Abstract
Here, we developed a prostate cancer (PCa) risk nomogram including lymphocyte-to-monocyte ratio (LMR) for initial prostate biopsy, and internal and external validation were further conducted. A prediction model was developed on a training set. Significant risk factors with P < 0.10 in multivariate logistic regression models were used to generate a nomogram. Discrimination, calibration, and clinical usefulness of the model were assessed using C-index, calibration plot, and decision curve analysis (DCA). The nomogram was re-examined with the internal and external validation set. A nomogram predicting PCa risk in patients with prostate-specific antigen (PSA) 4–10 ng ml−1 was also developed. The model displayed good discrimination with C-index of 0.830 (95% confidence interval [CI]: 0.812–0.852). High C-index of 0.864 (95% CI: 0.840–0.888) and 0.871 (95% CI: 0.861–0.881) was still reached in the internal and external validation sets, respectively. The nomogram exhibited better performance compared to the nomogram with PSA only (C-index: 0.763, 95% CI: 0.746–0.780, P < 0.001) and the nomogram with LMR excluded (C-index: 0.824, 95% CI: 0.804–0.844, P < 0.010). The calibration curve demonstrated good agreement in the internal and external validation sets. DCA showed that the nomogram was useful at the threshold probability of >4% and <99%. The nomogram predicting PCa risk in patients with PSA 4–10 ng ml−1 also displayed good calibration and discrimination performance (C-index: 0.734, 95% CI: 0.708–0.760). This nomogram incorporating age, PSA, digital rectal examination, abnormal imaging signals, PSA density, and LMR could be used to facilitate individual PCa risk prediction in initial prostate biopsy.
Keywords: lymphocyte-to-monocyte ratio, nomogram, prostate biopsy, prostate cancer
INTRODUCTION
Prostate cancer (PCa) is the second most commonly diagnosed cancer (13.5%) and the fifth most common cause of cancer-related death (6.7%) in men worldwide.1 In China, the incidence rate of PCa is rapidly increasing, likely caused by longer life expectancy and westernized lifestyles.2 In 2015, it was responsible for 40 300 new cases and 26 600 cancer-related deaths, making it the most lethal urological cancer in Chinese men.3 Despite much progress in detection and treatment, PCa is still a major health burden, especially among elderly men.
The use of prostate-specific antigen (PSA) has revolutionized PCa diagnosis.4 The need for prostate biopsy, which is still the gold standard for PCa diagnosis, is based on PSA level and/or suspicious digital rectal examination (DRE) and/or imaging. However, PSA, as an organ- but not cancer-specific serum marker, lacks sufficient sensitivity and specificity in PCa detection.5 Approximately 1 out of 5 men with PCa might be misdiagnosed at initial prostate biopsy.6 Patients with PSA between 4 ng ml−1 and 10 ng ml−1 (known as the diagnostic gray zone) present a challenge to clinicians. The PCa detection rate within this PSA range was only 16%–39%, and the rate of high-grade PCa ranged between 4.1% and 25%.7,8 Although several biomarkers have been investigated in PCa detection, for example, prostate health index (PHI), the four kallikrein (4K) score, and prostate cancer antigen 3 (PCA3), they are still not widely accepted by clinicians.9 Thus, new valid but simple prediction tools are needed to detect PCa and avoid unnecessary biopsy.
It has been recognized over the last decade that systemic inflammation plays a significant role in onset and development of cancer. This idea has led to the investigation of inflammatory biomarkers in cancer detection and prognosis, including lymphocyte-to-monocyte ratio (LMR), neutrophil-to-lymphocyte ratio, platelet-to-lymphocyte ratio, C-reactive protein, and albumin. LMR, an index integrating circulating lymphocytes and monocytes, has been investigated in various malignancies, including gastric cancer, lung cancer, bladder cancer, and renal cell carcinoma.10,11,12 Nevertheless, there is limited evidence for the use of LMR in diagnosis and prognosis of PCa. One study in a small cohort has investigated the utility of LMR as a potential biomarker for PCa detection.13 However, the diagnostic value of LMR in addition to PSA and other indicators for PCa needs to be validated. Therefore, we performed this double-center study in China to develop a nomogram including LMR to predict the result of initial prostate biopsy.
PATIENTS AND METHODS
Patients
Patients who underwent prostate needle biopsy at the Department of Urology, the Affiliated Hospital of Qingdao University (AHQDU, Qingdao, China), and Fudan University Shanghai Cancer Center (FUSCC, Shanghai, China), between January 2012 and December 2018 were included in this study. Transrectal ultrasound-guided 12-core systematic prostate needle biopsy was performed in both centers. Another two samples were obtained from the suspicious lesions under the transrectal ultrasound. The need for prostate biopsy was based on PSA level and/or suspicious DRE and/or imaging. Patients with elevated PSA >10 ng ml−1, or PSA 4–10 ng ml−1 with abnormal free-to-total (f/t) PSA, or PSA 4–10 ng ml-1 with abnormal PSA density (PSAD) were candidates for biopsy. We excluded patients with systemic or urinary tract infection (including symptomatic prostatitis), hematological diseases, autoimmune diseases, or medical history of anti-inflammatory or systemic steroid drug use within 2 weeks before biopsy. Patients with history of malignancy or any other disease that affected white blood cell count and differential leukocyte count were also excluded. Moreover, patients with atypical small acinar proliferation or high-grade intraepithelial neoplasia were excluded because of the small number of cases. For patients who underwent prostate biopsy twice or more, only the initial biopsy data were analyzed.
Clinical parameters such as age, body weight and height, alcohol or tobacco consumption, imaging record (including B-ultrasound and magnetic resonance imaging [MRI]), and Gleason score (GS) were recorded. Peripheral blood samples were obtained 3 days before biopsy. Blood index and PSA value were examined. LMR was calculated by dividing the lymphocyte count by the monocyte count. The study protocol was approved by the Ethics Committee of AHQDU (AYFYWZLL25772) and FUSCC (050432-4-1212B), respectively.
