Abstract
Coronavirus disease 2019 (COVID-19) is a highly contagious infectious disease. Similar to H7N9 infection, pneumonia and cytokine storm are typical clinical manifestations of COVID-19. Our previous studies found that H7N9 patients had intestinal dysbiosis. However, the relationship between the gut microbiome and COVID-19 has not been determined. This study recruited a cohort of 57 patients with either general (n = 20), severe (n = 19), or critical (n = 18) disease. The objective of this study was to investigate changes in the abundance of ten predominant intestinal bacterial groups in COVID-19 patients using quantitative polymerase chain reaction (q-PCR), and to establish a correlation between these bacterial groups and clinical indicators of pneumonia in these patients. The results indicated that dysbiosis occurred in COVID-19 patients and changes in the gut microbial community were associated with disease severity and hematological parameters. The abundance of butyrate-producing bacteria, such as Faecalibacterium prausnitzii, Clostridium butyricum, Clostridium leptum, and Eubacterium rectale, decreased significantly, and this shift in bacterial community may help discriminate critical patients from general and severe patients. Moreover, the number of common opportunistic pathogens Enterococcus (Ec) and Enterobacteriaceae (E) increased, especially in critically ill patients with poor prognosis. The results suggest that these bacterial groups can serve as diagnostic biomarkers for COVID-19, and that the Ec/E ratio can be used to predict death in critically ill patients.
Keywords: Intestinal microbiota, COVID-19, SARS-CoV-2
1. Introduction
Coronavirus disease 2019 (COVID-19) is a highly contagious infectious disease. The etiological agent has been designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the coronavirus study group of the International Committee on Taxonomy of Viruses. The most common clinical manifestation of COVID-19 is pneumonia.
H7N9 is a viral infectious disease that causes pulmonary damage. A previous study found that intestinal dysbiosis occurred in H7N9, especially in severe and critical patients [1]. Qin et al. [2] analyzed the composition of the intestinal microbiota of 26 H7N9 patients and found that the number of Bacteroides decreased while the number of Proteobacteria increased at the phylum level. At the genus level, the number of Eubacterium rectale and Bifidobacterium dramatically decreased, while the number of Salmonella and Enterococcus (Ec) increased in H7N9 patients. Beneficial intestinal bacteria can modulate immune function [3]. A prospective study in intensive care unit (ICU) patients showed that the serum levels of interleukin-6 (IL-6) decreased as systemic inflammation decreased using probiotics [4]. Imbalance in the intestinal microbiota seems to be related to the inflammatory response in humans [5], and changes in the composition of the gut microbiome are associated with secondary infections induced by bacterial translocation from the gastrointestinal tract [5]. Bacterial translocation is an important contributor to multiple organ dysfunction syndrome (MODS) [6], [7]. Clinical studies have shown that intestinal bacteria are influenced by hypoxia, ischemia, and the use of antibiotics in critical patients. Modulators of gut microbiota help prevent infections, pyemia, and MODS [8].
COVID-19 is an acute respiratory viral infection that leads to cytokine release syndrome in severe and critical cases. The serum levels of IL-6 increase significantly in severe patients [9]. Acute respiratory distress syndrome is the most common organ dysfunction in COVID-19 [10]. According to the abundance and clinical significance of bacteria in the intestinal tract, we selected ten kinds of main bacterial groups for detection by quantitative polymerase chain reaction (q-PCR), including probiotics (Lactobacillus and Bifidobacterium), conditionally pathogenic bacteria (Ec, Enterobacteriaceae (E), and Atopobium), and other effective symbiotic bacteria (Faecalibacterium prausnitzii (F. prausnitzii), Clostridium butyricum (C. butyricum), Clostridium leptum (C. leptum), Eubacterium rectale (E. rectale), and Bacteroides), and explored the relationship of these groups with the severity of COVID-19.
2. Materials and methods
2.1. Study design
Ten predominant intestinal bacterial groups were detected in fresh stool specimens of 57 COVID-19 patients with pneumonia. All patients were classified into one of three groups—general, severe, or critical—according to the diagnostic criteria of the Diagnosis and treatment protocol for novel coronavirus pneumonia (trail version 7) [11].
According to these criteria, cases with fever, respiratory symptoms, and lung imaging evidence of pneumonia were considered general. Cases with a respiratory rate (RR) ≥ 30 per minite, oxygen saturation ≤ 93% at rest, arterial partial pressure of oxygen (PaO2)/fraction of inspiration oxygen (FiO2) ≤ 300 mmHg (1 mmHg = 133.3 Pa), or significant improvement in lung imaging lesions (> 50%) within 24–48 h were considered severe. Cases with respiratory failure requiring mechanical ventilation, shock, or other organ failure requiring ICU care were considered critical [11]. All clinical data, including biochemical and immunological indexes were collected from electronic medical records and are shown in Table 1 .
Table 1.
