Abstract
The pandemic of COVID-19 caused by SARS-CoV-2 has made serious threats to the public health. Antibodies have been considered as promising therapeutics for the prevention and treatment of pathogens. So far, effectors that can influence the sustainability of SARS-CoV-2 specific antibodies in COVID-19 patients are still unclear. In this paper, we attempted to find potential key factors correlated with SARS-CoV-2 specific antibodies. Transcriptional analysis with the peripheral blood mononuclear cells (PBMCs) revealed proportional changes of immune cell subsets in COVID-19 convalescent patients, including a substantial decrease of monocytes and evident increase of dendritic cells (DCs). Moreover, we found that the gene expressions of chemokines associated with monocyte/macrophage were significantly up-regulated during the COVID-19 recovery phase. Most importantly, we found a set of 27 immune genes corresponding to a comparatively lower amount of SARS-CoV-2 specific antibodies, and identified two hub genes, IL1β and IL6, the protein expressions of which exhibited negative correlation with the immunoglobulin G (IgG) levels in COVID-19 convalescent sera. In addition, we found that high expressions of these 2 hub genes during the convalescent stage were negatively associated with the plasma cell marker CD138. Our study presented two key inflammatory factors correlated to the low level of SARS-CoV-2 specific antibodies, which indicated the potential regulatory process of plasmatic antibodies levels in some COVID-19 convalescent patients.
Keywords: Antibodies, Convalescent stage, COVID-19, Cytokines, IL1β and IL6
Introduction
Pandemic outbreak of COVID-19, caused by severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), has become a global threat to the society with its intense infectivity.1, 2, 3 Antibody responses will provide protection from SARS-CoV-2 infection,4 which have aroused extensive attention of researchers. Recently, the validity duration of SARS-CoV-2 specific antibodies in COVID-19 patients have sparked widespread controversy. Several studies showed that the levels of IgG and neutralizing antibodies in a high proportion of individuals who recovered from SARS-CoV-2 infection started to decrease within 2–3 months after infection.5, 6, 8, 7 Moreover, cases of reinfection with SARS-CoV-2 also raised the concerns of how long the antibodies could provide effective protection.9 However, some other studies indicated that the levels of antibodies against SARS-CoV-2 sustained for at least 4 months post-infection.10,11 More information is needed to understand the validity duration of effective protection of neutralizing antibodies against SARS-CoV-2. Therefore, it is important to search for the key factors correlated with the levels of SARS-CoV-2 specific neutralizing antibodies in COVID-19 patients. Some studies focused on the duration of antibodies responses to influenza A virus or hepatitis E virus, however, no definite regulatory mechanism of antibodies was proposed.12,13 Recently, interleukin 6 (IL6) was showed to have impact on antibodies-secreting B cells in Salmonella infected mice, which shed some light on potential correlation between cytokines and antibody production.14
In this study, we compared the proportions of various immune cell subsets, the expressions of immune genes and the amount of SARS-CoV-2 specific antibodies in the PBMCs of COVID-19 patients. With WGCNA (Weighted correlation network analysis) and Gene Ontology (GO) enrichment analyses, we identified 2 hub genes associated with the low level of SARS-CoV-2 specific antibodies. The findings of this study provided updated knowledge for understanding the regulation process of functional antibodies (Abs) against SARS-CoV-2 in patients, and for potential optimizations of the vaccine development for COVID-19.
Materials and methods
Study design and ethics statement
This study was designed to explore potential factors associated with the amount of functional antibodies against SARS-CoV-2 in COVID-19 convalescent patients. A total of 15 peripheral blood samples from COVID-19 patients of various ages and disease severity, from Chongqing Medical University affiliated Yongchuan Hospital, were enclosed for the bulk ribonucleic acid sequencing (RNA-Seq). The detailed patients’ information was described in Table S1. For comparison, blood samples from 3 healthy donors were collected in Chongqing Medical University. The informed consent was obtained from all patients and healthy donors in this study.
PBMCs preparation and RNA extraction
The PBMCs were separated by Ficoll density gradient centrifugation method. Each peripheral blood sample (approximately 5 ml) was diluted with Phosphate-Buffered Saline (PBS) at 1:1 ratio, and then transferred to a Ficoll tube which contained 15 ml Ficoll buffer (PBS with 2% Fetal Bovine Serum). Samples were centrifuged at 1200×g for 10 min at room temperature (RT), and the supernatants were quickly transferred to a new 15 ml centrifuge tube, with fresh PBS added up to 15 ml. Then, samples were centrifuged at 300×g for 10 min at RT, and the pellets were washed twice with 10 ml PBS and again centrifuged at 300×g for 10 min to collect PBMCs. Total RNAs of PBMCs were extracted using TRIzol reagent (Life Technologies) according to the manufacturer's instruction.