To develop an individualized biopsy prediction model including LMR, patients from FUSCC were randomly split into the training set and internal verification set according to the random number table. Data from AHQDU were used as the external verification set to verify the predictive capability and accuracy of the nomogram in an independent population. Similarly, a nomogram for prediction of clinically significant PCa (csPCa, defined as GS ≥7) was also developed with training set and internal verification set from FUSCC and external verification set from AHQDU. Patients with PSA higher than 100 ng ml−1 were excluded when developing the nomogram, since the clinical use of nomogram with this range seems unnecessary, and the contribution of other biomarkers might be concealed by PSA. The definite cutoff value of LMR was set as the median level of LMR in the training set, and accordingly, the patients were divided into high-LMR and low-LMR groups. When the population was restricted to patients with PSA 4–10 ng ml−1, patients from FUSCC were labeled as the training set and patients from AHQDU as the verification set.
Statistical analyses
Univariate and multivariate logistic regression analyses were performed to determine the factors affecting PCa diagnosis. Significant risk factors with P < 0.10 in multivariate logistic regression models were used to generate a nomogram predicting the probability of positive pathology in prostate biopsy. Discrimination was assessed using C-index. The calibration was examined by the calibration curves. The nomogram was subjected to bootstrapping validation (1000 bootstrap re-samples) to calculate a relative corrected C-index. The nomogram was re-examined with the internal and external validation sets. Decision curve analysis (DCA) was performed to assess the clinical usefulness of the nomogram by calculating the net benefits. Statistical analyses were conducted using SPSS version 19.0 software (IBM Corporation, Armonk, NY, USA) and R version 3.5.2 (The R Foundation, Vienna, Austria).
RESULTS
Patient characteristics
We included 1170 patients from FUSCC (820 in the training set and 350 in the internal verification set) and 1339 patients from AHQDU (external verification set). Among the patients in the training set, the median LMR was 4.800, and accordingly, the patients were divided into the high-LMR and low-LMR groups. The patient characteristics are shown in Table 1. PCa was diagnosed by initial biopsy in 550 (53.4%) patients in FUSCC and 590 (44.1%) patients in AHQDU. Patients with positive biopsy exhibited older age, higher PSA level, higher rates of abnormal DRE findings and imaging signals, higher PSAD, and lower LMR in both centers. History of alcohol consumption reached statistical significance only in FUSCC (P = 0.030). However, f/t PSA level (FUSCC: P = 0.409; AHQDU: P = 0.194), history of tobacco consumption (FUSCC: P = 0.276; AHQDU: P = 0.439), and body mass index (FUSCC: P = 0.804; AHQDU: P = 0.071) showed no difference between PCa and benign prostatic hyperplasia (BPH). Moreover, depending on the cutoff value, we assessed clinical usefulness of LMR. The result revealed that LMR got an accuracy rate of 53.3% (1338/2509) with a sensitivity of 66.1% (753/1140), a specificity of 42.7% (585/1369), a positive predictive value of 49.0% (753/1537), and a negative predictive value of 60.2% (585/972).
Table 1.
Baseline characteristics in the Fudan University Shanghai Cancer Center (Shanghai, China), and Affiliated Hospital of Qingdao University (Qiangdao, China)
| Variables | FUSCC (n=1170) | AHQDU (n=1339) | ||||
|---|---|---|---|---|---|---|
| BPH | PCa | P | BPH | PCa | P | |
| Patients, n (%) | 620 (46.6) | 550 (53.4) | 749 (56.0) | 590 (44.1) | ||
| Age (year), mean±s.d. | 64.38±9.69 | 69.17±8.29 | 0.000 | 67.24±7.85 | 71.14±8.25 | 0.000 |
| PSA (ng ml−1), median (IQR) | 8.56 (5.87–12.59) | 31.08 (12.10–100.00) | 0.000 | 11.99 (7.25–18.99) | 63.60 (20.04–100.00) | 0.000 |
| DRE (n), −/+ | 563/57 | 306/244 | 0.000 | 637/112 | 260/330 | 0.000 |
| Abnormal imaging (n), −/+ | 484/136 | 331/219 | 0.000 | 576/173 | 344/246 | 0.000 |
| PSAD (n), low/high | 133/220 | 26/286 | 0.000 | 169/226 | 28/244 | 0.000 |
| LMR (n), low/high | 268/352 | 317/233 | 0.000 | 516/233 | 436/154 | 0.045 |
| f/t PSA (n), low/high | 330/290 | 306/244 | 0.409 | 384/223 | 270/185 | 0.194 |
| Tobacco (n), −/+ | 197/156 | 246/166 | 0.276 | 438/307 | 355/228 | 0.439 |
| Alcohol (n), −/+ | 297/57 | 241/71 | 0.030 | 557/181 | 449/131 | 0.411 |
| BMI (kg m−2), mean±s.d. | 23.80±2.97 | 23.75±2.89 | 0.804 | 24.29±3.21 | 23.95±3.59 | 0.071 |
FUSCC: Fudan University Shanghai Cancer Center; AHQDU: the Affiliated Hospital of Qingdao University; PSA: prostate-specific antigen; IQR: interquartile range; DRE: digital rectal examination; PSAD: prostate-specific antigen density; LMR: lymphocyte-to-monocyte ratio; f/t PSA: free-to-total prostate-specific antigen; BMI: body mass index; PCa: prostate cancer
LMR value stratified by PSA level and GS
Compared with BPH patients, PCa patients exhibited decreased LMR in FUSCC (P < 0.001) and AHQDU (P = 0.045). Reduced LMR (mean ± standard deviation [s.d.]) was observed among patients with PSA >20 ng ml−1 compared with 4–10 ng ml−1 (FUSCC: 4.74 ± 1.98 ng ml−1 vs 5.31 ± 1.93 ng ml−1, P < 0.001; AHQDU: 3.88 ± 1.73 ng ml−1 vs 4.24 ± 1.46 ng ml−1, P = 0.003; Supplementary Figure 1 (213.7KB, tif) ). Similarly, LMR (mean ± s.d.) was significantly lower in men with PSA >20 ng ml−1 than in those with PSA 10–20 ng ml−1 (FUSCC: 4.74 ± 1.98 ng ml-1 vs 5.31 ± 2.0 ng ml−1, P < 0.001; AHQDU: 3.88 ± 1.73 ng ml-1 vs 4.31 ± 1.79 ng ml−1, P < 0.001). LMR value (mean ± s.d.) in the high-GS group was 4.27 ± 1.82, which was significantly lower than in the biopsy-negative group (mean ± s.d.: 4.66 ± 1.94, P < 0.001) and low-GS group (mean ± s.d.: 4.62 ± 1.96, P = 0.033). Given that LMR had a close relationship with PCa, we tried to develop an individualized biopsy prediction model including LMR.
