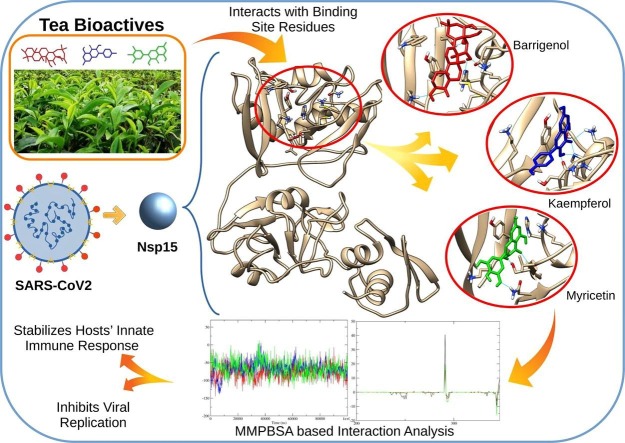
Graphical abstract
Abbreviations: Nsp15, Non-structural protein 15; MDS, Molecular dynamics simulations; RMSD, Root mean square deviation; RMSF, Root mean square fluctuations; MMPBSA, Molecular Mechanics Poisson Boltzmann Surface Area
Keywords: COVID-19, SARS-CoV-2, Nsp15, Bioactive molecules, In-silico
Abstract
Immensely aggravated situation of COVID-19 has pushed the scientific community towards developing novel therapeutics to fight the pandemic. Small molecules can possibly prevent the spreading infection by targeting specific vital components of the viral genome. Non-structural protein 15 (Nsp15) has emerged as a promising target for such inhibitor molecules. In this investigation, we docked bioactive molecules of tea onto the active site of Nsp15. Based on their docking scores, top three molecules (Barrigenol, Kaempferol, and Myricetin) were selected and their conformational behavior was analyzed via molecular dynamics simulations and MMPBSA calculations. The results indicated that the protein had well adapted the ligands in the binding pocket thereby forming stable complexes. These molecules displayed low binding energy during MMPBSA calculations, substantiating their strong association with Nsp15. The inhibitory potential of these molecules could further be examined by in-vivo and in-vitro investigations to validate their use as inhibitors against Nsp15 of SARS-CoV2.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) formerly known as the 2019-novel CoV reported to have spread from the Huanan market in China has ultimately led to a pandemic called the coronavirus disease 2019 (COVID-19) (Organization, 2020, Sinha et al., 2020). Gradually increasing its severity spectrum from mild respiratory tract infections in the initial days to acute pneumonia and currently having advanced to asymptomatic carriage, SARS-CoV2 has taken the globe by a storm in the past couple of months (Singhal, 2020).
Genetically it is a non-segmented positive sense RNA virus hailing from the Coronaviridae family of the order Nidovirales (Kim et al., 2020, Shang et al., 2020, Shannon et al., 2020, Yuan et al., 2020). SARS-CoV2 genome is one of the largest known RNA virus genomes (~30 kb in size), encoding for four structural proteins (spike protein, envelope protein, membrane protein, and nucleocapsid protein) and five accessory proteins (ORF3a, ORF6, ORF7, ORF8, and ORF9) (Kim et al., 2020, McDonald, 2013, Shannon et al., 2020, Sinha et al., 2020). Once the virus is inside the host cell, the ORFs are translated into polypeptides pp1a and pp1b comprising 4382 and 7073 amino acids, respectively (Cui et al., 2019, Sinha et al., 2020). These polypeptides are further proteolytically divided into 16 non-structural polyproteins (Nsps) (Báez-Santos et al., 2015, Gao et al., 2020, Sinha et al., 2020, Sinha et al., 2020, Ziebuhr, 2005). The Nsps congregate together to develop a large membrane bound replication-transcription complex known to perform several enzymatic activities (Báez-Santos et al., 2015, Pillaiyar et al., 2020, Sinha et al., 2020). The current investigation is based on Nsp15, one of the fifteenth members of the Nsp family.
Nsp15, a member of the EndoU family of enzymes, is nidoviral RNA uridylate-specific endoribonuclease (NendoU) with a catalytic domain at the C-terminal and has been observed to be conserved in various virus families (Elfiky, 2020, Kim et al., 2020). Earlier, it was thought to have direct involvement in only viral replication, recent research on Nsp15 also unraveled its interference with the innate immune response, hence proclaiming its biological importance (Bhardwaj et al., 2008, Deng et al., 2017, Kim et al., 2020, Sinha et al., 2020, Sinha et al., 2020). It is also responsible for snipping the double stranded RNA substrate via the Mn2+ dependent endoribonuclease activity that shows specificity towards uridylate in unpaired regions (Bhardwaj et al., 2008, Kim et al., 2020, Sinha et al., 2020). The active site of Nsp15 is shaped by the six critical amino acids (His235, His250, Lys290, Thr341, Tyr343, and Ser294), where His235 and His250 act as a general acid and a general base respectively. A catalytic triad is formed by the former three amino acids, and the latter two amino acids administer the uridine specificity (Kim et al., 2020, Sinha et al., 2020). The middle domain also offers a number of interaction sites (Kim et al., 2020). Lastly, the N-terminal domain stabilizes the complete hexamer conformation (Kim et al., 2020).
