Abstract
The main binding site for SARS-COV-2 spike protein in human body is human Angiotensin converting enzyme 2 (ACE2) protein receptor. Herein we present the effect of chloroquine (CLQ) on human ACE2 receptor. Molecular docking studies showed that chloroquine have a docking score is quite high compare to other well known drugs. Furthermore, molecular dynamics (MD) studies with CLQ docked ACE2 results in large fluctuations on RMSD up to 2.3 ns, indicating conformational and rotational changes due to the presence of drug molecule in the ACE2 moiety. Analysis of results showed that CLQ can effect the conformation of human ACE2 receptor. We believed that this work will help researchers to understand better the effect of CLQ on ACE2.
Keywords: Chloroquine (CLQ), Angiotensin converting enzyme 2 (ACE2) protein, Molecular docking, MD-simulation
1. Introduction
From December 2019 a massive social and economical pandemic [1] has been spread internationally due to COVID-19 (SARS-CoV-2) which creates public health emergency. SARS-CoV in not a new at all [2], [3], [4], [5]. SARS-CoV are mainly classified into four categories viz. α-COV, β-COV, γ-COV and δ-COV, of which the latest, classified as SARS-CoV-2, belongs to β-CoV category. It has four different proteins like spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins. SARS-CoV-2 has single stranded, positive-sense RNA and there are total seven genes. Most of the part of RNA genomes are coated by ORF while rest part is coated by mRNA and other proteins. PP1a and PP1ab replicase proteins are generated by ribosomal frame shifting and these proteins generates total sixteen non-structural proteins (NSPs) which are responsible for carrying on the viral reproduction of the replicase transcriptase compound process. The structural proteins (S, E, M and N) are formed by the mRNA [6]. The outer surface of SAR-COV-2 contains spike glycoprotein which binds with human ACE2 receptor. After binding, it creates channel to transfer its genetic material (RNA) inside the host cell for duplication. It is therefore important to block the human ACE2 receptor or prevent further modification of it by the virus. There are some drugs which are under clinical trials and which showed some positive results on COVID-19 [7]. But the main mechanism of potential drugs are still under investigation. Remdesivir shows some positive results on COVID-19 patients at the very primary stages but does not show any potential effect on the seriously affected patients [8]. Favipiravir shows inhibitory effect on viral RNA polymerase [9] but does not have any remarkable effect on COVID-19 patients. A widely used drug for the treatment of Zika virus is azithromycine [10], which has been used with hydroxychloroquine [11] COVID-19. Again there are some anticancer, antiviral, CNS stimulants approved by FDA [12, 13], while some are under clinical trials [14], but the results against COVID-19 are not uniformly satisfactory. Some positive results against COVID-19 patients have been shown by Chloroquine phosphate [15, 16]. Chloroquine itself is an anti-malarial drug also, having some anti-viral effect [17]. It must be mentioned that higher doses of both chloroquine and hydroxychloroquine can pose risks to the patients [18].
From the above, it must be clear that though numerous attempts have been made to combat the lethal effect of SARS-COV-2, till date there are no suitable drugs or vaccines which can cure COVID-19 affected patients with certainty. Most of the suspected drugs are now in clinical trials and there are no in-vivo potentially active established drugs are present for the proper treatment of COVID-19 patients. We considered docking studies with main protease of SARS-CoV-2 and with human ACE2 of 16 drugs, and found CLQ to be outperforming the others. This was followed by more detailed analysis, including MD studies on CLQ and ACE2.
2. Methodology
2.1. Docking of potential drugs with COVID-19 main protease and human ACE2 receptor
Structures of the main protease (Mpro) and human ACE2 receptor were obtained from Protein Data Bank, with PDB IDs 6LU7 and 6LZG respectively. The “sdf” files of 16 drugs were obtained from the PubChem (National Library of Medicine) and were converted to pdb files using UCSF Chimera to check the ligand-protein binding interaction. Both the structures of Mpro and ACE2 receptor were cleaned by removing hetero-atoms and water molecules using UCSF Chimera package [19]. Only chain-A of ACE2 receptor was chosen for performing the binding affinity with the drugs. Docking was done between the best binding pocket of the protein and the drugs using Autodock Vina package [20]. Necessary files for docking simulation were made by using Autodock Tools.