RNA-seq NEB library
The first strand of cDNA was synthesized using the M-MULV reverse transcriptase system. Endogenous RNAs were degraded by RNaseH, and the second strand was synthesized by DNA polymerase I with dNTPs. After purification, double-stranded cDNA was repaired at the end, followed by the addition of a tail and the connection of sequencing adapters. AMPure XP beads were used to screen cDNAs. PCR amplification was performed and the PCR products of AMPure XP beads were again used to purify each cDNA. The shot-gun libraries were sequenced on an Illumina Novaseq 6000 and paired-end reads with 150 bp were generated.
Enzyme-linked immunosorbent assay (ELISA)
Recombinant spike protein 1 (S1) of SARS-CoV-2 (2 μg/ml, Sino Biological, Beijing, China) were coated on 384-well plates (Corning Costar), and incubated at 4 °C ovnernight. Then the plates were blocked with the blocking buffer (PBS with 5% BSA) at 37 °C for 1 h, and subsequently added with serially diluted monoclonal antibodies (10-fold dilutions, from 10 μg/ml to 10 pg/ml). After 30 min incubation at 37 °C, the plates were washed 5 times and incubated with goat anti-human IgG (H + L) antibody conjugated with ALP (Thermo Fisher, a18808, 1:5000) for 30 min at 37 °C. For the quantification of bound IgG, 1 mg/ml PNPP (Thermo Fisher) was added and the absorbance at 405 nm was measured by the MultiSkan GO fluoro-microplate reader (Thermo Fisher).
Data analysis
Trimmomatic was used for the pre-processing of the raw RNA-Seq reads. Clean reads were mapped to human reference genome sequences (GRCh38) using HISAT2 with the GRCh38/V34 annotation file on gencode (https://www.gencodegenes.org/human/). Reads count matrix was obtained by using featureCount, then differentially expression gene analysis was completed with R packages DEseq2 in R v3.6.3, following the standard analysis process (|log (FC)| >2 and false discovery rate P value < 0.05). WGCNA was finished with R package WGCNA, which was used to describe the gene association modes among different samples. Gene Ontology (GO) enrichment analysis was implemented by the clusterProfiler R package, in which gene length bias was corrected. GO terms with corrected P value less than 0.05 were considered significantly enriched by differential expressed genes. Correlation analysis was implemented with R package psych and corplot. CIBERSORTx (https://cibersortx.stanford.edu) is used to impute gene expression profiles and provide an estimation of the abundances of various cell types in a mixed cell population, using gene expression data.
Data access
Raw RNA-Seq data of the PBMCs from 3 COVID-19 patients, 15 COVID19 convalescent patients and 3 healthy donors were already deposited in the Genome Sequence Archive in BIG Data Center (https://bigd.big.ac.cn/) under the accession number: CRA002390 and HRA000520. Raw data of the PBMCs from 6 influenza A convalescent patients and those from 6 HCV convalescent patients were download from the NCBI GEO datasets under the accession number: GSE114588 and GSE119117.
