsynonymous mutations. This mutational pattern is similar to that induced by the cytidine deaminases of the apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) family of proteins. The ability of this enzyme family to modify retroviruses, retroelements, and some DNA viruses is well established, while their potential to modify RNA viruses is exploratory. This review will provide an overview of the current knowledge of APOBEC function and evolution, the evidence for this mutational phenomenon in coronaviruses, and its potential evolutionary consequences for RNA viruses.
2. Discovery of the APOBEC family
In 1987, two groups identified that tissue-specific expression of ApoB100 and ApoB48 from the ApoB gene depended on post-transcriptional modifications of messenger RNA rather than differences in their genomic content (Chen et al., 1987; Powell et al., 1987). In both human and rat enterocytes, a cytosine to uracil modification (C6666U) generated a UAA stop codon and translation of a truncated version of ApoB48 with a modified biological function. In 1993, the protein catalyzing this reaction was isolated from rat enterocytes and designated apo B mRNA editing protein (REPR) (Teng et al., 1993). Shortly thereafter, the existence of other mRNA editing enzymes functioning through a similar catalytic subunit was hypothesized, and the enzyme responsible for ApoB48 was renamed Apolipoprotein B mRNA Editing Catalytic Polypeptide 1 (APOBEC1) (Davidson et al., 1995). Since that time, many APOBEC1-related genes and splice variants have been identified in the genomes and transcriptomes of fish, mammals, non-human primates, and humans, which collectively are referred to as the APOBEC family of proteins. In humans, the family of cytidine deaminases has 11 members: APOBEC1 (A1), APOBEC2 (A2), seven APOBEC3s (A3A, A3B, A3C, A3D, A3F, A3G, A3H), APOBEC4 (A4), and activation induced deaminase (AID) (Jarmuz et al., 2002; Rogozin et al., 2005). The proteins possess a shared, highly conserved zinc-dependent deaminase domain (ZDD) but are otherwise diverse in function, having been implicated in a plethora of diverse biological pathways including lipid metabolism, antibody diversification, restriction of viruses and transposable elements, and cancer (Burns et al., 2015; Green et al., 2016; Henderson and Fenton, 2015; Roberts et al., 2013; Smith, 2017; Taylor et al., 2013).
3. Conservation of structure
Members of the APOBEC family of proteins are readily recognized through their possession of a highly conserved ZDD motif His-X-Glu-X23-28-Pro-Cys-X2-4-Cys that is present in the cytidine deaminase fold (CD) (Salter et al., 2016). In those maintaining enzymatic activity, the active site of the ZDD coordinates a zinc ion that activates a water for nucleophilic attack of C4 of the pyrimidine ring of C, which in conjunction with the glutamic acid residue leads to the deamination of the base to U. The CD fold is nested in a conserved super-secondary structure of a five-stranded β-sheet surrounded by six α-helices, with minor variations in the X residues potentially leading to the diverse functions and targets of this family of proteins (Salter et al., 2016). A3B, A3F, and A3G contain double deaminase domains where the N-terminal domain is no longer catalytically active for deamination of ssDNA (Salter et al., 2016). In humans, all members of the family have multiple exons and some have been shown to produce splice variants (Harari et al., 2009; Lee et al., 1998; van Maldegem et al., 2009).
4. Diversity of function in humans
A1 primarily functions to increase diversity in host-expressed RNAs in a tissue-specific manner (Smith, 2017). The RNA binding and catalytic activity of A1 is dependent on the binding of one of two cofactors - A1 complementation factor (ACF) or RBM47 (Fossat et al., 2014; Mehta et al., 2000) – and, in addition to ApoB, edits NF1, eIF4G, and the 3′UTRs of other host mRNAs (Rosenberg et al., 2011; Skuse et al., 1996; Yamanaka et al., 1997). ACF binds an 11-nt ‘mooring sequence’ 3′ of the target cytidine residue that partially, but not completely, determines the specificity of the nucleotide target (Backus and Smith, 1991; Blanc et al., 2014; Rosenberg et al., 2011). A1 deaminates cytidines in the context of 5′-AC-3′ with the mooring sequence 4–6 nt downstream of the target and is primarily found in the cytoplasm but can traffic to the nucleus. However, A1 is the exception and not the rule to this family, as most APOBEC proteins are regarded for their ability to deaminate C to dU in ssDNA rather than RNA.
AID and AID-like enzymes are an essential component of the adaptative immune system in all jawed vertebrate species (Flajnik and Kasahara, 2010; Rogozin et al., 2007). In humans, AID drives somatic hypermutation and class-switch recombination in B cells through recruitment of DNA repair enzymes following introduction of dU into immunoglobulin V and C genes (Chaudhuri et al., 2003; Dickerson et al., 2003; Muramatsu et al., 1999, 2000; Pham et al., 2003). The introduced dU can lead to transitions upon DNA replication or stochastic introduction of insertions or deletions during mismatch repair, leading to the diversity that is needed to expand the antibody repertoire of the host. AID preferentially deaminates in the 5′-WRC-3′ context (W = A or T, R = A or G) and is basally present in the cytoplasm but traffics to the nucleus to perform immunoglobulin gene editing.
A2 has roles in skeletal and cardiac muscle differentiation (Etard et al., 2010; Liao et al., 1999; Sato et al., 2010; Vonica et al., 2011). Transcripts of eukaryotic translation initiation factor 4 gamma 2 (Eif4g2) and phosphatase and tensin homolog (PTEN) have been suggested as mRNA targets in A2-overexpressing hepatocytes (Okuyama et al., 2012). However, transgene expression of A1 in hepatocytes also leads to Eif4g2 editing (Yamanaka et al., 1995, 1997), so it is unclear if this represents the in vivo function or is an artifact of the overexpression model. A4 was discovered in humans through bioinformatic identification and contains orthologs in other mammals, chickens, and frogs (Rogozin et al., 2005). The function of the protein is as of yet unknown but expression is upregulated in the testes of mice, leading the authors to suggest a potential role in spermatogenesis. An A4 homolog was recently cloned from chicken cells and showed in vitro antiviral activity against Newcastle disease virus, even though editing of the viral genome was not described (Shi et al., 2020). Neither A2 nor A4 have described target sequences. A2 shows a distributed intracellular distribution while the cellular localization of A4 remains poorly characterised.
The principal activity of the A3 subfamily of proteins is the restriction of viruses and mobile genomic elements, which is covered extensively below. A3A, A3B, A3C, A3D, A3F, and A3H target sequences with a 5′-TC-3′ motif, while A3G targets 5′-CC-3’. Recently, McDaniel et al. investigated the relevance of sequence context adjacent to a TC motif and secondary structure elements to A3 activity on DNA oligonucleotides (McDaniel et al., 2020). Flanking sequence preference varied between subfamily members and within difference structural contexts. In general, A3A, A3F, and A3H preferred A or T flanking the target motif and all members of the subfamily preferred the edited site to be presented outside of defined secondary structure elements. A3A, A3C, and A3H show a broadly distributed intracellular distribution, while A3B is predominantly nuclear, and A3D, A3F, and A3G are primarily localized in the cytoplasm.
Intriguingly, both A3A and A3G have recently been experimentally shown to edit host mRNAs, with A3A-driven editing in activated innate immune cells and A3G-driven editing in hypoxic natural killer and CD8+ T cells and overexpressing HEK293T cells (Sharma et al., 2015, 2016, 2019). As all A3s have demonstrated RNA binding activity (Salter et al., 2016), it does open up the intriguing possibility of deamination of RNA in specific cellular contexts by members of the A3 subfamily. Perhaps reflecting the structure dependence of A3-meditated editing of DNA oligonucleotides (McDaniel et al., 2020), it had been shown that editing of host mRNA in activated macrophages and monocytes by A3A occurred preferentially at sites flanked by short palindromic sequences (Sharma et al., 2015). Using bioinformatic approaches, the authors further demonstrated that 72% of edited sites in monocytes and 67% of edited sites in macrophages were present in the loop of a predicted stem-loop structure. In both cell lines, the most common flanking sequence was an upstream A and a downstream A/G, consistent with context preferences for ssDNA targets (McDaniel et al., 2020). C was also commonly seen in the −3 position.
4.1. Deaminase-dependent restriction of viruses and mobile genetic elements
Human Immunodeficiency Virus (HIV-1). The inhibition of HIV-1 replication through the function of A3D, A3F, A3G, and A3H is the most extensively studied of the viral restriction phenotype of the APOBEC family and has been well reviewed elsewhere (Aydin et al., 2014; Delviks-Frankenberry et al., 2020; Desimmie et al., 2014; Haché et al., 2006; Harris and Dudley, 2015; Xu et al., 2020). During production of new virions in a productively infected host cell, A3 proteins form ribonucleoprotein complexes with HIV-1 RNA and the nucleocapsid domain of the Gag protein. The A3 proteins are subsequently packaged and encapsidated in the mature HIV-1 virions. The major hypermutation events take place during subsequent cellular infection. After viral entry and uncoating, A3 proteins co-localize with HIV-1 RNA during reverse transcription of nascent negative-strand viral cDNA. This single-stranded cDNA acts as the template for plus-strand genomic DNA synthesis and is highly susceptible to the cytidine deamination activities of the packaged A3 proteins. The C→U transitions in the template strand (negative-sense strand) result in G→A transitions in the genomic strand (positive-sense strand) (Mangeat et al., 2003). Many hypermutated proviruses are degraded during or after generation of the cDNA template strand, potentially through a pathway mediated by uracil DNA glycosylase-2, and do not integrate in the host cell genome (Kaiser and Emerman, 2006; Langlois and Neuberger, 2008; Weil et al., 2013; Yang et al., 2007). A large portion of those proviruses that do successfully integrate are rendered defective due to an abundance of mutated start codons and nonsense mutations in several open reading frames (Bruner et al., 2016; Janini et al., 2001; Kieffer et al., 2005).