2.2. MD simulation of ACE2-CLQ complex
The minimum energy configurations obtained from molecular docking studies were considered for molecular dynamics (MD) simulation studies using GROMACS (Version 2018.5) [21] with the CHARMM36-mar2019 force-field [22] using TIP3P model [23]. The ligand (CLQ) topology and parameters were generated using CHARMM General Force Field server. A cubical box was generated where the ACE2-CLQ complex was at least 1 nm from the edges of the box to maintain at least 2 nm distance between two successive images of the complex using periodic boundary conditions. Adequate number of Na+ ions was added to maintain charge neutrality of the system. First, energy minimization was carried out until the maximum force became less than 10 kJmol−1nm−1. The steepest descent algorithm was used followed by conjugate gradient protocol. Then the system was equilibrated for 100 ps using isochoric-isothermal (NVT) equilibration at 300 K. The time step was 2 fs. This was followed by equilibration at isothermal-isobaric or NPT ensemble at 300 K for 100 ps. Modified Berendsen thermostat was used for the NPT ensemble. Here also the time step was 2 fs. For both NVT and NPT equilibration, cut-offs for electrostatic and van der Waals interactions were kept at 1.0 nm. Long range interactions were calculated using smooth particle mesh Ewald (PME) method [24]. The equilibrated ensembles were finally subjected to MD simulation for 10 ns, with electrostatic and van der Waals cut off as before. PME method was used to calculate long range electrostatic interactions. A modified Berendsen thermostat and a Parinello-Rahman barostat were used with reference temperature and pressure at 300 K and 1 bar respectively. Snapshots of the trajectory were saved every 1 ns for each case.
2.3. Analysis of md simulations
For ACE2-CLQ systems, structural trajectories were calculated using trjconv tool. The trjconv tool of GROMACS was used to re-center the protein and other molecules within the cubical box. van der Waals interaction, electrostatic energy, interaction energy values etc. were calculated using gmx energy tool of GROMACS. RMSD plots, RMSF plot, Radius of gyration and Solvent Accessible Surface Area (SASA) plots were performed using xmgrace plotting tool.
3. Results and discussion
3.1. Molecular docking studies