Currently, there are no treatment measures or vaccination against SARS-CoV2, and the requirement of a prophylactic and therapeutic intervention technique is critical (Shannon et al., 2020, Sinha et al., 2020, Walls et al., 2020). Targeting the conserved Nsp15 active site via potent inhibitor molecules will not only hinder its involvement in virus replication activity but also prohibit the protein from interfering with the host’s innate immune response, enabling it to fight the viral invasion (Chandra et al., 2020, Khan et al., 2020, Surti et al., 2020). The current investigation was performed with the aim of finding potent inhibitor molecules that could strongly bind to the active site of Nsp15.
2. Material and methods
2.1. Datasets
The three dimensional crystal structure of Nsp15 (PDB ID: 6W01) (Kim et al., 2020) having a resolution of 1.90 Å was retrieved from the Protein Data Bank for this study (Berman et al., 2000). The bioactive molecules of tea were used as ligand molecules (Bhardwaj et al., 2020, Nakai et al., 2005, Namal Senanayake, 2013) that were obtained from pubchem (Kim, Chen, Cheng, Gindulyte, He, He, Li, & Bolton, 2019) in sdf file format. The geometry optimization of each ligand molecule was attained by Gaussian16 DFT minimization protocols (Zheng & Frisch, 2017).
2.2. Molecular docking
Molecular docking of the ligand molecules onto the active site of Nsp15 was achieved by utilizing the CDOCKER docking application of Accelrys Discovery Studio software (Hockney, Goel, & Eastwood, 1974). This application follows the protein-fixed ligand-flexible type of docking algorithm (Chen, Zhou, & Meng, 2018) during which Nsp15 is kept constant in a position, and the ligand molecules are free to move, setting the rest of the parameters to default. Before docking, structure of the protein was prepared via “prepare protein” wizard of the Discovery studio package, which checks the protein and for absent hydrogen atoms, irregular bond formations, and other anomalies.
The active site residues were selected as the binding site for the ligand molecules and it was defined to be of 10 Å radius, containing the six critical amino acids that shape it. Protein ligand interactions simulated during the docking process depends largely on the correct geometry of this ligand binding site. CDOCKER follows a grid based approach towards docking (Puratchikody et al., 2016, Wu et al., 2003), thus we defined a grid box with a grid angle of 90° and spacing of 0.50. The co-ordinates of the ligand binding site of the input sphere were 10.959, −0.514, 20.8275, 12, with the ligand molecule (citric acid) as the center point (Chandra et al., 2020, Kim et al., 2020, Surti et al., 2020). The number of starting random conformations and the number of rotated ligand orientations to refine for each of the conformations for 1000 dynamics steps were set to ten. Moreover, for annealing refinement, the number of heating steps were 2000 while the number of cooling sets were set to 5000. The distance to consider Pi-cation, Pi-Pi, and Pi-alkyl interactions was set to 5 Å, 6 Å, and 5.5 Å respectively. Docking was performed on this site with the bioactive molecules for generating poses of the protein–ligand complexes. This docking methodology follows the principles of CHARMm (Chemistry at Harvard Macromolecular Mechanics Energy) (Brooks et al., 1983), where the water molecules, heteroatoms, as well as any bound inhibitors in the protein–ligand complex are extracted since they are liable to influence the protein–ligand complex formation. CHARMm also minimizes the energy of the complexes to a gradient of 0.01 kcal/mol/Å. Lastly, hydrogen atoms were added to the proteins conformation. The ligands binding to the protein’s binding site obtained unique conformational poses and were analyzed via high-temperature kinetics. Successfully docked complexes were interpreted and ranked based on their total binding energies calculated after docking, and their interaction patterns were visualized by discovery studio.
