Abstract
Background
Bacterial co-pathogens are commonly identified in viral respiratory infections and are important causes of morbidity and mortality. The prevalence of bacterial infection in patients infected with SARS-CoV-2 is not well understood.
Aims
To determine the prevalence of bacterial co-infection (at presentation) and secondary infection (after presentation) in patients with COVID-19.
Sources
We performed a systematic search of MEDLINE, OVID Epub and EMBASE databases for English language literature from 2019 to April 16, 2020. Studies were included if they (a) evaluated patients with confirmed COVID-19 and (b) reported the prevalence of acute bacterial infection.
Content
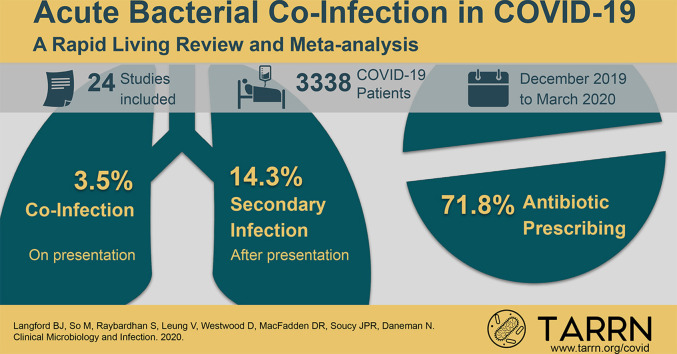
Data were extracted by a single reviewer and cross-checked by a second reviewer. The main outcome was the proportion of COVID-19 patients with an acute bacterial infection. Any bacteria detected from non-respiratory-tract or non-bloodstream sources were excluded. Of 1308 studies screened, 24 were eligible and included in the rapid review representing 3338 patients with COVID-19 evaluated for acute bacterial infection. In the meta-analysis, bacterial co-infection (estimated on presentation) was identified in 3.5% of patients (95%CI 0.4–6.7%) and secondary bacterial infection in 14.3% of patients (95%CI 9.6–18.9%). The overall proportion of COVID-19 patients with bacterial infection was 6.9% (95%CI 4.3–9.5%). Bacterial infection was more common in critically ill patients (8.1%, 95%CI 2.3–13.8%). The majority of patients with COVID-19 received antibiotics (71.9%, 95%CI 56.1 to 87.7%).
Implications
Bacterial co-infection is relatively infrequent in hospitalized patients with COVID-19. The majority of these patients may not require empirical antibacterial treatment.
Keywords: Bacterial infections, Co-infection, COVID-19, Living review, Secondary infection
Graphical abstract
Introduction
Bacterial co-pathogens are commonly identified in viral respiratory tract infections such as influenza and are an important cause of morbidity and mortality, necessitating timely diagnosis and antibacterial therapy [[1], [2], [3]]. Although highly variable, bacterial co-infection in patients with severe influenza has been reported to be as high as 20–30% [3,4] and is associated with a greater severity of illness, greater use of healthcare resources, and increased risk of death [5]. The prevalence, incidence and characteristics of bacterial infection in patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is not well understood and has been raised as an important knowledge gap [6,7].
While antibiotics are ineffective for treatment of COVID-19, they are prescribed in patients with suspected or documented COVID-19 for a variety of reasons. This includes difficulty in ruling out bacterial co-infection on presentation, but also the possibility of bacterial secondary infection during the course of the illness. Extrapolating from concerns of an increase in mortality in patients with bacterial superinfection during influenza pandemics, several guidelines advocate the use of empirical antibiotics for patients with severe COVID-19 [8,9]. However, this assumption raises concerns of antibiotic overuse and subsequent harm associated with bacterial resistance.
Understanding the proportion of COVID-19 patients with acute respiratory bacterial co-infection, and the culprit pathogens, is crucial for treating patients with COVID-19 and to help ensure responsible use of antibiotics and to minimize negative consequences of overuse [6]. In addition, this knowledge could have a significant impact in refining empirical antibiotic management guidelines for patients with COVID-19.
We performed a rapid review to determine the prevalence of bacterial infection in patients with COVID-19 and to identify the most common co-infecting respiratory organisms in these individuals.
Methods
We conducted a rapid systematic review guided by recommendations from the Cochrane Rapid Reviews Methods Group [10] to determine the prevalence of respiratory bacterial infections among individuals with confirmed active COVID-19 infection. A rapid review was selected as the ideal methodology to synthesize knowledge in a timely fashion for this emergent issue, so that learnings could impact clinical care during the ongoing pandemic.
We included studies of humans with laboratory-confirmed SARS-CoV-2 infection across all healthcare settings (i.e. hospital, community, long-term care) and age groups (paediatric and adult patients). Bacterial infection was defined as an acute infection including either (a) co-infection on presentation, or (b) secondary infection emerging during the course of illness or hospital stay. Cases where bacteria were detected in an isolate not from a respiratory tract or blood culture sample were excluded.
We included randomized controlled trials, cohort studies and case series with more than 10 patients, but excluded reviews, editorials, letters and case studies. We considered studies to be eligible regardless of experimental or observational design, and irrespective of their primary objective. However, we excluded studies that did not report data on bacterial infection or exclusively reported data on chronic co-infection or non-bacterial pathogens. This protocol was registered under PROSPERO, the international registry of systematic reviews (ID CRD42020180229).
Our aim is to ensure that this is a living review [11] which we will update on our website (https://www.tarrn.org/covid) every 3 months as new information becomes available throughout the course of the COVID-19 pandemic.