HIV-1 and other retroviruses contain a potent counteractor of APOBEC restriction – the accessory protein viral infectivity factor (Vif). The early 2000s contained a cascade of information regarding the interaction between Vif and A3G (Conticello et al., 2003; Kao et al., 2003; Marin et al., 2003; Mehle et al., 2004a, 2004b; Sheehy et al., 2002, 2003; Stopak et al., 2003; Yu et al., 2003). Per the current model, Vif recruits an E3 ubiquitin ligase complex composed of Elongin B/C, cullin 5, RING-box subunit 2 (Rbx2), and core-binding factor (CBFβ) to polyubiquitinate A3G, leading to degradation via the host proteasome (Fribourgh et al., 2014; Guo et al., 2014; Jäger et al., 2011; Zhang et al., 2011). Vif-like proteins have been described for other lentiviruses including simian immunodeficiency virus, feline immunodeficiency virus, bovine immunodeficiency virus, jembrana disease virus, visna/maedi virus, and caprine arthritis encephalitis virus (Kristbjörnsdóttir et al., 2004; Nakano et al., 2017; Wang et al., 2018; Zhang et al., 2014; Zhao et al., 2019; Zielonka et al., 2010). Species-specific spumaretrovirinae, another subfamily of retroviruses, infect a diverse number of mammals and encode the accessory protein Bet, which impairs the activity of A3G through direct binding and sequestration rather than targeted degradation (Jaguva Vasudevan et al., 2013; Löchelt et al., 2005; Russell et al., 2005). The presence of APOBEC-antagonism by protein-protein interactions in lentiviruses that infect diverse placental mammals strongly suggests active antiviral APOBEC activity in these representative species. As an alternative mechanism, human T-Cell leukemia virus (HTLV-1) avoids APOBEC activity simply through exclusion of A3G from the packaged virus (Derse et al., 2007). The diverse approaches of retroviruses to avoid A3 activity highlight the importance of this subfamily for restricting this class of viruses.
Retrotransposons and Mobile Genetic Elements. Transposable elements (TEs) are selfish genetic elements that are able to move within the genome via one of two methods (Boeke et al., 1985; Bourque et al., 2018; Rubin et al., 1982). Class I TEs (retrotransposons) replicate through a “copy and paste” mechanism wherein reverse transcription of an RNA intermediate produces a cDNA copy of the genetic element that integrates elsewhere in the genome. Class II TEs (DNA transposons) utilize a “cut and paste” mechanism via the activity of an encoded transposase enzyme and do not require RNA intermediaries. The evolution of the human genome has been shaped by mobile genetic elements, and estimates suggest that 46% of it may be comprised of inactivated TEs (Biémont and Vieira, 2006; Lander et al., 2001). In humans, only a subset of long-interspersed element-1 (LINE-1) retrotransposons and short interspersed nuclear elements (SINEs) remain retrotransposition-competent (Brouha et al., 2003; Richardson et al., 2015; Schumann, 2007). The majority of APOBEC family activity on retroelements has been demonstrated on LINE-1 elements in vitro. A3A inhibits LINE-1 retrotransposition through a deaminase-dependent mechanism (Richardson et al., 2014) while AID, A1, A3B, A3C, and A3F act through a deaminase-independent pathway (Ikeda et al., 2011; MacDuff et al., 2009; Pizarro and Cristofari, 2016). It has been hypothesized that the massive reduction in active LINE-1 elements in the primate taxon compared to rodentia could be the result of extensive duplication and expansion of the A3 gene (Ostertag and Kazazian, 2001; Schumann, 2007). However, analysis of the depletion of A3 targeted sequence motifs in human retroelements only demonstrated the footprint of A3 activity on a subset of endogenous retroviruses and not in LINE-1 or SINE elements (Anwar et al., 2013).
DNA viruses. Beyond retroviruses and TEs, APOBEC-driven C→U/G→A mutations have been demonstrated in a diverse set of ssDNA and dsDNA viruses (Moris et al., 2014). A3B, A3C, A3F, and A3G have been demonstrated to deaminate both the ssDNA negative-sense and positive-sense strand of the pararetrovirus Hepatitis B Virus (HBV) in vitro and in vivo resulting in a low proportion of G→A hypermutated genomes (Suspène et al., 2005). In vitro work has also demonstrated that AID and A1 can edit HBV DNA and HBV DNA and RNA, respectively (Gonzalez et al., 2009; Liang et al., 2013). Murine A1 (mA1) is canonically expressed in hepatic cells and, in a mouse model of HBV infection, was shown to edit transgenic HBV genomes (Renard et al., 2010). The low endogenous expression of AID and A1 in human cells canonically infected with HBV limits the interpretability of their contribution to hypermutation, although AID been shown to be upregulated in human hepatocytes upon infection with hepatitis C virus in vitro (Endo et al., 2007; Seeger and Mason, 2000; Uhlén et al., 2015). Human papillomaviruses (HPV), a family of dsDNA viruses, have also been shown to be edited by A3A, A3C, and A3H in A3-transfected 293T cells (Vartanian et al., 2008), while overexpressed A3A, but not A3B or A3C, was able to restrict HPV replication in immortalized keratinocytes (Warren et al., 2015b). Bioinformatic analysis of the HPV genome suggests that the preferred target of A3 enzymes (5′-TC-3′) is significantly depleted, consistent with long-term adaptation to infection of A3 expressing tissues (Warren et al., 2015a). Other DNA viruses shown to be hypermutated by A3 proteins included transfusion-transmitted virus, herpes simplex virus 1, and Epstein-Barr virus (Suspène et al., 2011; Tsuge et al., 2010).
RNA Viruses. Although the first description of APOBEC editing activity was on the cellular ApoB mRNA template, direct evidence of APOBEC-driven deamination in RNA viruses has been problematic to demonstrate. The earliest evidence came in 2005 when the rat A1 homolog was shown to edit HIV-1 RNA in vitro (Bishop et al., 2004). Recently, it was shown that the restriction of HCoV-NL63, a human-infective alphacoronavirus, by A3C, A3F, and A3H was deaminase-dependent, but, in this case, the authors were unable to demonstrate that this resulted from virus genome editing even after serial passaging of HCoV-NL63 in A3 overexpressing cells (Milewska et al., 2018). Furthermore, while bioinformatic analysis of a large dataset of human-infective viruses demonstrated significant depletion of A3 target motifs (5′-TC-3′) in the DNA viruses parvoviruses, herpesviruses, and papillomaviruses, there was little evidence for this in RNA virus genomes with the exception of the seasonal coronaviruses HCoV-NL63, HCoV-OC43, HCoV-HKU1, and HCoV-229E (although oddly this signature was not detected in the recently emerged MERS-CoV, SARS-CoV-1 and SARS-CoV-2 or their zoonotic source viruses) (Poulain et al., 2020). However, analysis of immunodeficiency-related vaccine-derived rubella virus in the granulomas of infected children did demonstrate a preponderance of C→U transitions and depletion of 5′-TC-3′, accounting for 30% of the transitions when compared to the virus strain used in vaccine manufacture (Perelygina et al., 2019).
Deaminase-Independent Restriction and Insensitivity. Deaminase-independent restriction has been demonstrated primarily using overexpression models of catalytically inactive A3 mutants. This phenomenon has been reported for many viruses, including HIV-1, measles, mumps, respiratory syncytial virus, HCoV-NL63, adeno-associated virus, enterovirus A71, and minute virus of mice (Chen et al., 2006; Fehrholz et al., 2012; Li et al., 2018; Narvaiza et al., 2009). Long-interspersed element 1 is also inhibited by A3B and A3F in a deaminase-independent manner (Stenglein and Harris, 2006). In HIV-1, the prevailing model for this restriction is binding of genomic RNA inhibiting the initiation of reverse transcription, which may also explain the phenotype in LINE-1. The mechanism in other RNA and DNA viruses is not well described. Influenza virus and vaccinia virus are not restricted by A3 overexpression and appear to avoid targeting through unknown mechanisms (Kremer et al., 2006; Pauli et al., 2009).
4.2. APOBEC origins, diversification, and evolution
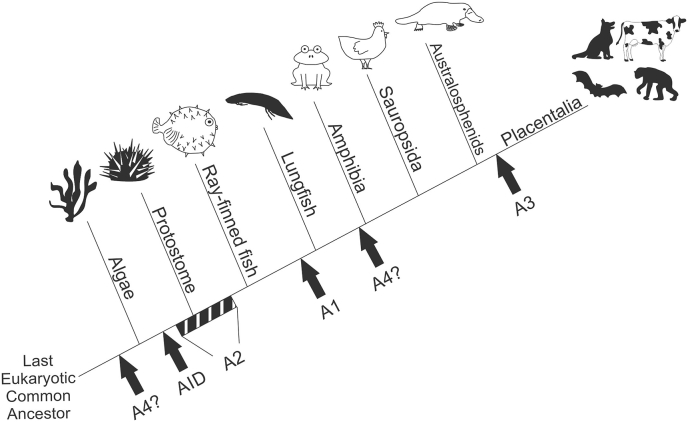
Origin. The APOBEC family is related to bacterial, yeast, and plant deaminases all possessing highly conserved amino acid motifs responsible for coordination of zinc in the active site (Conticello et al., 2005; Gerber and Keller, 2001; Iyer et al., 2011) (Fig. 1). Bioinformatic analysis suggests that A4 may be the ancient member of the APOBEC family, as homologs have been identified in the phylum cnidaria and occasionally in algal lineages (Krishnan et al., 2018). Another recent study identified, cloned, and demonstrated deaminase activity in AID homologs from sea urchins and branchiopods (Liu et al., 2018). The AID homologs showed increased expression in barrier tissues and were upregulated in response to bacterial challenge, suggesting a potential role in antimicrobial defense. Ancestral members of the family at the point of vertebrate speciation include both AID and A2, and homologs of the two proteins have been identified bioinformatically in birds, amphibians, and ray-finned fish (zebrafish and pufferfish), with an AID homolog responsible for diversity in variable lymphocyte receptors of jawless vertebrates (Conticello et al., 2005; Hsu, 2016). A1 and A3 both arose from independent duplication events of AID (Conticello et al., 2005). A1 has been identified in birds, reptiles, amphibians, and lungfish, suggesting it arose prior to the tetrapod-lungfish divergence (Krishnan et al., 2018). A3 emerged from AID duplication in placental mammals and has undergone frequent duplication, fusion, and loss in different branches of the clade (Krishnan et al., 2018; Munk et al., 2012).