Development of a nomogram for PCa prediction
Logistic regression analyses were performed in the training set to determine the factors affecting PCa diagnosis. Age (P < 0.001), PSA (P < 0.001), DRE (P < 0.001), abnormal imaging signals (P < 0.001), PSAD (P < 0.001), and LMR (P < 0.001) were associated with positive biopsy in univariate logistic regression model, and multivariate analysis revealed that age (P = 0.001), PSA (P = 0.001), DRE (P < 0.001), abnormal imaging signals (P = 0.019), PSAD (P < 0.001), and LMR (P = 0.009) were independent predictors of positive biopsy (Table 2). Based on this six-risk factor logistic regression model, the nomogram for positive biopsy prediction in initial prostate biopsy patients was developed (Figure 1).
Table 2.
Logistic regression analysis of predictors for positive biopsy in the training cohort
| Variables | Univariate regression | Multivariate regression | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |
| Age | 1.063 | 1.045–1.081 | 0.000 | 1.046 | 1.018–1.076 | 0.001 |
| PSA | 1.040 | 1.031–1.048 | 0.000 | 1.014 | 1.006–1.022 | 0.001 |
| DRE | 9.223 | 6.169–13.788 | 0.000 | 5.544 | 3.136–9.801 | 0.000 |
| Abnormal imaging | 2.498 | 1.842–3.388 | 0.000 | 1.839 | 1.105–3.060 | 0.019 |
| PSAD | 5.265 | 3.201–8.662 | 0.000 | 3.273 | 1.790–5.986 | 0.000 |
| LMR | 0.532 | 0.403–0.702 | 0.000 | 0.533 | 0.332–0.856 | 0.009 |
| Tobacco | 0.725 | 0.514–1.023 | 0.067 | 1.070 | 0.656–1.745 | 0.787 |
| Alcohol | 1.561 | 0.982–2.482 | 0.060 | 1.384 | 0.748–2.563 | 0.301 |
| BMI | 0.988 | 0.960–1.017 | 0.423 | 0.997 | 0.977–1.017 | 0.789 |
PSA: prostate-specific antigen; DRE: digital rectal examination; PSAD: prostate-specific antigen density; LMR: lymphocyte-to-monocyte ratio; BMI: body mass index; OR: odds ratio; CI: confidence interval
Figure 1.
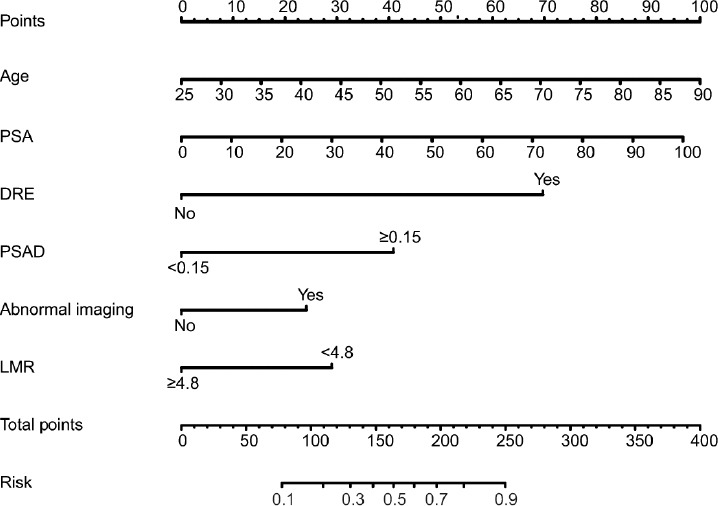
Nomogram for predicting PCa based on the training cohort. The prostate biopsy nomogram was developed in the training cohort, with age, PSA, abnormal DRE, PSAD, abnormal imaging findings, and LMR incorporated. PSA: prostate-specific antigen; DRE: digital rectal examination; PSAD: prostate-specific antigen density; LMR: lymphocyte-to-monocyte ratio.
Calibration and discrimination of the nomogram for PCa detection in the training set
The calibration curve of the nomogram for prediction of positive biopsy demonstrated good agreement between prediction and observation in the training set (Supplementary Figure 2a (383.3KB, tif) ). The C-index of the training set was 0.830 (95% confidence interval [CI]: 0.812–0.852) and confirmed to be 0.820 through bootstrapping validation. When LMR was excluded from the model, the C-index decreased to 0.824 (95% CI: 0.804–0.844, P < 0.010). When only PSA was included in this nomogram, the C-index was 0.763 (95% CI: 0.746–0.780, P < 0.001).
Internal and external validation of the nomogram for PCa detection
Using 30% of the Fudan cohort as the internal validation data set, acceptable calibration was also observed (Supplementary Figure 2b (383.3KB, tif) ), and the C-index of the nomogram was 0.864 (95% CI: 0.840–0.888). The calibration curve demonstrated good agreement in the independent external validation set (Supplementary Figure 2c (383.3KB, tif) ), and the C-index for the prediction of positive biopsy was 0.871 (95% CI: 0.861–0.881).
Clinical use of the nomogram for PCa detection
DCA for the nomogram is presented in Supplementary Figure 3 (272.4KB, tif) . The decision curve showed that if the threshold probability of a patient and doctor was >4% and <99%, respectively, which is a wide range, using this nomogram to predict positive biopsy would add more benefit than the intervention-all-patients scheme or the intervention-none scheme. Within this range, net benefit was comparable with several overlaps, on the basis of the nomogram.