Data sources
We performed a systematic search of MEDLINE, OVID Epub and EMBASE databases for published literature in the English language from January 1, 2019 to April 16, 2020 with assistance from a medical library information specialist. The search was structured to include COVID-19 terms and co-infection or bacterial infection or respiratory infection or epidemiology or descriptive cohort study terms. The complete search strategy is described in the Appendix. The results of the search were imported into Covidence (Covidence, Melbourne, Australia), an online software tool for systematic reviews. Duplicate records were removed using Covidence.
Study selection
All titles and abstracts were independently screened by two authors (BL, VL) to determine whether studies met the inclusion criteria; all full-text studies meeting these criteria were then reviewed by two authors (BL and MS or SR) for final inclusion in the rapid review. Disagreements that could not be resolved via consensus were reviewed independently by an additional author. The reference section of articles selected for inclusion were hand-searched by an author (BL) to identify any additional studies for potential inclusion.
Data extraction
Two authors performed data extraction from included studies using a spreadsheet, and cross-checked for accuracy and completeness (DW, BL). The following variables were extracted: country of study, start and end date of study, healthcare site name(s), study design (retrospective versus prospective), healthcare setting, sample size, age group, patient population, mean or median age, proportion of female patients, disease severity, proportion of patients requiring mechanical ventilation, proportion of patients that were smokers, reported comorbidities (i.e., chronic obstructive pulmonary disease, cardiovascular disease, hypertension), bacteriological testing methods, patients with acute respiratory co-infection, proportion of patients with secondary bacterial infection, respiratory organisms identified and their proportions, bacterial susceptibility, viral testing methods, proportion of patients receiving antibiotics and class of antibiotic. Studies potentially describing overlapping data were noted (e.g. same hospital and population during an overlapping time period).
Data synthesis
The main outcome of interest was the overall proportion of confirmed acute bacterial infections in patients with COVID-19 stratified by co-infection on initial presentation and secondary infection during the course of the illness. We also stratified bacterial infection rates by patient population as an estimate of the severity of COVID-19 illness, as: (a) all hospitalized patients or (b) critically ill patients (admitted to intensive care unit). Studies were categorized as reporting co-infection (bacterial infection on presentation) unless they explicitly stated that they were capturing secondary infection data by using the term ‘secondary infection’ or indicating that infection data were captured after admission. To avoid duplication of patient data, each study was captured only once, reporting either co-infection or secondary infection data, whichever component was larger. We pooled proportion data across studies using a random effects meta-analysis with the DerSimonian–Laird method [12]. Results were illustrated using forest plots. Heterogeneity was assessed by I2 statistic, with <40% considered low heterogeneity, 30–60% considered moderate heterogeneity, 50–90% considered substantial heterogeneity, and 75–100% considered considerable heterogeneity [13]. All analyses were carried out using R version 3.6.0 with the packages metafor and meta.
Assessment of bias
A formal assessment for risk of bias was deemed to have limited utility given the lack of an appropriate assessment tool. Although a risk-of-bias tool has been developed for meta-analyses of disease prevalence [14], many aspects of the tool are not directly relevant to our research question. Therefore, we incorporated study quality into our sensitivity analysis based on three key factors: whether bacterial diagnostic method was reported, whether co-infection was explicitly subcategorized (i.e. co-infection versus secondary infection), and with removal of studies with potentially overlapping patient cohorts.
Results
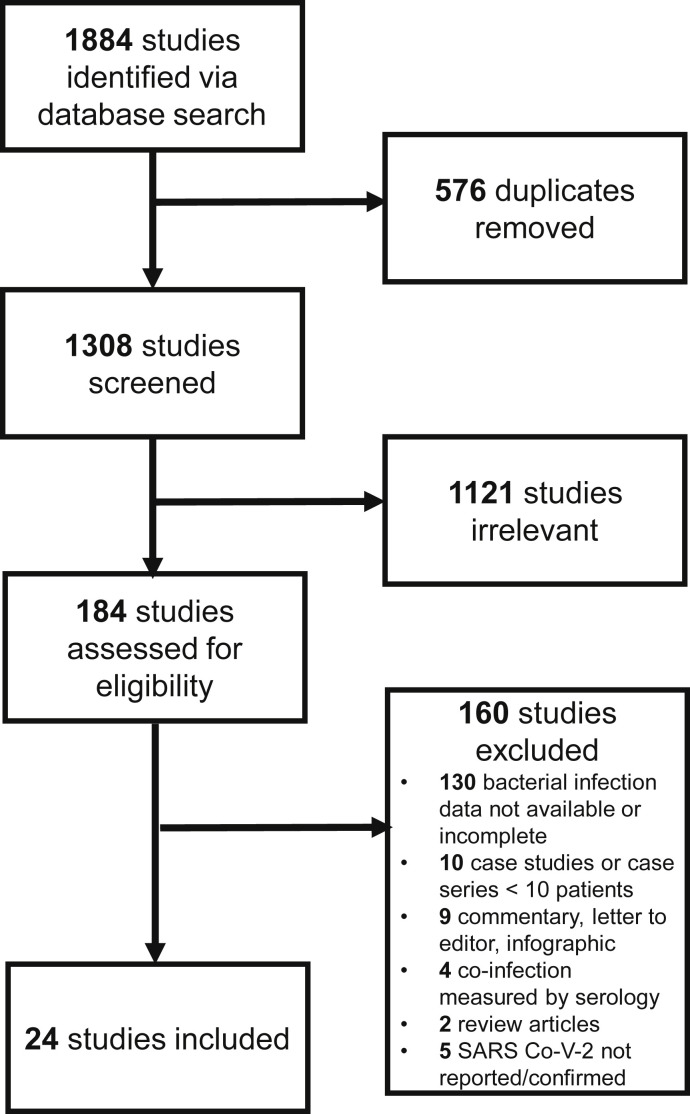
We screened a total of 1308 publications and included 24 studies [[15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38]] in the final analysis. The main reason for study exclusion was the lack of reporting of bacterial co-infection or bacterial secondary infection data (Fig. 1 ). Among the 24 studies, 3506 patients with laboratory-confirmed COVID-19 were evaluated. All studies (n = 24) employed retrospective cohort designs and most took place in Asia (n = 21). Studies were carried out between December 25, 2019 and March 31, 2020.