Fig. 1.
Emergence of APOBEC proteins. Branch length is not drawn to timescale. Question marks for A4 protein represent a lack of concrete evidence at the divergence of amphibians (Rogozin et al., 2005) and putative evidence in algae species (Krishnan et al., 2018). Block for A2 represents uncertainty on emergence prior to protostome - deuterostome split. A1 and A3 both arose from duplication of AID.
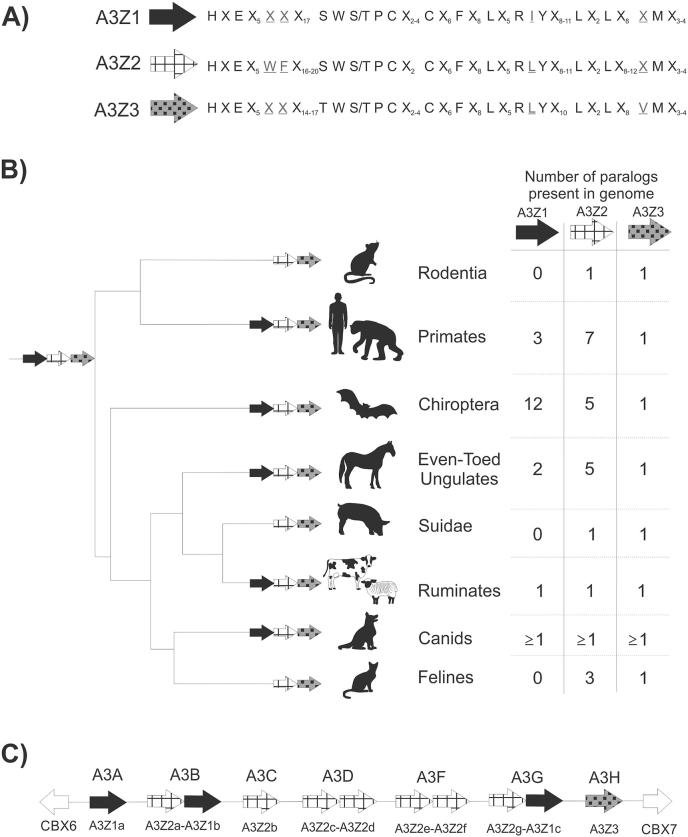
A3 paralog expansion. A3 homologs are not present in marsupials or monotreme genomes and appear to be exclusive to placental mammals (Ito et al., 2020; Mikkelsen et al., 2007; Warren et al., 2008). The A3 proteins contains three paralogs (A3Z1, A3Z2, A3Z3) that are designated according to conservation of amino acid residues within the zinc-coordinating domain (LaRue et al., 2009) (Fig. 2a). From the most recent common ancestor of the three A3 paralogs, A3Z3 may be the most ancient having diverged approximately 250 million years ago with A3Z1 and A3Z2 diverging from each other 50 million years later (Munk et al., 2012). It is hypothesized that the most recent common ancestor of placental mammals contained a gene set of A3Z1-A3Z2-A3Z3 that, through multiple processes of differential evolution, have led to the diversity seen in extant species (Fig. 2b). Mapping and identification of A3 genes takes advantage the presence of genes CBX6 and CBX7 upstream and downstream of the gene locus, respectively. For the purpose of this article, particular attention will be played to species known to be hosts for coronaviruses – bats and humans – but we draw attention to an extensive review of the activity and characteristics of feline A3s (Zhang et al., 2018).
Fig. 2.
Expansion of A3 paralogs. (A) Amino acid motifs within the zinc deaminase domain of the three A3 paralogs. Differences highlighted in grey and underlined. (B) Expansion of paralogs in different branches of placental mammals. A3Z1 has been independently lost in several taxonomic classes. Branch length is not drawn to timescale. Table lists number of paralogs and not number of gene products. Taxonomic relationships designed in consultation with the Tree of Life web project. (C) Diagram of the A3 gene locus in humans and primates. Adjacent paralogs (e.g. A3Z2a-A3Z1b) represent fused paralogs in a single gene product.
The complexity of APOBEC expression in bats has recently been revealed (Hayward et al., 2018). Within the pteropid family, eighteen A3 proteins were identified, of which twelve are single deaminase A3Z1, one is a single deaminase A3Z2, four are composed of a single deaminase of a novel paralog termed ‘A3Z2B’, and one is a double deaminase of A3Z2B-Z3. Active expression of 13 of these proteins was identified in spleen tissue extracted from a black flying fox. The large expansion of Z1 is particularly unusual as this paralog has been independently lost in the mammalian clades Rodentia, Felidae, and Suidae (domestic and wild pigs) (LaRue et al., 2008) (Fig. 2b).
The A3 locus and its evolution in humans compared to other mammals is thoroughly described in Münk et al. (Munk et al., 2012) (Fig. 2c). A3A-G form the seven A3 proteins in the human A3 locus, and the dual-deaminase proteins form a complicated interrelationship of the A3Z1, A3Z2, and A3Z3 paralogs. The current organization likely arose through as many as 6 gene duplication events. In all known species, there is no evidence for duplication of the A3Z3 gene.
APOBEC protein evolution. The long-term evolution of the APOBEC proteins is of particular interest as it may indicate which of the proteins perform essential activities for host survival (and thus show low rates of evolution, referred to as purifying or negative selection) and which may be involved in host-pathogen interactions (and thus show rapid fixation of nonsynonymous mutations, referred to as positive or directional selection). Current evidence for the overall evolutionary trajectory is summarized in Table 1. When evaluating the evolution of AID using representatives from seven mammalian clades, the protein was shown to be under strict purifying evolution (Munk et al., 2012). Repeating the analysis for A3Z1, A3Z2, and A3Z2 demonstrated multimodal evolution and sliding-window analysis indicated that certain regions of the gene are under purifying selection and others are under positive selection. This multimodal relationship for A1 was also present but to a lesser degree than the A3 paralogs. It is likely that the regions under purifying selection are necessary for the direct function of the protein but the regions under positive selection allow for diversity in targets or are the targets of antagonizing viral proteins. Similar analysis with a more limited subset of clades indicated that A3Z1, A3Z2, and A3Z3 all showed dN/dS ratios greater than 1, consistent with overall positive selection (LaRue et al., 2008). When comparing whole genes between humans and chimpanzees, AID and A3A were under purifying selection, A3B, A3D, and A3G were under positive selection, and A3F was under positive selection in a sliding window analysis only (Sawyer et al., 2004). Comparing humans to a broader range of primates, A2 was under purifying selection and A3C was under positive selection in a sliding window analysis. A1 was under purifying selection in whole gene comparisons and positive selection under sliding window, consistent with the multimodal results seen above. Curiously, the sites under positive selection for A3G were not isolated to the regions targeted by the HIV-1 Vif protein as one may expect if the core function of positive selection was to limit retroviral antagonism escape. Altogether, the evidence implicates the A3 proteins and potentially some functionality of A1 as being involved in the pathogens arms race and being suitable targets for further investigation in an RNA virus context.
Table 1.
Overview of APOBEC family proteins.
| Name | Taxonomic Emergence | Evolutionary Pressure | Deaminase Domains (active) | Cellular Localizationa | Preferred target Sequence (principally defined in ssDNA) |
|---|---|---|---|---|---|
| A1 | Chordata | Multimodal (all mammals), purifying (whole gene), positive (sliding window) | 1 | n/C | 5′-AC-3′ |
| AID | Deuterostomes | Purifying | 1 | n/C | 5′-WRC-3′ |
| A2 | Bony vertebrates (Euteleostomi) | Positive | ? | N/C | ? |
| A4 | LECA? | ? | ? | ? | ? |
| A3Z1 | Placental Mammals | Multimodal | 1 | ? | ? |
| A3Z2 | Placental Mammals | Multimodal | 1 | ? | ? |
| A3Z3 |
Placental Mammals |
Multimodal |
1 |
? |
? |
| A3A | Primates | Purifying | 1 (N-A3Z1-C) | N/C | 5′-TC-3′ |
| A3B | Primates | Positive | 2 (N-A3Z2-A3Z1-C) | N | 5′-TC-3′ |
| A3C | Primates | Positive (sliding window) | 1 (N-A3Z2-C) | N/C | 5′-TC-3′ |
| A3D | Primates | Positive | 2 (N-A3Z2-A3Z2-C) | C | 5′-TC-3′ |
| A3F | Primates | Positive (sliding window) | 2 (N-A3Z2-A3Z2-C) | C | 5′-TC-3′ |
| A3G | Primates | Positive | 2 (N-A3Z2-A3Z1-C) | C | 5′-CC-3′ |
| A3H | Primates | Unclear | 1 (N-A3Z3-C) | N/C | 5′-TC-3′ |
Adapted from Salter et al. (2016). C, predominantly cytoplasmic; n/C, can traffic to nucleus but predominantly cytoplasmic; N/C, even localization; N, predominantly nuclear.
