Abstract
Background
The early clinical course of COVID-19 can be difficult to distinguish from other illnesses driving presentation to hospital. However, viral-specific PCR testing has limited sensitivity and results can take up to 72 h for operational reasons. We aimed to develop and validate two early-detection models for COVID-19, screening for the disease among patients attending the emergency department and the subset being admitted to hospital, using routinely collected health-care data (laboratory tests, blood gas measurements, and vital signs). These data are typically available within the first hour of presentation to hospitals in high-income and middle-income countries, within the existing laboratory infrastructure.
Methods
We trained linear and non-linear machine learning classifiers to distinguish patients with COVID-19 from pre-pandemic controls, using electronic health record data for patients presenting to the emergency department and admitted across a group of four teaching hospitals in Oxfordshire, UK (Oxford University Hospitals). Data extracted included presentation blood tests, blood gas testing, vital signs, and results of PCR testing for respiratory viruses. Adult patients (>18 years) presenting to hospital before Dec 1, 2019 (before the first COVID-19 outbreak), were included in the COVID-19-negative cohort; those presenting to hospital between Dec 1, 2019, and April 19, 2020, with PCR-confirmed severe acute respiratory syndrome coronavirus 2 infection were included in the COVID-19-positive cohort. Patients who were subsequently admitted to hospital were included in their respective COVID-19-negative or COVID-19-positive admissions cohorts. Models were calibrated to sensitivities of 70%, 80%, and 90% during training, and performance was initially assessed on a held-out test set generated by an 80:20 split stratified by patients with COVID-19 and balanced equally with pre-pandemic controls. To simulate real-world performance at different stages of an epidemic, we generated test sets with varying prevalences of COVID-19 and assessed predictive values for our models. We prospectively validated our 80% sensitivity models for all patients presenting or admitted to the Oxford University Hospitals between April 20 and May 6, 2020, comparing model predictions with PCR test results.
Findings
We assessed 155 689 adult patients presenting to hospital between Dec 1, 2017, and April 19, 2020. 114 957 patients were included in the COVID-negative cohort and 437 in the COVID-positive cohort, for a full study population of 115 394 patients, with 72 310 admitted to hospital. With a sensitive configuration of 80%, our emergency department (ED) model achieved 77·4% sensitivity and 95·7% specificity (area under the receiver operating characteristic curve [AUROC] 0·939) for COVID-19 among all patients attending hospital, and the admissions model achieved 77·4% sensitivity and 94·8% specificity (AUROC 0·940) for the subset of patients admitted to hospital. Both models achieved high negative predictive values (NPV; >98·5%) across a range of prevalences (≤5%). We prospectively validated our models for all patients presenting and admitted to Oxford University Hospitals in a 2-week test period. The ED model (3326 patients) achieved 92·3% accuracy (NPV 97·6%, AUROC 0·881), and the admissions model (1715 patients) achieved 92·5% accuracy (97·7%, 0·871) in comparison with PCR results. Sensitivity analyses to account for uncertainty in negative PCR results improved apparent accuracy (ED model 95·1%, admissions model 94·1%) and NPV (ED model 99·0%, admissions model 98·5%).
Interpretation
Our models performed effectively as a screening test for COVID-19, excluding the illness with high-confidence by use of clinical data routinely available within 1 h of presentation to hospital. Our approach is rapidly scalable, fitting within the existing laboratory testing infrastructure and standard of care of hospitals in high-income and middle-income countries.
Funding
Wellcome Trust, University of Oxford, Engineering and Physical Sciences Research Council, National Institute for Health Research Oxford Biomedical Research Centre.
Research in context.
Evidence before this study
A detailed systematic review identified 91 diagnostic models for COVID-19 as of July 1, 2020; however, all were appraised to be at “high risk of bias”. Existing early detection models overwhelmingly consider radiological imaging (60 of 91 models), such as CT, which is less readily available than blood tests and involves exposure of patients to ionising radiation. Few studies assessed routine laboratory tests, with the scarce literature considering small numbers of patients with confirmed COVID-19 (<180), labelling patients as negative by use of the imperfectly sensitive PCR test and thereby failing to ensure disease freedom, inadequately accounting for breadth of alternative disease, and not being prospectively validated. No published studies considered whether laboratory artificial intelligence models can be applied to a clinical population as a screening test for COVID-19.
Added value of this study
To our knowledge, this was the largest laboratory artificial intelligence study on COVID-19 to date, training with clinical data from more than 115 000 patients presenting to hospital, and the first to integrate laboratory blood tests with point-of-care measurements of blood gases and vital signs. The breadth of our pre-pandemic control cohort exposed our classifiers to a wide variety of alternative illnesses and offered confidence that control patients did not have COVID-19. Here, we developed context-specific models for patient populations attending the emergency department and being admitted to hospital, and we showed clinically minded calibration by selecting for high negative predictive values at high classification performance. In doing so, we developed an effective screening test for COVID-19 using clinical data that are routinely acquired for patients presenting to hospital in the UK and typically available within 1 h. By simulating performance of our screening test at different stages of a pandemic, we showed high negative predictive values (>98·5%) when disease prevalence is low (≤5%), safely and rapidly excluding COVID-19. We prospectively validated our models by applying them to all patients presenting and admitted to the Oxford University Hospitals in a 2-week test period, achieving high accuracy (>92%) compared with PCR results.
Implications of all the available evidence
Rapid and accurate detection of COVID-19 in hospital admissions is essential for patient safety. Well described limitations of the current gold-standard test include turnaround times up to 72 h (as of July, 2020), limited sensitivity of about 70%, and shortages of skilled operators and reagents. The benefits of our artificial intelligence screening test are that it is immediately deployable at low cost, fits within existing clinical pathways and laboratory testing infrastructure, gives a result within 1 h that can safely exclude COVID-19, and ensures that patients can receive upcoming treatments rapidly.