Fig. 1.
Study flow diagram.
The median or mean patient age ranged from 2 to 71 years across studies; however, the majority (18/24, 75%) included only adult patients. The median proportion of female patients was 45.8% (interquartile range (IQR) 37.6–50.0%). Among studies reporting specific patient characteristics, a median of 6.4% (IQR 3.9–9.7%, n = 11 studies) were smokers, 3.2% (IQR 0.4–5.5%, n = 18 studies) had chronic obstructive pulmonary disease, 9.7% (IQR 4.9–19.2%, n = 18 studies) had cardiovascular disease, and 11.5% (IQR 7.7–18.3%, n = 20 studies) had diabetes.
Most studies focused on all hospitalized patients (n = 19), but some were limited to critically ill patients (n = 5). Seven studies exclusively reported secondary bacterial infections, the remaining 17 studies were categorized as reporting bacterial co-infection. The main characteristics of the included studies are shown in Table 1 .
Table 1.
Study, patient, and infection characteristics
| Author, year | Country | Setting | Sample size | Age | Female (n, %) |
Bacteria diagnostic method | Infection type | Bacterial infection (n, %) |
Antibiotic use (%) |
|---|---|---|---|---|---|---|---|---|---|
| Arentz M, 2020 | USA | ICU (critically Ill) | 21 | 70 (mean) | 10 (47.6) | Unspecified | Co-infection | 1 (4.8) | Unspecified |
| Barrasa H, 2020 | Spain | ICU (critically ill) | 48 | 63 (median) | 21 (43.8) | Unspecified | Co-infection | 6 (12.5) | 87.5 |
| Bhatraju P, 2020 | USA | ICU (critically ill) | 15 | 64 (mean) | 9 (37.5) | Respiratory, blood culture | Co-infection | 0 (0.0) | Unspecified |
| Cai Q, 2020 | China | Hospital | 298 | 48 (median) | 153 (51.3) | Unspecified | Secondary | 30 (10.1) | 12.4 |
| Chen N, 2020 | China | Hospital | 99 | 56 (mean) | 32 (32.3) | Respiratory culture | Co-infection | 1 (1.0) | 70.7 |
| Chen T, 2020 | China | Hospital | 203 | 54 (median) | 95 (46.8) | Unspecified | Co-infection | 2 (1.0) | Unspecified |
| Feng Y, 2020 | China | Hospital | 410 | 53 (median) | 205 (43.1) | Respiratory culture | Secondary | 35 (8.5) | 67.0 |
| Lian J, 2020 | China | Hospital | 788 | 46 (mean) | 381 (48.4) | Unspecified | Secondary | 0 (0.0) | Unspecified |
| Ling L, 2020 | China | ICU (critically ill) | 8 | 64.5 (mean) | 4 (50.0) | Unspecified culture | Co-infection Secondary |
0 (0.0) 2 (25.0) |
100.0 |
| Liu W, 2020 | China | Hospital | 78 | 38 (median) | 39 (50.0) | Respiratory NAAT | Co-infection | 0 (0.0) | Unspecified |
| Liu Y, 2020 | China | Hospital | 12 | 54 (mean) | 4 (33.3) | Unspecified | Co-infection | 2 (16.7) | Unspecified |
| Mo P, 2020 | China | Hospital | 155 | 54 (median) | 69 (44.5) | Unspecified | Co-infection | 2 (1.3) | Unspecified |
| Pongpirul W, 2020 | Thailand | Hospital | 11 | 56 (mean) | 5 (45.5) | Respiratory NAAT | Co-infection | 5 (45.4) | 55.5 |
| Tan Y, 2020 | China | Hospital (children) | 10 | 7 (mean) | 7 (70.0) | Respiratory culture | Co-infection | 0 (0.0) | 10.0 |
| Wang L, 2020 | China | Hospital (adults >60) | 339 | 71 (mean) | 173 (51.0) | Unspecified | Secondary | 143 (42.2) | Unspecified |
| Wang Z, 2020 | China | Hospital | 29 | 42 (median) | 37 (53.6) | Respiratory culture | Co-infection | 3 (10.3) | 98.6 |
| Wu C, 2020 | China | Hospital | 148 | 51 (median) | 73 (36.3) | Respiratory culture | Co-infection | 0 (0.0) | 97.5 |
| Wu J, 2020 | China | Hospital | 280 | 43 (median) | 129 (46.1) | Respiratory culture | Co-infection | 6 (2.1) | 67.1 |
| Wu J, 2020 | China | Hospital | 80 | 46 (mean) | 41 (51.3) | Respiratory culture | Co-infection | 0 (0.0) | 91.3 |
| Xia W, 2020 | China | Hospital (children) | 20 | 2 (median) | 7 (35.0) | Unspecified | Co-infection | 4 (20.0) | Unspecified |
| Yang X, 2020 | China | ICU (critically Ill) | 52 | 60 (mean) | 17 (32.7) | Respiratory, blood culture | Secondary | 7 (13.5) | 94.2 |
| Young B, 2020 | Singapore | Hospital | 18 | 47 (median) | 9 (50.0) | Unspecified | Co-infection | 0 (0) | Unspecified |
| Zheng F, 2020 | China | Hospital (children) | 25 | 3 (median) | 11 (44.0) | Unspecified | Co-infection | 4 (16.0) | 52.0 |
| Zhou F, 2020 | China | Hospital | 191 | 56 (median) | 72 (37.7) | Respiratory, blood culture | Secondary | 28 (14.7) | 94.8 |
NAAT, nucleic acid amplification.