2.3. Molecular dynamics simulations
Subsequently, the top three ligand molecules having the best total binding energies were put through 100 ns of molecular dynamic simulation (MDS) via version 5.1.4 of GROningen MAchine for Chemical Simulations (GROMACS) (Abraham et al., 2015, Hess et al., 2008, Van Der Spoel et al., 2005), to ensure flexibility and stability of complex developed after docking. In MDS, firstly the protein and ligand topologies were obtained via the ‘pdb2gmx’ script and the GlycoBioChem PRODRG1 server respectively, and joined together into a conformation followed by its energy minimization via the GROMOS96 43a1 force field. The PRODRG2 server was given the PDB coordinates of the molecules, which were converted by it to topologies compatible with GROMACS and other programs. These conformations were then put into a solvated cubic box comprising water molecules (30232 for Barrigenol, and 30,227 for Kaempferol and Myricetin) which was electrically stabilized by addition of 12Na+ ions using the ‘gmx genion’ script. Inside the simulation box the protein was kept at a distance of 1.4 nm from the walls of the box to comply with the minimum image conventions. This system was equilibrated under NVT and NPT ensembles and put through position restrained MDS for the time period of 100 ns, which keeps the backbone C-α atoms fixed and allows the movement of solvent molecules. Linear constraint solver algorithm (Hess, Bekker, Berendsen, & Fraaije, 1997) was used to obtain the covalent interactions and the particle mesh Eshwald method for electrostatic interactions (Essmann et al., 1995). The temperature and pressure of the system was equilibrated to 310 K and 1 atm respectively by V-rescale weak coupling method (Berendsen, Postma, van Gunsteren, DiNola, & Haak, 1984) and Parrinello-Rahman method respectively (Parrinello & Rahman, 1981). Motion equation was integrated at a time step of 2 fs via the leap frog algorithm, updating the list of neighbors after every 5 steps. Finally these solvated and equilibrated complexes (three protein–ligand complexes) were put to production phase to generate trajectories of root mean square deviation (RMSD), root mean square fluctuation (RMSF) and hydrogen bond (Hbond) graphs by the in-built GROMACS scripts.
2.4. MMPBSA calculations
Lastly, the Molecular Mechanics Poisson Boltzmann Surface Area (MMPBSA) calculations were performed on the simulated protein–ligand complexes to obtain the final binding free energies and residue contribution energies of the complexes, by using the gmmpbsa module of GROMACS (Kumari, Kumar, & Lynn, 2014). This module is based on an end point method that applies the following equations for the calculation of binding free energy (BFE):
Here ΔGbinding is the BFE, ΔEelectrostatic is the electrostatic contribution, ΔEvdW+ is the vander Waals contribution, and ΔGpolar and ΔGnon-polar are the polar and non polar solvation terms respectively. The non polar solvation term is more commonly referred as the SASA contribution. Since the study involves the binding of similar ligands to the protein repeatedly, thus the entropic contribution is ignored here. MMPBSA brings together continuum solvent approaches with molecular mechanics and generalized born electrostatics to calculate the binding as well as contribution energies (Ghosh et al., 2020, Kumar et al., 2020).
3. Results and discussion
3.1. Interaction analysis
Molecular docking is regarded as one of the primary methods for identifying favourable protein–ligand interactions based on affinity parameters (Bhardwaj, Singh, Das, & Purohit, 2021). In the current investigation, we screened a library of bioactive molecules of tea and docked them onto the active site of Nsp15, which is shaped by the six aforementioned critical residues. We calculated the total binding energy of all the docked molecules, as illustrated in Table S1. The binding energy score was used for assessing the strength of protein–ligand interactions and rank them accordingly. A more negative binding energy score suggests more favourable binding between protein and ligand. Our molecular docking results showed that the molecules Barrigenol, Kaempferol, Myricetin, Theanine, Methyl Salicylate, Linalool Oxide, Epicatechin, Quercetin, and Apigenin could be considered as potential inhibitors of NSp15. To narrow down the investigation, the top three ligand molecules with the best total binding energies were selected. Barrigenol showed the maximum affinity, followed by Kaempferol and Myricetin.
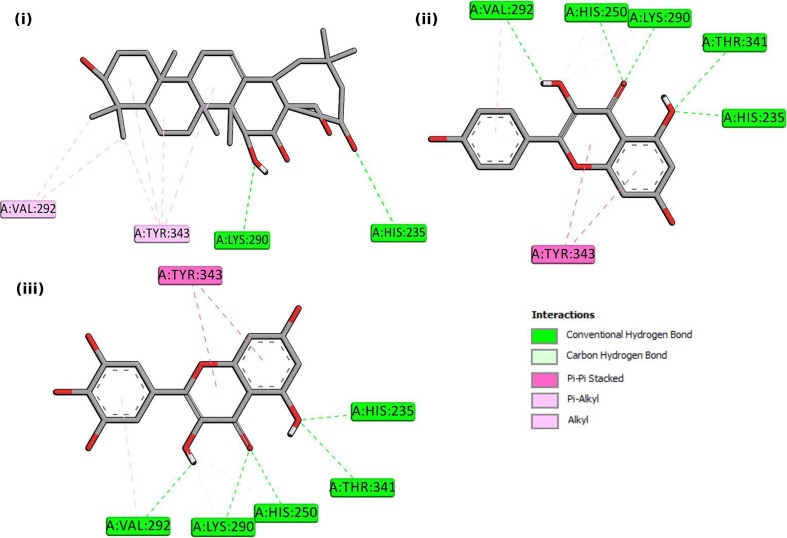
The interactions of these three molecules with Nsp15 were explored in-depth by analyzing their binding patterns. This exploration revealed that out of the six critical residues of the Nsp15 active site, Barrigenol interacted with three of them, namely His235, Lys290, and Tyr343. It formed two conventional H bonds with His235, one with Lys290, and four pi-alkyl bonds with Tyr343. Apart from these residues, it has also interacted with Val292, forming two alkyl bonds. Similarly, Kaempferol interacted with five of the critical residues of Nsp15 active site, namely His235, His250, Lys290, Thr341, and Tyr343, forming one conventional H bond with His235, one conventional H bond, and two carbon H bonds with His250; one conventional H bond and one carbon H bond with Lys290; one conventional H bond with Thr341, and two pi-pi stacked bonds with Tyr343. Apart from these residues, it has also interacted with Val292, forming one conventional H bond and one pi-alkyl bond. In the case of Myricetin, we noticed that it followed the exact similar interaction pattern, as shown by Kaempferol, forming the same bonds with the Nsp15 active site residues.