Result
Changes of immune cell subsets in COVID-19 patients
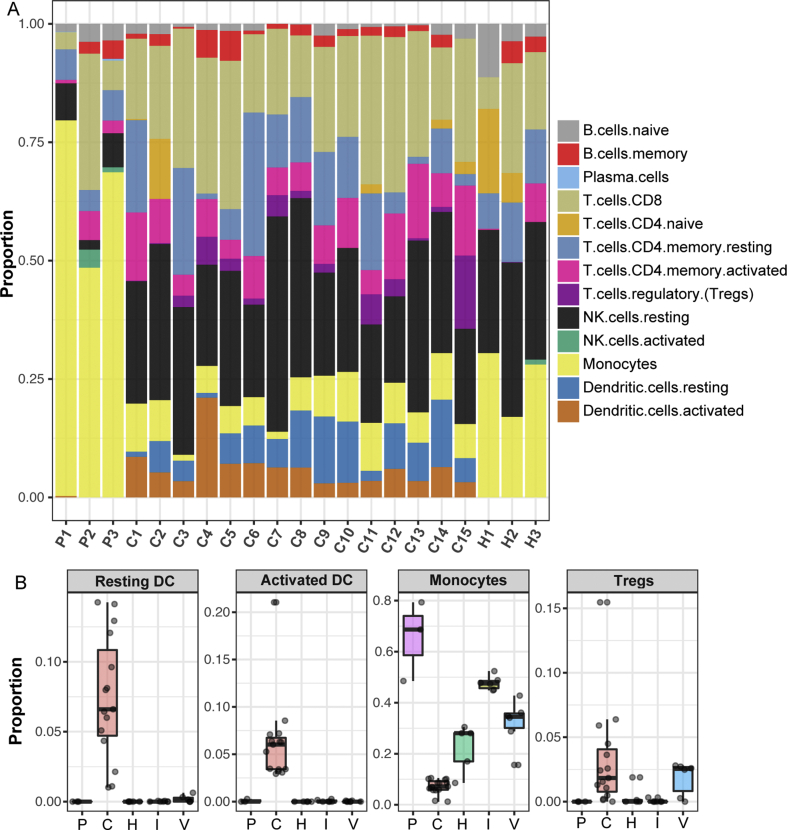
PBMCs samples from 15 COVID-19 convalescent patients (C1–C15) and 3 healthy donors (H1–H3) were separately applied for RNA-Seq. The detailed patients’ information was described in Table S1. For comparison, RNA-Seq data from the PBMCs samples of 3 COVID-19 acute patients (P1–P3) were downloaded from the BIG Data Center.15 After the differential gene expression analysis, we adapted a digital cytometry method16,17 (CIBRSORTx) to delineate the transcriptome matrix and translated it into the abundances of immune cell subsets in the PBMCs of different groups. As shown in Fig. 1A, the proportion of monocytes in the PBMCs from COVID-19 convalescent patients was lower than that from COVID-19 acute patients or from health donors, whereas the proportions of dendritic cells (DCs) increased drastically in the COVID-19 convalescent group, compared with the other two groups. In order to determine whether it was a unique immune response to SARS-CoV-2 infection, we enclosed additional RNA-Seq data from the PBMCs of influenza A convalescent patients (I1–I6) and hepatitis C Virus (HCV) convalescent patients (V1–V6), from the NCBI GEO datasets.18,19 The RNA-Seq data of 3 additional health donors (H4–H6) from the BIG Data Center15 was included. Comparative analysis revealed that the increase of the DC proportions, including both activated DCs and resting DCs, and the sharp decrease of the monocyte proportion were specific in COVID-19 convalescent patients (Fig. 1B). Meanwhile, we found that the proportion of regulatory T cells (Tregs) was elevated in the convalescent PBMCs of the SARS-CoV-2 and the HCV sets, indicating an induction of suppressive responses to these infections during the convalescent stages.
Figure 1.
Comparison of different immune cell subsets responding to COVID-19. (A) Proportions of different immune cell populations in the PBMCs of COVID-19 patients and health donors. (B) Four immune cell subsets with distinct proportional alterations in COVID-19 convalescent patients. P: COVID-19 acute patients; C: COVID-19 convalescent patients; H: health donors; I: influenza A convalescent patients; V: HCV convalescent patients.
The specific clusters of up-regulated immune genes in COVID-19 convalescent patients
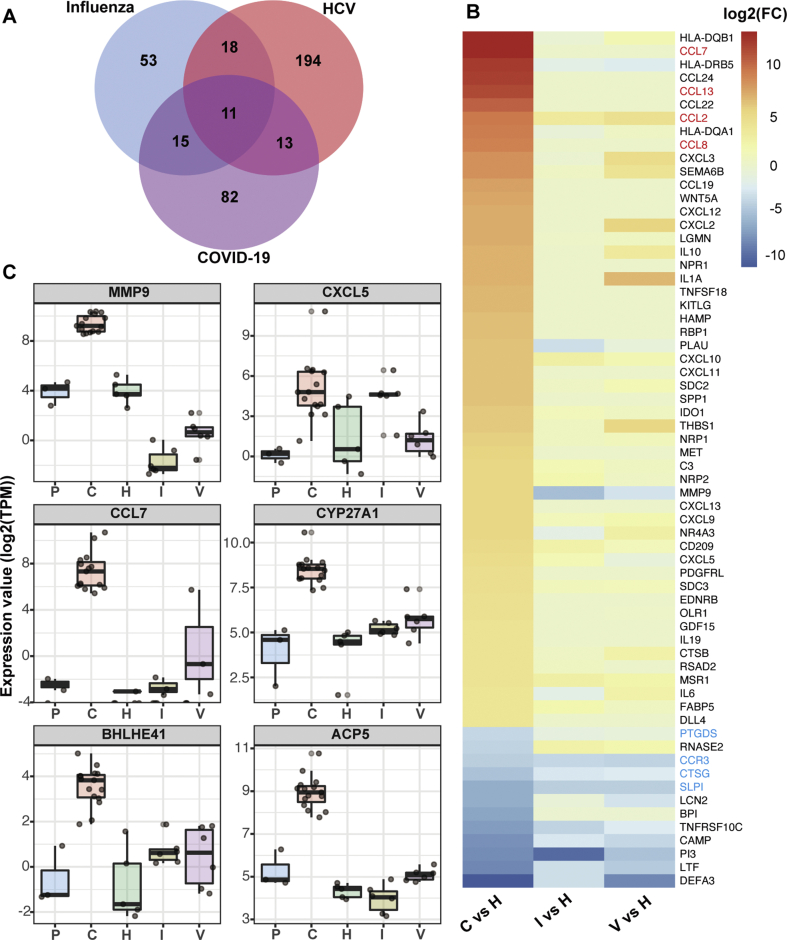
Following the standardized process of differential gene expressions analysis with R package DESeq2, we performed paired comparison of the immune gene profiles between the health donor (HD) group and the groups of COVID-19, influenza or HCV convalescent patients. Specially, we selected immune genes, as listed in the Immport database immune gene set,20 from differentially expressed genes (DEGs) of individual disease group. Compared to the HD group, the numbers of immune DEGs were 97, 121 and 236 in the influenza, COVID-19 and HCV group, respectively (Fig. 2A). We found that only 11 immune DEGs were shared by all three groups, and the majority of differentially expressed immune genes were rather specific to the type of infection (Fig. 2A). Next, we screened the 64 immune DEGs with significantly altered expressions in the PBMCs of COVID-19 convalescents (|log (FC)| > 4 and P value < 0.01), and ranked them according to the log (FC) values. Consistently, the heatmap results showed that a few down-regulated immune DEGs were common in all 3 groups, while most of the up-regulated genes were not overlapped between different groups (Fig. 2B).
Figure 2.
Assessment of immune gene expressions in COVID-19 convalescent patients. (A) Venn diagram showing overlaps and differences between immune DEGs in influenza, HCV and COVID-19 convalescent PBMCs. (B) Heatmap showing the fold change value of 64 immune genes in 3 disease groups. Immune DEGs shared in 3 groups were marked in blue, 4 MCP genes were labeled in red. (C) Comparison of 6 M0 stage macrophage-associated genes among different disease groups.
Interestingly, among the above 64 immune DEGs in the PBMCs of COVID-19 convalescents, 10 out of the top 15 up-regulated DEGs were chemokines (Fig. 2B). Furthermore, we found that 4 of these chemokines (CCL7, CCL2, CCL13 and CCL8) were monocyte chemotactic proteins (MCPs) that could recruit monocytes or macrophages to infected tissues (Fig. 2B). Six immune genes exhibited high expressions, including MMP9, CXCL5, CCL7, CYP27A, BHLHE41 and ACP5, which were found to be specifically associated with the early stage of macrophages (M0), according to the database of cell type-specific genes.16 Importantly, the high expressions of all these 6 genes were only found in the convalescent patients with SARS-CoV-2 infection, but not in those infected with influenza virus or HCV (Fig. 2C). These findings reflected the possibility that exceptionally large amount of monocytes were recruited to infected tissues and differentiated to macrophages during the COVID-19 convalescent stage.
A negative correlation between the expressions of specific cytokines and the amount of SARS-COV-2 specific antibodies
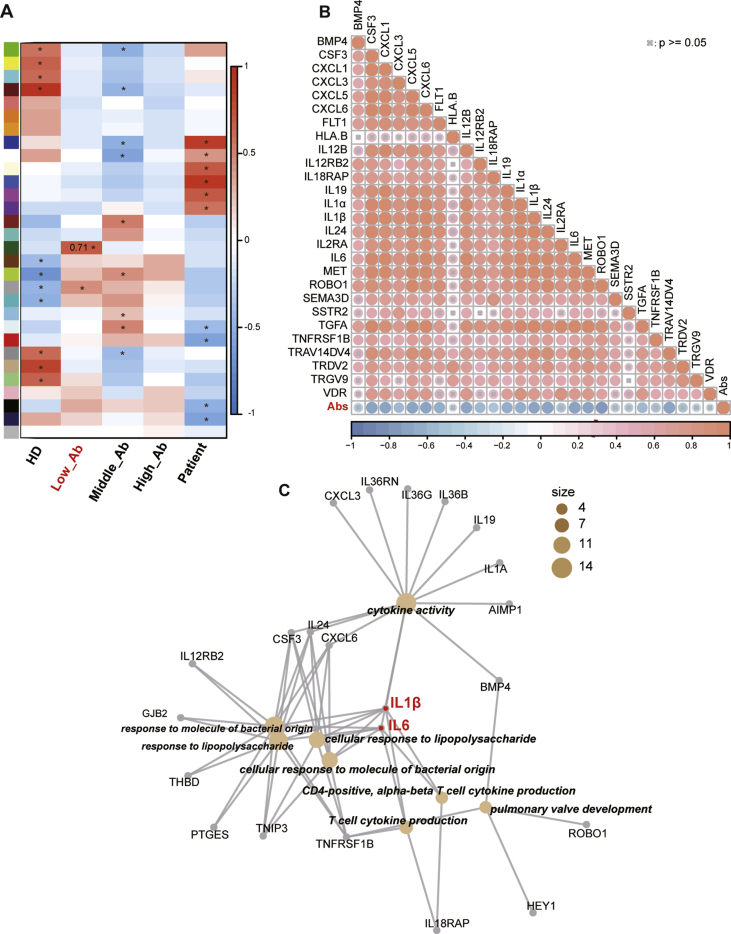
To detect the relative expression levels of the SARS-CoV-2 specific antibodies, the 15 COVID-19 convalescent patients’ plasma samples were subjected for the ELISA experiment against the SARS-CoV-2 recombinant Spike 1 protein (S1), with sequential dilutions (Fig. S1). And based on the levels of the SARS-CoV-2 specific antibodies in each plasma sample, we divided the 15 convalescent patients into 3 groups, termed as Low, Middle and High Abs, for the correlation assessment (Fig. S1). The weighted correlation network analysis (WGCNA) was performed with the RNA-Seq data, in order to find potential molecular candidates correlated with the amount of SARS-CoV-2 specific antibodies (Fig. S2). Specifically, we focused on gene sets associated with the group of Low Abs, and found that a specific cluster of 27 immune genes exhibited a significantly higher correlation coefficient with low antibody levels, compared to the other gene sets (Fig. 3A). The detailed information of these 27 immune genes was described in Table S3.