Meta-analysis of bacterial infection in patients with COVID-19
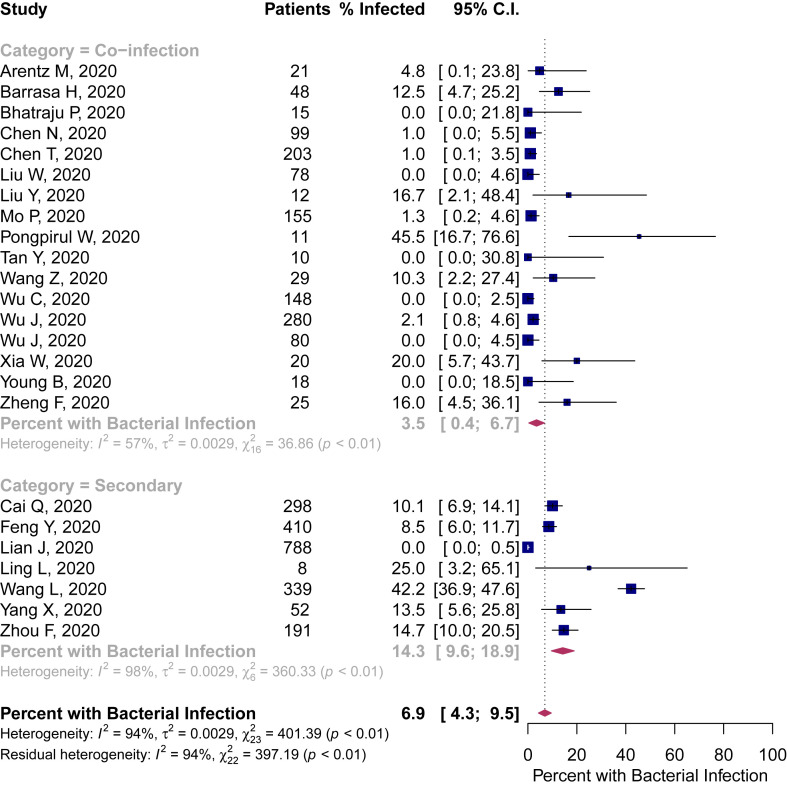
Bacteriological testing methods were reported as respiratory culture with or without blood culture in ten studies, respiratory nucleic acid amplification in two studies, and not specified in 12 studies. Of 3506 patients, 3338 were evaluated for bacterial infection, of which 281 had a reported bacterial infection. In the random effects meta-analysis bacterial co-infection was identified in 3.5% of patients (95%CI 0.4–6.7%) and bacterial secondary infection was identified in 14.3% of patients (95%CI 9.6–18.9%). When pooling all included studies, the proportion of COVID-19 patients with bacterial infection was 6.9% (95%CI 4.3–9.5%) (Fig. 2 ).
Fig. 2.
Percentage of patients with COVID-19 and bacterial co-infection or secondary infection.
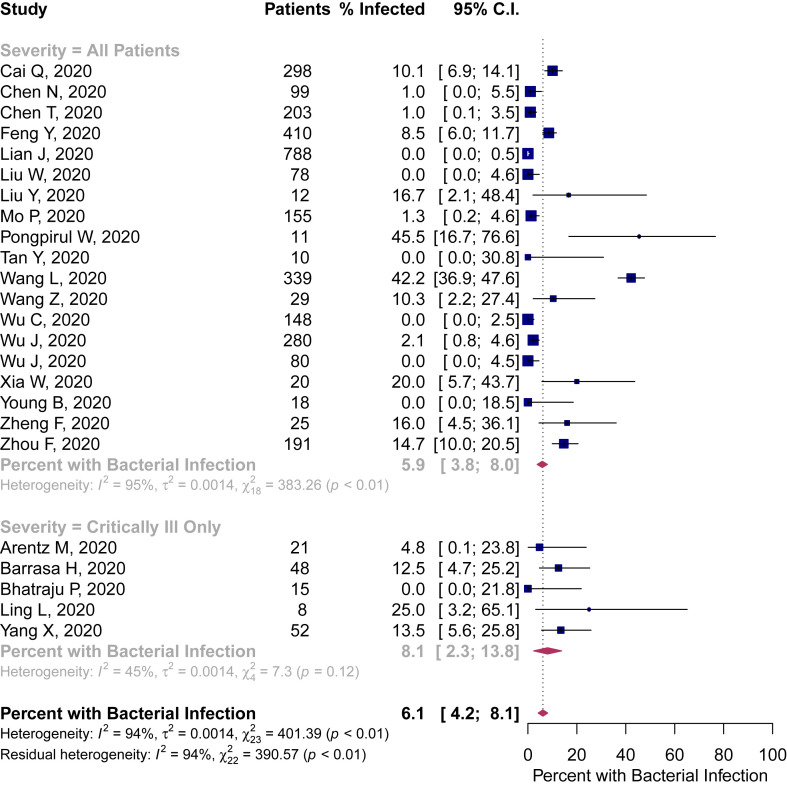
When stratified by patient population (an estimate of COVID-19 illness severity), bacterial infection ranged from 5.9% (95%CI 3.8–8.0%) in all hospitalized patients to 8.1% in critically ill patients (95%CI 2.3–13.8) (Fig. 3 ).