Development of a nomogram for prediction of significant prostate cancer
To investigate the predictive value of LMR in high-risk PCa, a nomogram for prediction of csPCa was developed. Similarly, logistic regression analyses revealed that age (P = 0.002), PSA (P < 0.001), DRE (P < 0.001), abnormal imaging signals (P = 0.001), PSAD (P < 0.001), and LMR (P = 0.015) were independent predictors of csPCa (Supplementary Table 1), and the nomogram is displayed in Figure 2.
Supplementary Table 1.
Logistic regression analysis of predictors for clinically significant prostate cancer in the training cohort
| Variables | Univariate regression | Multivariate regression | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |
| Age | 1.063 | 1.045–1.082 | 0.000 | 1.047 | 1.017–1.078 | 0.002 |
| PSA | 1.042 | 1.033–1.051 | 0.000 | 1.017 | 1.009–1.026 | 0.000 |
| DRE | 7.779 | 5.338–11.337 | 0.000 | 4.360 | 2.480–7.667 | 0.000 |
| Abnormal imaging | 2.625 | 1.937–3.557 | 0.000 | 2.335 | 1.388–3.929 | 0.001 |
| PSAD | 7.482 | 4.236–13.214 | 0.000 | 4.581 | 2.331–9.000 | 0.000 |
| LMR | 0.640 | 0.454–0.902 | 0.000 | 0.546 | 0.334–0.891 | 0.015 |
| Tobacco | 0.725 | 0.514–1.023 | 0.011 | 0.932 | 0.561–1.546 | 0.784 |
| Alcohol | 1.470 | 0.928–2.328 | 0.101 | 1.243 | 0.658–2.350 | 0.503 |
| BMI | 0.994 | 0.940–1.051 | 0.821 | 1.051 | 0.971–1.139 | 0.216 |
PSA: prostate-specific antigen; DRE: digital rectal examination; PSAD: prostate-specific antigen density; LMR: lymphocyte-to-monocyte ratio; BMI: body mass index; OR: odds ratio; CI: confidence interval
Figure 2.
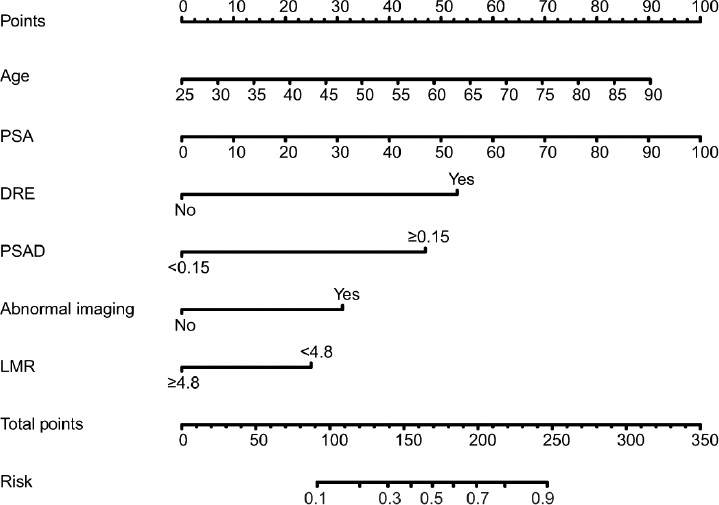
Nomogram for predicting csPCa based on the training cohort. The prostate biopsy nomogram was developed in the training cohort, with age, PSA, abnormal DRE, PSAD, abnormal imaging findings, and LMR incorporated. PSA: prostate-specific antigen; DRE: digital rectal examination; PSAD: prostate-specific antigen density; LMR: lymphocyte-to-monocyte ratio; csPCa: clinically significant prostate cancer.
Performance of the nomogram for prediction of csPCa
Good calibration was also observed for the probability of csPCa in the training set, as well as internal and external validation set (Supplementary Figure 4 (378.4KB, tif) ). The C-index of the training set was 0.848 (95% CI: 0.829–0.867) and confirmed to be 0.840 through bootstrapping validation. In the internal and external validation set, the C-index was 0.882 (95% CI: 0.858–0.906) and 0.882 (95% CI: 0.869–0.895), respectively. DCA revealed that threshold probability was >5% and <100%, respectively (Supplementary Figure 5 (270KB, tif) ).
Development of a nomogram for positive biopsy prediction with PSA 4–10 ng ml−1
To investigate further the predictive value of LMR accompanied with other factors in patients with PSA 4–10 ng ml−1, logistic regression analyses were performed in the training set. Univariate logistic regression showed that age (P < 0.001), abnormal DRE findings (P < 0.001) and imaging signals (P < 0.001), f/t PSA (P < 0.001), and LMR (P = 0.045) were associated with high probability of diagnosis of PCa, and PSA lost its predictive value (Supplementary Table 2). On multivariate analysis, DRE findings (P < 0.001), f/t PSA (P < 0.001), and LMR (P = 0.010) were still significant predictors, and age reached marginal significance (P = 0.060). Thus, age, DRE, f/t PSA, and LMR were finally included in the nomogram (Figure 3).
Supplementary Table 2.