Personal and laboratory findings of 57 cases of COVID-19 with pneumonia in the general, severe, and critical groups.
| Variable | Disease subtype |
|||
|---|---|---|---|---|
| General (n = 20) | Severe (n = 19) | Critical (n = 18) | P value | |
| Age (year), median (IQR) | 59 (53–63) | 66 (61–74) | 68 (55–70) | 0.026* |
| Sex (male), (proportion) | 8 (40.0%) | 9 (47.4%) | 12 (66.7%) | 0.242 |
| Length of stay in hospital (d), median (IQR) | 20 (14–24) | 23(19–27) | 22(13–28) | 0.221 |
| Underlying disease, (proportion) | 10 (50.0%) | 12 (63.2%) | 14 (77.8%) | 0.208 |
| Hypertension, (proportion) | 7 (35.0%) | 8 (42.1%) | 12 (66.7%) | 0.02* |
| Diabetes mellitus, (proportion) | 3 (15.0%) | 0 (0%) | 6 (33.3%) | 0.021* |
| WBC (×109 L−1), median (IQR) | 4.96 (4.60–6.60) | 6.40 (4.90–9.00) | 8.00 (6.60–11.60) | 0.002** |
| Neutrophil ratio (%), median (IQR) | 56.2 (60.0–66.7) | 66.5 (59.7–74.0) | 88.7 (75.4–91.5) | < 0.0001*** |
| Lymphocyte ratio (%), median (IQR) | 30.95 (21.50–36.10) | 20.50 (13.70–28.50) | 6.30 (3.20–13.90) | < 0.0001*** |
| Monocyte ratio (%), median (IQR) | 9.4 (6.7–11.8) | 9.0 (7.9–10.5) | 3.7 (2.5–6.6) | 0.001** |
| Platelet count (×109 L−1), median (IQR) | 198.5 (141.0–269.5) | 260.0 (213.0–322.0) | 145.0 (98.3–288.0) | 0.08 |
| CRP (mg·L−1), median (IQR) | 5.0 (5.0–10.9) | 6.0 (4.0–25.7) | 47.1 (23.1–100.1) | 0.001** |
| PCT (ng·mL−1), median (IQR) | 0.033 (0.027–0.054) | 0.057 (0.033–0.076) | 0.147 (0.099–0.336) | < 0.0001*** |
| D-dimer (mg·L−1), median (IQR) | 0.87 (0.50–1.30) | 1.60 (0.60–3.20) | 5.10 (3.10–19.50) | < 0.0001*** |
| IL-6 (pg·mL−1), median (IQR) | 2.47 (1.50–3.70) | 5.00 (1.60–17.10) | 12.40 (6.50–55.20) | 0.002** |
| ALT (U·L−1), median (IQR) | 23.0 (17.3–51.8) | 35.0 (32.0–73.0) | 28.0 (17.5–40.8) | 0.149 |
| AST (U·L−1), median (IQR) | 20.5 (17.0–25.0) | 34.0 (21.0–40.0) | 30.5 (19.3–48.0) | 0.05 |
| ALB (g·L−1), median (IQR) | 36.5 (32.5–41.2) | 36.7 (33.9–40.7) | 31.9 (28.7–34.7) | 0.001** |
| Creatinine (μmol·L−1), median (IQR) | 59.5 (50.8–67.8) | 52.0 (42.0–64.0) | 56.0 (41.0–66.3) | 0.362 |
| CK (U·L−1), median (IQR) | 39.0 (24.0–58.0) | 26.0 (17.0–42.0) | 39.5 (22.8–56.5) | 0.204 |
| LDH (U·L−1), median (IQR) | 204.0 (177.8–250.5) | 219.0 (183.0–253.0) | 377.0 (350.0–592.8) | < 0.0001*** |
| Myoglobin (μg·L−1), median (IQR) | 28.4 (19.0–41.8) | 28.5 (21.2–34.8) | 60.0 (30.8–139.0) | 0.006** |
Significant differences among general, severe, and critical groups are indicated by asterisks (*: P < 0.05; **: P < 0.01; ***: P < 0.001).
IQR: interquartile range; WBC: white blood cell; CRP: C-reactive protein; PCT: procalcitonin; ALT: alanine aminotransferase; AST: aspartate aminotransferase; ALB: albumin; CK: creatine kinase; LDH: lactate dehydrogenase.
2.2. Use of q-PCR to detect predominant intestinal bacterial population
The primers used are shown in Appendix A Table S1. All oligonucleotide primers were synthesized by GenScript (China). A ViiA™ 7 real-time PCR system (Applied Biosystems, USA) was used to perform q-PCR. Amplification reactions contained 10 μL of SYBR™ green PCR master mix (TongChuang, China), 8 μL of primer (0.2–0.6 μmol·L−1), and 2 μL of crude template DNA or 2 μL of water (negative control), for a final volume of 20 μL. Each reaction was performed in triplicate, and a Δcycle threshold (ΔCt) < 0.5 between duplicates was required. Amplifications were performed with the following temperature profiles: one cycle at 95 °C for 3 min, followed by 40 cycles at 95 °C for 15 s and at 60 °C for 30 s. The annealing and plate-reading temperatures for each primer pair are shown in Table S1. The copy number of ribosomal DNA (rDNA) operons of targeted bacteria in crude DNA templates was determined by comparison with serially diluted plasmid DNA standards run on the same plate. Plasmid DNA standards were made from known concentrations of plasmid DNA that contained the respective amplicon for each set of primers.