Fig. 3.
Percentage of patients with COVID-19 and bacterial infection stratified by estimated severity of illness.
There was considerable heterogeneity across all included studies (I2 = 94%), with the lowest, albeit moderate, heterogeneity in the critically ill subgroup (I2 = 45%).
Sensitivity analyses
The proportion of COVID-19 patients with bacterial infection differed based on the reported method of testing: 4.6% (95%CI 0.9–8.2) for culture, 4.4% (95%CI 0.0–14.0) for nucleic acid amplification and 9.6% (95%CI 5.9–13.3) when the method was not specified (Appendix Fig. A).
Studies reporting concomitant bacterial pathogens in COVID-19 were assumed to represent co-infection rather than secondary infection. In a sensitivity analysis where studies were stratified by those explicitly and those not explicitly distinguishing co-infection from secondary infection, there was negligible difference in the reported prevalence of bacterial infection: 4.6% (95%CI 0.2–9.0%) and 2.5% (95%CI 0.0–7.9%), respectively (Appendix Fig. B).
Of 24 studies included, nine contained potential duplicate data due to overlapping hospital site, patient age group and study time period. Removing any potential duplicate study cohorts did not significantly change the estimate of bacterial co-infection (3.8%, 95%CI 0.0–8.0%), secondary infection (14.3%, 95%CI 8.9–19.7%), or the overall proportion with bacterial infection (7.7%, 95%CI 4.4–11.1%) compared to original estimates (Appendix Fig. C).
Bacterial pathogens
Specific species of bacterial co-pathogens were reported in 11/24 studies (45.8%), representing less than 14% of patients with reported infections. The most common organisms reported were Mycoplasma species (n = 11 patients, n = 3 reported as M. pneumoniae), Haemophilus influenzae (n = 5 patients) and Pseudomonas aeruginosa (n = 5 patients) (Appendix Table A).
Antibiotic prescribing
The proportion of patients receiving antibiotic agents was reported in 14 studies (58%). In these studies, the majority of patients received antibiotics (71.8%, 95%CI 56.1–87.7%). Antibiotic use was generally broad in spectrum with fluoroquinolones and third-generation cephalosporins comprising 74% of the antibiotics prescribed (Appendix Table B).
Discussion
To our knowledge, this is the first living meta-analysis and systematic review focusing on bacterial co-infections in patients hospitalized for COVID-19; 24 studies were included. Co-infection was reported in 3.5% (95%CI: 0.4–6.7%) of patients and secondary infection in 14.3% (95%CI: 9.6–18.9%) of patients with COVID-19. Overall, reported bacterial infection was 6.9% (95% CI 4.3–9.5%) but varied slightly by patient population, ranging from 5.9% in hospitalized patients to 8.1% in critically ill patients. Despite an overall low rate of bacterial infections, over 70% of patients received antibiotics, with the majority constituting broad-spectrum agents such as fluoroquinolones and third-generation cephalosporins.
Our findings are consistent with those of a recent systematic review and meta-analysis that included 30 studies evaluating co-infections among patients infected with COVID-19. Similarly, the authors reported that 7% of patients had a bacterial co-infection with a high degree of heterogeneity (I2 = 92.2%) and higher prevalence in the ICU compared to mixed inpatient settings [39]. Our study adds to this literature by estimating co-infection versus secondary infection prevalence and by updating the evaluation throughout the pandemic.
Among studies reporting on other coronaviruses, 11% of patients were estimated to have co-infections, predominantly secondary infections in the largest SARS-CoV-1 patient series [40], and a limited role for bacterial infections in MERS [41]. A narrative review reported the proportion of COVID-19 patients with co-infections to vary widely, from no co-infections to 100% in those who died, and also wide variability of antimicrobial use by severity of illness, ranging from 20% to 100% for antibiotics [42]. As the authors did not generate an overall effect estimate in their narrative review, we are unable to make comparisons with their findings.
Previous epidemic and pandemic outbreaks of viral respiratory infections have reported bacterial infections complicating initial viral illness. During the 2009 A(H1N1) influenza pandemic, bacterial co-infection was reported in up to 30% of critically ill patients [3,43,44] and in 12% of hospitalized patients not requiring ICU admission [45]. When reported, the most commonly identified bacterial co-pathogens were Staphylococcus aureus and Streptococcus pneumoniae [3,43,45]. In contrast, our review found those pathogens to be uncommonly reported amongst patients with COVID-19. There are scant data on bacterial co-infections from patients infected with SARS-CoV-1 and MERS-Co-V [41,46,47]. Given that the current knowledge of the pathophysiology of SARS-CoV-2 is evolving, our understanding of the pathogenesis of bacterial co-infection is also incomplete. For influenza, it is postulated that viral damage of epithelial cells in the lower airway, coupled with mucociliary dysfunction, facilitate binding to cell surfaces of pathogenic bacteria aspirated from the nasopharynx [48]. Consequently, bacterial infection is established, which causes further damage by inhibiting repair and regeneration of the epithelial cell layer [48]. Whether this mechanism applies to SARS-CoV-2 remains to be determined.