Logistic regression analysis of predictors for positive biopsy in patients with prostate-specific antigen 4–10 ng ml−1 in the training cohort
| Variables | Univariate regression | Multivariate regression | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |
| Age | 1.057 | 1.029–1.086 | 0.000 | 1.039 | 0.998–1.082 | 0.060 |
| PSA | 1.076 | 0.946–1.223 | 0.265 | 1.155 | 0.924–1.443 | 0.205 |
| DRE | 5.819 | 3.355–10.092 | 0.000 | 4.936 | 2.217–10.99 | 0.000 |
| Abnormal Imaging | 2.486 | 1.541–4.010 | 0.000 | 1.154 | 0.513–2.597 | 0.729 |
| PSAD | 1.634 | 0.917–2.914 | 0.096 | 1.283 | 0.621–2.65 | 0.501 |
| f/tPSA | 0.400 | 0.248–0.645 | 0.000 | 0.099 | 0.035–0.278 | 0.000 |
| LMR | 0.644 | 0.419–0.991 | 0.045 | 0.390 | 0.19–0.798 | 0.010 |
| Tobacco | 0.758 | 0.457–1.258 | 0.284 | 1.399 | 0.673–2.910 | 0.369 |
| Alcohol | 1.142 | 0.535–2.438 | 0.731 | 1.225 | 0.464–3.238 | 0.682 |
| BMI | 0.981 | 0.910–1.058 | 0.619 | 0.995 | 0.968–1.022 | 0.690 |
PSA: prostate-specific antigen; DRE: digital rectal examination; PSAD: prostate-specific antigen density; f/t PSA: free-to-total prostate-specific antigen; LMR: lymphocyte-to-monocyte ratio; BMI: body mass index
Figure 3.
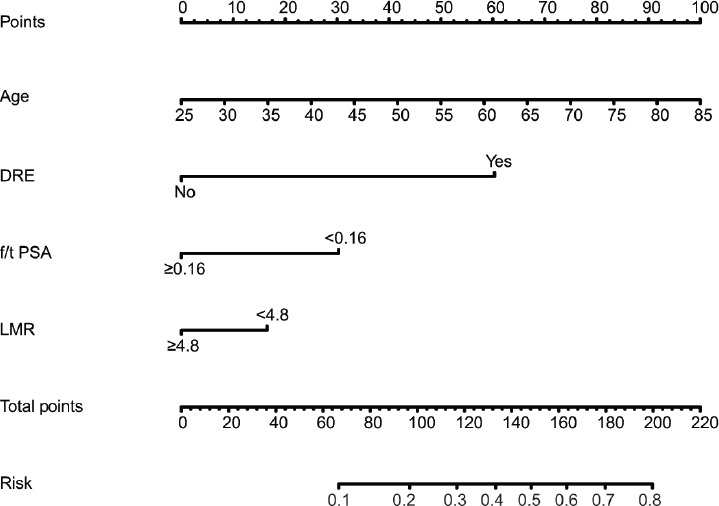
Nomogram for predicting PCa in patients with PSA 4–10 ng ml−1 based on the training cohort. The biopsy nomogram was developed in the training cohort, with age, abnormal DRE, f/t PSA, and LMR incorporated. DRE: digital rectal examination; f/t PSA: free-to-total prostate-specific antigen; LMR: lymphocyte-to-monocyte ratio.
Performance of the nomogram for patients with PSA 4–10 ng ml−1
Good calibration was observed for the probability of positive biopsy in the training set and validation set (Supplementary Figure 6 (554.6KB, tif) ). The C-index of the training set was 0.734 (95% CI: 0.708–0.760) and confirmed to be 0.725 through bootstrapping validation. In the validation set, the C-index was 0.711 (95% CI: 0.670–0.752). DCA revealed that threshold probability was >2% and <81%, respectively (Supplementary Figure 7 (277.1KB, tif) ).
DISCUSSION
PCa screening with PSA has allowed for early detection of PCa at the cost of diagnosing large numbers of men with false-positive results. As a baseline test, PSA screening with a cutoff value of 4.0 ng ml−1 has a specificity of approximately 60% and sensitivity of 56%.14 In order to avoid unnecessary biopsy and overtreatment, several biomarkers have been studied to increase the accuracy of PCa diagnosis, including the PHI test (integrating the value of total and free PSA and the [−2]pro PSA isoform in a formula) and the 4K score (combining free, intact, and total PSA and kallikrein-like peptidase 2).15 These two PSA-based serum tests are intended to distinguish PCa from benign diseases and might be considered as secondary tests with potential utility to determine the need for biopsy.16,17 PCA3, a long noncoding mRNA in urine, exhibited better performance than total and f/t PSA, and increased the area under the receiver operating characteristic curve for positive biopsy.18,19 The use of multiparametric MRI (mpMRI) for the detection of PCa has been steadily increasing over the past few years. Plenty of studies have revealed its great role in detecting PCa, as well as in predicting tumor aggressiveness.20,21 Multiparametric MRI fusion-guided biopsy for the diagnosis of PCa has also been explored recently, revealing its improved accuracy in PCa detection.22,23 However, these technologies were not universally performed due to lack of availability or high cost. Thus, there is interest in developing a new biomarker that is cheap, effective, and widely accessible to avoid overdiagnosis of PCa.
The interaction between tumors and inflammation has been studied in various cancers, including PCa.24,25,26 Chronic inflammation can promote PCa onset and progression through disruption of the immune response and regulation of the tumor microenvironment. Inflammatory cells are involved in stimulating DNA mutation, promoting resistance to apoptosis, regulating the cell cycle, and inducing angiogenesis, cancer invasion, and metastasis through various molecules.27 In particular, anti-inflammatory drugs, such as aspirin, can reduce PCa risk.28,29 For example, lymphocytes play a significant role in antitumor immunity. CD8+ T cells can induce tumor cell apoptosis and reduce tumor growth by cytotoxic activity, with the help of CD4+ T cells. Meanwhile, monocytes can product reactive oxygen species, reactive nitrogen species, and other cytokines, leading to DNA mutation and tumor progression.30 Therefore, decreased lymphocytes and increased monocytes, that is, lower LMR is associated with elevated cancer risk. LMR has also been studied regarding the diagnosis and prognosis of PCa in several clinical studies. A meta-analysis revealed that elevated LMR was associated with favorable overall survival and progression-free survival in PCa.31 Caglayan et al.13 examined the prognostic value of LMR in PCa and found that it was significantly decreased in PCa in the entire cohort and in patients with PSA 4–10 ng ml−1. In the diagnostic gray zone, the combination of f/t PSA ratio and LMR improves the diagnostic accuracy more than that of f/t PSA ratio alone. However, the prognostic value of LMR in prostate biopsy needs to be validated. In the current study, we chose LMR, together with age, PSA, DRE, abnormal imaging signals, and PSAD, to develop a new nomogram to predict PCa risk.