2.3. Statistical analysis
SPSS software version 22.0 and GraphPad Prism version 8 were used for statistical analyses. Normally distributed data were expressed as means and standard deviations, whereas non-normally distributed data were presented as median and interquartile range (IQR). Intergroup differences in clinical data were analyzed by one-way analysis of variance (ANOVA). Non-normally distributed data were analyzed using the Mann–Whitney U test or the Kruskal–Wallis test. Categorical data were compared by means of the χ 2 test. Correlations between the microbiota and laboratory findings were established using Pearson correlation analysis. Receiver operating characteristic (ROC) curves were constructed to determine the optimal cutoff values. The area under the curve (AUC) was used to compare the diagnostic value of the microbial composition. P values of less than 0.05 were considered to be statistically significant.
2.4. Ethics
This study was approved by the Clinical Research Ethics Committee of Renmin Hospital of Wuhan University (WDRY2020-K153). The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki [12].
3. Results
3.1. Demographic and clinical characteristics
All patients with pneumonia were classified as general, severe, or critical, according to the Diagnosis and treatment protocol for novel coronavirus pneumonia (trail version 7) [11]. Ten predominant intestinal bacterial groups were identified in the study population. Twenty patients presented general disease; the median age was 59 years (IQR: 53–63), and the male-to-female ratio was 2:3 (Table 1). Nineteen patients presented severe disease; the median age was 66 years (IQR: 61–74), and the male-to-female ratio was 9:10. Eighteen patients presented critical disease; the median age was 68 years (IQR: 55–70), and the male-to-female ratio was 2:1. The median age of the severe and critical groups was higher than that of the general group. There was no significant difference in the sex ratio among the three groups. The time interval from admission to bacterial identification in the three groups was 20, 23, and 22 d, respectively, without significant difference between the groups. Many patients had more than one chronic disease, including hypertension, diabetes, liver disease, and kidney disease. The proportion of patients with chronic diseases in the three groups was 50.0%, 63.2%, and 77.8%, respectively. There was a significant difference in the rate of hypertension among the groups (P < 0.05), corresponding to 35.0% (7), 42.1% (8), and 66.7% (12), respectively, with an upward trend in all groups. There was no significant difference in the rates of liver and kidney disease because the sample size was small.
3.1.1. Laboratory findings
There was a significant difference in white blood cell (WBC) count (P < 0.05) among the three groups. WBC count was significantly higher in the critical group than in the general group (P < 0.05), but there was no significant difference in this parameter between the general and severe groups. The number of neutrophils, lymphocytes, and monocytes was significantly different among the three groups (P < 0.05). There was no significant difference in the serum levels of C-reactive protein (CRP), procalcitonin (PCT), and IL-6 between the general and severe groups, whereas the levels of these markers were significantly higher in the critical group than in the severe group (P < 0.05). There was a significant difference in the levels of D-dimer, myoglobin, and lactate dehydrogenase (LDH) between critical and severe patients (P < 0.05), but no significant difference between general and severe patients. There was no significant intergroup difference in alanine aminotransferase (ALT), aspartate aminotransferase (AST), and creatinine. Serum albumin (ALB) level was significantly lower in critical patients.
3.1.2. Treatment and outcomes
A total of 50.9%, 5.3%, and 12.3% of our patients received antibiotics, antifungal drugs, and probiotics, respectively. Antibiotics and antifungal drugs were used more frequently in critical patients. Probiotics were not used regularly. One patient in the general group (5.0%), four patients in the severe group (21.1%), and two patients in the critical group (11.1%) were given probiotics. Eight of the 18 patients (44.4%) from the critical group died.
3.2. Changes in the composition of the gut microbiome in COVID-19 patients with pneumonia
The abundance of probiotic bacteria Lactobacillus and Bifidobacterium and of anti-inflammatory bacteria F. prausnitzii, C. butyricum, C. leptum, and E. rectale decreased in all patients. The abundance of conditional pathogenic bacteria E decreased as the concentration of Ec increased with disease severity. The concentration of Atopobium did not change significantly. The abundance of Bacteroides was not significantly different between the three groups, corresponding to 55.0%, 47.4%, and 61.1% decreased, respectively, but was close to the lower limit of the normal range.