4.3. Bioinformatic evidence for RNA editing in SARS-CoV-2 and other coronavirus genomes
C→U Mutational Asymmetry in SARS-CoV-2. Coronaviridae are a family of single-stranded, positive-sense RNA viruses, of which there are seven known to infect and transmit in human populations (Table 2). HCoV-NL63 and HCoV-229E are seasonal alphacoronaviruses and HCoV-HKU1 and HCoV-OC43 are seasonal betacoronaviruses that all cause cold-like symptoms and circulate during the winter months (Gaunt et al., 2010). MERS-CoV, SARS-CoV-1, and SARS-CoV-2 are recently emergent betacoronaviruses that are responsible for large outbreaks of viral disease associated with high levels of morbidity and mortality. Coronaviruses, in common with other members of the Nidovirales order, encode a 3′-5′ exonuclease that dramatically reduces the effective error rate of the viral RNA-dependent RNA polymerase (Eckerle et al., 2007, 2010; Smith et al., 2013). Consequently, coronaviruses demonstrate a low rate substitution rate and minor differences in sequence diversity despite a genome size of nearly 30,000 bases.
Table 2.
Characteristics of human-infective coronaviruses.
| Virus | Classification | Year of Discovery or Emergence | |
|---|---|---|---|
| Alphacoronavirus | 229E | Seasonal | 1965 (Hamre and Procknow, 1966) |
| NL63 | Seasonal | 2004 (van der Hoek et al., 2004) | |
| Betacoronavirus | HKU1 | Seasonal | 2005 (Woo et al., 2005) |
| OC43 | Seasonal | 1967 (McIntosh et al., 1967) | |
| MERS-CoV | Emergent | 2012 (Zaki et al., 2012) | |
| SARS-CoV-1 | Emergent | 2003 (Ksiazek et al., 2003) | |
| SARS-CoV-2 | Emergent | 2019 (Wu et al., 2020; Zhou et al., 2020; Zhu et al., 2020) |
Potential editing effects of APOBECs on RNA virus genomes is facilitated by the availability of an unprecedentedly large, expertly assembled and curated dataset of SARS-CoV-2 complete genome sequences that has accumulated since the start of the COVID-19 pandemic. The accuracy of the sequences obtained by a range of NGS technologies, combined with minimal evolutionary diversification in the few months since the start of its global spread provides a unique opportunity to investigate mutational events at very high resolution. All analyses of SARS-CoV-2 genomic sequence data report a preponderance of C→U transitions in the viral genome. Our early analysis (Simmonds, 2020b) of 1001 SARS-CoV-2 sequences indicated that 38–42% of all mutations in the queried datasets were due to C→U changes and that the ratio of C→U to U→C transitions was nearly six times higher than expected. Demonstrating that this was not an artifact of the sequencing or bioinformatics methods used, a parallel analysis of datasets of Ebola virus from historical strains and the 2014-16 West African outbreak demonstrated no such mutational asymmetry. Analysis of single nucleotide variations (SNVs) in a small collection of 34 SARS-CoV-2 sequences using the Wuhan-Hu-1 strain (MN908947) as a reference genome revealed 85 SNVs (Matyášek and Kovařík, 2020). Within these 85 SNVs, C→U changes showed an observed/expected ratio of 5.19 while U→C had a markedly lower ratio of 1.32. In Kilmczak et al. (Klimczak et al., 2020), a larger analysis of 32,115 isolates deposited on GenBank by late June tabled 251,273 mutations when compared to the reference MN908947 genome. Once duplicate mutations were removed and 12,156 mutational event identified, C→U and A→G mutations were shown to be the most prevalent single base substitutions. However, the removal of duplicate mutations may have biased the analysis against detection of independently occurring (homoplastic) C→U transitions at context-favored sites, as documented extensively in SARS-CoV-2 mutational analyses (Simmonds, 2020b). Additionally, a 3.5-fold asymmetry in C→U/U→C mutations was noted in samples from infected minks that had arisen during its transmission within mink farms in the Netherlands (Oude Munnink et al., 2021).
Most studies of mutation frequencies have been based upon the characterization of substitutions away from the SARS-CoV-2 global consensus sequence in individual genome sequences. Di Giorgio et al. (Di Giorgio et al., 2020) have contrastingly investigated the accumulation of mutations in the viral transcriptome via processing of RNA sequencing data from bronchoalveolar lavage fluid (BALF) samples of COVID-19 patients. Through this method, mutations occurring before fitness selection can be observed and potentially provide a better snapshot of the mutational spectrum of SARS-CoV-2 genomic sequences; for example, mutations that were likely to be lethal were readily identified. The authors state that the most common mutations in their datasets, namely A→G and T→C, were likely due to the action of adenosine deaminases that act on RNA (ADARs) while the proportion of C→U and G→A mutations was greater than that typically seen in the human transcriptome, suggesting an additional mechanism leading to the diversity.
The locations of C→U transition were loosely associated with the base and structure context of RNA deamination favored by APOBEC3 proteins. Base context in our analysis and Di Giorgio et al. indicated high preference for C→U deamination in the presence of upstream and downstream A or U. Klimczak et al. analyzed preference in trinucleotide motifs and found strong preference for uCn with total ablation of the phenotype in the context of an upstream C. Intriguingly, this extended context compared to the standard preference of several APOBECs for uC was similar to the A3A-driven editing of cellular mRNA sequences (Sharma et al., 2015) and in A3A-mediated mutations in ssDNA templates (Chan et al., 2015).
The other factor appearing to influence site preference in RNA sequences is the context of the target C in surrounding RNA secondary structure elements. Over half of the sites in human mRNA that were edited by A3A were flanked by short palindromic sequences and typically located in the unpaired termini of stem-loops (Sharma et al., 2016). Supporting this, an analysis of RNA structural contexts of edited sites in SARS-CoV-2 and rubella identified preferential C→U mutations in terminal loop compared to stem sequences (Klimczak et al., 2020), or in predicted paired compared to unpaired regions (Simmonds, 2020a).
4.4. Functional evidence for a role for APOBEC in editing and antiviral restriction of coronaviruses
The observed excess of C→U transitions, the nature of the 5′ and 3’ contexts where such transitions occur and the previously documented ability of several APOBEC paralogs to edit RNA templates make a compelling case for a role of APOBEC in editing of coronavirus genomes. However, functional studies have not, to date, unequivocally demonstrated the introduction of C→U transitions in viral RNA templates by APOBEC. In the most relevant functional study to date, interactions between human A3 proteins and HCoV-NL63 were demonstrated but without clear evidence of systematically biased RNA editing (Milewska et al., 2018). Infection of human airway epithelium culture upregulated A3A, A3C, A3D, and A3F. Transfection of LLC-Mk2 cells (rhesus macaque kidney) revealed deaminase-dependent virus restriction by A3C, A3F, and A3H but not A3A, A3E, or A3G. C→U and G→A point mutations were identified in virus serially passaged in A3A, A3C, A3E, A3F, A3G, and A3H overexpressing cells but did not result in hypermutation of progeny virus. However, potentially recapitulating interactions of APOBEC during assembly and consequent incorporation into retroviral particles, A3C, A3F, and A3H co-localized with the coronavirus nucleocapsid (N) protein through a catalytically-independent mechanism, while A3A, A3D, and A3G did not co-localize, associated with their differences in virus restriction.
Protein-protein interactions between A3 proteins and coronaviruses have furthermore been previously demonstrated in chimeric HIV-1 expressing SARS-CoV or HCoV-229E N (Wang and Wang, 2009). In these experiments, A3G packaged in the chimeric virions and was dependent on the carboxy-terminal self-association domain of the coronavirus N protein and not through binding to genomic RNA. Viral-like particles composed of SARS-CoV N and M proteins also co-packaged A3G.
A role of APOBEC in coronavirus restriction is further although indirectly supported by observations of the upregulation of A4, A3A, A3G, and A3H and downregulation of A3C in BALF samples from patients hospitalized with COVID-19 although there were no significant changes in APOBEC expression in peripheral blood mononuclear cells (PBMCs) (Xiong et al., 2020). Analysis of immune cells in BALF samples from nine patients demonstrated upregulation of A3A and A3B in macrophages and A3G in NK cells (Liao et al., 2020). Tissue expression in two post-mortem lung biopsies demonstrated upregulation of A3A only (Blanco-Melo et al., 2020). While these findings have been advanced as supporting evidence for a role of APOBECs in controlling infections, it should be noted that their expression, as ISGs, is primarily dependent on IFN-induced signaling, a process that activates hundreds of cellular genes that play roles in innate immunity, irrespective of pathway. Demonstration of specific interactions of individual APOBEC proteins with coronaviruses and functional effects of their expression on virus replication is required to substantiate these associations.
Evolutionary consequences of C→U mutational bias in coronaviruses. Irrespective of the underlying mutational mechanisms, we have reviewed the substantial evidence for a potent editing process that drives C→U mutations into the genomes of coronaviruses and potentially other RNA viruses in certain favored sequence and RNA structural contexts. In evolutionary terms, this driver of virus sequence change is quite distinct from the occurrence of random mutations through misincorporation errors during transcription that is assumed in standard evolutionary models. Apart from the restriction of mutations to C→U changes (or G→A if the negative-sense strand is edited), sequence changes are also primarily distributed in mutational hotspots that may lead to the appearance of convergent changes between otherwise genetically unlinked strains. For example, several C→U mutations occurred independently among variants of SARS-CoV-2 that were distributed into different parts of the tree (Simmonds, 2020b), and therefore conflicted with relationships reconstructed from phylogenetically informative sites. A high frequency of homoplastic mutations were similarly described in a larger analysis of 7666 genome sequences of SARS-CoV-2 (van Dorp et al., 2020). Conflicts between the distribution of C→U changes with underlying phylogenetic relationships hampers evolutionary reconstructions and rate calculations (Mavian et al., 2020).