Initial screening of the drug molecules were performed by applying molecular docking studies. The drugs used for docking have either anti-malerial, anti HIV or anti-viral agents. Results of the docking studies of all the drugs with both the Mpro and ACE2 receptor are shown in Table 1 below.
Table 1.
Screening of drugs by molecular docking studies with Mpro and ACE2 receptor.
| PUBCHEM CID | Compound Name | MW (g/mol) | Molecular Formula | Docking Score (kcal/mol) | |
|---|---|---|---|---|---|
| Main protease | ACE2-receptor | ||||
| 2791 | Chloroquine | 319.90 | C18H26ClN3 | −5.9 | −7.5 |
| 6475 | Chlorphenoxamine | 303.80 | C18H22ClNO | −6.1 | −6.7 |
| 2726 | Chlorpromazine | 318.90 | C17H19ClN2S | −6.3 | −6.0 |
| 2801 | Clomipramine | 314.90 | C19H23ClN2 | −5.9 | −6.9 |
| 5,284,550 | Dosulepin | 295.40 | C19H21NS | −6.5 | −6.8 |
| 60,877 | Emtricitabine | 247.25 | C8H10FN3O3S | −5.8 | −6.7 |
| 3461 | Gemcitabin | 263.20 | C9H11F2N3O4 | −6.2 | −6.5 |
| 3652 | Hydroxychloroquine | 335.90 | C18H26ClN3O | −6.3 | −7.3 |
| 3658 | Hydroxyzine | 374.90 | C21H27ClN2O2 | −6.6 | −6.9 |
| 60,825 | Lamivudin | 229.26 | C8H11N3O3S | −5.7 | −6.5 |
| 104,762 | Mizoribine | 259.22 | C9H13N3O6 | −6.4 | −6.7 |
| 4756 | Phenazopyridine | 213.24 | C11H11N5 | −6.4 | −6.7 |
| 4927 | Promethazine | 284.40 | C17H20N2S | −6.0 | −6.5 |
| 37,542 | Ribavirin | 244.20 | C8H12N4O5 | −6.2 | −6.5 |
| 5568 | Triflupromazine | 352.40 | C18H19F3N2S | −7.0 | −7.1 |
| 35,370 | Zidovudine | 267.24 | C10H13N5O4 | −6.6 | −6.6 |
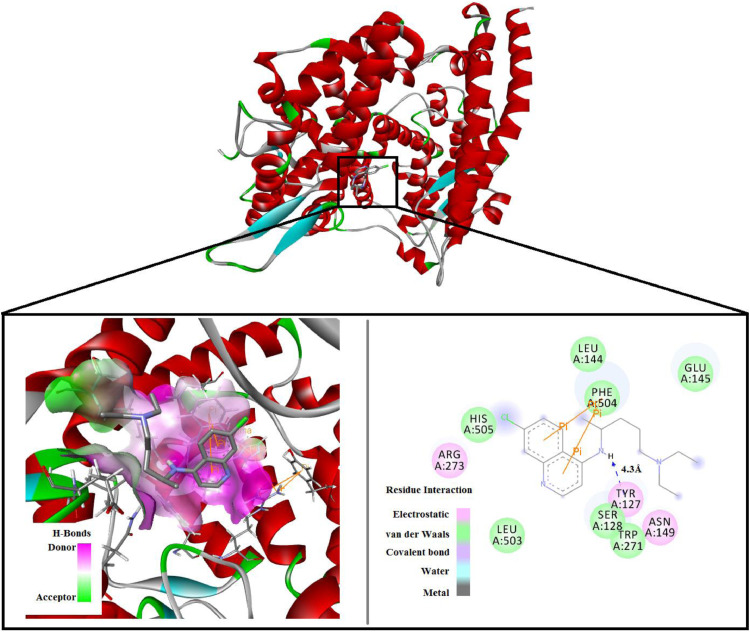
All the drugs showed good binding affinities with both main protease and ACE2. Our previous study showed that HCQ has an inhibitory effect against Mpro of SARS-CoV-2. But chloroquine (CLQ) showed the best binding affinity with ACE2 receptor with a score of −7.5 kcal/mol. It showed greater binding affinity vis-a-vis its derivative hydroxychloroquine (HCQ) (−7.3 kcal/mol) which is used for COVID-19 treatment [25]. However, CLQ has less binding affinity (−5.9 kcal/mol) with the Mpro compared to HCQ (−6.3 kcal/mol). Docking studies showed that CLQ has the highest docking score with ACE2. From Fig. 1 , it is seen that there are H-bonding and electrostatic interactions between CLQ and ACE2. The left panel of Fig. 1 shows the donor and acceptor sites of the docked structure for the H-bond formation. The neighbouring residues of CLQ in the docked structure are ARG 273, HIS 505, LEU 144, PHE 504, TYR 127, SER 128, TRP 271, ASN 149, LEU 503, GLU 145 as shown in the right panel in Fig. 1.
Fig. 1.
Docked structure of CLQ with ACE2 receptor.