These interaction patterns of the complexes were depicted in Fig. 1 . The docked complexes revealed that the molecules were extensively involved in the binding with the residues of the active site of Nsp15 and attained high docking scores. The information about the binding site residues of Nsp15 and ligand molecules were presented in Table S2. The three complexes were thus proceeded to MDS experiments to further deepen our knowledge about their interaction profiles and to examine their stability in a dynamic 3D conformational space.
Fig. 1.
2-D interactions of Nsp15 of SARS-CoV2 with the selected bioactive molecules (i) Barrigenol (ii) Kaempferol (iii) Myricetin.
3.2. Molecular dynamics simulation
Since, the protein–ligand interactions are entity dynamic, therefore MDS is considered as an imperative part of any computational analysis. It supplements with a detailed data regarding the protein–ligand interaction with a dynamic perspective. So intending to achieve molecular insight into the self conformational perturbations brought by the protein to attain conformational flexibility and stability with the ligand molecules, we allowed the three protein–ligand complexes to undergo MDS for a time period of 100 ns. The successful completion of the simulation experiments provided us with trajectories for RMSD, RMSF, and H bonds of the complexes. Moreover, the clusters of complexes formed during simulation were produced and analyzed. Subsequently, we also performed MMPBSA calculations to diversify our analysis by obtaining the binding free energies (BFE) and residue contribution energies (CE).
3.3. Root mean square deviations
One of the critical parameters to analyze a protein–ligand complex is the RMSD of the protein backbone C-α atoms, which characterizes its overall conformational stability in a dynamic state during the simulation. The system is equilibrated and stabalized when it obtains low levels of RMSD with consistent fluctuations for the entire simulation; on the other hand, higher fluctuations indicate low stability. Highly deviated RMSD graphs can also imply major conformational transitions occurring in the protein to obtain stable conformation with the ligand.
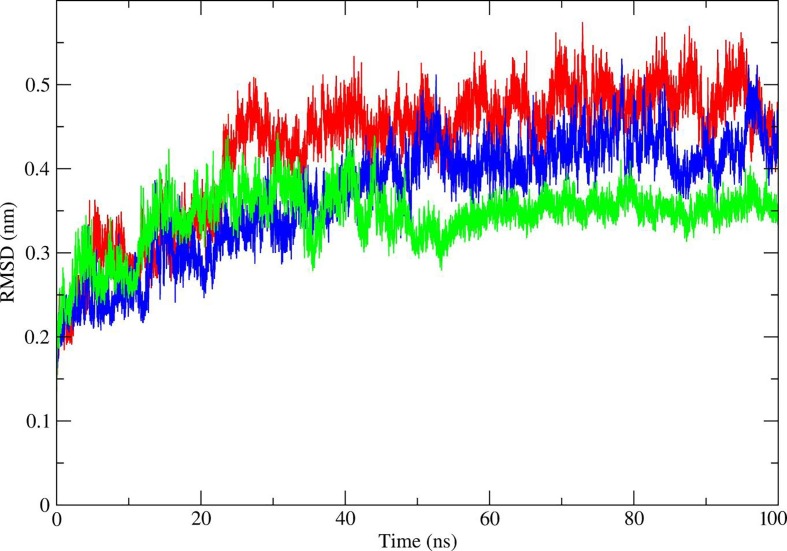
The RMSDs for the protein backbone C-α atoms in complex with the three ligand molecules were calculated, and produced as a graph depicted in Fig. 2 . Here we noticed that the minimum and maximum RMSDs obtained by Nsp15-Barrigenol, Nsp15-Kaempferol, and Nsp15-Myricetin were of very low magnitude ranging from 0.2 nm to 0.5 nm. This was a strong indication of conformational stability that the protein had achieved with the ligand molecules. The plot depicted stable trajectories with consistent and minor fluctuations implying that the protein backbone underwent minor structural perturbations. Also a few major fluctuations were observed among the trajectories at different time periods. These fluctuations were first observed at 10 ns where the RMSDs of the three complexes first gradually decreased from 0.3 nm to 0.25 nm and then suddenly increased to 0.35 nm. Other than these, for Myricetin we have observed fluctuations at 35 ns and 42 ns, whereas for Barrigenol and Kaempferol fluctuations were observed at 87 ns and 90 ns respectively. These observations indicated that the protein–ligand complex obtained a stable conformation during the simulations with a few conformational transitions.