The occurrence of C→U hypermutation in SARS-CoV-2 also has a substantial skewing effect on relative frequencies of non-synonymous (dN - amino acid changing) to synonymous substitutions (dS) occurring during virus evolution. Most viral sequence diversity is generated through fixation of neutral substitution that do not influence the fitness of the virus. These typically are synonymous, and comparative analyses of virus sequence datasets typically reveals low dN/dS ratios indicative of purifying selection, such as the 0.1 ratio found in a dataset of Ebolavirus genome sequences during its recent epidemic spread in Central and West Africa (Simmonds, 2020b). Contrastingly, the high dN/dS ratio between strains (Simmonds, 2020b; van Dorp et al., 2020) or between SARS-CoV-2 and the most closely related bat sarbecovirus, RaTG13 (Wang et al., 2020) indicates that much of the sequence diversity in SARS-CoV-2 is driven by mutational mechanisms distinct from a process of random mutation and fixation assumed in models of neutral evolution.
Furthermore, a substantial proportion of amino acid changes that evolve within SARS-CoV-2 populations originate from non-synonymous changes induced by C→U transitions (Matyášek and Kovařík, 2020; Simmonds, 2020b), rather than necessarily representing evolutionarily selected phenotypic changes that might be expected to occur as the virus adapts to replication and transmission in a new host (MacLean et al., 2020). The occurrence of C→U driven amino acid changes with their likely often mildly deleterious phenotypes and likelihood of reversion considerably complicates analyses of the critical protein changes associated with adaptation to a human host. The occurrence of large numbers of homoplastic sites similarly complicates analyses of sites under positive selection and recombination analysis. Finally, the sheer number of excess C→U changes observed in SARS-CoV-2 genomes (>40% of observed substitutions in some datasets) has a distorting effect on estimates of SARS-CoV-2 substitution rates, particularly as many sites may rapidly revert over time (van Dorp et al., 2020).
The long-term effects of the (C→U)/(U→C) transition asymmetry on virus diversification are difficult to predict. However, the compositional abnormalities of other coronaviruses, particularly human seasonal coronaviruses, might plausibly be attributed to the long term outcome of prolonged C→U editing (Berkhout and van Hemert, 2015; Simmonds, 2020b; Woo et al., 2007). For example, the enrichment of U (40%), depletion of C (13%) and intermediate frequencies of A and G bases (28% and 19%) in HCoV-HKU1 has been proposed to have been driven by APOBEC associated mutations (Woo et al., 2007). Of potential relevance to exploring host differences in APOBEC activity in different hosts, there is no compositional asymmetry and a relatively higher G + C contents in bat sarbecoviruses from which SARS-CoV-2 derives. The intensity of C→U mutational pressure observed in human SARS-CoV-2 sequences since its zoonotic emergence may therefore reflect a less permissive, hostile internal cellular environment in human cells than might be found in a better co-adapted, virus-tolerized immune system of the average bat (Baker et al., 2013).
Downstream induction of inflammatory cytokine and adaptive immune system responses may contribute the exaggerated and damaging inflammatory pathology observed in COVID-19 cases. The pronounced activity of APOBEC in human cells may derive from an inability of a bat-adapted virus to counteract the human APOBEC editing pathway. The varying degrees of base asymmetry and G + C contents of human seasonal coronaviruses may then be indicative of the varying times they have circulated in humans following their acquisition from bats (Corman et al., 2018). SARS-CoV-2 may follow their evolutionary trajectory if infections become permanently established in human populations.
5. A perspective on future functional investigations of APOBEC-mediated editing of SARS-CoV-2 and other RNA viruses
APOBECs have been shown to be important for host and retroviral function, and the possibility that they play a role in RNA viral evolution would substantially expand the scope of their antiviral activities. However, part of the difficulty in studying the process functionally is the plethora of different APOBECs in human with likely different virus, RNA, and DNA targeting and biological activities. Furthermore, the number and identities of APOBECs in high variable in mammals; for example, the mouse which might otherwise represent a suitable in vivo model for APOBEC function in whole host defense possesses only one A3 gene compared to seven or more in primates and bats.
Functional studies using human cell lines are similarly hampered by the current lack of information of which APOBEC protein might mediate the observed RNA editing in coronaviruses and in other RNA viruses, or whether their might exist currently uncharacterized defense pathways. Among the members of the diverse family, A1 and A3, with their evidence for extensive positive selection during mammalian evolution, represent the frontrunners for deaminase-dependent mutation of RNA viruses. A1 is an unlikely candidate for SARS-CoV-2 editing in humans, as its target specificity and restricted hepatic expression of the protein limits its ability to mutate what is principally a respiratory pathogen. Among the A3 proteins, it may be possible to exclude A3G from contributing to SARS-CoV-2 deamination as its upstream base preference for C preference for G does not match the observed contexts of editing sites in SARS-CoV-2 at. Beyond that, one may consider intracellular distributions of this proteins as limiting factors, as SARS-CoV-2 and other coronaviruses spend their entire intracellular life cycles in the host cytoplasm, but first the intracellular distributions of the A3 proteins in different activation contexts should be better defined. Further, it is conceivable that, similar to HIV-1, any editing of SARS-CoV-2 RNA may be enhanced by packaging of A3 proteins in the viral capsid with the bulk of deleterious editing taking place in the subsequent cellular infection. In this way, experiments aimed at identifying protein-protein interactions between the viral capsid and host proteins may assist in narrowing the breadth of proteins that should be investigated. In all cases, trans complementation of the editing phenotype by gene transfection may be difficult to achieve in vitro if there are functionally required interacting cofactors or activated intracellular states. Further research into the protein family is warranted (Box 1).
Box 1. Outstanding research questions.
A more thorough understanding of the general evolution and role of the APOBEC family proteins will benefit from addressing the following research themes:
-
•APOBEC emergence and evolution
-
oIdentification of the presence of A4 homologs in ray-finned fish or protostomes.
-
oEvolutionary relationship between AID and A2.
-
oSignificance of the loss of A3Z1 in multiple branches of placental mammals.
-
oStudy of the activity of APOBECS in the Carnivora genus motivated by the activity of feline and potentially mink homologs.
-
o
-
•APOBEC function
-
oFunction of A2 or A4 in vitro and in vivo.
-
oNecessary cofactors for deaminase activity in a variety of cell and activation contexts. Tissue and cellular distribution of these cofactors.
-
oFunctional importance of dual-deaminase domains in A3 proteins in placental mammals.
-
oThe role of base context and secondary structure motifs on APOBEC deamination activity on ssRNA.
-
oIntracellular localization of APOBEC proteins at rest and in activated contexts.
-
o
-
•APOBEC-virus interactions
-
oDependence of deamination phenotype on protein-protein interactions with viral proteins.
-
oMechanisms of escape of APOBEC targeting for influenza and vaccinia virus.
-
oCellular location and virus replication stage where APOBEC-mediated RNA virus genome editing occurs
-
oPackaging of APOBEC in non-retrovirus virions?
-
o
Alt-text: Box 1
Whether or not the C→U phenotype is driven by APOBEC or by currently uncharacterized intracellular mechanism does not mitigate the functional significance of the observed hypermutation phenotype. Homoplastic transitions in the viral genome impair attempts to construct phylogenetic trees used for molecular epidemiology investigations and may result in spurious associations between unrelated lineages. Were these mutations to be driven by a host element rather than misincorporation errors leading to selection, it would also undermiuneunder the assumptions of neutral evolution underpinning much of the conceptual basis of virus evolutionary models. Lastly, the overrepresentation of these transitions in nonsynonymous mutations underpins its role in generating protein diversity. The long term effect of these potentially driven mutations to viral fitness is uncertain but will contribute to the overall population diversity.
Funding
The work was supported by a Wellcome Investigator Award Grant (WT103767MA).
CRediT authorship contribution statement
Jeremy Ratcliff: Conceptualization, Writing - original draft, preparation, Writing - review & editingWriting – review & editing. Peter Simmonds: Writing - original draft, Writing - Draft extension, Writing - review & editing, Funding acquisition.