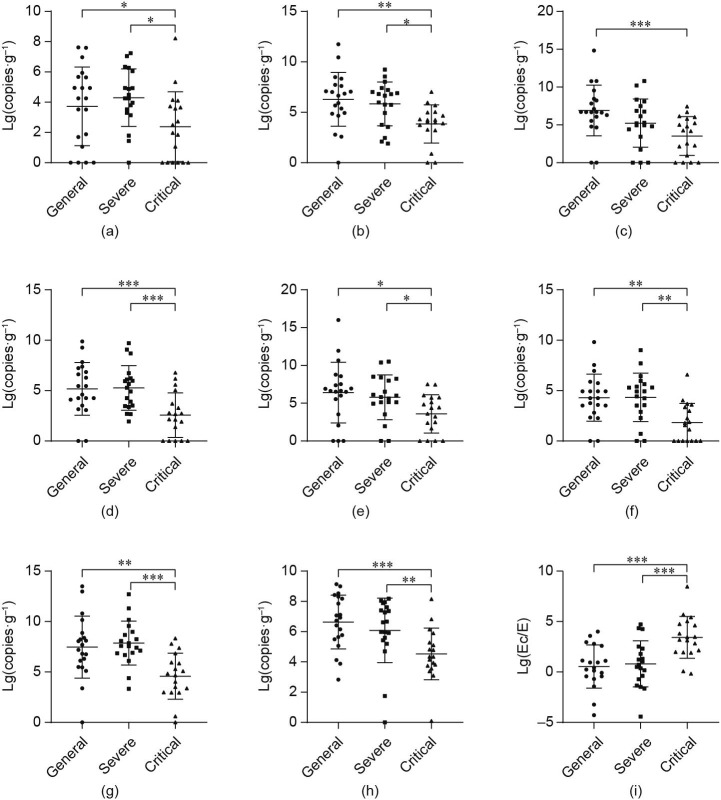
Common logarithm (lg) was used for the bacterial population values (copy number of fecal bacteria per gram) (Fig. 1 ). The number of probiotic and anti-inflammatory bacteria were significantly different between the three groups. The number of beneficial bacterial decreased as disease severity increased. The Ec/E ratio was calculated as lg(1000 × Ec/E). Bacterial counts were shown as lg and were multiplied by 1:1000, according to the Ec/E ratio in healthy individuals as reported in our previous study. This ratio was significantly increased in critical patients.
Fig. 1.
Lg values of copy number of fecal bacteria per gram in general, severe, and critical patients with COVID-19 pneumonia, respectively, and the normal reference range for each bacterial strain. (a) Lactobacillus: the median value was 1.8 × 104 in the general group, 1.8 × 104 in the severe group, and 200 in the critical group; and the normal reference range was 1.0 × 106–9.0 × 108. (b) Bifidobacterium: the median value was 5.7 × 106 in the general group, 4.4 × 106 in the severe group, and 1.4 × 104 in the critical group; and the normal reference range was 1.0 × 105–9.0 × 108. (c) F. prausnitzii: the median value was 5.0 × 106 in the general group, 1.6 × 105 in the severe group, and 1.7 × 104 in the critical group; and the normal reference range was 1.0 × 106–9.0 × 109. (d) C. butyricum: the median value was 1.6 × 105 in the general group, 1.9 × 105 in the severe group, and 310 in the critical group; and the normal reference range was 1.0 × 105–9.0 × 108. (e) C. leptum: the median value was 4.3 × 106 in the general group, 6.4 × 105 in the severe group, and 1.4 × 104 in the critical group; and the normal reference range was 1.0 × 106–9.0 × 108. (f) E. rectale: the median value was 1.8 × 104 in the general group, 8.2 × 104 in the severe group, and 49 in the critical group; and the normal reference range was 1.0 × 105–9.0 × 106. (g) E: the median value was 1.1 × 104 in the general group, 8.6 × 104 in the severe group, and 240 in the critical group; and the normal reference range was 1.0 × 105–9.0 × 106. (h) Atopobium: the median value was 6.9 × 106 in the general group, 4.3 × 106 in the severe group, and 2.2 × 104 in the critical group; and the normal reference range was 1.0 × 103–9.0 × 106. (i) Intestinal Ec/E ratio: the median value in the general, severe, and critical groups was 0.4, 0.7, and 3.3, respectively. Statistical analysis was performed with one-way ANOVA. Values are mean ± standard deviation. *: P < 0.05; **: P < 0.01; ***: P < 0.001. Lg(copies·g−1): lg values of copies of microbial DNA per gram of feces.
3.3. Correlation between bacterial abundance and clinical indicators in COVID-19 with pneumonia
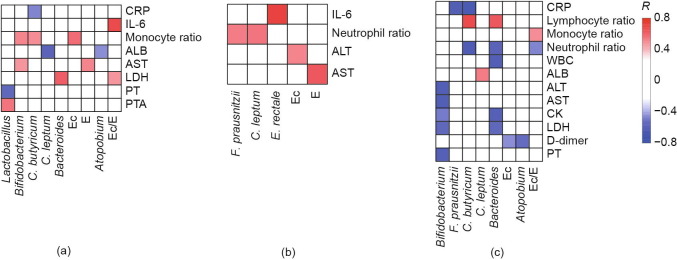
A Pearson correlation analysis was performed between ten bacterial groups and clinical indicators of COVID-19 to determine the clinical value of the intestinal microbiota (Fig. 2 ). There was a significant correlation between butyrate-producing bacteria (BPB) and inflammatory markers (CRP, WBC, lymphocyte ratio, neutrophil ratio, and IL-6).