Fig. 2.
RMSD of the backbone Cα atoms of Nsp15 of SARS-CoV2 in complex with bioactive molecules: Nsp15-Barrrigenol (Red), Nsp15-Kaempferol (Blue), and Nsp15-Myricetin (Green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Root mean square fluctuations
The residues of the protein play a vital role in achieving a stable conformation for a protein-ligand complex, which can be gauged by using the RMSF as a parameter. RMSF of the residues basically analyses a particular segment of the protein that is deviating from its mean structure which generally happens upon ligand interaction. The fluctuations observed for every residue signify the levels of flexibility obtained by them. Thus residues or a group of residues showing higher levels of RMSF imply increased flexibility which in turn indicates their increased potential to interact with the ligand molecule. Similarly lower RMSF fluctuations imply lesser flexibility, hence diminished interaction potential.
RMSF for the residues of the protein towards the three ligands were calculated and produced onto a graph, depicted in Fig. S1. The graph for RMSF depicted the residues on the x-axis whereas fluctuation values on the y-axis. Significant fluctuations were observed in the active site region with residues obtaining significant peaks. Moreover, several significant peaks of increased fluctuations were also observed by the residues other than the active site residues, indicating their increased interaction potential implying that the ligands were able to adapt well in the binding pocket of the protein.
3.5. Cluster description
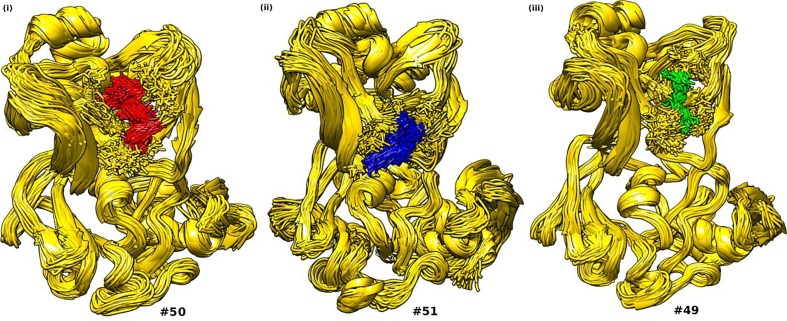
To further elucidate the noticeable flexibility of the dynamic protein receptor towards the ligand molecules as well as the degree of stability achieved by them as a complex, we performed cluster analysis over the MD trajectories of these complexes for the final 5 ns of the simulation (Fig. 3 ). This analysis provided us with a significant number of clusters for a protein–ligand complex accounted for the flexibility of the protein, along with their average RMSDs and energy matrix accounted for the conformational stability of the complex. Greater the number of clusters generated, higher the flexibility of the protein, whereas lower RMSD and lower energy of the matrix implied greater stability of the complex. The results of this analysis were shown in Table 1 . It was evident from the table that a significant number of clusters were formed for all the three complexes confirming the higher flexibility of the protein towards the ligand molecules which also supported our RMSF analysis. Moreover, the values for RMSDs and matrix energies were also of very low degrees. These low values pertain to compactness of the mean conformation of the complexes in 3D space with very less deviation among the clusters, suggesting conformational stability of the clusters.
Fig. 3.
Pictorial representation of the central conformations representative of the average structure of each clusters of (i) Nsp15-Barrrigenol (ii) Nsp15-Kaempferol (iii) Nsp15-Myricetin. #: Number of clusters.
Table 1.
Cluster analysis results depicting number of clusters formed for a protein–ligand complex, their average RMSDs, and the energy of their cluster matrix.
| Complexes | Number of Clusters | Average RMSD (nm) | Energy of the Matrix (nm) |
|---|---|---|---|
| Nsp15-Barrigenol | 50 | 0.187876 | 3.39181 |
| Nsp15-Kaempferol | 51 | 0.224762 | 1.2038. |
| Nsp15-Myricetin | 49 | 0.170349 | 0.934839. |
3.6. Hydrogen bond analysis
Although, the aforementioned analyses already indicated a stable and flexible conformation of the protein with the three ligands, we also performed H bond analysis to second our previous results. Hydrogen bonds formed between a ligand and the protein are responsible for maintaining a compact and a properly oriented structure, where the flexibility of the protein’s residues also comes into play as they are the ones that will be forming bonds with the ligand molecules.