Figure 3.
Identification of hub immune genes associated with the low level of SARS-CoV-2 specific antibodies in COVID-19 convalescent patients. (A) WGCNA of gene expressions and the levels of SARS-CoV-2 specific antibodies (each color in left bar represent a gene set, ∗ means P < 0.05). (B) Correlation analysis between the expressions of 27 immune genes and the concentrations of SARS-CoV2 S1 specific antibodies. (C) GO enrichment analysis of 27 immune genes from the gene set with the most significant correlation to Low_Abs group. (Low_Ab/Middle_Ab/High_Ab: low/middle/high level group of IgG in convalescent PBMCs; HD: health donor group; Patient: COVID-19 patient group).
Furthermore, another correlation analysis was performed between these 27 immune genes and the S1-specific antibody expression, with 100-fold dilution. We found pan positive correlations among the gene expressions of these 27 cytokines with each other (Fig. 3B). And we confirmed that majority of them were negatively correlated with the SARS-CoV-2 S1 antibody levels (Fig. 3B). With the GO enrichment analysis, we found that these 27 genes participated in a variety of immune functions, such as cytokine productions, cytokine activities, the responses to external stimulus process, etc. Essentially, IL1β and IL6 were involved in almost all of these functional processes, thus defined as hub genes associated with the expression of SARS-CoV-2 specific antibodies, as listed in Figure 3C.
The expressions and correlations of the two hub genes with the plasma cell marker and MCPs
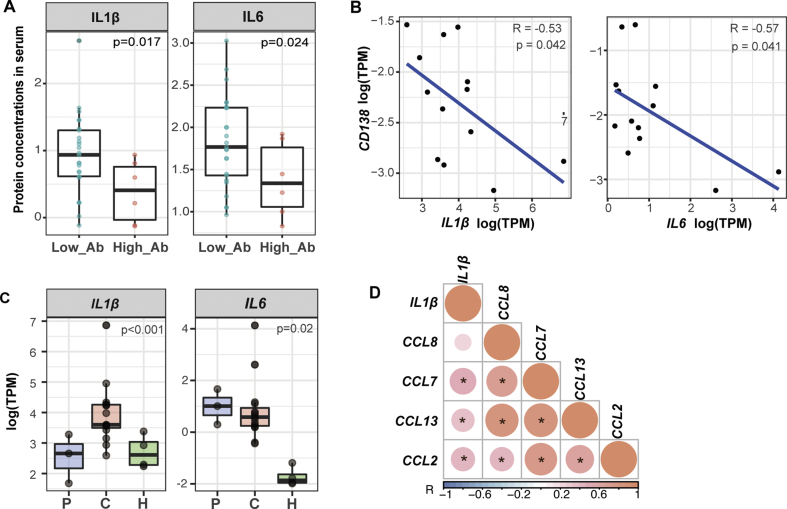
To confirm the association of the high expressions of hub genes with the low antibody levels, we collected additional publicly available data from COVID-19 convalescent patients (Table S4).5 Consistently, evidences from another cohort of 31 patients showed higher concentrations of both IL1β and IL6 in low Abs group than in high Abs group, which indicated a similar negative correlation between IL1β or IL6 gene expressions and the virus-specific IgG levels from the COVID-19 convalescent serum (Fig. 4A). These results suggested that high levels of IL1β and IL6 might imply a low level of SARS-CoV-2 specific antibodies in convalescent patients (Fig. S3). Furthermore, the relationships between these 2 hub genes and the antibody-secreting plasma cells were addressed by correlation analysis. Given the difficulty of detecting plasma cells in PBMCs samples, the plasma cell marker CD138 was used instead. We found that the gene expressions of both IL1β and IL6 were also negatively correlated with that of CD138 (Fig. 4B). Collectively, these results confirmed a close relationship between the two hub genes and SARS-CoV-2 specific antibodies.