Introduction
An outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) led to the COVID-19 pandemic of 2020.1 The early clinical course of COVID-19, which often includes common symptoms such as fever and cough, can be challenging for clinicians to distinguish from other respiratory illnesses.2, 3, 4
Testing for SARS-CoV-2, most commonly by real-time RT-PCR assay of nasopharyngeal swabs, has been widely adopted, but has limitations.3, 5, 6 These include limited sensitivity,5, 7 a long turnaround time of up to 72 h, and requirements for specialist laboratory infrastructure and expertise.8 Studies have shown a significant proportion of asymptomatic carriage and limited specificity for common symptoms (fever and cough), hampering symptom-guided hospital triage.9 Therefore, an urgent clinical need exists for rapid, point-of-care identification of COVID-19 to support expedient delivery of care and to assist front-door triage and patient streaming for infection control purposes.10
The increasing use of electronic health-care record (EHR) systems has improved the richness of clinical datasets available to study COVID-19. High-throughput electronic data extraction and processing can enable curation of rich datasets, incorporating all clinical data available on presentation, and might combine with advanced machine learning techniques to produce a rapid screening tool for COVID-19 that fits within existing clinical pathways.11, 12
Approaches to produce a rapid screening tool, with utility during the early phase of hospital presentations, should use only clinical data available before the point of prediction.13 Basic laboratory blood tests and physiological clinical measurements (vital signs) are among the routinely collected health-care data typically available within the first hour of presentation to hospitals in high-income and middle-income countries, and patterns of changes have been described in retrospective observational studies of patients with COVID-19 (variables including lymphocyte count, and alanine aminotransferase, C-reactive protein [CRP], D-dimer, and bilirubin concentrations).3, 4, 14, 15 Moreover, previous health-care data available in the EHR might be useful in identifying risk factors for COVID-19 or underlying conditions that might cause alternative, but similar, presentations.
In this study, we applied artificial intelligence methods to a rich clinical dataset to develop and validate a rapidly deployable screening model for COVID-19. Such a tool would facilitate rapid exclusion of COVID-19 in patients presenting to hospital, optimising patient flow and serving as a pretest where access to confirmatory molecular testing is limited.
Methods
Data collection
Linked deidentified demographic and clinical data for all patients presenting to emergency and acute medical services at Oxford University Hospitals (Oxford, UK) between Dec 1, 2017, and April 19, 2020, were extracted from EHR systems. Oxford University Hospitals consist of four teaching hospitals, serving a population of 600 000 and providing tertiary referral services to the surrounding region.
For each presentation, data extracted included presentation blood tests, blood gas testing, vital signs, results of RT-PCR assays for SARS-CoV-2 (Abbott Architect [Abbott, Maidenhead, UK], and Public Health England-designed RNA-dependent RNA polymerase) from nasopharyngeal swabs, and PCR for influenza and other respiratory viruses. Where available, the following baseline health data were included: the Charlson comorbidity index, calculated from comorbidities recorded during a previous hospital encounter since Dec 1, 2017 (if any existed); and changes in blood test values relative to pre-presentation results. Patients who had opted out of EHR research, did not receive laboratory blood tests, or were younger than 18 years were excluded from analysis. Analyses were confined to clinical, laboratory, and historical data routinely available within the first hour of presentation to hospital.
Adult patients presenting to hospital before Dec 1, 2019, and thus before the global outbreak, were included in the COVID-19-negative cohort. A subset of this cohort was admitted to hospital and included in the COVID-19-negative admissions cohort. Patients presenting to hospital between Dec 1, 2019, and April 19, 2020, with PCR-confirmed SARS-CoV-2 infection were included in the COVID-19-positive cohort, with the subset admitted to hospital included in the COVID-19-positive admissions cohort. Because of incomplete penetrance of testing during early stages of the pandemic and limited sensitivity of the PCR swab test, there is uncertainty in the viral status of patients presenting during the pandemic who were untested or tested negative. Therefore, these patients were excluded from the analysis.
The study protocol, design and data requirements were approved by the National Health Service (NHS) Health Research Authority (IRAS ID 281832) and sponsored by the University of Oxford.
Feature sets
The five sets of clinical variables investigated are shown in table 1 . We considered presentation blood tests and blood gas results from the first blood draw on arrival to hospital. Routine blood tests were determined to include the full blood count, urea and electrolytes, liver function tests, and CRP. We selected these tests because they are widely done within existing care pathways in emergency departments, and results are typically available within 1 h.16 We computed changes in blood tests from previous laboratory samples taken at least 30 days before presentation to hospital (available from Dec 1, 2017, onwards).
Table 1.
Clinical parameters included in each feature set
| Clinical parameters | |
|---|---|
| Feature sets of data routinely acquired on presentation to hospital | |
| Presentation blood tests | Haemoglobin, haematocrit, mean cell volume, white cell count, neutrophil count, lymphocyte count, monocyte count, eosinophil count, basophil count, platelets, prothrombin time, INR, APTT, sodium, potassium, creatinine, urea, eGFR, CRP, albumin, alkaline phosphatase, ALT, bilirubin |
| Presentation point-of-care blood gas results | Actual base excess, standard base excess, bicarbonate, calcium, chloride, estimated osmolality, fraction of carboxyhaemoglobin, glucose, haemoglobin, haematocrit, potassium, methaemoglobin, sodium, oxygen saturation, calculated lactate, calculated oxygen content, calculated p5O, partial pressure of carbon dioxide at point of care, pH, partial pressure of oxygen |
| Presentation vital signs | Heart rate, respiratory rate, oxygen saturation, systolic blood pressure, diastolic blood pressure, temperature, oxygen flow rate |
| Feature sets of previous health data | |
| Change (Δ) in blood test results from baseline | Δ albumin, Δ alkaline phosphatase, Δ ALT, Δ basophil count, Δ bilirubin, Δ creatinine, Δ eosinophil count, Δ haematocrit, Δ haemoglobin, Δ lymphocyte count, Δ mean cell volume, Δ monocyte count, Δ neutrophil count, Δ platelets, Δ potassium, Δ sodium, Δ urea, Δ white cell count, Δ eGFR |
| Baseline comorbidity data | Charlson comorbidity index |
ALT=alanine aminotransferase. APTT=activated partial thromboplastin time. CRP=C-reactive protein. eGFR=estimated glomerular filtration rate. INR=international normalised ratio. p50=pressure at which haemoglobin is 50% bound to oxygen.