Our results show that there is currently insufficient evidence to support widespread empirical use of antibiotics in most hospitalized patients, as the overall proportion of bacterial infections in patients with COVID-19 was low. For critically ill patients the proportion is higher, although this was reported in only five studies. As antibiotics likely provide minimal benefit as empirical treatment in COVID-19 and are associated with unintended consequences—including adverse events, toxicity, resistance, and Clostridioides difficile infections—it is prudent for clinicians to prescribe them judiciously [6,40,42,[49], [50], [51]]. For those with suspected bacterial infections, antibiotic selection should be based on local epidemiology and patient factors, and early discontinuation should be considered if there is no evidence of bacterial infection. Our findings reinforce current clinical guidelines that recommend use of antibiotics in patients with COVID-19 who have suspected or proven bacterial co-infection, with re-evaluation based on clinical signs and symptoms, laboratory and imaging findings [49].
Our rapid review and meta-analysis has several strengths. We followed a robust literature search strategy with guidance from a healthcare information specialist and we used a dual-reviewer process to screen and select appropriate studies meeting the inclusion criteria. In addition, we performed stratified analyses according to estimated severity of illness to better inform clinical practice. However, this review and meta-analysis also has important limitations. While we identified 24 studies representing over 3000 patients, bacterial infections in patients with COVID-19 disease may be under- or over-represented, as there was a lack of consistent bacteriological diagnostic method and a specific testing method was not reported in half of all the studies. Further, distinguishing bacterial colonization from infection presents a challenge, particularly in the context of COVID-19 infection. We only examined the subset of COVID-19 studies that reported the presence or absence of bacterial infections, and the temporal relationship between bacterial and viral infection was not explicit; hence differentiating co-infection from secondary infection was challenging. In addition, we did not perform a formal assessment for risk of bias. However, we attempted to address these limitations through our three sensitivity analyses, which remained robust to our initial estimates. The generalizability of our estimates may be limited as the majority of studies were from Asia during the start of the pandemic. Regional variations in patient population, access to care, and infection prevention and control are all potential variables that could impact the likelihood of bacterial co-infection and secondary infection. To overcome this limitation, we are planning a living meta-analysis with regular systematic searches to better inform our co-infection and secondary infection estimates, which will be hosted on the Toronto Antimicrobial Resistance Research Network (TARRN) website (https://www.tarrn.org/covid). Lastly, the estimates in our meta-analysis were primarily based on hospitalized adult patients, and may not reflect the overall bacterial infection rates as the vast majority of COVID-19 patients experience mild disease and do not require hospitalization [52].
Opportunities for improvement in future research reporting on concomitant bacterial infection in COVID-19 include indicating the temporal relationship of bacterial infection relative to patient presentation, specifying the bacterial testing method, and clearly differentiating colonization from infection when possible. Ideally, prospective cohort studies could include systematic blood and respiratory specimen collection from all patients with COVID-19.
Conclusion
Among patients with COVID-19, the overall proportion of bacterial co-infection was low, but usage of antibiotics was high. There is insufficient evidence to support widespread use of empirical antibiotics in patients hospitalized for COVID-19, particularly those without critical illness.
Author contributions
Concept and design: all authors. Acquisition, analysis, or interpretation of data: BJL, MS, SR, VL, DW, J-PS. Drafting of the manuscript: BJL, MS, SR, VL. Critical revision of the manuscript for important intellectual content: all authors. Statistical analysis: BJL. Administrative, technical, or material support: all authors.
Transparency declaration
The authors have no conflicts of interest to declare. The University of Toronto, Department of Medicine, Network Seed Funding Grant supported the role of a research coordinator (DW) to provide research project management. The University was not involved in study concept, analysis or synthesis of evidence, nor in the decision to publish.
Acknowledgements
We thank Ashley Farrell, BA, MLIS, AHIP, Information Specialist, for her support with formulating and executing the search strategy for this systematic review. We thank the Toronto Antimicrobial Resistance Research Network (TARRN), an interprofessional forum of clinicians and researchers that acted as a venue for bringing together this research team, and a repository for our living review.