Figure 4.
Correlations of IL1B or IL6 to different functional elements of COVID-19 convalescent stage. (A) IL1β or IL6 protein expressions and the SARS-CoV-2-S-specific IgG levels from the convalescent serum. (Low_Ab: low level group of IgG; High_Ab: high level group of IgG) (B) Correlation analysis between the gene expressions of IL1β or IL6 and CD138. (C) The expressions of IL1β and IL6, in COVID-19 acute patients, convalescent patients and health donors. P values among 3 groups were calculated by ANOVA test. (D) Correlation analysis between the gene expressions of IL1β and all 4 MCP family members (∗ means P < 0.05).
Given the importance of IL1β and IL6, group comparison analysis was applied with their gene expression levels in the PBMCs of COVID-19 acute patients, convalescent patients and health donors. Surprisingly, we found that IL1β was expressed at a higher level during the convalescent stage of COVID-19 than the acute phase, in contrast to other types of viral infections (Fig. 4C). Meanwhile, IL6 was found to be intensively expressed in the acute group, which was sustained during the convalescent phase, probably subsequent to the elevated level of IL1β.21 As IL1β mainly be produced by macrophages post stimulation,22 we examined the gene expressions of chemotactic proteins of macrophages. We found positive correlations between the gene expression of IL1β and CCL7, CCL13, CCL2 or CCL8, all of which are MCP family members (Fig. 4D). These results suggested that the previously speculated induction and activation of macrophages were highly likely to be the source of the elevated expression of IL1β during the COVID-19 convalescent stage.
Discussion
Our study revealed the proportional changes of immune cells in the PBMCs of COVID-19 convalescent patients, and the immune gene expressions correlated to different stages of this disease. Specifically, we found the gene set corresponding to a comparatively lower amount of SARS-CoV-2 specific antibodies, and identified two candidate hub genes, IL1β and IL6, which might be linked to the low level of antibodies in COVID-19 convalescent patients. Additionally, we found that IL1β and IL6 were highly expressed during the COVID-19 convalescent stage, with negative correlation to the plasma cell marker CD138.
As the first line of defense against infections, the innate immune cells, especially monocytes, are often rapidly expanded.9,23 The proportion of monocytes increased sharply in the PBMCs of COVID-19 patients,24, 25, 26 similar to the immune responses to other viral infections (Fig. 1B). However, different from the influenza or the HCV convalescent stages, the monocytes’ expansion was completely reversed during the COVID-19 convalescent stage, to a level even lower than that from healthy donors. This was accompanied by the increase of other immune cell populations. Meanwhile, many cytokines and chemokines were up-regulated with viral infections. Among them, all members of the MCP family, CCL7, CCL2, CCL13 and CCL8, were induced to relatively higher levels in the convalescent PBMCs (Fig. 2B). These findings supported the speculation that monocytes might undergo differentiations into macrophages during the recovery stage of COVID-19 progression.