We used three imputation strategies—population mean, population median, and age-based imputation—to impute missing data. We report mean and SD across imputation strategies. A full description of the data processing pipeline is available in the appendix (pp 3–5).
Model training, calibration, and testing
Linear (logistic regression) and non-linear ensemble (random forest and XGBoost) classifiers17, 18 were trained to distinguish patients presenting or admitted to hospital with confirmed COVID-19 from pre-pandemic controls. We developed separate models to predict COVID-19 in all patients attending the emergency department (ED model) and then in the subset of those who were subsequently admitted to hospital (admissions model).
Models were trained and tested with use of data from Dec 1, 2017, to April 19, 2020, (table 2 ). We did an 80:20 stratified split to generate a training set and held-out test set. Using the training set, we first trained models with each independent feature set (table 1) to distinguish presentations of COVID-19 from pre-pandemic controls. Next, we started model training using the presentation blood results set and sequentially added additional sets. The area under the receiver operating characteristic curve (AUROC) achieved during training with stratified 10-fold cross-validation is reported alongside SDs. During training, controls were matched for age, gender, and ethnicity. Model thresholds were calibrated to achieve sensitivities of 70%, 80%, and 90% for identifying patients with COVID-19 in the training set before evaluation. The selection of 70%, 80%, and 90% sensitivity thresholds was a pragmatic decision to allow clear presentation of the data.
Table 2.
Population characteristics for the study cohorts and the prospective validation set
|
Study population |
Prospective validation cohorts |
||||||
|---|---|---|---|---|---|---|---|
| Presenting to hospital |
Admitted to hospital |
Presenting to hospital (n=3326) | Admitted to hospital (n=1715) | ||||
| COVID-19 negative (n=114 957) | COVID-19 positive (n=437) | COVID-19 negative (n=71 927) | COVID-19 positive (n=383) | ||||
| Patients positive for COVID-19 | 0 | 437 | 0 | 383 | 107 | 91 | |
| Age, years | 60 (38) | 69 (26) | 65 (33) | 71 (26) | 56 (37) | 64 (34) | |
| Sex | |||||||
| Men | 53 570 (46·6%) | 246 (56·3%) | 34 381 (47·8%) | 211 (55·1%) | 1513 (45·5%) | 832 (48·5%) | |
| Women | 61 387 (53·4%) | 191 (43·7%) | 37 546 (52·2%) | 172 (44·9%) | 1813 (54·5%) | 883 (51·5%) | |
| Previous EHR encounter | 85 183 (74·1%) | 367 (84·0%) | 53 370 (74·2%) | 33 091 (86·4%) | 2671 (80·3%) | 1367 (79·7%) | |
| Ethnicity | |||||||
| White British | 76·0% | 65·4% | 78·5% | 68·4% | 66·3% | 68·2% | |
| Not stated | 11·8% | 17·4% | 11·0% | 16·2% | 19·5% | 20·5% | |
| Any other White background | 5·0% | 3·7% | 4·0% | 3·4% | 6·5% | 4·7% | |
| Pakistani | 1·3% | 1·1% | 1·1% | 1·0% | 1·2% | 1·0% | |
| Any other Asian background | 0·9% | 2·5% | 0·8% | 1·8% | 1·4% | 1·2% | |
| Indian or British Indian | 0·8% | 1·1% | 0·7% | 0·8% | 0·9% | 0·8% | |
| White Irish | 0·7% | 0·7% | 0·7% | 0·8% | 0·7% | 0·8% | |
| African | 0·6% | 3·0% | 0·6% | 2·9% | 0·6% | 0·8% | |
| Any other Black background | 0·3% | 0·9% | 0·3% | 0·5% | 0·5% | 0·3% | |
| Bangladeshi | 0·2% | 0·7% | 0·2% | 0·8% | 0·3% | 0·3% | |
| Chinese | 0·2% | 0·2% | 0·2% | 0·3% | 0·4% | 0·3% | |
| Any other ethnic group | 2·0% | 3·2% | 1·8% | 3·2% | 1·6% | 1·3% | |
| Patients positive for influenza | 484 (<0·1%) | 0 | 466 (<0·1%) | 0 | 0 | 0 | |
Data are n (%) or median (IQR). EHR=electronic health-care record.
We assessed performance of each configuration using the held-out test set. First, we configured the test set with equal numbers of COVID-19 cases and pre-pandemic controls, ensuring that performance was assessed in conditions free of class imbalance, and reported AUROC alongside sensitivity and specificity at each threshold. Second, to simulate model performance at varying stages of the pandemic, we generated a series of test sets with various prevalences of COVID-19 (1–50%) by use of the held-out set. We report positive and negative predictive values for each model at the 70% and 80% sensitivity thresholds. AUROC, sensitivity, specificity, and precision are reported for candidate models at the described thresholds. Positive predictive values (PPVs) and negative predictive values (NPVs) are reported for the simulated test sets. To understand the contribution of individual features to model predictions, we queried importance scores and did SHAP (Shapley additive explanations) analysis.