Fig. 2.
Correlation between intestinal bacteria and clinical indexes in COVID-19 with pneumonia. (a) General patients; (b) severe patients; (c) critical patients. PT: prothrombin time; PTA: prothrombin activity; R: Pearson correlation coefficient.
In the general group, C. butyricum was negatively correlated with CRP levels (Pearson correlation coefficient (R) = −0.5). In the severe group, F. prausnitzii and C. leptum were positively associated with neutrophil levels (R = 0.5), and E. rectale was positively correlated with IL-6 levels (R = 0.7). In the critical group, there was a negative correlation between C. butyricum and CRP (R = −0.7) or neutrophil levels (R = −0.6), and between F. prausnitzii and CRP (R = −0.6).
Shifts in specific bacterial groups were related to changes in the concentration of liver function markers ALT and AST. In general and severe patients, the conditional pathogenic E was positively correlated with AST (R = 0.5 and 0.6, respectively). Moreover, the abundance of Ec was positively correlated with abnormal serum concentrations of ALT (R = 0.5) in severe patients. In the critical group, the probiotic Bifidobacterium was negatively correlated with ALT and AST (both R = −0.6).
Some bacterial species were linked to markers of organ dysfunction (D-dimer, LDH, and creatine kinase (CK)). In the general group, Lactobacillus was negatively associated with prothrombin time (PT) (R = −0.6). In the critical group, there was a negative correlation between Bifidobacterium and PT and LDH (R = −0.6 and −0.5, respectively). Furthermore, there was a negative relationship between Atopobium and D-dimer levels (R = −0.5) and between Bacteroides and LDH and CK levels (R = −0.6) in the critical group.
3.4. Intestinal microecological failure occurred in critical patients
Seven beneficial bacteria, including probiotic bacteria (Lactobacillus and Bifidobacterium), anti-inflammatory bacteria (F. prausnitzii, C. butyricum, C. leptum, and E. rectale), and Bacteroides, were chosen for further analysis. If we chose 1000 fold under the lower limit of the normal range as the threshold of obvious disorder, more than one specie disorder accounted for 80.0%, 84.2%, and 94.4% of general, severe, and critical patients, respectively. The abundance of beneficial bacteria decreased significantly in critical patients, and ten patients (55.6%) appeared to present intestinal microecological failure with more than three kinds of bacterial disorder. The bacterial count of these groups was decreased in 15 patients, and the mortality rate of these patients in 7 and 14 d was 26.7% and 40.0%, respectively.
Both Ec and E are conditional pathogens. The mean Ec/E ratio was 1.3 ± 2.5 in survivors (49 patients) and 3.3 ± 1.4 in non-survivors (8 patients) (P < 0.05). The Ec/E ratio increased in 17 out of 18 critical patients.
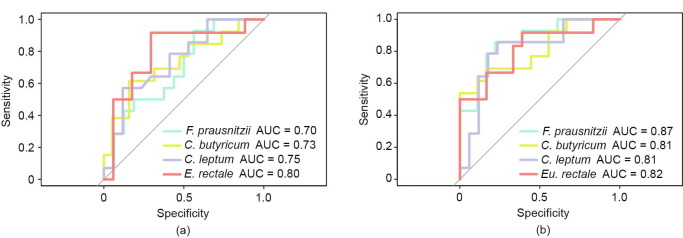
3.5. Prognostic value of the decrease in the abundance of BPB in critical patients
Four BPB—F. prausnitzii, C. butyricum, C. leptum, and E. rectale—were identified. ROC analysis was used to compare the microbiota of severe and critical patients. The AUC for F. prausnitzii, C. butyricum, C. leptum, and E. rectale was 0.70 (95% confidence interval (CI): 0.50–0.89; specificity (SPE): 0.44; sensitivity (SEN): 0.93), 0.73 (95% CI: 0.54–0.92; SPE: 0.84; SEN: 0.62): 0.75 (95% CI: 0.57–0.92; SPE: 0.88; SEN: 0.57), and 0.80 (95% CI: 0.61–0.98; SPE: 0.71; SEN: 0.92), respectively, and the cutoff values were 6.4 × 104, 2.2 × 103, 7.8 × 104, and 2.8 × 103, respectively (Fig. 3 (a)). ROC analysis was performed to assess the prognostic value of BPB in general and critical patients. The AUC for F. prausnitzii, C. butyricum, C. leptum, and E. rectale was 0.87 (95% CI: 0.74–0.99; SPE: 0.78; SEN 0.86), 0.81 (95% CI: 0.64–0.97; SPE: 1.00; SEN 0.54), 0.81 (95% CI: 0.65–0.97; SPE 0.77; SEN: 0.86), and 0.82 (95% CI: 0.65–0.98: SPE, 0.61; SEN: 0.92), respectively, and the cutoff values were 1.7 × 106, 880, 8.0 × 105, and 180, respectively (Fig. 3(b)).