References
- Anwar F., Davenport M.P., Ebrahimi D. Footprint of APOBEC3 on the genome of human retroelements. J. Virol. 2013;87:8195–8204. doi: 10.1128/JVI.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin H., Taylor M.W., Lee J.E. Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure. 2014;22:668–684. doi: 10.1016/j.str.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Backus J.W., Smith H.C. Apolipoprotein B mRNA sequences 3' of the editing site are necessary and sufficient for editing and editosome assembly. Nucleic Acids Res. 1991;19:6781–6786. doi: 10.1093/nar/19.24.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.L., Schountz T., Wang L.F. Antiviral immune responses of bats: a review. Zoonoses and public health. 2013;60:104–116. doi: 10.1111/j.1863-2378.2012.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., van Hemert F. On the biased nucleotide composition of the human coronavirus RNA genome. Virus Res. 2015;202:41–47. doi: 10.1016/j.virusres.2014.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biémont C., Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- Bishop K.N., Holmes R.K., Sheehy A.M., Malim M.H. APOBEC-mediated editing of viral RNA. Science. 2004;305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- Blanc V., Park E., Schaefer S., Miller M., Lin Y., Kennedy S., Billing A.M., Ben Hamidane H., Graumann J., Mortazavi A., Nadeau J.H., Davidson N.O. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol. 2014;15:R79. doi: 10.1186/gb-2014-15-6-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D., Garfinkel D.J., Styles C.A., Fink G.R. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Bourque G., Burns K.H., Gehring M., Gorbunova V., Seluanov A., Hammell M., Imbeault M., Izsvák Z., Levin H.L., Macfarlan T.S., Mager D.L., Feschotte C. Ten things you should know about transposable elements. Genome Biol. 2018;19:199. doi: 10.1186/s13059-018-1577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner K.M., Murray A.J., Pollack R.A., Soliman M.G., Laskey S.B., Capoferri A.A., Lai J., Strain M.C., Lada S.M., Hoh R., Ho Y.C., Richman D.D., Deeks S.G., Siliciano J.D., Siliciano R.F. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2016;22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M.B., Leonard B., Harris R.S. APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed. J. 2015;38:102–110. doi: 10.4103/2319-4170.148904. [DOI] [PubMed] [Google Scholar]
- Chan K., Roberts S.A., Klimczak L.J., Sterling J.F., Saini N., Malc E.P., Kim J., Kwiatkowski D.J., Fargo D.C., Mieczkowski P.A., Getz G., Gordenin D.A. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 2015;47:1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F.W. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Chen H., Lilley C.E., Yu Q., Lee D.V., Chou J., Narvaiza I., Landau N.R., Weitzman M.D. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Current biology. CB. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chen S.H., Habib G., Yang C.Y., Gu Z.W., Lee B.R., Weng S.A., Silberman S.R., Cai S.J., Deslypere J.P., Rosseneu M. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238:363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- Conticello S.G., Harris R.S., Neuberger M.S. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. : CB. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Conticello S.G., Thomas C.J., Petersen-Mahrt S.K., Neuberger M.S. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson N.O., Innerarity T.L., Scott J., Smith H., Driscoll D.M., Teng B., Chan L. Proposed nomenclature for the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme: APOBEC-1. Rna. 1995;1:3. [PMC free article] [PubMed] [Google Scholar]
- Delviks-Frankenberry K.A., Desimmie B.A., Pathak V.K. Structural insights into APOBEC3-mediated lentiviral restriction. Viruses. 2020;12 doi: 10.3390/v12060587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D., Hill S.A., Princler G., Lloyd P., Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2915–2920. doi: 10.1073/pnas.0609444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desimmie B.A., Delviks-Frankenberrry K.A., Burdick R.C., Qi D., Izumi T., Pathak V.K. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J. Mol. Biol. 2014;426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio S., Martignano F., Torcia M.G., Mattiuz G., Conticello S.G. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci. Advances. 2020 doi: 10.1126/sciadv.abb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson S.K., Market E., Besmer E., Papavasiliou F.N. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., Scherbakova S., Graham R.L., Baric R.S., Stockwell T.B., Spiro D.J., Denison M.R. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Marusawa H., Kinoshita K., Morisawa T., Sakurai T., Okazaki I.M., Watashi K., Shimotohno K., Honjo T., Chiba T. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene. 2007;26:5587–5595. doi: 10.1038/sj.onc.1210344. [DOI] [PubMed] [Google Scholar]
- Etard C., Roostalu U., Strähle U. Lack of Apobec2-related proteins causes a dystrophic muscle phenotype in zebrafish embryos. J. Cell Biol. 2010;189:527–539. doi: 10.1083/jcb.200912125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrholz M., Kendl S., Prifert C., Weissbrich B., Lemon K., Rennick L., Duprex P.W., Rima B.K., Koning F.A., Holmes R.K., Malim M.H., Schneider-Schaulies J. The innate antiviral factor APOBEC3G targets replication of measles, mumps and respiratory syncytial viruses. J. Gen. Virol. 2012;93:565–576. doi: 10.1099/vir.0.038919-0. [DOI] [PubMed] [Google Scholar]
- Flajnik M.F., Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nature reviews. Genetics. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat N., Tourle K., Radziewic T., Barratt K., Liebhold D., Studdert J.B., Power M., Jones V., Loebel D.A., Tam P.P. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep. 2014;15:903–910. doi: 10.15252/embr.201438450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourgh J.L., Nguyen H.C., Wolfe L.S., Dewitt D.C., Zhang W., Yu X.F., Rhoades E., Xiong Y. Core binding factor beta plays a critical role by facilitating the assembly of the Vif-cullin 5 E3 ubiquitin ligase. J. Virol. 2014;88:3309–3319. doi: 10.1128/JVI.03824-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A.P., Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- Gonzalez M.C., Suspène R., Henry M., Guétard D., Wain-Hobson S., Vartanian J.P. Human APOBEC1 cytidine deaminase edits HBV DNA. Retrovirology. 2009;6:96. doi: 10.1186/1742-4690-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A.M., Landry S., Budagyan K., Avgousti D.C., Shalhout S., Bhagwat A.S., Weitzman M.D. APOBEC3A damages the cellular genome during DNA replication. Cell Cycle. 2016;15:998–1008. doi: 10.1080/15384101.2016.1152426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Dong L., Qiu X., Wang Y., Zhang B., Liu H., Yu Y., Zang Y., Yang M., Huang Z. Structural basis for hijacking CBF-β and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- Haché G., Mansky L.M., Harris R.S. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS Rev. 2006;8:148–157. [PubMed] [Google Scholar]
- Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Harari A., Ooms M., Mulder L.C., Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Dudley J.P. APOBECs and virus restriction. Virology. 2015;479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward J.A., Tachedjian M., Cui J., Cheng A.Z., Johnson A., Baker M.L., Harris R.S., Wang L.F., Tachedjian G. Differential evolution of antiretroviral restriction factors in pteropid bats as revealed by APOBEC3 gene complexity. Mol. Biol. Evol. 2018;35:1626–1637. doi: 10.1093/molbev/msy048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S., Fenton T. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol. Med. 2015;21:274–284. doi: 10.1016/j.molmed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Hsu E. Assembly and expression of shark ig genes. J. Immunol. 2016;196:3517–3523. doi: 10.4049/jimmunol.1600164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T., Abd El Galil K.H., Tokunaga K., Maeda K., Sata T., Sakaguchi N., Heidmann T., Koito A. Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Res. 2011;39:5538–5554. doi: 10.1093/nar/gkr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Gifford R.J., Sato K. Retroviruses drive the rapid evolution of mammalian APOBEC3 genes. Proc. Natl. Acad. Sci. U. S. A. 2020;117:610–618. doi: 10.1073/pnas.1914183116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L.M., Zhang D., Rogozin I.B., Aravind L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 2011;39:9473–9497. doi: 10.1093/nar/gkr691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Kim D.Y., Hultquist J.F., Shindo K., LaRue R.S., Kwon E., Li M., Anderson B.D., Yen L., Stanley D., Mahon C., Kane J., Franks-Skiba K., Cimermancic P., Burlingame A., Sali A., Craik C.S., Harris R.S., Gross J.D., Krogan N.J. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature. 2011;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaguva Vasudevan A.A., Perkovic M., Bulliard Y., Cichutek K., Trono D., Häussinger D., Münk C. Prototype foamy virus Bet impairs the dimerization and cytosolic solubility of human APOBEC3G. J. Virol. 2013;87:9030–9040. doi: 10.1128/JVI.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janini M., Rogers M., Birx D.R., McCutchan F.E. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J. Virol. 2001;75:7973–7986. doi: 10.1128/JVI.75.17.7973-7986.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Kaiser S.M., Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J. Virol. 2006;80:875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S., Khan M.A., Miyagi E., Plishka R., Buckler-White A., Strebel K. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer T.L., Kwon P., Nettles R.E., Han Y., Ray S.C., Siliciano R.F. G-->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J. Virol. 2005;79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak L.J., Randall T.A., Saini N., Li J.L., Gordenin D.A. Similarity between mutation spectra in hypermutated genomes of rubella virus and in SARS-CoV-2 genomes accumulated during the COVID-19 pandemic. PloS One. 2020;15 doi: 10.1371/journal.pone.0237689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer M., Suezer Y., Martinez-Fernandez Y., Münk C., Sutter G., Schnierle B.S. Vaccinia virus replication is not affected by APOBEC3 family members. Virol. J. 2006;3:86. doi: 10.1186/1743-422X-3-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A., Iyer L.M., Holland S.J., Boehm T., Aravind L. Diversification of AID/APOBEC-like deaminases in metazoa: multiplicity of clades and widespread roles in immunity. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E3201–e3210. doi: 10.1073/pnas.1720897115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristbjörnsdóttir H.B., Andrésdóttir V., Svansson V., Torsteinsdóttir S., Matthíasdóttir S., Andrésson O.S. The vif gene of maedi-visna virus is essential for infectivity in vivo and in vitro. Virology. 