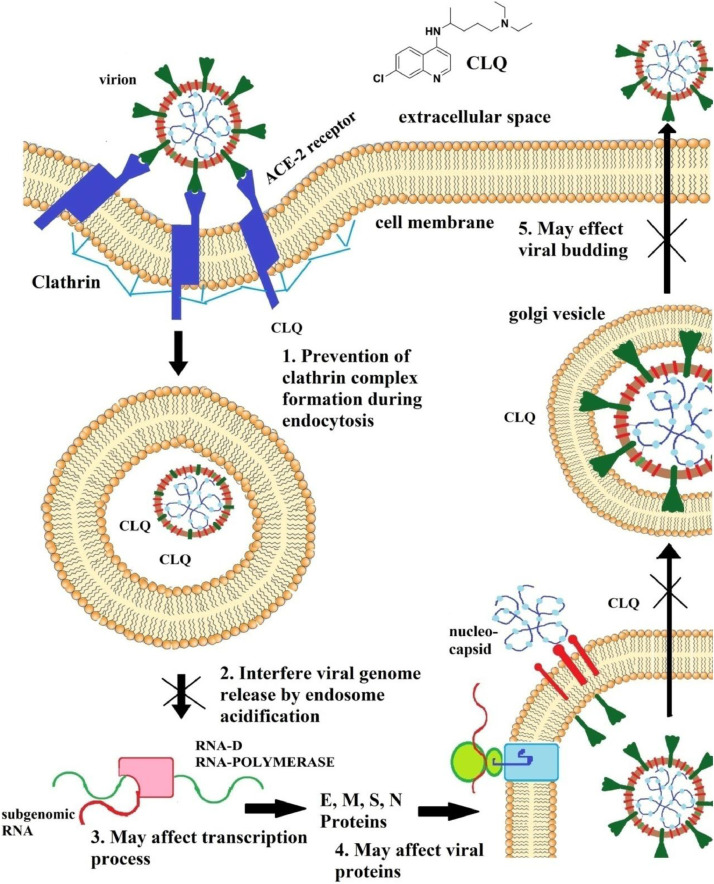
Chloroquine causes multiple effects on cellular functions viz. 1. Immunomoddulatory effect, 2. Prevents endocytosis as well as lysosomal function and fusion [26], 3. zinc ionophore [27] and 4. prevents binding with sialic acid and ACE-2 receptor [28]. These effects may make CLQ as a preventer against SARS-CoV-2. CLQ is alkaline in nature which causes the increase in alkalinity of endosome and lysosome that leads to increase the endosomal and lysosomal pH. SARS-CoV interacts with ACE-2 receptor in the host cell by spike (S) proteins and endocytosed into the endosome. Two functional domains of spike proteins are there (a) S1, binds with receptor and (b) S2, for viral and cellular membrane fusion with specified fusion peptide region, interacting with lipid bilayer. After virion uptake, the S protein is specifically cleaved to expose the fusion peptide region. In presence of CLQ the process is blocked due to its alkalinity increase the pH of endosome and lysosome. CLQ may also inhibit the proton pump present in the lysosome though the mechanism is still not clear. Hence the process of endocytosis i.e. the fusion of endosome into lysosome also be stopped or slowed down in presence of chloroquine [26]. Enhancing the lysosomal pH, chloroquine inactivates pH-dependent lysosomal proteases like cathepsin L necessary for S-protein cleavage, finally preventing the membrane fusion and viral genome release. Previous research also showed that Zn2+ inhibits the corona virus or RNA viruses by blocking the replication process. The viral RNA i.e. RdRp which is responsible for making viral RNA and responsible of replication process inhibited by Zn2+ ionophore [29]. Zinc ions can not enter into host cells through the cell membrane. CLQ allows zinc ion to bind with it which allows to enter into the cell membrane of the host cell [27]. Then by increasing the zinc concentration, Zn can inhibit RdRp and it prevents for making the viral RNA. These results by the application of CLQ zinc concentration can be increased and viral RNA can be decreased [29]. Again sialic acid is attached with the cell membrane. Spike proteins can also bind with sialic acid. Chloroquine can directly bind with sialic acid and block the binding site of spike protein of SARS-CoV-2 and also CLQ also binds with the ACE-2 receptor which prevents the proper binding SARS-CoV-2 spike protein with the ACE-2 as well as sialic acid [28].
From the above discussion it is showed that CLQ interacts with ACE-2 to reduce the glycosylation of the ACE-2 receptor and the viral penetration into the cell. The endocytosis process is inhibited due to the alkaline nature of CLQ by lowering the affinity of viral spike protein against the receptor which results the inhibition of infection development [30]. Again significant reduction of the development of infection is administrated by CLQ which has a immense therapeutic but also prophylactic activity [31]. Both CLQ and HCQ caused immunomodulating effect and it is assumed that the cytokine storm of SARS-CoV-2 infected patients suffering from COVID-19 will be inhibited [32]. The probable mechanism of action of CLQ on SARS-CoV-2 is shown in Fig. 2 .
Fig. 2.
Mechanism of action of chloroquine (CLQ) against SARS-CoV-2.
To gain inside how chloroquine affect the conformational and structural changes of ACE-2 receptor we have done MD-simulation with the stable docked structure of ACE-2 and CLQ.