Our recent reports have shown that most of the SARS-CoV-2 S1-specific antibodies exhibited neutralizing capability, which indicated that the expression levels of S1-specific IgG in the serum can represent the amount of neutralizing Abs.27,28 To explore factors associated with SARS-CoV-2 specific antibodies in COVID-19 convalescent sera, we detected the relative expression levels of Abs using recombinant Spike 1 protein. By WGCNA and GO enrichment analyses, we identified IL1β and IL6 as hub factors which were closely associated with the low level of SARS-CoV-2 specific antibodies (Fig. 3D). And we found both IL1β and IL6 were highly expressed during COVID-19 convalescent stage, with a negative correlation to the plasma cell marker CD138 (Fig. 4B). Therefore, we speculated that the negative correlations between the two hub genes and the low level of SARS-CoV-2 specific antibodies might be a result of reduced abundancy and/or inhibited functions of the plasma cells. As IL6 signaling is known to be crucial in plasma cell differentiation and long-term survival, excess IL6 may directly inhibit the functions of antibody-secreting plasma cells.29 Also, it has been reported that the combination of IL6 and IL1β can provide the necessary commitment signals for naïve B cells to regulatory B cells (Bregs),21,30 thereby limiting the probable number of plasma cells differentiated from the same naïve B cell reservoir. In addition, Bregs function through suppressing the immune responses by secreting inhibitory cytokines,21 such as IL35 and IL10. In turn, IL10 has been specifically linked to inhibit antibody productions, by reducing the expression of B cell activating factor (BAFF) and by impairing B cell differentiation upon mitogenic stimuli.31 In parallel, IL10 may induce the expansion of the Treg population, which can aggravate the immune inhibitory reactions.32, 33, 34 As a collective consequence, these factors and the chain of events can probably lead to a decrease of antibody-secreting plasma cells, which might be a possible explanation for the reduced amount of SARS-CoV-2 specific antibodies in COVID-19 convalescent patients.
Moreover, we demonstrated a potential expansion of the macrophage population in COVID-19 convalescent patients. We also identified a positive correlation between the gene expression of IL1β and 4 MCPs, which suggested that the induction or activation of macrophages were highly likely to be the source of the elevated expression of IL1β during the COVID-19 convalescent stage. Furthermore, the increasing of IL1β might, in turn, contribute to the sustained high levels of its downstream effector IL6 in the convalescents.21,35,36 Therefore, monocytes/macrophages might also be involved in the down-regulation of SARS-CoV-2 specific antibodies in COVID-19 convalescents, probably through the two hub genes.
Taken together, our study identified two hub genes which indicated that a potential transition of monocytes to macrophages during the COVID-19 convalescent stage, which was accompanied by drastically shifted expressions of correlated cytokines and chemokines. Such interconnectivity of the cellular and molecular components in the innate immune system might contribute to the reduction of SARS-CoV-2 specific antibodies. Our findings might shed some light on the potential mechanisms and the development of corresponding interventions to amend the validity duration of neutralizing antibodies in some COVID-19 patients.
Author contributions
A.J., S.L. and J.W. conceived and designed the study. L.D. and Y.N. offered help on collection of convalescent patient blood samples and preparation of PBMCs. T.L. completed the ELISA experiments, X.H., C.H., Y.M. and M.S. helped on data analysis. S.L., W.W. and J.W. generated figures and tables, and also wrote the manuscript.
Conflict of interests
We declare no competing financial interest.
Acknowledgements
We acknowledge the work and contribution of blood sample providers from Chongqing Medical University affiliated Yongchuan Hospital and the third affiliated Hospital of Chongqing Medical University. This study was sponsored by Natural Science Foundation of Chongqing, China (No. cstc2020jcyj-bshX0027), with the donation from Mr Yuling Feng.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.12.007.
Contributor Information
Jianwei Wang, Email: jianweiwangcn@cqmu.edu.cn.
Aishun Jin, Email: aishunjin@cqmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chen N., Zhou M., Dong X., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummings M.J., Baldwin M.R., Abrams D., et al. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet. 2020;395(10239):1763–1770. doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long Q.X., Liu B.Z., Deng H.J., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 5.Long Q.X., Tang X.J., Shi Q.L., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 6.Seow J., Graham C., Merrick B., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5(12):1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X., Guo X., Xin Q., et al. Neutralizing antibody responses to severe acute respiratory syndrome coronavirus 2 in coronavirus disease 2019 inpatients and convalescent patients. Clin Infect Dis. 2020;71(10):2688–2694. doi: 10.1093/cid/ciaa721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckerle I., Meyer B. SARS-CoV-2 seroprevalence in COVID-19 hotspots. Lancet. 2020;396(10250):514–515. doi: 10.1016/S0140-6736(20)31482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tillett R.L., Sevinsky J.R., Hartley P.D., et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21(1):52–58. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudbjartsson D.F., Norddahl G.L., Melsted P., et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. 2020;383(18):1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripperger T.J., Uhrlaub J.L., Watanabe M., et al. Orthogonal SARS-CoV-2 serological assays enable surveillance of low prevalence communities and reveal durable humoral immunity. Immunity. 2020;53(5):925–933. doi: 10.1016/j.immuni.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride J.M., Lim J.J., Burgess T., et al. Phase 2 randomized trial of the safety and efficacy of MHAA4549A, a broadly neutralizing monoclonal antibody, in a human influenza A virus challenge model. Antimicrob Agents Chemother. 2017;61(11) doi: 10.1128/AAC.01154-17. e01154–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyuregyan K.K., Potemkin I.A., Lopatukhina M.A., et al. The duration of preservation of anamnestic antibodies to hepatitis E virus. Klin Lab Diagn. 2018;63(5):310–314. doi: 10.18821/0869-2084-2018-63-5-310-314. [DOI] [PubMed] [Google Scholar]
- 14.Barr T.A., Brown S., Mastroeni P., Gray D. TLR and B cell receptor signals to B cells differentially program primary and memory Th1 responses to Salmonella enterica. J Immunol. 2010;185(5):2783–2789. doi: 10.4049/jimmunol.1001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong Y., Liu Y., Cao L., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman A.M., Liu C.L., Green M.R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenberg B.R., Depla M., Freije C.A., et al. Longitudinal transcriptomic characterization of the immune response to acute hepatitis C virus infection in patients with spontaneous viral clearance. PLoS Pathog. 2018;14(9) doi: 10.1371/journal.ppat.1007290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Forst C.V., Gordon A., et al. Characterization of antibiotic resistance and host-microbiome interactions in the human upper respiratory tract during influenza infection. Microbiome. 2020;8(1):39. doi: 10.1186/s40168-020-00803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya S., Andorf S., Gomes L., et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58(2–3):234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 21.Rosser E.C., Oleinika K., Tonon S., et al. Regulatory B cells are induced by gut microbiota-driven interleukin-1β and interleukin-6 production. Nat Med. 2014;20(11):1334–1339. doi: 10.1038/nm.3680. [DOI] [PubMed] [Google Scholar]
- 22.Dinarello C.A. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281(1):8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouayad A. Innate immune evasion by SARS-CoV-2: comparison with SARS-CoV. Rev Med Virol. 2020;30(6):1–9. doi: 10.1002/rmv.2135. [DOI] [PubMed] [Google Scholar]
- 24.Yan Q., Li P., Ye X., et al. Longitudinal peripheral blood transcriptional analysis of COVID-19 patients captures disease progression and reveals potential biomarkers. MedRxiv. 2020 doi: 10.1101/2020.05.05.20091355. Submitted for publication. [DOI] [Google Scholar]
- 25.Jingyi Y., Zhong M., Zhang E., et al. Broad phenotypic alterations and potential dysfunctions of lymphocytes in COVID-19 recovered individuals. MedRxiv. 2020 doi: 10.1101/2020.07.01.20144030. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han X.J., Wang Y., Li S., Tang N., Jin A. A rapid and efficient screening system for neutralizing antibodies and its application for the discovery of potent neutralizing antibodies to SARS-CoV-2 S-RBD. BioRxiv. 2020 doi: 10.1101/2020.08.19.253369. Submitted for publication. [DOI] [Google Scholar]
- 28.Li T., Han X., Wang Y., Gu C., Jin A. A key linear epitope for a potent neutralizing antibody to SARS-CoV-2 S-RBD. BioRxiv. 2020 doi: 10.1101/2020.09.11.292631. Submitted for publication. [DOI] [Google Scholar]
- 29.Nguyen D.C., Joyner C.J., Sanz I., Lee F.E. Factors affecting early antibody secreting cell maturation into long-lived plasma cells. Front Immunol. 2019;10:2138. doi: 10.3389/fimmu.2019.02138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter C.A., Jones S.A. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16(5):448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 31.Palmer V.L., Worth A.N., Scott R.L., et al. IL10 restrains autoreactive B cells in transgenic mice expressing inactive RAG1. Cell Immunol. 2018;331:110–120. doi: 10.1016/j.cellimm.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Garra A., Chang R., Go N., Hastings R., Haughton G., Howard M. Ly-1 B (B-1) cells are the main source of B cell-derived interleukin 10. Eur J Immunol. 2010;22(3):711–717. doi: 10.1002/eji.1830220314. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto M., Baba A., Yokota T., et al. Interleukin-10-producing plasmablasts exert regulatory function in autoimmune inflammation. Immunity. 2014;41(6):1040–1051. doi: 10.1016/j.immuni.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 34.Sattler S., Ling G.S., Xu D., et al. IL-10-producing regulatory B cells induced by IL-33 (BregIL-33) effectively attenuate mucosal inflammatory responses in the gut. J Autoimmun. 2014;50(100):107–122. doi: 10.1016/j.jaut.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose-John S. Interleukin-6 family cytokines. Cold Spring Harb Perspect Biol. 2018;10(2) doi: 10.1101/cshperspect.a028415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taniguchi K., Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin Immunol. 2014;26(1):54–74. doi: 10.1016/j.smim.2014.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.