Two independent cohorts from FUSCC and AHQDU were enrolled in the current study and formed the training set and validation set. The cutoff for LMR was 4.800, while in Caglayan et al.'s study13, the cutoff was 3.05. We attributed this difference to different study populations. We found that PCa patients exhibited older age, higher PSA level, higher rates of abnormal DRE findings and imaging signals, higher PSAD, and lower LMR. When stratified by pathological features, LMR differed by PSA value and GS, suggesting the close relationship between LMR and PCa. Therefore, developing a nomogram including LMR value seemed to be reasonable.
In the entire cohort, our nomogram included six variables: age, PSA, DRE, abnormal imaging signals, PSAD, and LMR. All of these predictors were easy to measure clinically before surgery. Our nomogram demonstrated good discrimination and calibration power, and it was confirmed by the internal and independent external validation. The C-index of the nomogram was high, exhibiting moderate accuracy in predicting PCa in the biopsy cohort. Compared with PSA alone and without LMR, the nomogram demonstrated more accuracy in biopsy prediction. The C-index increased only a little in the entire cohort when LMR was added with basic factors to the nomogram (0.824–0.830, P < 0.010). However, in individual patients who presented with suspicion of PCa, LMR still added 30 points to the total score, that is, an increase of approximately 20% to the PCa risk. DCA showed that the threshold probability was 4% and 99%, which demonstrated a high net benefit across a wide range of threshold probabilities. In the nomogram for csPCa detection, age, PSA, DRE, abnormal imaging signals, PSAD, and LMR were included. Similar result was obtained in this nomogram with even better discrimination performance. Moreover, we investigated the prognostic value of LMR when PSA level was located in the gray zone. Age, DRE, f/t PSA, and LMR were included in the nomogram. The nomogram also exhibited good performance, with a C-index of 0.734 and a threshold probability between 2% and 81%, providing an efficient tool for PCa prediction in patients within the gray zone.
The current study had several strengths. First, to our knowledge, this is the first nomogram including a systemic inflammatory marker for prostate biopsy, offering an opportunity to increase diagnostic accuracy without additional cost in clinical practice. Second, the nomogram was based on two independent cohorts from FUSCC and AHQDU with a large number of patients, exhibiting a high degree of stability and flexibility. Third, the cutoff of LMR might vary in different populations. In the current study, we set the definite cutoff value of LMR as 4.800, and the rationality was verified with the independent cohort. This cutoff value might be applied and examined in further studies.
However, our research had several limitations. First, our study was derived from a retrospective cohort, which might have introduced recall bias. Second, a wide variety of exogenous factors were associated with PCa risk, such as metabolic syndrome, dietary factors, family history, and hormonally active medication. These known or unknown factors were not taken into consideration, which might have resulted in some bias in multivariate regression analyses. Third, the two centers were in China. The different ethnicities may have contributed to the difference in PCa diagnosis between different geographical areas. The nomogram should be tested for its applicability in other races and ethnicities.
CONCLUSIONS
We performed a large double-center retrospective study to construct a nomogram for predicting the probability of PCa and csPCa in initial prostate biopsy based on age, PSA, DRE, abnormal imaging signals, PSAD, and LMR. Moreover, a nomogram including age, DRE, f/t PSA, and LMR for PCa detection when PSA level was located in the gray zone was developed. This is the first nomogram to include a systemic inflammatory marker. This model may help clinicians to make clinical decisions when patients present with suspicion of PCa.
AUTHOR CONTRIBUTIONS
GMZ, LJS, YZ and DWY designed the study, collected, analyzed, and interpreted the clinical data, and wrote the manuscript. ZHZ, FL, and WJW collected part of the patients' clinical data. ZHZ, FL, and XL analyzed part of the data. GMZ and DWY supervised the project and revised the manuscript. All authors vouch for the respective data and analysis, read and approved the final version, and agreed to publish the manuscript.
COMPETING INTERESTS
All authors declared no competing interests.
LMR stratified by pathological results. (a) LMR in the 4 groups with different PSA range in FUSCC. (b) LMR in the 4 groups with different PSA range in AHQDU. (c) LMR in the 3 groups with different pathological features: BPH, PCa with GS lower than 3 + 4 and higher than 4 + 3. *P < 0.05; **P < 0.01; ***P < 0.001. LMR: lymphocyte-to-monocyte ratio; BPH: benign prostatic hypertrophy; PSA: prostate-specific antigen; PCa: prostate cancer; GS: Gleason score; FUSCC: Fudan University Shanghai Cancer Center.
Calibration curves of the nomogram for PCa detection in the training, and internal and external validation cohorts. (a) Calibration curves of the nomogram in the training cohort. (b) Calibration curves of the nomogram in the internal validation cohort. (c) Calibration curves of the nomogram in the external validation cohort. The observed rate of positive biopsy is represented on the Y axis, and nomogram predicted probability is plotted on the X axis. Perfect prediction would correspond to a slope of 1 (diagonal 45° dashed line). The histogram on the top side of the figure shows the frequency counts of predicated probability. In the training cohort, the dotted line represents the apparent accuracy of this nomogram without correction for overfit. The solid line indictates bootstrap-corrected nomogram performance. In the validation cohort, the solid line represents the logistic calibration of the model, and the dotted line shows the result of the validation cohort, of which a closer fit to the solid line represents a better prediction. PCa: prostate cancer.
Decision curve analysis of the nomogram for PCa detection. The Y axis measures the net benefit. The blue line represents the prostate positive biopsy prediction nomogram. The thin solid line represents the assumption that all patients have positive biopsy. The thick solid line represents the assumption that all patients have negative biopsy. The decision curve showed that if the threshold probability of a patient and a doctor were 4% and 99%, respectively, using this nomogram in the current study to predict biopsy result would add more benefit than the intervention-all-patients scheme or the intervention-none scheme. PCa: prostate cancer.