3.2. MD simulation studies
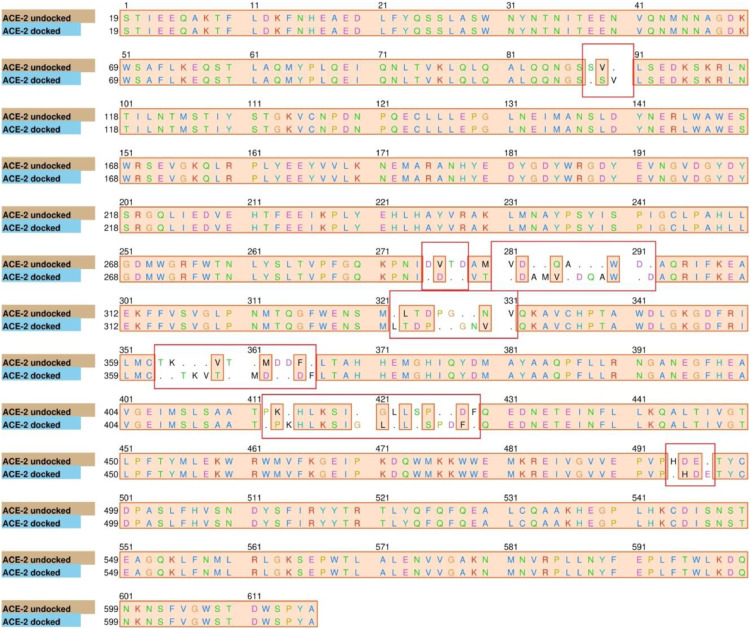
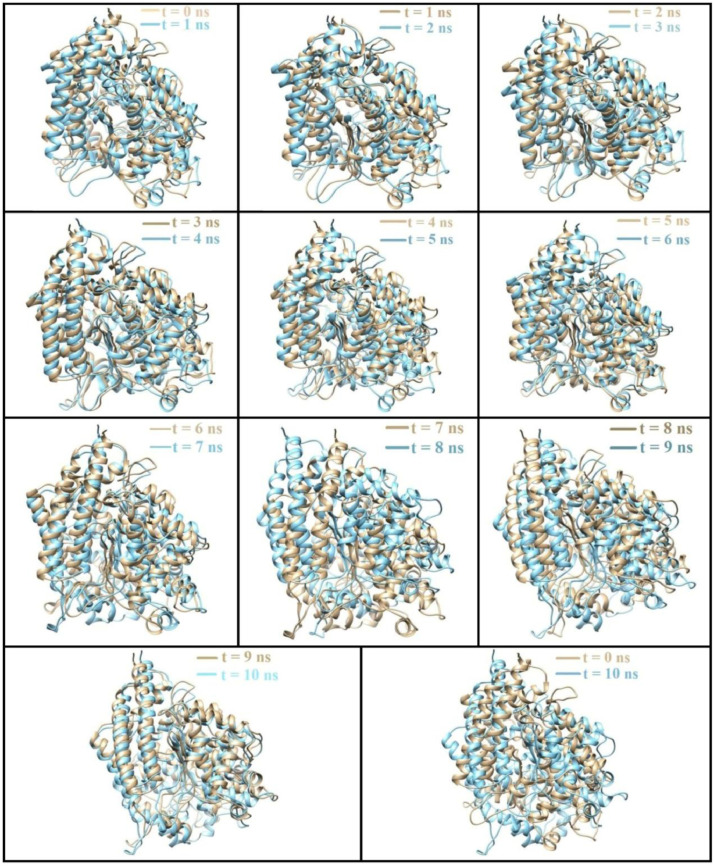
MD simulation studies for a period 10 ns were performed with the energy minimised docked structure. Due to translational and rotational motion of the drug molecule, remarkable changes were observed during the simulation, which may be seen in the snapshots taken during simulation process. Fig. 3 shows the structural changes in the ACE2 sequence after MD.
Fig. 3.
Structural changes in the sequence of ACE2 after MD, with CLQ.