Validation
Models were validated independently by use of data for all adult patients presenting or admitted to Oxford University Hospitals between April 20 and May 6, 2020, by direct comparison of model prediction against SARS-CoV-2 PCR results. Because of incomplete penetrance of testing and limited sensitivity of the PCR swab test, we did a sensitivity analysis to ensure disease freedom in patients labelled as COVID-19 negative during validation, replacing patients who tested negative by PCR assay or who were not tested with truly negative pre-pandemic patients matched for age, gender, and ethnicity. Accuracy, AUROC, NPV, and PPV were reported during validation. We assessed rates of misclassification by characteristics of age, gender, and ethnicity; comparison between groups was done with Fishers’ exact test. We used the SciPy library for Python, version 1.2.3.
Model development and reporting followed TRIPOD (transparent reporting of a multivariable prediction model for individual prediction or diagnosis) guidelines.13
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the manuscript. All authors had full access to all the data in the study. AASS and SK guarantee the data and analysis. The corresponding author had final responsibility for the decision to submit for publication.
Results
We assessed 155 689 adult patients presenting to hospital between Dec 1, 2017, and April 19, 2020 (appendix p 2). We included 114 957 patients who presented to hospital before Dec 1, 2019, of whom 71 927 were admitted to hospital, in the COVID-19-negative cohorts. 437 patients had a diagnosis of COVID-19 confirmed by RT-PCR between Dec 1, 2019, and April 19, 2020, of whom 383 were admitted to hospital, and were included in the COVID-19-positive cohorts. 40 295 patients who presented to hospital during the pandemic and either were not tested for SARS-CoV-2 by PCR or were tested and had negative results were excluded from analysis due to uncertainty in viral status. Therefore, the full study population comprised 115 394 patients.
Patients presenting to hospital with COVID-19 had a higher median age than pre-pandemic controls (69 years, IQR 26, for the COVID-19-positive cohort vs 60 years, 38, for the COVID-19-negative cohort; Kruskal-Wallis test p<0·0001; table 2). Similarly, patients admitted to hospital with COVID-19 were older than those in the COVID-19-negative admissions cohort (median 71 years, SD 26, for COVID-19-positive admissions vs 65 years, 33, for COVID-19-negative admissions; p<0·0001). Summary statistics of vital signs for the COVID-19-positive cohort showed median oxygen saturation at presentation of 95·3% (IQR 94–98), median systolic blood pressure of 132 mm Hg (115–147), and median diastolic blood pressure of 74 mm Hg (65–84; appendix p 5). 85 183 (74·1%) of 114 957 presenting to hospital before the COVID-19 pandemic had had a previous clinical encounter at the Oxford University Hospitals.
We assessed the relative performance of models trained with each independent feature set at identifying presentations due to COVID-19, reported as AUROC (SD) achieved during training with stratified 10-fold cross-validation (table 3 ). Both ensemble methods outperformed logistic regression, possibly due to their intrinsic ability to detect non-linear effects of the feature sets. XGBoost classifiers trained on laboratory blood tests and vital signs done at presentation showed the highest predictive performance for COVID-19 (table 3). The narrow SDs showed model stability.
Table 3.
AUROCs achieved for each independent feature set and for increasing feature sets using stratified 10-fold cross-validation during training
|
Independent feature sets |
Sets routinely performed on presentation |
Sets integrating previous health data |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Presentation blood tests | Blood gas results | Vital signs | Δ blood tests | Presentation blood tests | Presentation blood tests plus blood gas results | Presentation blood tests plus blood gas results plus vital signs | Sets performed on presentation plus Δ blood tests | Sets performed on presentation plus Δ blood tests plus CCI | |
| Logistic regression | 0·897 (0·003) | 0·730 (0·001) | 0·810 (0·003) | 0·805 (0·008) | 0·897 (0·003) | 0·898 (0·003) | 0·919 (0·002) | 0·920 (0·004) | 0·920 (0·004) |
| Random forest | 0·901 (0·004) | 0·780 (0·000) | 0·815 (0·005) | 0·835 (0·006) | 0·901 (0·004) | 0·907 (0·003) | 0·922 (0·002) | 0·941 (0·004) | 0·937 (0·002) |
| XGBoost | 0·904 (0·000) | 0·770 (0·000) | 0·823 (0·005) | 0·808 (0·050) | 0·904 (0·000) | 0·916 (0·003) | 0·929 (0·003) | 0·942 (0·002) | 0·942 (0·002) |
Data are AUROC (SD). Δ=change in results from baseline. AUROC=area under the receiver operating characteristic curve. CCI=Charlson comorbidity index.
The stepwise addition of routinely collected clinical data improved model performance to a peak AUROC of 0·929 (SD 0·003), achieved with 10-fold cross-validation during training using the XGBoost classifier (table 3). Incorporating previous blood results further improved model performance to an AUROC of 0·942 (0·002); however, having added previous blood results, the addition of the Charlson comorbidity index did not further improve performance.
Our preliminary results suggest that a non-linear approach with clinical data routinely available on presentation achieves high classification performance (table 3). Although incorporating previous health data supports a small increment in performance, missingness could limit generalisability. Therefore, we developed and optimised context-specific models with use of the XGBoost classifier, using only clinical datasets routinely available on presentation, training separate models to predict COVID-19 in patients attending the emergency department (ED model) and in the subset admitted to hospital (admissions model). This approach has the advantage of being applicable to all patients, and is specific to the clinical contexts in which model use is intended. Detailed performance metrics for all feature-set combinations, at each threshold, are available in the appendix (pp 6–7).