Fig. 3.
ROC curves of the prediction of gut microbiota signatures with AUC values. (a) ROC curve of severe versus critical patients. (b) ROC curve of general versus critical patients.
4. Discussion
Patients with COVID-19 were classified into three groups according to disease severity. The results showed that critical patients were slightly older than general and severe patients. Moreover, critical patients had more underlying diseases, and WBC, PCT, IL-6, D-dimer, LDH, and CRP in peripheral blood were significantly higher in this group than in the other two groups, whereas the levels of these markers did not differ significantly between general and severe patients. However, there were no significant differences in the serum levels of ALT, AST, and creatinine between the three groups.
In our study, eight patients died (all with very severe disease), 50.9% of patients were treated with antibiotics, especially critical patients, and seven patients received probiotics. For severe and critical patients, antibiotic use was high because of the need to prevent and control secondary infections. Detecting changes in the composition of the intestinal microbiota can reveal intestinal microflora disorder in patients with COVID-19, especially severe and critical patients, to provide clues for empirical antibiotic treatment.
As the largest immune organ of the human body, the intestine harbors approximately 1014 bacteria [13]. The gut microbiome is an indispensable functional organ and participates in immune response regulation, nutrient absorption and metabolism, and the control of infection [14], [15], [16]. However, the homeostasis between the intestinal microbiome and the host immune system is impaired as viral infections, antibiotic use, and chemotherapeutic applications increase [1], [3]. Microbiota dysbiosis is the abnormal shift in the number, species, and proportion of normal microbial populations in the intestine, which results in an abnormal physiological and pathological combination. There were significant changes in the intestinal microbiota in our cohort of patients with COVID-19, as was observed in H7N9 patients, including a decrease in the number of Lactobacillus, Bifidobacterium, and BPB, and an overgrowth of opportunistic pathogens such as Ec and E, which are involved in disease severity [2].
Mounting evidence has shown that the predominant bacteria we selected play a vital role in intestinal homeostasis in the intestinal tract. It is known that Lactobacillus and Bifidobacterium produce lactic acid and play an important role in regulating immunity and maintaining intestinal barrier function [17], [18]. F. prausnitzii, C. butyricum, C. leptum, and E. rectale produce short-chain fatty acids (SCFAs) and contribute to host resistance to enteric pathogen colonization and immunomodulation. E, particularly Escherichia coli (E. coli), have been noted for their various virulence factors. Therefore, we hypothesized that measuring the abundance of this bacterial group during COVID-19 would reveal the dysbiosis of intestinal microecology, including bacterial community composition and function.
The abundance of Lactobacillus and Bifidobacterium was below or close to the lower limit of the normal range (Fig. 1), especially in the critical group, which agrees with the findings in severe H7N9 patients [2]. In addition, Lactobacillus was not detected in some patients. The abundance of Bifidobacterium was negatively correlated with ALT, AST, and LDH in critical patients, suggesting that Bifidobacterium is associated with the maintenance of liver and heart function.
The abundance of BPB, such as F. prausnitzii, C. butyricum, C. leptum, and E. rectale, was decreased in most COVID-19 patients. The median count of these species was close to the lower limit of the normal range in the general and severe groups, and below the lower limit of the normal range in the critical group. Butyrate plays an important role in inhibiting the overgrowth of opportunistic pathogens, maintaining the integrity of the intestinal mucosal barrier, activating the adaptive immune response, and enhancing antiviral immunity [19], [20]. An in vitro study has shown that butyrate can regulate the number and function of regulatory T cells (Tregs) and promote the activation of T helper 17 (Th17) and T helper 1 (Th1) cells [21]. Moreover, it has been shown that the intensity of the inflammatory response—especially of a cytokine storm—is related to the aggravation of COVID-19 [22]. It has been suggested that BPB play an important role in SARS-CoV-2 infections. The lower the number of anti-inflammatory bacteria is, the worse the disease severity will be. The reduced abundance of BPB in critical patients may be related to decreased immunity and inflammation against viral pneumonia. The Pearson correlation coefficient between the number of BPB and the levels of peripheral inflammatory markers was negative for the association between C. butyricum and CRP level (R = −0.5) in the general group, as well as for the association between C. butyricum and CRP (R = −0.7), C. butyricum and neutrophils (R = −0.6), and F. prausnitzii and CRP (R = −0.6) in the critical group. Therefore, the decrease in BPB may aggravate the inflammatory response to viral pneumonia and may be positively linked to disease severity. Thus, shifts in the populations of F. prausnitzii, C. butyricum, C. leptum, and E. rectale can be used to predict disease severity. Changes in the abundance of BPB could accurately discriminate critical patients from general patients, and microbial signatures may be a powerful tool to predict disease severity.