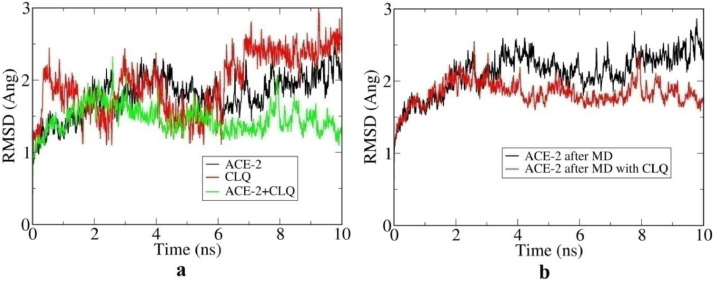
Fig. 4a represents the RMSD plot for the individual ACE2, CLQ and the combined system while Fig. 4b represents the RMSD plot for docked and undocked ACE2. From Fig. 4a it is clear that compare to individual ACE2 and CLQ the RMSD fluctuation of the combined system become progressively lower, while that of undocked ACE2 fluctuates much more and progressively increases. This suggests stabilization of the combined system. After 8 ns, the RMSD fluctuation is minimum and equilibration is expected to have occurred.
Fig. 4.
Root mean square deviation (RMSD) plot of ACE2 receptor, CLQ and ACE2+CLQ docked structure (a) RMSD plot of ACE2 receptor alone before and after docking (b).
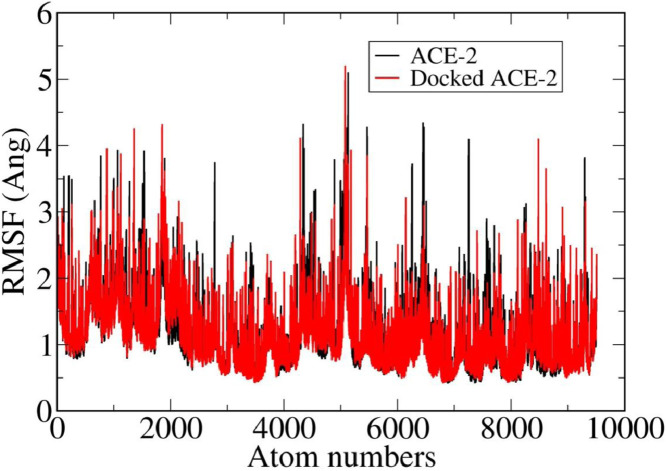
Furthermore Fig. 4b reveals the fact that ACE2 shows a continuous fluctuation but after docking this fluctuation is reduced to 1.7 Å from 2.3 Å. Root mean square fluctuation (RMSF) behavior of atoms, shown in Fig. 5 , also shows a similar trend and support the above conclusion.
Fig. 5.
RMSF of the ACE2 and its docked form with the CLQ.
From the RMSF it is clear that in undocked ACE2, residues that fluctuate more are: ASN33, HIS34, GLN42, ALA46, ASN49, ASN51, LEU97, GLN98, LYS112, THR118, LEU144, MET152, ARG177, TRP203, ASP206, ILE358, ALA550, GLY575 and MET579. On the other hand, in docked ACE2, the fluctuation is more near THR129, LYS131, HIS241, LYS247, SER545 residues only. Table 2 displays the conformational energy, total energy and interaction energy of ACE2 and ACE2+CLQ complex.
Table 2.
Average conformational, total and interaction energy of ACE2 and ACE2+CLQ.
| System | Conformational Energy (kJ/mol) | Total Energy (kJ/mol) | Total interaction energy (kJ/mol) |
|---|---|---|---|
| ACE2 | 34,593.48 | −1.36 × 106 | – |
| ACE2+CLQ | 34,825.45 | −1.37 × 106 | −157.93 |
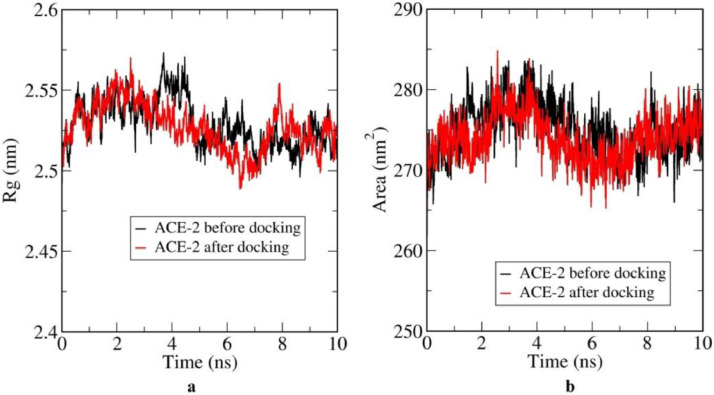
More conformational energy in the composite system leads to more flexibility within the same system. On the other hand, higher value of total energy of the composite system makes the system more stabilized. A considerably higher value of interaction energy suggests that there is strong interaction between CLQ and the ACE2 receptor. Fig. 6 shows the radius of gyration of the docked and undocked ACE2 and Solvent Accessible Surface Area (SASA) plot of docked and undocked ACE2.