Performance of our ED and admissions models was assessed on a held-out test set, generated using a stratified 80:20 train-test split of cases and configured initially with equal numbers of patients with COVID-19 and pre-pandemic controls (ie, 50% prevalence; table 4 ). Our ED and admissions models, calibrated during training to sensitivity of 80%, achieved an AUROC of 0·939 (ED model) and 0·940 (admissions model), sensitivity of 77·4% (for both models), and specificity of 95·7% (ED) and 94·8% (admissions).
Table 4.
Assessment of performance of the ED and admissions models, calibrated to 70%, 80%, and 90% sensitivities during training, in identifying COVID-19 in patients presenting to or admitted to hospital in the held-out test set
|
Calibrated threshold during training |
|||
|---|---|---|---|
| Sensitivity 0·70 | Sensitivity 0·80 | Sensitivity 0·90 | |
| ED model performance on test set | |||
| Sensitivity | 0·697 (0·009) | 0·774 (0·019) | 0·847 (0·014) |
| Specificity | 0·986 (0·005) | 0·957 (0·009) | 0·917 (0·018) |
| Precision (PPV) | 0·979 (0·007) | 0·944 (0·012) | 0·905 (0·018) |
| NPV | 0·777 (0·005) | 0·820 (0·013) | 0·866 (0·011) |
| AUROC | 0·939 (0·003) | 0·939 (0·003) | 0·939 (0·003) |
| Admissions model performance on test set | |||
| Sensitivity | 0·663 (0·029) | 0·774 (0·013) | 0·854 (0·007) |
| Specificity | 0·973 (0·000) | 0·948 (0·005) | 0·891 (0·009) |
| Precision (PPV) | 0·950 (0·002) | 0·922 (0·006) | 0·861 (0·010) |
| NPV | 0·785 (0·014) | 0·841 (0·007) | 0·886 (0·005) |
| AUROC | 0·940 (0·001) | 0·940 (0·001) | 0·940 (0·001) |
Data are performance (SD). The test set was generated from an 80:20 stratified train-test split of the dataset and balanced equally with controls (50% assumed prevalence). AUROC=area under the receiver operating characteristic curve. ED=emergency department. NPV=negative predictive values. PPV=positive predictive values.
Relative feature importance analysis showed that all feature sets contributed to the most-informative variables for model predictions (figure ). In the ED model, three laboratory blood markers (eosinophils, basophils, and CRP) were among the highest-ranking variables. Blood gas measurements (calcium and methaemoglobin) and vital signs (oxygen requirement and respiratory rate) were additionally among the variables most informative to model predictions. Similar top-ranking features are seen in the admissions model, but with greater relative weights for CRP and white cell counts and lesser weights for blood gas measurements. Results of SHAP analysis confirmed that CRP, eosinophil counts, and basophil counts had the greatest effect on predictions of both models (appendix p 7).
Figure.
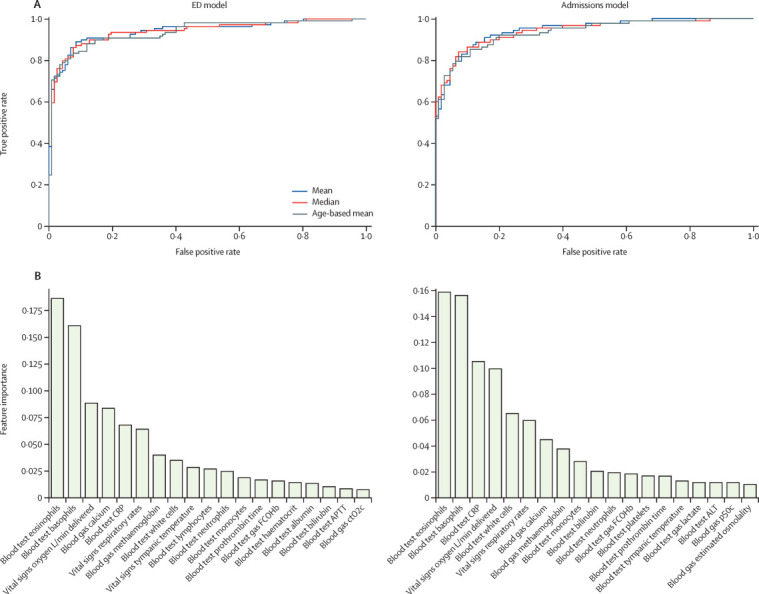
Receiver operating characteristic curves (A) and relative importance of features (B) for the ED and admissions models
ALT=alanine aminotransferase. APTT=activated partial thromboplastin time. CRP=C-reactive protein. ctO2c=calculated oxygen content. ED=emergency department. FCOHb=fraction of carboxyhaemoglobin. p50c=calculated pressure at which haemoglobin is 50% bound to oxygen.
To reflect performance at varying stages of an epidemic, we assessed positive and negative predictive values on test sets configured to various prevalences of COVID-19, with results calibrated to two sensitivity thresholds (70% and 80%; table 5 ). For both ED and admissions models, the higher sensitivity configuration (80%) achieved high NPV (>98·5%) where COVID-19 is uncommon (≤5% prevalence), supporting safe exclusion of the disease. At high disease prevalences (≥20%), the 70% sensitivity configuration optimises for high PPV (>83%) at good NPV (>92%; table 5). The 70% sensitivity configurations achieved high PPV (76·3% in the ED model and 83·0% in the admissions model) and NPV (95·3% in the ED model and 92·6% in the admissions model) at the prevalence of COVID-19 observed in patients presenting and admitted to hospital at the study hospitals during April 1–8, 2020 (table 5).
Table 5.