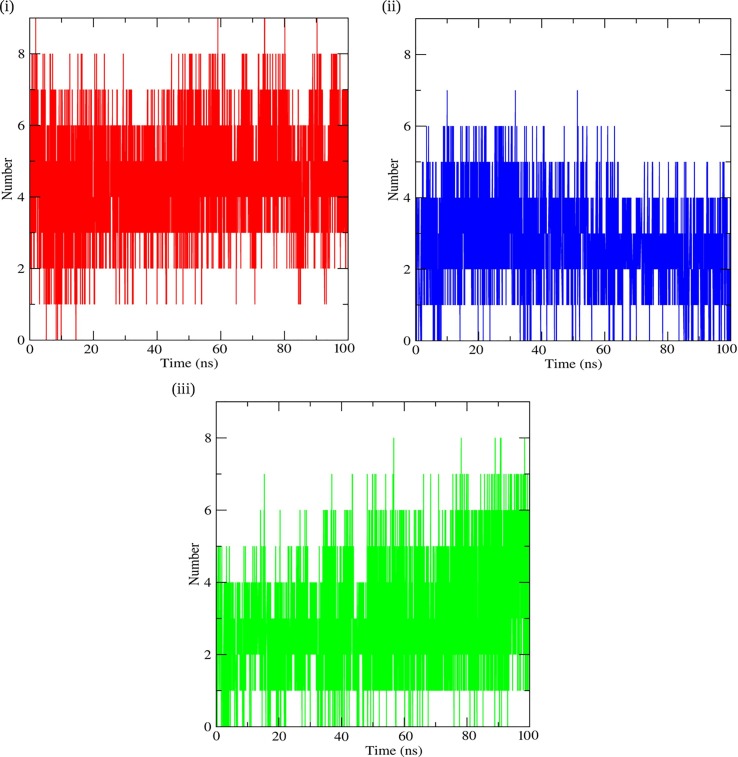
The number of H bonds formed by the protein with the ligand molecules were calculated and depicted on a graph, as shown in Fig. 4 . The H bond graph for Barrigenol depicted that the protein formed a minimum of 0 H bonds whereas a maximum of ~9 bonds during the simulation, fluctuating from time to time which narrowed down to an average of ~4 H bonds that remained almost constant for the complete simulation of 100 ns. Kaempferol on the other hand formed a maximum of ~7 H bonds with the protein and a minimum of 0 H bonds, whereas the average number of H bonds decreased form ~4 to ~3 as the simulation progressed. In the case of Myricetin a maximum of ~8 H bonds and a minimum of 0H bonds were observed, whereas the average number of H bonds gradually increased from ~ 3 to ~ 5 during the simulation. The H bond formation between the protein and the ligand molecules emphasized that they had a strong and stable binding during the entire simulation period.
Fig. 4.
Hydrogen bond profiles of Nsp15 in complex with bioactive molecules: (i) Nsp15-Barrrigenol (Red) (ii) Nsp15-Kaempferol (Blue) (iii) Nsp15-Myricetin (Green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Prior to further investigation, we extracted the simulated conformations of the complexes at the beginning (5 ns), middle (50 ns), and the end (100 ns) of the simulation. The interaction poses of each conformation were shown in Fig. S2. The protein–ligand poses confirmed that the ligand molecules stayed inside the active site of Nsp15 during the entire simulation. In this time period, these molecules showed extensive interactions with various residues of the binding site. These results suggested that the ligand molecules formed stable complexes with Nsp15 and that by tightly and stably binding to it, they would be able to prevent the protein from conducting its activities.
3.6.1. Molecular mechanics Poisson Boltzmann surface area calculations
Results obtained via MMPBSA calculations included BFE as well as residue CEs, considered to be one of the most accurate results in computational analysis for a protein–ligand complex under simulation. The BFE computed for a complex comprises various constituents, namely, Van der waals energy, electrostatic energy, polar solvation energy, and SASA energy, as illustrated in Table 2 , where a positive value is unfavourable for interaction and a negative value is favourable. All the constituent energies summed up to form the total BFE for a simulated complex in that time period. Among the complexes under observation we found Barrigenol (−76.073 kJ/mol) to have the most favourable BFE with Nsp15, followed by Kaempferol (−66.259 kJ/mol) and Myricetin (−65.663 kJ/mol) implying that Barrigenol has the best association with the protein receptor and formed a stable complex. Van der waals energy was observed to be the primary benefactor for interaction of these molecules with Nsp15. These results were also in accordance with the total binding energy scores obtained in docking analysis.
Table 2.