Calibration curves of the nomogram for csPCa detection in the training, internal and external validation cohort. (a) Calibration curves of the nomogram in the training cohort. (b) Calibration curves of the nomogram in the internal validation cohort. (c) Calibration curves of the nomogram in the external validation cohort. csPCa: clinically significant prostate cancer.
Decision curve analysis of the nomogram for csPCa detection. The decision curve showed that if the threshold probability of a patient and a doctor were 5% and 100%, respectively, using this nomogram to predict biopsy result would add more benefit than the intervention-allpatients scheme or the intervention-none scheme. csPCa: clinically significant prostate cancer.
Calibration curves of the nomogram for PCa detection in patients with PSA 4–10 ng ml−1 in the training and validation cohorts. (a) Calibration curves of the nomogram in the training cohort. (b) Calibration curves of the nomogram in the validation cohort. PSA: prostate-specific antigen; PCa: prostate cancer.
Decision curve analysis of the nomogram for PCa detection in patients with PSA 4-10 ng ml−1. The decision curve showed that if the threshold probability of a patient and a doctor were 2% and 81%, respectively, using this nomogram to predict biopsy result would add more benefit than the intervention-all-patients scheme or the intervention-none scheme. PSA: prostate-specific antigen; PCa: prostate cancer.
ACKNOWLEDGMENTS
This study was partly funded by the National Natural Science Foundation of China (No. 81502195).
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen R, Sjoberg DD, Huang Y, Xie L, Zhou L, et al. Prostate specific antigen and prostate cancer in Chinese men undergoing initial prostate biopsies compared with Western cohorts. J Urol. 2017;197:90–6. doi: 10.1016/j.juro.2016.08.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Stamey TA, Yang N, Hay AR, McNeal JE, Freiha FS, et al. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N Engl J Med. 1987;317:909–16. doi: 10.1056/NEJM198710083171501. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi T, Fujita K, Tanigawa G, Kawashima A, Nagahara A, et al. Serum monocyte fraction of white blood cells is increased in patients with high Gleason score prostate cancer. Oncotarget. 2017;8:35255–61. doi: 10.18632/oncotarget.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang L, Li X, Wang B, Luo G, Gu L, et al. Prognostic value of neutrophil-to-lymphocyte ratio in localized and advanced prostate cancer: a systematic review and meta-analysis. PLoS One. 2016;11:e0153981. doi: 10.1371/journal.pone.0153981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aminsharifi A, Howard L, Wu Y, De Hoedt A, Bailey C, et al. Prostate specific antigen density as a predictor of clinically significant prostate cancer when the prostate specific antigen is in the diagnostic gray zone: defining the optimum cutoff point stratified by race and body mass index. J Urol. 2018;200:758–66. doi: 10.1016/j.juro.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 8.Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, et al. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–83. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kearns JT, Lin DW. Improving the specificity of PSA screening with serum and urine markers. Curr Urol Rep. 2018;19:80. doi: 10.1007/s11934-018-0828-6. [DOI] [PubMed] [Google Scholar]
- 10.Nishijima TF, Muss HB, Shachar SS, Tamura K, Takamatsu Y. Prognostic value of lymphocyte-to-monocyte ratio in patients with solid tumors: a systematic review and meta-analysis. Cancer Treat Rev. 2015;41:971–8. doi: 10.1016/j.ctrv.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Chan JC, Chan DL, Diakos CI, Engel A, Pavlakis N, et al. The lymphocyte-to-monocyte ratio is a superior predictor of overall survival in comparison to established biomarkers of resectable colorectal cancer. Ann Surg. 2017;265:539–46. doi: 10.1097/SLA.0000000000001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y, Li M, Bo C, Liu X, Zhang J, et al. Prognostic significance of the lymphocyte-to-monocyte ratio and the tumor-infiltrating lymphocyte to tumor-associated macrophage ratio in patients with stage T3N0M0 esophageal squamous cell carcinoma. Cancer Immunol Immunother. 2017;66:343–54. doi: 10.1007/s00262-016-1931-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caglayan V, Onen E, Avci S, Sambel M, Kilic M, et al. Lymphocyte-to-monocyte ratio is a valuable marker to predict prostate cancer in patients with prostate specific antigen between 4 and 10 ng/dl. Arch Ital Urol Androl. 2019;90:270–5. doi: 10.4081/aiua.2018.4.270. [DOI] [PubMed] [Google Scholar]
- 14.Xiao LH, Chen PR, Gou ZP, Li YZ, Li M, et al. Prostate cancer prediction using the random forest algorithm that takes into account transrectal ultrasound findings, age, and serum levels of prostate-specific antigen. Asian J Androl. 2017;19:586–90. doi: 10.4103/1008-682X.186884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kretschmer A, Tilki D. Biomarkers in prostate cancer – current clinical utility and future perspectives. Crit Rev Oncol Hematol. 2017;120:180–93. doi: 10.1016/j.critrevonc.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Heijnsdijk EA, Denham D, de Koning HJ. The cost-effectiveness of prostate cancer detection with the use of prostate health index. Value Health. 2016;19:153–7. doi: 10.1016/j.jval.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Vedder MM, de Bekker-Grob EW, Lilja HG, Vickers AJ, van Leenders GJ, et al. The added value of percentage of free to total prostate-specific antigen, PCA3, and a kallikrein panel to the ERSPC risk calculator for prostate cancer in prescreened men. Eur Urol. 2014;66:1109–15. doi: 10.1016/j.eururo.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filella X, Foj L, Mila M, Auge JM, Molina R, et al. PCA3 in the detection and management of early prostate cancer. Tumour Biol. 2013;34:1337–47. doi: 10.1007/s13277-013-0739-6. [DOI] [PubMed] [Google Scholar]
- 19.Fenner A. Prostate cancer: PCA3 as a grade reclassification predictor. Nat Rev Urol. 2017;14:390. doi: 10.1038/nrurol.2017.70. [DOI] [PubMed] [Google Scholar]