Qin et al. [2] found that populations of F. prausnitzii and E. rectale significantly decreased in H7N9 patients, suggesting that the decrease in BPB in COVID-19 patients may be linked to the inflammatory pathogenesis of respiratory viral infections and, consequently, to cytokine release syndrome. Microbiota-targeted therapies administered at the early stage of COVID-19 may help decrease the number of severe and critical cases. However, it is notable that the abundance of C. butyricum decreased in COVID-19 patients but increased in H7N9 patients. Therefore, more studies are necessary to elucidate the effect of viral infections on the dynamics of beneficial intestinal bacteria.
Opportunistic pathogens such as Ec and E can enter the bloodstream through the intestinal barrier and cause infection. Moreover, the overgrowth of opportunistic pathogens causes further intestinal dysbiosis, impairment of the intestinal epithelial barrier, and secondary infections [5], [23], [24]. In the present study, the Ec/E ratio increased in 73.7% (42/57) of patients, especially in the critical group, and was significantly higher in severe patients and non-survivors than in general patients and survivors. The abundance of E was lowest in critical patients, which may be due to the use of antibacterial drugs. The exposure of critical patients to antibiotics such as carbapenem and cephalosporins significantly decreased the population of E, which increased Ec/E ratio and caused further dysbiosis [1]. Moreover, the abundance of Ec was positively correlated with abnormal serum concentrations of ALT (R = 0.5) in severe patients. In addition to secondary fungal infections, attention should be paid to the occurrence of vancomycin-resistant Ec bloodstream infections during the treatment of late complications [25]. Preventing bacterial translocation from the gastrointestinal tract is a priority in the treatment of COVID-19 and associated complications. However, empiric prophylaxis at the expense of maintaining a balance in the gut microbiota is a risk factor for secondary bacterial and fungal infections. Microbiota-targeted therapies have not currently received enough attention in the study of acute infectious diseases [26]. A small number of patients in our cohort received probiotic supplements to improve the gut microbiota and maintain the integrity of the intestinal barrier.
Qin et al. [2] compared the intestinal microbiome of 40 H7N9 patients with healthy controls and found that the abundance of Bacteroides decreased in the former, while the number of pathogens such as E. coli and Ec increased. Similarly, the abundance of Bacteroides decreased in severe and critical cases of COVID-19. Bacteroides is beneficial to the host when located in the gut, but can cause significant pathology when translocated to other sites [27]. Moreover, Bacteroides can produce SCFAs, which improve intestinal function, microbial balance, and the host immune system [28]. However, some Bacteroides species may cause infection. Nevertheless, the role of this bacterial group in COVID-19 is unclear [29].
Changes in bacterial abundance were associated with abnormal serum concentrations of markers of organ dysfunction, such as D-dimer, LDH, and CK. For example, the abundance of lactic acid bacteria was negatively correlated with PT (R = −0.6) in general patients. There was a negative correlation between Bifidobacterium and PT (R = −0.6) and LDH (R = −0.5) in the critical group, as well as the correlation of Atopobium and D-dimer, and Bacteroides and LDH/CK. Therefore, intestinal dysbiosis may lead to changes in hematological parameters [30].
In our clinical experience, we monitored the shifts in ten predominant intestinal bacterial groups by q-PCR in order to achieve complementary personalized infection control strategies and maintain intestinal microbial balance. q-PCR is fast, easy to use, reliable, and less laborious than second-generation sequencing [31]. Promptly detecting microbial imbalance helps clinicians to implement effective diagnostic and therapeutic interventions, reduce the severity of diseases caused by intestinal dysbiosis and even failure, and improve treatment. The receptor used by SARS-CoV-2 to enter host cells is angiotensin-converting enzyme 2 (ACE2) [32]. ACE2 receptors are present in almost all human cells, and it is reported that SARS-CoV-2 can be detected in feces [9], [33]. We postulate that intestinal dysbiosis increases the risk of viral infections because the microbiome helps maintain the integrity of the intestinal mucosa. Given the key role of the gut microbiome in COVID-19 patients, including antiviral response and prevention of infections, it is crucial to identify the interaction between viruses and the gut microbiota and its effect on host response to SARS-CoV-2 infection.
5. Conclusions
Our results demonstrated a potential correlation between changes in intestinal bacterial populations and hematological parameters in COVID-19 patients. The present study is the first to use Ec/E ratio to predict death in critically ill patients.
Acknowledgments
Acknowledgements
This study was funded by the Zhejiang Key Research and Development Plan Emergency Project (2020C03123), the National Science and Technology Major Project (2017ZX10204401), and the Zhejiang Provincial Natural Science Foundation of China (LED20H190001).