MMPBSA based total binding free energies along with its constituent energies for the selected bioactive molecules.
| Complex | Total Binding Free Energy (kJ/mol) | Van der Waals Energy (kJ/mol) | Electrostatic energy (kJ/mol) | Polar solvation energy (kJ/mol) | SASA energy (kJ/mol) |
|---|---|---|---|---|---|
| Barrigenol | −76.073 | −161.943 | −103.552 | 205.191 | −15.769 |
| Kaempferol | −66.259 | −123.574 | −51.059 | 120.275 | −11.902 |
| Myricetin | −65.663 | −145.387 | −52.664 | 145.288 | −12.900 |
Finally, the CEs for the residues involved in interaction with the ligand molecules were calculated via MMPBSA, and produced on to a graph, as depicted in Fig. S3, where higher the contribution of a residue towards favourable interaction more negative is its CE value, whereas unfavourable contribution attains positive CE value. Five residues of Nsp15 (His235, His250, Ser294, Thr341, and Tyr343) were observed to play a key role in interaction with the ligand molecules. The obtained energies were depicted in Table S3. Apart from these, there were also a fairly good number of other residues showing favourable contribution towards interaction with the ligands. The residues with the most favourable CE were labeled in the Figure. Cys291 had unfavourable contribution for all the ligand molecules which might be due to some steric hindrance in the conformation obtained by these molecules inside the active site of Nsp15. But the unfavourable effect of Cys291 was immensely overpowered by the favourable effect of the other residues.
The study carried out on the three ligand molecules led us to believe that they had a strong affinity for Nsp15 and were tightly bound to it. The close association between the bioactive molecules and the binding site could essentially avoid the infection by inhibiting the functioning of the Nsp15 protein. These results, however, needs further validation by in-vitro and in-vivo experiments.
4. Conclusion
Bearing its involvement in important virus survival functions, Nsp15 acted as a promising target, whose inhibition could possibly control the pandemic by not only ceasing its replication inside the host but also activating the hosts immune response against the virus. With this objective, we tried to explore the binding patterns of the bioactive molecules of tea by docking them against the active site of Nsp15. This exploration uncovered the bioactive molecules Barrigenol, Kaempferol, Myricetin, Theanine, Methyl Salicylate, Linalool Oxide, Epicatechin, Quercetin, and Apigenin to have achieved good binding with Nsp15 based on their docking scores. To further elucidate their affinity towards the protein, we carried out molecular dynamics simulations on the top three molecules (Barrigenol, Kaempferol, and Myricetin) in complex with Nsp15 and analyzed the obtained trajectories of RMSD, RMSF, and H bond. These trajectories exposed not only the increased flexibility (through RMSF) of Nsp15 towards the molecules but also the higher stability (through RMSD and H bond) of the complexes formed by them. Conformational stability and flexibility were also verified by cluster analysis of the complexes. Lastly, low binding energies and residue contribution energies obtained via MMPBSA calculations provided the final proof of strong binding between the molecules and Nsp15. This investigation provided evidence of strong interaction between three bioactive molecules and Nsp15 of SARS-CoV2, therefore they could be used as a potent inhibitor against the protein. Inhibitory potential of these molecules could further be examined and verified by in-vivo and in-vitro investigations. Exploitation of the backbone structures of these molecules could further help in developing more molecules with higher affinity and specificity for NSP15. Moreover, the currently examined active site of Nsp15 could also be explored further for drug discovery.
5. Author contribution statement
RP conceived of and designed the study. RP, JS, VKB and RS analyzed and interpreted the data. VKB, VR, RP and SK critically revised it for important intellectual content. All the authors gave final approval of the version to be published.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
RP gratefully acknowledges the Department of Science and Technology, New Delhi (SERB File No: ECR/2016/000031) and Board of Research in Nuclear Sciences, Department of Atomic Energy, Mumbai, India for financial support (Letter No: 37(1)/14/26/2015/BRNS). VKB acknowledges the Academy of Scientific & Innovative Research (AcSIR) India for providing junior research fellowship. VR gratefully acknowledges the Department of Science and Technology, New Delhi (SERB File No: PDF/2019/000607) for the awarding National Post Doctoral Fellowship. We also acknowledge the CSIR-Institute of Himalayan Bioresource Technology, Palampur for providing the facilities to carry out this work. This manuscript represents CSIR-IHBT communication no. 4717.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.foodchem.2020.128933.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- Báez-Santos Y.M., St. John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Research. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics. 1984;81(8):3684–3690. [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H.…Bourne P.E. The protein data bank. Nucleic Acids Research. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Palaninathan S., Alcantara J.M.O., Yi L.L., Guarino L., Sacchettini J.C., Kao C.C. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. Journal of Biological Chemistry. 2008;283(6):3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure and Dynamics. 2020 doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Computers in Biology and Medicine. 2021;128:104117. doi: 10.1016/j.compbiomed.2020.104117. (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. Journal of Computational Chemistry. 1983;4(2):187–217. doi: 10.1002/jcc.540040211. [DOI] [Google Scholar]