Fig. 6.
Radius of gyration (a) and SASA plot (b) ACE2 receptor before and after docking.
Fig. 6a shows variation of radius of gyration of ACE2, which measures its compactness, before and after docking,. From the plot it is clear that up to 3 ns both the docked and undocked ACE2 receptor had similar size. But after that the undocked structure suddenly expands and remains larger in size. This indicates a destabilization of undocked ACE2, whereas the docked structure stabilises more smoothly, leaving aside smaller fluctuations around 8 ns. This agrees with the RMSD plot more or less. Fig. 6b indicates the easy incorporation of the drug molecule within the protein binding site. With increased value of SASA for ACE2 indicates its expansion of volume and lower fluctuation with time. After
Fig. 7 indicates the structural changes that occur during 1–10 ns of MD-simulation. During 1 to 3 ns there is a robust change in structure which is in accordance with the RMSD plot. After 4 ns the system becomes stabilised with little overall structural change. This continues till 7 ns. Between 7 to 8 ns there is huge change in the structure which is also reflected in the RMSD plot. Again, after 8 ns, there is comparative stability of the structure indicating equilibration.
Fig. 7.
Structural change in ACE2 receptor at 1–10 nanosecond during MD-simulation docking of CLQ the SASA value reached a maxima at 3 ns which reveals that the drug stabilize the ACE2 receptor.
4. Conclusion
16 drug molecules were screened against the main protease of SARS-CoV-2 and human ACE2 receptor by molecular docking. Chloroquine (CLQ) showed the highest binding affinity with ACE2 via formation of H-bonding and electrostatic interactions. The docked ACE2-CLQ structure was further studied with molecular dynamics. MD study also shows formation of stable complex between CLQ and ACE2. RMSD plot revealed the docked ACE2-CLQ structure to be more stable than ACE2 alone. ACE2-CLQ composite system is found to equilibrate after about 8 ns. The same conclusion was reached from RMSF, radius of gyration and SASA plots. Hence we conclude that CLQ binds with human ACE2 receptor reasonably strongly and the stable ACE2-CLQ may prevent further binding of ACE2 with spike protein of SARS-CoV-2.
CRediT authorship contribution statement
Nabajyoti Baildya: Conceptualization, Visualization, Writing - original draft, Methodology. Narendra Nath Ghosh: Methodology, Conceptualization, Visualization, Writing - original draft. Asoke P. Chattopadhyay: Methodology, Supervision, Writing - original draft.
Declaration of Competing Interest
The authors declare no conflicting interest in the present work.
Acknowledgements
The authors wish to acknowledge infrastructural support from Department of Chemistry, University of Kalyani, Kalyani, Nadia, India and from Department of Chemistry, University of Gour Banga, Mokdumpur, Malda, India.
Data availability
Data is available upon request to the corresponding author.