PPV and NPV of the ED and admissions models, calibrated during training to 70% and 80% sensitivities, for identifying COVID-19 in test sets with various prevalences
|
Prevalence of COVID-19 in test set |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1% | 2% | 5% | 10%* | 20%† | 25% | 33% | 50% | ||
| ED model | |||||||||
| Sensitivity 0·70 | |||||||||
| PPV | 0·203 | 0·383 | 0·613 | 0·763 | 0·834 | 0·902 | 0·888 | 0·979 | |
| NPV | 0·996 | 0·990 | 0·985 | 0·953 | 0·932 | 0·871 | 0·886 | 0·778 | |
| Sensitivity 0·80 | |||||||||
| PPV | 0·133 | 0·282 | 0·493 | 0·638 | 0·767 | 0·831 | 0·823 | 0·944 | |
| NPV | 0·997 | 0·993 | 0·991 | 0·962 | 0·946 | 0·909 | 0·908 | 0·820 | |
| Admissions model | |||||||||
| Sensitivity 0·70 | |||||||||
| PPV | 0·175 | 0·304 | 0·513 | 0·595 | 0·830 | 0·859 | 0·876 | 0·950 | |
| NPV | 0·996 | 0·992 | 0·982 | 0·969 | 0·926 | 0·905 | 0·881 | 0·785 | |
| Sensitivity 0·80 | |||||||||
| PPV | 0·098 | 0·211 | 0·390 | 0·509 | 0·755 | 0·797 | 0·812 | 0·922 | |
| NPV | 0·998 | 0·994 | 0·986 | 0·977 | 0·942 | 0·920 | 0·907 | 0·841 | |
ED=emergency department. NPV=negative predictive values. PPV=positive predictive values.
The 10% scenario approximates the observed prevalence of COVID-19 in patients presenting to the study hospitals during April 1–8, 2020.
The 20% scenario approximates the observed prevalence of COVID-19 in patients admitted to the study hospitals during April 1–8, 2020.
We prospectively validated our ED and admissions models, calibrated during training to 80% sensitivity, for all patients presenting or admitted to Oxford University Hospitals between April 20 and May 6, 2020. 3326 patients presented to hospital and 1715 were admitted during the validation period. Prevalences of COVID-19 were 3·2% (107 of 3326) in patients presenting to hospital and 5·3% (91 of 1715) in those admitted to hospital. Our ED model performed with 92·3% accuracy (AUROC 0·881) and the admission model performed with 92·5% accuracy (0·871) on the validation set, assessed against results of laboratory PCR testing. PPVs were 46·7% (ED model) and 40·0% (admissions model) and NPVs were 97·6% (ED) and 97·7% (admissions).
We did a sensitivity analysis to account for uncertainty in the viral status of patients testing negative by PCR or who were not tested. Our ED model showed an apparent improvement in accuracy to 95·1% (AUROC 0·960) and our admission model improved to 94·1% accuracy (0·937) on the adjusted validation set. NPVs achieved were also improved to 99·0% (ED model) and 98·5% (admissions model).
To assess model performance on clinically important subgroups, we assessed performance of our admissions model on patients presenting during prospective validation who went on to require admission to the intensive care unit (ICU) or who died. The model performed highest on the subpopulation admitted to ICU (AUROC 0·930, accuracy 93·5%, NPV 98·3%, PPV 37·8%) and also achieved high performance for patients who died during admission (0·916, accuracy 93·0%, NPV 98·3%, PPV 37·6%). Additionally, we investigated model performance for the subset of patients presenting with respiratory symptoms, showing high performance for this key group (0·895, accuracy 92·8%, NPV 98·0%, PPV 35·6%).
To evaluate for biases in model performance, we assessed rates of patient misclassification during validation of our ED and admissions models. We observed that rates of misclassification were similar between White British (ED model 9%, admissions model 10%) and Black, Asian, and minority ethnic group patients (ED 11%, admissions 13%; Fishers’ exact test p=0·37 for ED and p=0·36 for admissions), and between men (11% for both models) and women (8% for both models; p=0·15 for ED and p=0·091 for admissions). We also found no difference between misclassification of patients older than 60 years (10% for both models) and patients aged 18–60 years (9% ED model and 8% admissions model; p=0·19 for ED and admissions).
Discussion
The limitations of the gold-standard PCR test for COVID-19 have challenged health-care systems across the world. Because COVID-19 can be difficult to distinguish clinically from other illnesses, there remains an urgent need for rapid and accurate screening of patients on arrival to hospitals, with the available diagnostic test limited by long turnaround times,19 shortages of specialist equipment and operators, and relatively low sensitivity.8 NHS guidelines require testing of all emergency admissions,20 irrespective of clinical suspicion, highlighting the demand for rapid and accurate exclusion of COVID-19 in the acute care setting.6
In this study, we developed and assessed two context-specific artificial intelligence-driven screening tools for COVID-19. Our ED and admissions models effectively identified patients with COVID-19 among all patients presenting and admitted to hospital, using data typically available within the first hour of presentation. Simulation on test sets with varying prevalences of COVID-19 showed that our models achieved clinically useful NPVs (>98·5%) at low prevalences (≤5%). On validation, using prospective cohorts of all patients presenting or admitted to the Oxford University Hospitals, our models achieved high accuracies and NPVs compared with PCR test results. A sensitivity analysis to account for uncertainty in negative PCR results improved apparent accuracy and NPVs.