Editor: J. Rodriguez-Baño
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.07.016.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Esper F.P., Spahlinger T., Zhou L. Rate and influence of respiratory virus co-infection on pandemic (H1N1) influenza disease. J Infect. 2011;63:260–266. doi: 10.1016/j.jinf.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein E.Y., Monteforte B., Gupta A., Jiang W., May L., Hsieh Y.-H., et al. The frequency of influenza and bacterial coinfection: a systematic review and meta-analysis. Influenza Other Respir Viruses. 2016;10:394–403. doi: 10.1111/irv.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rice T.W., Rubinson L., Uyeki T.M., Vaughn F.L., John B.B., Miller R.R., et al. Critical illness from 2009 pandemic influenza A virus and bacterial coinfection in the United States. Crit Care Med. 2012;40:1487–1498. doi: 10.1097/CCM.0b013e3182416f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah N.S., Greenberg J.A., McNulty M.C., Gregg K.S., Riddell J., Mangino J.E., et al. Bacterial and viral co-infections complicating severe influenza: incidence and impact among 507 US patients, 2013–14. J Clin Virol. 2016;80:12–19. doi: 10.1016/j.jcv.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martín-Loeches I., Sanchez-Corral A., Diaz E., Granada R.M., Zaragoza R., Villavicencio C., et al. Community-acquired respiratory coinfection in critically ill patients with pandemic 2009 influenza A(H1N1) virus. Chest. 2011;139:555–562. doi: 10.1378/chest.10-1396. [DOI] [PubMed] [Google Scholar]
- 6.Huttner B., Catho G., Pano-Pardo J.R., Pulcini C., Schouten J. COVID-19: don’t neglect antimicrobial stewardship principles! Clin Microbiol Infect. 2020;26:808–810. doi: 10.1016/j.cmi.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox M.J., Loman N., Bogaert D., O’Grady J. Co-infections: potentially lethal and unexplored in COVID-19. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30009-4. S2666524720300094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization . World Health Organization; Geneva, Switzerland: 2020. Clinical management of COVID-19 interim guidance [Internet]https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected Available from: [Google Scholar]
- 9.Alhazzani W., Møller M.H., Arabi Y.M., Loeb M., Gong M.N., Fan E., et al. Surviving sepsis campaign: guidelines on the management of critically ill adults with coronavirus disease 2019 (COVID-19) Crit Care Med. 2020;48:e440–e469. doi: 10.1097/CCM.0000000000004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garritty C., Gartlehner G., Kamel C., King V.J., Nussbaumer-Streit B., Stevens A., et al. Cochrane rapid reviews. Interim guidance from the Cochrane rapid reviews methods Group. 2020. Available at: https://methods.cochrane.org/rapidreviews/sites/methods.cochrane.org.rapidreviews/files/public/uploads/cochrane_rr_-_guidance-23mar2020-final.pdf. [DOI] [PMC free article] [PubMed]
- 11.Elliott J.H., Turner T., Clavisi O., Thomas J., Higgins J.P.T., Mavergames C., et al. Living systematic reviews: an emerging opportunity to narrow the evidence–practice gap. PLoS Med. 2014;11 doi: 10.1371/journal.pmed.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DerSimonian R., Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Deeks J., Higgins J., Altman D. 2019. Cochrane handbook for systematic reviews of interventions [Internet]https://training.cochrane.org/handbook Available from: [Google Scholar]
- 14.Hoy D., Brooks P., Woolf A., Blyth F., March L., Bain C., et al. Assessing risk of bias in prevalence studies: modification of an existing tool and evidence of interrater agreement. J Clin Epidemiol. 2012;65:934–939. doi: 10.1016/j.jclinepi.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Arentz M., Yim E., Klaff L., Lokhandwala S., Riedo F.X., Chong M., et al. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA [Internet] 2020 doi: 10.1001/jama.2020.4326. https://jamanetwork.com/journals/jama/fullarticle/2763485 [cited 2020 Apr 6]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrasa H., Rello J., Tejada S., Martin A., Balziskueta G., Vinuesa C., et al. SARS-Cov-2 in Spanish intensive care: early experience with 15-day survival in Vitoria. Anaesth Crit Care Pain Med. 2020 doi: 10.1016/j.accpm.2020.04.001. 101652401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhatraju P.K., Ghassemieh B.J., Nichols M., Kim R., Jerome K.R., Nalla A.K., et al. Covid-19 in critically ill patients in the Seattle region—case series. N Engl J Med. 2020 doi: 10.1056/NEJMoa2004500. NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai Q, Huang D, Ou P, Yu H, Zhu Z, Xia Z et al. COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy 2020 ((Su, Fu) School of Medicine, Southern University of Science and Technology, Shenzhen, Guangdong 518055, China). [DOI] [PubMed]
- 19.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen T, Dai Z, Mo P, Li X, Ma Z, Song S et al. Clinical characteristics and outcomes of older patients with coronavirus disease 2019 (COVID-19) in Wuhan, China (2019): a single-centered, retrospective study. J Gerontol A Biol Sci Med Sci 2020 (cba, 9502837). [DOI] [PMC free article] [PubMed]
- 21.Feng Y, Ling Y, Bai T, Xie Y, Huang J, Li J et al. COVID-19 with different severity: a multi-center study of clinical features. Am J Respir Crit Care Med 2020 (9421642, bzs). [DOI] [PMC free article] [PubMed]
- 22.Lian J., Jin X., Hao S., Cai H., Zhang S., Zheng L., et al. Analysis of epidemiological and clinical features in older patients with Corona Virus Disease 2019 (COVID-19) out of Wuhan. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa242. (Jin, Gao, Ren) Department of Gastroenterology, First Affiliated Hospital, College of Medicine, Zhejiang University. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling L., So C., Shum H.P., Chan P.K.S., Lai C.K.C., Kandamby D.H., et al. Critically ill patients with COVID-19 in Hong Kong: a multicentre retrospective observational cohort study. Crit Care Resusc J Australas Acad Crit Care Med. 2020 doi: 10.51893/2020.2.oa1. 100888170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W., Tao Z.