References
- 1.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S. ACS Publications; 2020. Research and Development On Therapeutic Agents and Vaccines For COVID-19 and Related Human Coronavirus Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.-.D., Rappuoli R. SARS—Beginning to understand a new virus. Nature Reviews Microbiology. 2003;1(3):209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peiris J., Lai S., Poon L., Guan Y., Yam L., Lim W., Nicholls J., Yee W., Yan W., Cheung M. Coronavirus as a possible cause of severe acute respiratory syndrome. The Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahir M., Shah S.I.A., Zaman G., Khan T. A Dynamic Compartmental Mathematical Model Describing The Transmissibility Of MERS-CoV Virus In Public. Punjab University Journal of Mathematics. 2019;51(4):57–71. (ISSN 1016-2526) [Google Scholar]
- 5.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. The Lancet. 2015;386(9997):995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McBride R., Van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gautret P., Lagier J.-.C., Parola P., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q. placebo-controlled, multicentre trial, The Lancet; 2020. Remdesivir in Adults With Severe COVID-19: a randomised, Double-Blind. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proceedings of the Japan Academy, Series B. 2017;93(7):449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Retallack H., Di Lullo E., Arias C., Knopp K.A., Laurie M.T., Sandoval-Espinosa C., Leon W.R.M., Krencik R., Ullian E.M., Spatazza J. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proceedings of the National Academy of Sciences. 2016;113(50):14408–14413. doi: 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosseboeuf E., Aubry M., Nhan T., De Pina J., Rolain J., Raoult D., Musso D. Azithromycin inhibits the replication of Zika virus. J Antivirals Antiretrovirals. 2018;10(1):6–11. [Google Scholar]
- 12.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6(1):16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in. The lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J., Tian Z., Yangbreakthrough X., Chloroquine Phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies, Biosci Trends 14 (1) (2020) 72–73, doi:10.5582/bst.2020.01047. Epub 2020 Feb 19. PMID: 32074550. [DOI] [PubMed]
- 16.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biot C., Daher W., Chavain N., Fandeur T., Khalife J., Dive D., De Clercq E. Design and synthesis of hydroxyferroquine derivatives with antimalarial and antiviral activities. J. Med. Chem. 2006;49(9):2845–2849. doi: 10.1021/jm0601856. [DOI] [PubMed] [Google Scholar]
- 18.Marmor M.F., Kellner U., Lai T.Y., Melles R.B., Mieler W.F. Recommendations on screening for chloroquine and hydroxychloroquine retinopathy (2016 revision) Ophthalmology. 2016;123(6):1386–1394. doi: 10.1016/j.ophtha.2016.01.058. [DOI] [PubMed] [Google Scholar]
- 19.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 20.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berendsen H.J., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91(1–3):43–56. [Google Scholar]
- 22.Lee S., Tran A., Allsopp M., Lim J.B., Hénin J.r.m., Klauda J.B. CHARMM36 united atom chain model for lipids and surfactants. The journal of physical chemistry B. 2014;118(2):547–556. doi: 10.1021/jp410344g. [DOI] [PubMed] [Google Scholar]
- 23.Boonstra S., Onck P.R., van der Giessen E. CHARMM TIP3P water model suppresses peptide folding by solvating the unfolded state. The journal of physical chemistry B. 2016;120(15):3692–3698. doi: 10.1021/acs.jpcb.6b01316. [DOI] [PubMed] [Google Scholar]
- 24.Abraham M.J., Gready J.E. Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5. J Comput Chem. 2011;32(9):2031–2040. doi: 10.1002/jcc.21773. [DOI] [PubMed] [Google Scholar]
- 25.Baildya N., Ghosh N.N., Chattopadhyay A.P. Inhibitory activity of hydroxychloroquine on COVID-19 main protease: an insight from MD-simulation studies. J Mol Struct. 2020 doi: 10.1016/j.molstruc.2020.128595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delvecchio R., Higa L.M., Pezzuto P., Valadão A.L., Garcez P.P., Monteiro F.L., Loiola E.C., Dias A.A., Silva F.J., Aliota M.T. Chloroquine, an endocytosis blocking agent, inhibits Zika virus infection in different cell models. Viruses. 2016;8(12):322. doi: 10.3390/v8120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue J., Moyer A., Peng B., Wu J., Hannafon B.N., Ding W.-.Q. Chloroquine is a zinc ionophore. PLoS ONE. 2014;9(10) doi: 10.1371/journal.pone.0109180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modeling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Te Velthuis A.J., van den Worm S.H., Sims A.C., Baric R.S., Snijder E.J., van Hemert M.J. Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6(11) doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devaux C.A., Rolain J.-.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. Journal of Microbiology, Immunology and Infection. 2020 doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases. The Lancet infectious diseases. 2003;3(11):722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felsenstein S., Herbert J.A., McNamara P.S., Hedrich C.M. COVID-19: immunology and treatment options. Clinical Immunology. 2020 doi: 10.1016/j.clim.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is available upon request to the corresponding author.