The high negative predictive performance of our models supports their use as a screening test to rapidly exclude a diagnosis of COVID-19 in emergency departments, assisting immediate care decisions, guiding safe patient streaming, and serving as a pretest for diagnostic molecular testing where availability is limited. In subgroup analyses during model validation, high accuracy and NPV were maintained for the subset of patients presenting with respiratory symptoms, showing superiority over a simple symptom-based triage strategy, and were also maintained for critically ill patients who required ICU admission or died. Key beneficiary populations include a majority of viral-free patients correctly predicted to be COVID-19 negative. In our clinically minded, safety-first approach, COVID-19 is ruled-in for an enriched subpopulation at higher risk of testing positive, for whom waiting for definitive testing is advisable. This screening paradigm is widely established in clinical practice after popularisation of the D-dimer test for suspected deep-vein thrombosis and pulmonary embolism.21
The strengths of our artificial intelligence approach include an ability to scale rapidly, taking advantage of cloud computing platforms and working with laboratory tests widely available and routinely done within the current standard of care. Moreover, we showed that our models can be calibrated to meet changing clinical requirements at different stages of the pandemic, such as a high PPV model.
To date, early-detection models have overwhelmingly focused on assessment of radiological imaging, such as CT,5, 19, 22, 23 which is less readily available and involves patient exposure to ionising radiation. Few studies have assessed routine laboratory tests, with studies to date including small numbers of patients with confirmed COVID-19, using PCR results for data labelling and thereby not ensuring disease freedom in so-called negative patients and not being validated in the clinical population that is the target for their intended use.11, 24, 25 A substantial limitation of existing works is the use of narrow control cohorts during training, inadequately exposing models to the breadth and variety of alternative infectious and non-infectious pathologies, including seasonal pathologies. Moreover, although the use of artificial intelligence techniques for early detection holds great promise, many published models to date have been assessed to be at high risk of bias.22
Our study includes the largest dataset of any laboratory artificial intelligence study on COVID-19 to date, considering over 115 000 hospital attendances and 5 million measurements, and it is prospectively validated with use of appropriate patient cohorts for the models’ intended clinical contexts. The breadth of our pre-pandemic control cohort gives exposure to a wide range of undifferentiated presentations, including other seasonal infectious pathologies (eg, influenza), and offers confidence in SARS-CoV-2 freedom. Additionally, to our knowledge, our study is the first to integrate laboratory blood results with blood gas and vital signs measurements taken at presentation to hospital, maximising the richness of the dataset available.
Although our results showed that integrating previous health data incrementally improved model performance, we did not include previous health data in our final models. As this data was missing for 29 844 (25·9%) of 115 394 patients (table 2), the cost of generalisability would outweigh the benefit of a marginal performance improvement.
We selected established linear and non-linear modelling approaches, achieving highest performance with XGBoost, an extreme gradient boosted tree method. Information variables from all sets were important in model predictions, including three measured biochemical quantities (eosinophils, basophils, and CRP), blood gas measurements (methaemoglobin and calcium), and vital signs (respiratory rate and oxygen delivery).
Existing literature has reported an association between lymphopenia and COVID-19.3, 15 We observed that lymphopenia was frequently absent on first-available laboratory tests done on admission (appendix pp 3–4) and was not a highly ranked feature in our models (figure 1). Univariate analysis identified that eosinopenia on presentation was more strongly correlated with COVID-19 diagnosis than lymphocyte count (appendix p 6; χ2 score 41·61 for eosinopenia and 31·56 for lymphocyte count).
Recognising concerns of biases within artificial intelligence models, we assessed cases misclassified during validation for evidence of ethnicity, age, and gender biases. Our results showed misclassification was not significantly different between White British and Black, Asian, and minority ethnic patients; men and women; and older (>60 years) and younger (18–59 years) patients.
Our study seeks to address limitations common to EHR research. We used multiple imputations for missing data, taking a mean of three strategies (age-based imputation, population mean, and population median). We queried whether our results were sensitive to the imputation strategy and found similar model performance across the three strategies.
A potential limitation of this study is the relatively limited ethnic diversity of patients included. 87 653 (76·0%) of 115 394 study patients reported their ethnicity to be White British (table 2). Although our models do not appear to be more likely to misclassify patients of an ethnic minority, integrating data from international centres where patients might attend hospital with different spectrums of complaints would increase confidence in model generalisability abroad. We excluded patients younger than 18 years from the analysis, noting that COVID-19 is a rare cause of hospital presentation in the paediatric population; however, this limits model applicability to adults.26 Additionally, as the first wave of COVID-19 cases in the UK largely followed the conclusion of the 2019–20 influenza season, data for patients who were co-infected were not available for this study.27, 28 Future work might examine a role for rapid screening in the paediatric population to reduce nosocomial transmission and assess model applicability in co-infection.
Our work shows that an artificial intelligence-driven screening test can effectively triage patients presenting to hospital for COVID-19 while confirmatory laboratory testing is pending. Our approach is rapidly scalable, fitting within the existing laboratory testing infrastructure and standard of care, and serves as proof of concept for a rapidly deployable software tool in future pandemics. Prospective clinical trials would further assess model generalisability and real-world performance.
Data sharing
The data studied are available from the Infections in Oxfordshire Research Database, subject to an application meeting the ethical and governance requirements of the Database (contact email iord@ndm.ox.ac.uk). Code and supplementary information for this paper are available online, alongside publication.
Acknowledgments
Acknowledgments
We express our sincere thanks to all patients, clinicians, and staff across Oxford University Hospitals NHS Foundation Trust. We additionally thank staff across the University of Oxford Institute of Biomedical Engineering, Research Services, and Clinical Trials and Research Group. In particular, we thank Dr Ravi Pattanshetty for clinical input and Jia Wei. This research was supported by grants from the Wellcome Trust (University of Oxford Medical and Life Sciences Translational Fund, award 0009350), the Engineering and Physical Sciences Research Council (EP/P009824/1 and EP/N020774/1), and the NIHR Oxford Biomedical Research Centre and NIHR Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance at the University of Oxford (NIHR200915), in partnership with Public Health England (PHE). AASS is an NIHR Academic Clinical Fellow. DWE is a Robertson Foundation Fellow and an NIHR Oxford Biomedical Research Centre Senior Fellow. The views expressed are those of the authors and not necessarily those of the NHS, NIHR, PHE, Wellcome Trust, or the Department of Health.