-W., Lei W., Ming-Li Y., Kui L., Ling Z., et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin Med J (Engl) 2020 doi: 10.1097/CM9.0000000000000775. (Ming) Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430014, China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mo P., Xing Y., Xiao Y., Deng L., Zhao Q., Wang H., et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa270. (Cheng) Department of Respiratory Medicine, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pongpirul W.A., Mott J.A., Woodring J.V., Uyeki T.M., MacArthur J.R., Vachiraphan A., et al. Clinical characteristics of patients hospitalized with coronavirus disease, Thailand. Emerg Infect Dis. 2020;26 doi: 10.3201/eid2607.200598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan Y.-P., Tan B.-Y., Pan J., Wu J., Zeng S.-Z., Wei H.-Y. Epidemiologic and clinical characteristics of 10 children with coronavirus disease 2019 in Changsha, China. J Clin Virol. 2020:127. doi: 10.1016/j.jcv.2020.104353. (Wu) Division of Pharmacy, College of Medicine, Hunan Normal University, Changsha 410005, China:104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, He W, Yu X, Hu D, Bao M, Liu H et al. Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. J Infect 2020 ((Liu, Zhou) Department of Cardiology, Renmin Hospital of Wuhan University, Wuhan 430060, China). [DOI] [PMC free article] [PubMed]
- 30.Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa272. (Yang) Department of Respiratory and Critical Care Medicine, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C., Chen X., Cai Y., Xia J., Xu S., Huang H., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med [Internet] 2020 doi: 10.1001/jamainternmed.2020.0994. http://archinte.jamanetwork.com/issues.aspx (Song) National Clinical Research Center for Aging and Medicine, Huashan Hospital, Fudan University, Shanghai, China. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J., Li W., Shi X., Chen Z., Jiang B., Liu J., et al. Early antiviral treatment contributes to alleviate the severity and improve the prognosis of patients with novel coronavirus disease (COVID-19) J Intern Med. 2020 doi: 10.1111/joim.13063. (Cao) Zhejiang Provincial Key Laboratory for Diagnosis and Treatment of Aging and Physic-chemical Injury Diseases, 79 Qingchun Rd, Hangzhou 310003, China. [DOI] [PubMed] [Google Scholar]
- 33.Wu J., Liu J., Zhao X., Liu C., Wang W., Wang D., et al. Clinical characteristics of imported cases of COVID-19 in Jiangsu Province: a multicenter descriptive study. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa199. (Cao) Zhejiang Provincial Key Laboratory for Diagnosis and Treatment of Aging and Physic-chemical Injury Diseases, Hangzhou, China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia W., Shao J., Guo Y., Peng X., Li Z., Hu D. Clinical and CT features in pediatric patients with COVID-19 infection: different points from adults. Pediatr Pulmonol. 2020 doi: 10.1002/ppul.24718. ppul.24718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med [Internet] 2020 doi: 10.1016/S2213-2600(20)30079-5. http://www.elsevier.com/journals/the-lancet-respiratory-medicine/2213-2600 (Yu) Department of Critical Care Medicine, Renmin Hospital of Wuhan University, Wuhan, China. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA [Internet] 2020 doi: 10.1001/jama.2020.3204. http://jama.jamanetwork.com/journal.aspx (Chen, Lee, Leo) Saw Swee Hock School of Public Health, Singapore, Singapore. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng F., Liao C., Fan Q.-H., Chen H.-B., Zhao X.-G., Xie Z.-G., et al. Clinical characteristics of children with coronavirus disease 2019 in Hubei, China. Curr Med Sci. 2020 doi: 10.1007/s11596-020-2172-6. (Zhong) Department of Pediatrics, Huazhong University of Science and Technology Hospital, Wuhan 430074, China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect 2020 (ig9, 7908424). [DOI] [PMC free article] [PubMed]
- 40.Rawson T.M., Moore L.S.P., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M., et al. Bacterial and fungal co-infection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa530. ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arabi Y.M., Deeb A.M., Al-Hameed F., Mandourah Y., Almekhlafi G.A., Sindi A.A., et al. Macrolides in critically ill patients with Middle East Respiratory Syndrome. Int J Infect Dis. 2019;81:184–190. doi: 10.1016/j.ijid.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clancy C.J., Nguyen M.H. COVID-19, superinfections and antimicrobial development: what can we expect? Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa524. ciaa524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar A. Critically ill patients with 2009 influenza A(H1N1) infection in Canada. JAMA. 2009;302:1872. doi: 10.1001/jama.2009.1496. [DOI] [PubMed] [Google Scholar]
- 44.Critical care services and 2009 H1N1 influenza in Australia and New Zealand. N Engl J Med. 2009;361:1925–1934. doi: 10.1056/NEJMoa0908481. [DOI] [PubMed] [Google Scholar]
- 45.MacIntyre C.R., Chughtai A.A., Barnes M., Ridda I., Seale H., Toms R., et al. The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza A(H1N1)pdm09. BMC Infect Dis. 2018;18:637. doi: 10.1186/s12879-018-3548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Memish Z.A., Perlman S., Van Kerkhove M.D., Zumla A. Middle East respiratory syndrome. Lancet. 2020;395:1063–1077. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozak R., Prost K., Yip L., Williams V., Leis J.A., Mubareka S. Severity of coronavirus respiratory tract infections in adults admitted to acute care in Toronto, Ontario. J Clin Virol. 2020;126:104338. doi: 10.1016/j.jcv.2020.104338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chertow D.S., Memoli M.J. Bacterial coinfection in influenza: a grand rounds review. JAMA. 2013;309:275. doi: 10.1001/jama.2012.194139. [DOI] [PubMed] [Google Scholar]
- 49.National Institute for Health and Care Excellence (NICE) 2020. COVID-19 rapid guideline: antibiotics for pneumonia in adults in hospital [Internet]https://www.nice.org.uk/guidance/ng173/chapter/4-Assessing-the-ongoing-need-for-antibiotics Available from: [PubMed] [Google Scholar]
- 50.Tamma P.D., Avdic E., Li D.X., Dzintars K., Cosgrove S.E. Association of adverse events with antibiotic use in hospitalized patients. JAMA Intern Med. 2017;177:1308. doi: 10.1001/jamainternmed.2017.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buetti N., Mazzuchelli T., Lo Priore E., Balmelli C., Llamas M., Pallanza M., et al. Early administered antibiotics do not impact mortality in critically ill patients with COVID-19. J Infect. 2020 doi: 10.1016/j.jinf.2020.06.004. S0163445320303819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.