- 20.Bittencourt LK, Hollanda ES, Oliveira RV. Multiparametric MR imaging for detection and locoregional staging of prostate cancer. Top Magn Reson Imaging. 2016;25:109–17. doi: 10.1097/RMR.0000000000000089. [DOI] [PubMed] [Google Scholar]
- 21.Lee CH, Ku JY, Park WY, Lee NK, Ha HK. Comparison of the accuracy of multiparametric magnetic resonance imaging (mpMRI) results with the final pathology findings for radical prostatectomy specimens in the detection of prostate cancer. Asia Pac J Clin Oncol. 2019;15:e20–7. doi: 10.1111/ajco.13027. [DOI] [PubMed] [Google Scholar]
- 22.Monni F, Fontanella P, Grasso A, Wiklund P, Ou YC, et al. Magnetic resonance imaging in prostate cancer detection and management: a systematic review. Minerva Urol Nefrol. 2017;69:567–78. doi: 10.23736/S0393-2249.17.02819-3. [DOI] [PubMed] [Google Scholar]
- 23.Wang R, Wang J, Gao G, Hu J, Jiang Y, Zhao Z, et al. Prebiopsy mp-MRI can help to improve the predictive performance in prostate cancer: a prospective study in 1,478 consecutive patients. Clin Cancer Res. 2017;23:3692–9. doi: 10.1158/1078-0432.CCR-16-2884. [DOI] [PubMed] [Google Scholar]
- 24.Taverna G, Pedretti E, Di Caro G, Borroni EM, Marchesi F, et al. Inflammation and prostate cancer: friends or foe? Inflamm Res. 2015;64:275–86. doi: 10.1007/s00011-015-0812-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhong Z, Sanchez-Lopez E, Karin M. Autophagy, inflammation, and immunity: a troika governing cancer and its treatment. Cell. 2016;166:288–98. doi: 10.1016/j.cell.2016.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sciarra A, Gentilucci A, Salciccia S, Pierella F, Del Bianco F, et al. Prognostic value of inflammation in prostate cancer progression and response to therapeutic: a critical review. J Inflamm (Lond) 2016;13:35. doi: 10.1186/s12950-016-0143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Vidal AC, Howard LE, Moreira DM, Castro-Santamaria R, Andriole GL, et al. Aspirin, NSAIDs, and risk of prostate cancer: results from the REDUCE study. Clin Cancer Res. 2015;21:756–62. doi: 10.1158/1078-0432.CCR-14-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang TB, Yan Y, Guo ZF, Zhang XL, Liu H, et al. Aspirin use and the risk of prostate cancer: a meta-analysis of 24 epidemiologic studies. Int Urol Nephrol. 2014;46:1715–28. doi: 10.1007/s11255-014-0703-4. [DOI] [PubMed] [Google Scholar]
- 30.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–8. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 31.Peng H, Luo X. Prognostic significance of elevated pretreatment systemic inflammatory markers for patients with prostate cancer: a meta-analysis. Cancer Cell Int. 2019;19:70. doi: 10.1186/s12935-019-0785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LMR stratified by pathological results. (a) LMR in the 4 groups with different PSA range in FUSCC. (b) LMR in the 4 groups with different PSA range in AHQDU. (c) LMR in the 3 groups with different pathological features: BPH, PCa with GS lower than 3 + 4 and higher than 4 + 3. *P < 0.05; **P < 0.01; ***P < 0.001. LMR: lymphocyte-to-monocyte ratio; BPH: benign prostatic hypertrophy; PSA: prostate-specific antigen; PCa: prostate cancer; GS: Gleason score; FUSCC: Fudan University Shanghai Cancer Center.
Calibration curves of the nomogram for PCa detection in the training, and internal and external validation cohorts. (a) Calibration curves of the nomogram in the training cohort. (b) Calibration curves of the nomogram in the internal validation cohort. (c) Calibration curves of the nomogram in the external validation cohort. The observed rate of positive biopsy is represented on the Y axis, and nomogram predicted probability is plotted on the X axis. Perfect prediction would correspond to a slope of 1 (diagonal 45° dashed line). The histogram on the top side of the figure shows the frequency counts of predicated probability. In the training cohort, the dotted line represents the apparent accuracy of this nomogram without correction for overfit. The solid line indictates bootstrap-corrected nomogram performance. In the validation cohort, the solid line represents the logistic calibration of the model, and the dotted line shows the result of the validation cohort, of which a closer fit to the solid line represents a better prediction. PCa: prostate cancer.
Decision curve analysis of the nomogram for PCa detection. The Y axis measures the net benefit. The blue line represents the prostate positive biopsy prediction nomogram. The thin solid line represents the assumption that all patients have positive biopsy. The thick solid line represents the assumption that all patients have negative biopsy. The decision curve showed that if the threshold probability of a patient and a doctor were 4% and 99%, respectively, using this nomogram in the current study to predict biopsy result would add more benefit than the intervention-all-patients scheme or the intervention-none scheme. PCa: prostate cancer.
Calibration curves of the nomogram for csPCa detection in the training, internal and external validation cohort. (a) Calibration curves of the nomogram in the training cohort. (b) Calibration curves of the nomogram in the internal validation cohort. (c) Calibration curves of the nomogram in the external validation cohort. csPCa: clinically significant prostate cancer.
Decision curve analysis of the nomogram for csPCa detection. The decision curve showed that if the threshold probability of a patient and a doctor were 5% and 100%, respectively, using this nomogram to predict biopsy result would add more benefit than the intervention-allpatients scheme or the intervention-none scheme. csPCa: clinically significant prostate cancer.
Calibration curves of the nomogram for PCa detection in patients with PSA 4–10 ng ml−1 in the training and validation cohorts. (a) Calibration curves of the nomogram in the training cohort. (b) Calibration curves of the nomogram in the validation cohort. PSA: prostate-specific antigen; PCa: prostate cancer.
Decision curve analysis of the nomogram for PCa detection in patients with PSA 4-10 ng ml−1. The decision curve showed that if the threshold probability of a patient and a doctor were 2% and 81%, respectively, using this nomogram to predict biopsy result would add more benefit than the intervention-all-patients scheme or the intervention-none scheme. PSA: prostate-specific antigen; PCa: prostate cancer.