Contributors
AASS, DAC, TZ, DWE, ZBH, and TP conceived of and designed the study. DWE extracted the data from EHR systems. TT, SK, and AASS pre-processed the data. DK, SK, AASS, TZ, TT, DWE, and DAC developed the models. AASS, SK, DK, TZ, AJB, DWE, and DAC validated the models. AASS, SK, and ZBH wrote the manuscript. DWE, TT, AASS, and SK had access to and verified the data. All authors revised the manuscript.
Declaration of interests
DWE reports personal fees from Gilead, outside the submitted work. DAC reports personal fees from Oxford University Innovation, BioBeats, and Sensyne Health, outside the submitted work. All other authors declare no competing interests.
Supplementary Material
References
- 1.WHO Rolling updates on coronavirus disease (COVID-19) 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen
- 2.Adhikari SP, Meng S, Wu Y. Novel coronavirus during the early outbreak period: epidemiology, causes, clinical manifestation and diagnosis, prevention and control. Infect Dis Poverty. 2020;9:1–12. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan WJ, Ni ZY, Hu Y. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Hu B, Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long C, Xu H, Shen Q. Diagnosis of the coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol. 2020;126 doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.UK National Health Service . National Health Service; London: 2020. Guidance and standard operating procedure: COVID-19 virus testing in NHS laboratories. [Google Scholar]
- 7.Long DR, Gombar S, Hogan CA. Occurrence and timing of subsequent SARS-CoV-2 RT-PCR positivity among initially negative patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa722. published online June 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Y, Schmitz JE, Persing DH, Stratton CW. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J Clin Microbiol. 2020;58:1–9. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen I, Phillips A. Three quarters of people with SARS-CoV-2 infection are asymptomatic: analysis of English household survey data. Clin Epidemiol. 2020;12:1039–1043. doi: 10.2147/CLEP.S276825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udugama B, Kadhiresan P, Kozlowski HN. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 11.Brinati D, Campagner A, Ferrari D, Locatelli M, Banfi G, Cabitza F. Detection of COVID-19 infection from routine blood exams with machine learning: a feasibility study. J Med Syst. 2020;44:135. doi: 10.1007/s10916-020-01597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menni C, Valdes AM, Freidin MB. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med. 2020;26:1037–1040. doi: 10.1038/s41591-020-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med. 2015;162:55–63. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 14.Docherty AB, Harrison EM, Green CA. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kermali M, Khalsa RK, Pillai K, Ismail Z, Harky A. The role of biomarkers in diagnosis of COVID-19—a systematic review. Life Sci. 2020;254 doi: 10.1016/j.lfs.2020.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kristensen M, Iversen AKS, Gerds TA. Routine blood tests are associated with short term mortality and can improve emergency department triage: a cohort study of >12,000 patients. Scand J Trauma Resusc Emerg Med. 2017;25:115. doi: 10.1186/s13049-017-0458-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T, Guestrin C. XGBoost: a scalable tree boosting system. KDD 19: The 25th ACM SIGKDD Conference on Knowledge Discovery and Data Mining; Anchorage, AK, USA; Aug 13–17, 2019.
- 18.Varoquaux G, Buitinck L, Louppe G, Grisel O, Pedregosa F, Mueller A. Scikit-learn. GetMobile Mob Comput Commun. 2015;19:29–33. [Google Scholar]
- 19.Mei X, Lee HC, Diao KY. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020;26:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.NHS England and NHS Improvement Healthcare associated COVID-19 infections—further action. 2020. https://www.england.nhs.uk/coronavirus/wp-content/uploads/sites/52/2020/06/Healthcare-associated-COVID-19-infections–further-action-24-June-2020.pdf
- 21.Wells PS, Anderson DR, Rodger M. Evaluation of D-dimer in the diagnosis of suspected deep-vein thrombosis. N Engl J Med. 2003;349:1227–1235. doi: 10.1056/NEJMoa023153. [DOI] [PubMed] [Google Scholar]
- 22.Wynants L, Van Calster B, Collins GS. Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. BMJ. 2020;369 doi: 10.1136/bmj.m1328. (accessed June 1, 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Zha Y, Li W. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur Respir J. 2020;56 doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng C, Huang Z, Wang L. A Novel triage tool of artificial intelligence assisted diagnosis aid system for suspected COVID-19 pneumonia in fever clinics. SSRN. 2020 doi: 10.21037/atm-20-3073. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3551355 published online March 19. (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Koh V, Marimuthu K. Epidemiological and clinical predictors of COVID-19. Clin Infect Dis. 2020;71:786–792. doi: 10.1093/cid/ciaa322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim L, Whitaker M, O'Halloran A. Hospitalization rates and characteristics of children aged <18 years hospitalized with laboratory-confirmed COVID-19—COVID-NET, 14 States, March 1-July 25, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1081–1088. doi: 10.15585/mmwr.mm6932e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.UK Government Coronavirus (COVID-19) in the UK: dashboard. 2020. https://coronavirus.data.gov.uk/
- 28.UK Government Weekly national flu reports: 2019 to 2020 season. 2020. https://www.gov.uk/government/statistics/weekly-national-flu-reports-2019-to-2020-season
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data studied are available from the Infections in Oxfordshire Research Database, subject to an application meeting the ethical and governance requirements of the Database (contact email iord@ndm.ox.ac.uk). Code and supplementary information for this paper are available online, alongside publication.