- Chandra A., Gurjar V., Qamar I., Singh N. Identification of potential inhibitors of SARS-COV-2 endoribonuclease (EndoU) from FDA approved drugs: A drug repurposing approach to find therapeutics for COVID-19. Journal of Biomolecular Structure and Dynamics. 2020 doi: 10.1080/07391102.2020.1775127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Zhou S., Meng Q. A molecular docking study of Rhizoma Atractylodis and Rhizoma Atractylodis Macrocephalae herbal pair with respect to type 2 diabetes mellitus. Journal of Traditional Chinese Medical Sciences. 2018;5(2):185–198. doi: 10.1016/j.jtcms.2018.05.004. [DOI] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hackbart M., Mettelman R.C., O’Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proceedings of the National academy of Sciences of the United States of America. 2017;114(21):E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sciences. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. The Journal of Chemical Physics. 1995;103(19):8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T.…Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science:abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in silico docking and molecular dynamics simulation study. Journal of Biomolecular Structure and Dynamics. 2020 doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1997;18:1463. http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.48.2727 [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hockney R.W., Goel S.P., Eastwood J.W. Quiet high-resolution computer models of a plasma. Journal of Computational Physics. 1974;14(2):148–158. doi: 10.1016/0021-9991(74)90010-2. [DOI] [Google Scholar]
- Khan R.J., Jha R.K., Singh E., Jain M., Amera G.M., Singh R.P.…Singh A.K. Identification of promising antiviral drug candidates against non-structural protein 15 (NSP15) from SARS-CoV-2: An in silico assisted drug-repurposing study. Journal of Biomolecular Structure and Dynamics. 2020 doi: 10.1080/07391102.2020.1814870. [DOI] [PubMed] [Google Scholar]
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., … Bolton, E. E. (2019). PubChem 2019 update: Improved access to chemical data. Nucleic Acids Research. DOI:10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed]
- Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A.…Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Science. 2020;29(7):1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Kumari K., Vishvakarma V.K., Jayaraj A., Kumar D., Ramappa V.K.…Singh P. Promising inhibitors of main protease of novel corona virus to prevent the spread of COVID-19 using docking and molecular dynamics simulation. Journal of Biomolecular Structure and Dynamics. 2020 doi: 10.1080/07391102.2020.1779131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R., Kumar R., Lynn A. g_mmpbsa —A GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- McDonald S.M. RNA synthetic mechanisms employed by diverse families of RNA viruses. Wiley Interdisciplinary Reviews: RNA. 2013:351–367. doi: 10.1002/wrna.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M., Fukui Y., Asami S., Toyoda-Ono Y., Iwashita T., Shibata H.…Kiso Y. Inhibitory effects of oolong tea polyphenols on pancreatic lipase in vitro. Journal of Agricultural and Food Chemistry. 2005;53(11):4593–4598. doi: 10.1021/jf047814+. [DOI] [PubMed] [Google Scholar]
- Namal Senanayake S.P.J. Green tea extract: Chemistry, antioxidant properties and food applications – A review. Journal of Functional Foods. 2013;5(4):1529–1541. doi: 10.1016/j.jff.2013.08.011. [DOI] [Google Scholar]
- Organization, W. H. (2020). Coronavirus disease 2019 (COVID-19) Situation Report-72 HIGHLIGHTS.
- Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics. 1981;52(12):7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discovery Today. 2020;25(4):668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puratchikody A., Sriram D., Umamaheswari A., Irfan N. 3-D structural interactions and quantitative structural toxicity studies of tyrosine derivatives intended for safe potent inflammation treatment. Chemistry Central Journal. 2016;10(1) doi: 10.1186/s13065-016-0169-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K.e., Wan Y., Luo C., Aihara H.…Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Le N.-T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C.…Canard B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Research. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian Journal of Pediatrics. 2020;87(4):281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S.K., Prasad S.K., Islam M.A., Gurav S.S., Patil R.B., AlFaris N.A.…Shakya A. Identification of bioactive compounds from Glycyrrhiza glabra as possible inhibitor of SARS-CoV-2 spike glycoprotein and non-structural protein-15: A pharmacoinformatics study. Journal of Biomolecular Structure and Dynamics. 2020 doi: 10.1080/07391102.2020.1779132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S.K., Shakya A., Prasad S.K., Singh S., Gurav N.S., Prasad R.S., Gurav S.S. An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics. 2020 doi: 10.1080/07391102.2020.1762741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surti M., Patel M., Adnan M., Moin A., Ashraf S.A., Siddiqui A.J.…Reddy M.N. Ilimaquinone (marine sponge metabolite) as a novel inhibitor of SARS-CoV-2 key target proteins in comparison with suggested COVID-19 drugs: Designing, docking and molecular dynamics simulation study. RSC Advances. 2020;10(62):37707–37720. doi: 10.1039/D0RA06379G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: Fast, flexible, and free. Journal of Computational Chemistry. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Robertson D.H., Brooks C.L., Vieth M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER?A CHARMm-based MD docking algorithm. Journal of Computational Chemistry. 2003;24(13):1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C., So R.T.Y., Lv H.…Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi: 10.1126/science:abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Frisch M.J. Efficient geometry minimization and transition structure optimization using interpolated potential energy surfaces and iteratively updated hessians. Journal of Chemical Theory and Computation. 2017 doi: 10.1021/acs.jctc.7b00719. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Current Topics in Microbiology and Immunology. 2005 doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.