2004;318:350–359. doi: 10.1016/j.virol.2003.09.044. [DOI] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J.P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann N., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J.C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R.H., Wilson R.K., Hillier L.W., McPherson J.D., Marra M.A., Mardis E.R., Fulton L.A., Chinwalla A.T., Pepin K.H., Gish W.R., Chissoe S.L., Wendl M.C., Delehaunty K.D., Miner T.L., Delehaunty A., Kramer J.B., Cook L.L., Fulton R.S., Johnson D.L., Minx P.J., Clifton S.W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J.F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R.A., Muzny D.M., Scherer S.E., Bouck J.B., Sodergren E.J., Worley K.C., Rives C.M., Gorrell J.H., Metzker M.L., Naylor S.L., Kucherlapati R.S., Nelson D.L., Weinstock G.M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D.R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H.M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R.W., Federspiel N.A., Abola A.P., Proctor M.J., Myers R.M., Schmutz J., Dickson M., Grimwood J., Cox D.R., Olson M.V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G.A., Athanasiou M., Schultz R., Roe B.A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W.R., de la B.M., Dedhia N., Blocker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J.A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D.G., Burge C.B., Cerutti L., Chen H.C., Church D., Clamp M., Copley R.R., Doerks T., Eddy S.R., Eichler E.E., Furey T.S., Galagan J., Gilbert J.G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L.S., Jones T.A., Kasif S., Kaspryzk A., Kennedy S., Kent W.J., Kitts P., Koonin E.V., Korf I., Kulp D., Lancet D., Lowe T.M., McLysaght A., Mikkelsen T., Moran J.V., Mulder N., Pollara V.J., Ponting C.P., Schuler G., Schultz J., Slater G., Smit A.F., Stupka E., Szustakowski J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y.I., Wolfe K.H., Yang S.P., Yeh R.F., Collins F., Guyer M.S., Peterson J., Felsenfeld A., Wetterstrand K.A., Patrinos A., Morgan M.J., Szustakowki J., de Jong P., Catanese J.J., Osoegawa K., Shizuya H., Choi S. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Langlois M.A., Neuberger M.S. Human APOBEC3G can restrict retroviral infection in avian cells and acts independently of both UNG and SMUG1. J. Virol. 2008;82:4660–4664. doi: 10.1128/JVI.02469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue R.S., Andrésdóttir V., Blanchard Y., Conticello S.G., Derse D., Emerman M., Greene W.C., Jónsson S.R., Landau N.R., Löchelt M., Malik H.S., Malim M.H., Münk C., O'Brien S.J., Pathak V.K., Strebel K., Wain-Hobson S., Yu X.F., Yuhki N., Harris R.S. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue R.S., Jónsson S.R., Silverstein K.A., Lajoie M., Bertrand D., El-Mabrouk N., Hötzel I., Andrésdóttir V., Smith T.P., Harris R.S. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.M., Hirano K., Anant S., Baunoch D., Davidson N.O. An alternatively spliced form of apobec-1 messenger RNA is overexpressed in human colon cancer. Gastroenterology. 1998;115:1096–1103. doi: 10.1016/s0016-5085(98)70080-0. [DOI] [PubMed] [Google Scholar]
- Li Z., Ning S., Su X., Liu X., Wang H., Liu Y., Zheng W., Zheng B., Yu X.F., Zhang W. Enterovirus 71 antagonizes the inhibition of the host intrinsic antiviral factor A3G. Nucleic Acids Res. 2018;46:11514–11527. doi: 10.1093/nar/gky840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Kitamura K., Wang Z., Liu G., Chowdhury S., Fu W., Koura M., Wakae K., Honjo T., Muramatsu M. RNA editing of hepatitis B virus transcripts by activation-induced cytidine deaminase. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2246–2251. doi: 10.1073/pnas.1221921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Liao W., Hong S.H., Chan B.H., Rudolph F.B., Clark S.C., Chan L. APOBEC-2, a cardiac- and skeletal muscle-specific member of the cytidine deaminase supergene family. Biochem. Biophys. Res. Commun. 1999;260:398–404. doi: 10.1006/bbrc.1999.0925. [DOI] [PubMed] [Google Scholar]
- Liu M.C., Liao W.Y., Buckley K.M., Yang S.Y., Rast J.P., Fugmann S.D. AID/APOBEC-like cytidine deaminases are ancient innate immune mediators in invertebrates. Nat. Commun. 2018;9:1948. doi: 10.1038/s41467-018-04273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löchelt M., Romen F., Bastone P., Muckenfuss H., Kirchner N., Kim Y.B., Truyen U., Rösler U., Battenberg M., Saib A., Flory E., Cichutek K., Münk C. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7982–7987. doi: 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDuff D.A., Demorest Z.L., Harris R.S. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res. 2009;37:1854–1867. doi: 10.1093/nar/gkp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean O.A., Lytras S., Singer J.B., Weaver S., Pond S.L.K., Robertson D.L. bioRxiv; 2020. Evidence of Significant Natural Selection in the Evolution of SARS-CoV-2 in Bats, Not Humans. [Google Scholar]
- Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Marin M., Rose K.M., Kozak S.L., Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Matyášek R., Kovařík A. 2020. Mutation Patterns of Human SARS-CoV-2 and Bat RaTG13 Coronavirus Genomes Are Strongly Biased towards C>U Transitions, Indicating Rapid Evolution in Their Hosts. Genes (Basel) p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavian C., Marini S., Manes C., Capua I., Prosperi M., Salemi M. medRxiv; 2020. Regaining Perspective on SARS-CoV-2 Molecular Tracing and its Implications. 2020.2003.2016.20034470. [Google Scholar]
- McDaniel Y.Z., Wang D., Love R.P., Adolph M.B., Mohammadzadeh N., Chelico L., Mansky L.M. Deamination hotspots among APOBEC3 family members are defined by both target site sequence context and ssDNA secondary structure. Nucleic Acids Res. 2020;48(3):1353–1371. doi: 10.1093/nar/gkz1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Becker W.B., Chanock R.M. Growth in suckling-mouse brain of "IBV-like" viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. U. S. A. 1967;58:2268–2273. doi: 10.1073/pnas.58.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A., Goncalves J., Santa-Marta M., McPike M., Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18:2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A., Strack B., Ancuta P., Zhang C., McPike M., Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- Mehta A., Kinter M.T., Sherman N.E., Driscoll D.M. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell Biol. 2000;20:1846–1854. doi: 10.1128/mcb.20.5.1846-1854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Jurka J., Kamal M., Mauceli E., Searle S.M., Sharpe T., Baker M.L., Batzer M.A., Benos P.V., Belov K., Clamp M., Cook A., Cuff J., Das R., Davidow L., Deakin J.E., Fazzari M.J., Glass J.L., Grabherr M., Greally J.M., Gu W., Hore T.A., Huttley G.A., Kleber M., Jirtle R.L., Koina E., Lee J.T., Mahony S., Marra M.A., Miller R.D., Nicholls R.D., Oda M., Papenfuss A.T., Parra Z.E., Pollock D.D., Ray D.A., Schein J.E., Speed T.P., Thompson K., VandeBerg J.L., Wade C.M., Walker J.A., Waters P.D., Webber C., Weidman J.R., Xie X., Zody M.C., Graves J.A., Ponting C.P., Breen M., Samollow P.B., Lander E.S., Lindblad-Toh K. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Milewska A., Kindler E., Vkovski P., Zeglen S., Ochman M., Thiel V., Rajfur Z., Pyrc K. APOBEC3-mediated restriction of RNA virus replication. Sci. Rep. 2018;8:5960. doi: 10.1038/s41598-018-24448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris A., Murray S., Cardinaud S. AID and APOBECs span the gap between innate and adaptive immunity. Front. Microbiol. 2014;5:534. doi: 10.3389/fmicb.2014.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk C., Willemsen A., Bravo I.G. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol. Biol. 2012;12:71. doi: 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Nakano Y., Aso H., Soper A., Yamada E., Moriwaki M., Juarez-Fernandez G., Koyanagi Y., Sato K. A conflict of interest: the evolutionary arms race between mammalian APOBEC3 and lentiviral Vif. Retrovirology. 2017;14:31. doi: 10.1186/s12977-017-0355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaiza I., Linfesty D.C., Greener B.N., Hakata Y., Pintel D.J., Logue E., Landau N.R., Weitzman M.D. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama S., Marusawa H., Matsumoto T., Ueda Y., Matsumoto Y., Endo Y., Takai A., Chiba T. Excessive activity of apolipoprotein B mRNA editing enzyme catalytic polypeptide 2 (APOBEC2) contributes to liver and lung tumorigenesis. Int. J. Canc. 2012;130:1294–1301. doi: 10.1002/ijc.26114. [DOI] [PubMed] [Google Scholar]
- Ostertag E.M., Kazazian H.H., Jr. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., Bouwmeester-Vincken N., Harders F., Hakze-van der Honing R., Wegdam-Blans M.C.A., Bouwstra R.J., GeurtsvanKessel C., van der Eijk A.A., Velkers F.C., Smit L.A.M., Stegeman A., van der Poel W.H.M., Koopmans M.P.G. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371(6525):172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli E.K., Schmolke M., Hofmann H., Ehrhardt C., Flory E., Münk C., Ludwig S. High level expression of the anti-retroviral protein APOBEC3G is induced by influenza A virus but does not confer antiviral activity. Retrovirology. 2009;6:38. doi: 10.1186/1742-4690-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelygina L., Chen M.H., Suppiah S., Adebayo A., Abernathy E., Dorsey M., Bercovitch L., Paris K., White K.P., Krol A., Dhossche J., Torshin I.Y., Saini N., Klimczak L.J., Gordenin D.A., Zharkikh A., Plotkin S., Sullivan K.E., Icenogle J. Infectious vaccine-derived rubella viruses emerge, persist, and evolve in cutaneous granulomas of children with primary immunodeficiencies. PLoS Pathog. 2019;15:e1008080. doi: 10.1371/journal.ppat.1008080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M.F. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Pizarro J.G., Cristofari G. Post-transcriptional control of LINE-1 retrotransposition by cellular host factors in somatic cells. Front Cell Dev Biol. 2016;4:14. doi: 10.3389/fcell.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain F., Lejeune N., Willemart K., Gillet N.A. Footprint of the host restriction factors APOBEC3 on the genome of human viruses. PLoS Pathog. 2020;16(8) doi: 10.1371/journal.ppat.1008718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L.M., Wallis S.C., Pease R.J., Edwards Y.H., Knott T.J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Renard M., Henry M., Guétard D., Vartanian J.P., Wain-Hobson S. APOBEC1 and APOBEC3 cytidine deaminases as restriction factors for hepadnaviral genomes in non-humans in vivo. J. Mol. Biol. 2010;400:323–334. doi: 10.1016/j.jmb.2010.05.029. [DOI] [PubMed] [Google Scholar]