Compliance with ethics guidelines
Lingling Tang, Silan Gu, Yiwen Gong, Bo Li, Haifeng Lu, Qiang Li, Ruhong Zhang, Xiang Gao, Zhengjie Wu, Jiaying Zhang, Yuanyuan Zhang, and Lanjuan Li declare that they have no conflict of interest or financial conflicts to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eng.2020.05.013.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lu H., Zhang C., Qian G., Hu X., Zhang H., Chen C. An analysis of microbiota-targeted therapies in patients with avian influenza virus subtype H7N9 infection. BMC Infect Dis. 2014;14:359. doi: 10.1186/1471-2334-14-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin N., Zheng B., Yao J., Guo L., Zuo J., Wu L. Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci Rep. 2015;5(1):14771. doi: 10.1038/srep14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belkaid Y., Harrison O.J. Homeostatic immunity and the microbiota. Immunity. 2017;46(4):562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanaie S., Ebrahimi-Mameghani M., Hamishehkar H., Mojtahedzadeh M., Mahmoodpoor A. Effect of a multispecies probiotic on inflammatory markers in critically ill patients: a randomized, double-blind, placebo-controlled trial. J Res Med Sci. 2014;19(9):827–833. [PMC free article] [PubMed] [Google Scholar]
- 5.Hanada S., Pirzadeh M., Carver K.Y., Deng J.C. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front Immunol. 2018;9:2640. doi: 10.3389/fimmu.2018.02640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deitch E.A. Gut-origin sepsis: evolution of a concept. Surgeon. 2012;10(6):350–356. doi: 10.1016/j.surge.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Q., Verne G.N. Intestinal hyperpermeability: a gateway to multi-organ failure? J Clin Invest. 2018;128(11):4764–4766. doi: 10.1172/JCI124366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders M.E., Merenstein D.J., Reid G., Gibson G.R., Rastall R.A. Author correction: probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16(10):642. doi: 10.1038/s41575-019-0199-6. [DOI] [PubMed] [Google Scholar]
- 9.Xu K., Cai H., Shen Y., Ni Q., Chen Y., Hu S. Management of corona virus disease-19 (COVID-19): the Zhejiang experience. J Zhejiang Univ, Med Sci. 2020;49(1):147–157. doi: 10.3785/j.issn.1008-9292.2020.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020;180(7):934–43. [DOI] [PMC free article] [PubMed]
- 11.National Health Commission of the People’s Republic of China, National Administration of Traditional Medicine. Diagnosis and treatment protocol for novel coronavirus pneumonia (trail version 7) [Internet]. Beijing: The State Council of the People’s Republic of China; 2020 Mar 3 [cited 2020 Mar 12]. Available from: http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989/files/ce3e6945832a438eaae415350a8ce964.pdf. Chinese.
- 12.Declaration of Helsinki—recommendations guiding medical doctors in biomedical research involving human subjects [Internet]. Ferney-Voltaire: The World Medical Assembly; c2020 [adopted 1964 Jun; revised 1975 Oct; cited 2020 Mar 12]. Available from: https://www.wma.net/wp-content/uploads/2018/07/DoH-Oct1975.pdf.
- 13.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Argenio V., Salvatore F. The role of the gut microbiome in the healthy adult status. Clin Chim Acta. 2015;451(Pt A):97–102. doi: 10.1016/j.cca.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Hooper L.V., Macpherson A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10(3):159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 16.O'Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Q., Yuan L., Deng J., Yang Q. Lactobacillus protects the integrity of intestinal epithelial barrier damaged by pathogenic bacteria. Front Cell Infect Microbiol. 2015;5:26. doi: 10.3389/fcimb.2015.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salazar N., Gueimonde M., de C.G., Los Reyes-Gavilán, Ruas-Madiedo P. Exopolysaccharides produced by lactic acid bacteria and bifidobacteria as fermentable substrates by the intestinal microbiota. Crit Rev Food Sci Nutr. 2016;56(9):1440–1453. doi: 10.1080/10408398.2013.770728. [DOI] [PubMed] [Google Scholar]
- 19.Andoh A., Tsujikawa T., Fujiyama Y. Role of dietary fiber and short-chain fatty acids in the colon. Curr Pharm Des. 2003;9(4):347–358. doi: 10.2174/1381612033391973. [DOI] [PubMed] [Google Scholar]
- 20.Louis P., Flint H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294(1):1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 21.Park J., Kim M., Kang S.G., Jannasch A.H., Cooper B., Patterson J. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8(1):80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swank G.M., Deitch E.A. Role of the gut in multiple organ failure: bacterial translocation and permeability changes. World J Surg. 1996;20(4):411–417. doi: 10.1007/s002689900065. [DOI] [PubMed] [Google Scholar]
- 24.Bäumler A.J., Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535(7610):85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ubeda C., Taur Y., Jenq R.R., Equinda M.J., Son T., Samstein M. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120(12):4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemon K.P., Armitage G.C., Relman D.A., Fischbach M.A. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4(137):137rv5. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wexler H.M. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20(4):593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 29.Choi V.M., Herrou J., Hecht A.L., Teoh W.P., Turner J.R., Crosson S. Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice. Nat Med. 2016;22(5):563–567. doi: 10.1038/nm.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haak B.W., Wiersinga W.J. The role of the gut microbiota in sepsis. Lancet Gastroenterol Hepatol. 2017;2(2):135–143. doi: 10.1016/S2468-1253(16)30119-4. [DOI] [PubMed] [Google Scholar]
- 31.Bartosch S., Fite A., Macfarlane G.T., McMurdo M.E. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Micro. 2004;70(6):3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.