- Richardson S.R., Doucet A.J., Kopera H.C., Moldovan J.B., Garcia-Perez J.L., Moran J.V. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0061-2014. Mdna3-0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S.R., Narvaiza I., Planegger R.A., Weitzman M.D., Moran J.V. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife. 2014;3 doi: 10.7554/eLife.02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G., Harris S., Shah R.R., Resnick M.A., Getz G., Gordenin D.A. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin I.B., Basu M.K., Jordan I.K., Pavlov Y.I., Koonin E.V. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Iyer L.M., Liang L., Glazko G.V., Liston V.G., Pavlov Y.I., Aravind L., Pancer Z. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat. Immunol. 2007;8:647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- Rosenberg B.R., Hamilton C.E., Mwangi M.M., Dewell S., Papavasiliou F.N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3' UTRs. Nat. Struct. Mol. Biol. 2011;18:230–236. doi: 10.1038/nsmb.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M., Kidwell M.G., Bingham P.M. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982;29:987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- Russell R.A., Wiegand H.L., Moore M.D., Schäfer A., McClure M.O., Cullen B.R. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 2005;79:8724–8731. doi: 10.1128/JVI.79.14.8724-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter J.D., Bennett R.P., Smith H.C. The APOBEC protein family: united by structure, divergent in function. Trends Biochem. Sci. 2016;41:578–594. doi: 10.1016/j.tibs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Probst H.C., Tatsumi R., Ikeuchi Y., Neuberger M.S., Rada C. Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J. Biol. Chem. 2010;285:7111–7118. doi: 10.1074/jbc.M109.052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S.L., Emerman M., Malik H.S. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G.G. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem. Soc. Trans. 2007;35:637–642. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]
- Seeger C., Mason W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Patnaik S.K., Taggart R.T., Baysal B.E. The double-domain cytidine deaminase APOBEC3G is a cellular site-specific RNA editing enzyme. Sci. Rep. 2016;6:39100. doi: 10.1038/srep39100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Patnaik S.K., Taggart R.T., Kannisto E.D., Enriquez S.M., Gollnick P., Baysal B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015;6:6881. doi: 10.1038/ncomms7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Wang J., Alqassim E., Portwood S., Cortes Gomez E., Maguire O., Basse P.H., Wang E.S., Segal B.H., Baysal B.E. Mitochondrial hypoxic stress induces widespread RNA editing by APOBEC3G in natural killer cells. Genome Biol. 2019;20:37. doi: 10.1186/s13059-019-1651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis N.C., Malim M.H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Shi M., Tan L., Zhang Y., Meng C., Wang W., Sun Y., Song C., Liu W., Liao Y., Yu S., Ren T., Ding Z., Liu X., Qiu X., Ding C. Characterization and functional analysis of chicken APOBEC4. Dev. Comp. Immunol. 2020;106:103631. doi: 10.1016/j.dci.2020.103631. [DOI] [PubMed] [Google Scholar]
- Simmonds P. Pervasive RNA secondary structure in the genomes of SARS-CoV-2 and other coronaviruses. mBio. 2020;11 doi: 10.1128/mBio.01661-20. e01661–01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. Rampant C→U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. mSphere. 2020;5 doi: 10.1128/mSphere.00408-20. e00408–00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse G.R., Cappione A.J., Sowden M., Metheny L.J., Smith H.C. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res. 1996;24:478–485. doi: 10.1093/nar/24.3.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Blanc H., Surdel M.C., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.C. RNA binding to APOBEC deaminases; Not simply a substrate for C to U editing. RNA Biol. 2017;14:1153–1165. doi: 10.1080/15476286.2016.1259783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein M.D., Harris R.S. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Stopak K., de Noronha C., Yonemoto W., Greene W.C. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- Suspène R., Aynaud M.M., Koch S., Pasdeloup D., Labetoulle M., Gaertner B., Vartanian J.P., Meyerhans A., Wain-Hobson S. Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo. J. Virol. 2011;85:7594–7602. doi: 10.1128/JVI.00290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspène R., Guétard D., Henry M., Sommer P., Wain-Hobson S., Vartanian J.P. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B.J., Nik-Zainal S., Wu Y.L., Stebbings L.A., Raine K., Campbell P.J., Rada C., Stratton M.R., Neuberger M.S. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2 doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng B., Burant C.F., Davidson N.O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- Tsuge M., Noguchi C., Akiyama R., Matsushita M., Kunihiro K., Tanaka S., Abe H., Mitsui F., Kitamura S., Hatakeyama T., Kimura T., Miki D., Hiraga N., Imamura M., Takahashi S., Hayses C.N., Chayama K. G to A hypermutation of TT virus. Virus Res. 2010;149:211–216. doi: 10.1016/j.virusres.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., Ortiz A.T., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol.: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Maldegem F., Scheeren F.A., Aarti Jibodh R., Bende R.J., Jacobs H., van Noesel C.J. AID splice variants lack deaminase activity. Blood. 2009;113:1862–1864. doi: 10.1182/blood-2008-08-175265. author reply 1864. [DOI] [PubMed] [Google Scholar]
- Vartanian J.P., Guétard D., Henry M., Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- Vonica A., Rosa A., Arduini B.L., Brivanlou A.H. APOBEC2, a selective inhibitor of TGFβ signaling, regulates left-right axis specification during early embryogenesis. Dev. Biol. 2011;350:13–23. doi: 10.1016/j.ydbio.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Pipes L., Nielsen R. bioRxiv; 2020. Synonymous Mutations and the Molecular Evolution of SARS-Cov-2 Origins. 2020.2004.2020.052019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Shaban N.M., Land A.M., Brown W.L., Harris R.S. Simian immunodeficiency virus vif and human APOBEC3B interactions resemble those between HIV-1 vif and human APOBEC3G. J. Virol. 2018;92 doi: 10.1128/JVI.00447-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.M., Wang C.T. APOBEC3G cytidine deaminase association with coronavirus nucleocapsid protein. Virology. 2009;388:112–120. doi: 10.1016/j.virol.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C.J., Van Doorslaer K., Pandey A., Espinosa J.M., Pyeon D. Role of the host restriction factor APOBEC3 on papillomavirus evolution. Virus evolution. 2015;1 doi: 10.1093/ve/vev015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C.J., Xu T., Guo K., Griffin L.M., Westrich J.A., Lee D., Lambert P.F., Santiago M.L., Pyeon D. APOBEC3A functions as a restriction factor of human papillomavirus. J. Virol. 2015;89:688–702. doi: 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren W.C., Hillier L.W., Marshall Graves J.A., Birney E., Ponting C.P., Grützner F., Belov K., Miller W., Clarke L., Chinwalla A.T., Yang S.P., Heger A., Locke D.P., Miethke P., Waters P.D., Veyrunes F., Fulton L., Fulton B., Graves T., Wallis J., Puente X.S., López-Otín C., Ordóñez G.R., Eichler E.E., Chen L., Cheng Z., Deakin J.E., Alsop A., Thompson K., Kirby P., Papenfuss A.T., Wakefield M.J., Olender T., Lancet D., Huttley G.A., Smit A.F., Pask A., Temple-Smith P., Batzer M.A., Walker J.A., Konkel M.K., Harris R.S., Whittington C.M., Wong E.S., Gemmell N.J., Buschiazzo E., Vargas Jentzsch I.M., Merkel A., Schmitz J., Zemann A., Churakov G., Kriegs J.O., Brosius J., Murchison E.P., Sachidanandam R., Smith C., Hannon G.J., Tsend-Ayush E., McMillan D., Attenborough R., Rens W., Ferguson-Smith M., Lefèvre C.M., Sharp J.A., Nicholas K.R., Ray D.A., Kube M., Reinhardt R., Pringle T.H., Taylor J., Jones R.C., Nixon B., Dacheux J.L., Niwa H., Sekita Y., Huang X., Stark A., Kheradpour P., Kellis M., Flicek P., Chen Y., Webber C., Hardison R., Nelson J., Hallsworth-Pepin K., Delehaunty K., Markovic C., Minx P., Feng Y., Kremitzki C., Mitreva M., Glasscock J., Wylie T., Wohldmann P., Thiru P., Nhan M.N., Pohl C.S., Smith S.M., Hou S., Nefedov M., de Jong P.J., Renfree M.B., Mardis E.R., Wilson R.K. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil A.F., Ghosh D., Zhou Y., Seiple L., McMahon M.A., Spivak A.M., Siliciano R.F., Stivers J.T. Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E448–E457. doi: 10.1073/pnas.1219702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Wong B.H., Huang Y., Lau S.K., Yuen K.Y. Cytosine deamination and selection of CpG suppressed clones are the two major independent biological forces that shape codon usage bias in coronaviruses. Virology. 2007;369:431–442. doi: 10.1016/j.virol.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.K., Byun H., Dudley J.P. The role of APOBECs in viral replication. Microorganisms. 2020;8 doi: 10.3390/microorganisms8121899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S., Balestra M.E., Ferrell L.D., Fan J., Arnold K.S., Taylor S., Taylor J.M., Innerarity T.L. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S., Poksay K.S., Arnold K.S., Innerarity T.L. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- Yang B., Chen K., Zhang C., Huang S., Zhang H. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J. Biol. Chem. 2007;282:11667–11675. doi: 10.1074/jbc.M606864200. [DOI] [PubMed] [Google Scholar]
- Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X.F. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang W., Du J., Evans S.L., Yu Y., Yu X.F. T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2011;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- Zhang W., Wang H., Li Z., Liu X., Liu G., Harris R.S., Yu X.F. Cellular requirements for bovine immunodeficiency virus Vif-mediated inactivation of bovine APOBEC3 proteins. J. Virol. 2014;88:12528–12540. doi: 10.1128/JVI.02072-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Gu Q., Marino D., Lee K.L., Kong I.K., Häussinger D., Münk C. Feline APOBEC3s, barriers to cross-species transmission of FIV? Viruses. 2018;10 doi: 10.3390/v10040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Li Z., Huan C., Wang H., Su X., Zhang W. CAEV vif hijacks ElonginB/C, CYPA and Cullin5 to assemble the E3 ubiquitin ligase complex stepwise to degrade oaA3Z2-Z3. Front. Microbiol. 2019;10:565. doi: 10.3389/fmicb.2019.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka J., Marino D., Hofmann H., Yuhki N., Löchelt M., Münk C. Vif of feline immunodeficiency virus from domestic cats protects against APOBEC3 restriction factors from many felids. J. Virol. 2010;84:7312–7324. doi: 10.1128/JVI.00209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]