Abstract
In the wake of COVID-19 disease, caused by the SARS-CoV-2 virus, we designed and developed a predictive model based on Artificial Intelligence (AI) and Machine Learning algorithms to determine the health risk and predict the mortality risk of patients with COVID-19. In this study, we used a dataset of more than 2,670,000 laboratory-confirmed COVID-19 patients from 146 countries around the world including 307,382 labeled samples. This study proposes an AI model to help hospitals and medical facilities decide who needs to get attention first, who has higher priority to be hospitalized, triage patients when the system is overwhelmed by overcrowding, and eliminate delays in providing the necessary care. The results demonstrate 89.98% overall accuracy in predicting the mortality rate. We used several machine learning algorithms including Support Vector Machine (SVM), Artificial Neural Networks, Random Forest, Decision Tree, Logistic Regression, and K-Nearest Neighbor (KNN) to predict the mortality rate in patients with COVID-19. In this study, the most alarming symptoms and features were also identified. Finally, we used a separate dataset of COVID-19 patients to evaluate our developed model accuracy, and used confusion matrix to make an in-depth analysis of our classifiers and calculate the sensitivity and specificity of our model.
Keywords: COVID-19, Coronavirus, Machine learning, Predictive analytics, Data analytics
1. Introduction
In late 2019, a novel form of Coronavirus, named SARS-CoV-2 (stands for Severe Acute Respiratory Syndrome Coronavirus 2), started spreading in the province of Hubei in China, and claimed numerous human lives (Li et al., 2020, Xu et al., 2020). In January 2020, the World Health Organization (WHO) declared the novel coronavirus outbreak a Public Health Emergency of International Concern (PHEIC) (Bogoch et al., 2020; World Health Organization). In February 2020, WHO selected an official name, COVID-19 (stands for Coronavirus Disease 2019), for the infectious disease caused by the novel coronavirus, and later in March 2020 declared a COVID-19 Pandemic (World Health Organization;WHO Director General).
Coronavirus is a family of viruses that usually causes respiratory tract disease and infections that can be fatal in some cases such as in SARS, MERS, and COVID-19. Some kinds of coronavirus can affect animals, and sometimes, on rare occasions, coronavirus jumps from animal species into the human population. The novel coronavirus might have jumped from an animal species into the human population, and then begun spreading. A recent study has shown that once the coronavirus outbreak starts, it will take less than four weeks to overwhelm the healthcare system. Once the hospital capacity gets overwhelmed, the death rate jumps (McConghy et al., 2020).
Artificial Intelligence (AI) has been shown to be an effective tool in predicting medical conditions and adverse events, and help caregivers with medical decision-making (Kalatzis et al., 2018; Chang, Chang, & Pourhomayoun, 2019, Kwon, 2018, Yoo et al., 2018). In this study, we proposed a data-driven predictive analytics algorithm based on Artificial Intelligence (AI) and machine learning to determine the health risk and predict the mortality risk of patients with COVID-19. The developed system can help hospitals and medical facilities decide who needs to get attention first, who has higher priority to be hospitalized, triage patients when the system is overwhelmed by overcrowding, and eliminate delays in providing the necessary care. The algorithm predicts the mortality risks based on patients’ physiological conditions, symptoms, and demographic information.
The proposed system includes a set of algorithms for preprocessing the data to extract new features, handling missing values, eliminating redundant and useless data elements, and selecting the most informative features. After preprocessing the data, we use machine learning algorithms to develop a predictive model to classify the data, predict the medical condition, and calculate the probability and risk of mortality. The processed dataset and code have been released in hope to benefit the research community1 .
The rest of this paper is organized as follows: in section 2, we will introduce the different methods and model architecture. Discuss each method by providing detailed information about the model, data preprocessing, and challenges that we encountered and the steps to mitigate these challenges, feature selection, and feature extraction. Section 3 provides the results with various approaches and metrics. Section 4, 5 includes the discussion and conclusion.
2. Methods
2.1. Dataset
In this paper, we used a dataset of more than 2,670,000 laboratory-confirmed COVID-19 patients from 146 countries around the world (Xu et al., 2020), including 307,382 labeled samples containing both male and female patients with an average age of 44.75 (Xu et al., 2020). The disease confirmed by detection of virus nucleic acid (Xu et al., 2020). The original dataset contained 32 data elements from each patient, including demographic and physiological data. At the data cleaning stage, we removed useless and redundant data elements such as data source, admin id, and admin name. We have also removed the unlabeled data samples. Then, data imputation techniques including mean/median/mode value replacement and KNN technique were used to handle missing values.
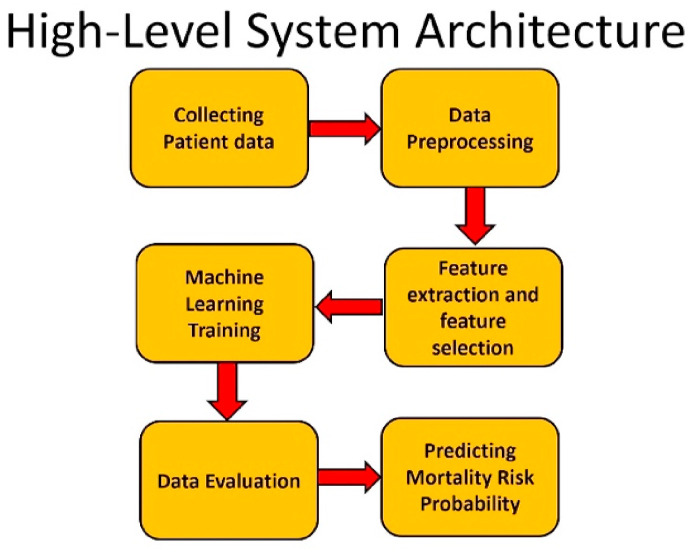
To have an accurate and unbiased model, we made sure that our dataset is balanced. A balanced dataset with equal number of observations for both recovered and deceased patients was created to train and test our model. The data samples (patients) in the training dataset have been selected randomly and they are completely separate from the testing data. Fig. 1 shows a high-level architecture of our system.
Fig. 1.
High-level system architecture.
2.2. Feature selection
The outcome label contained multiple values explaining the patient's health status. We considered patients that were discharged from hospital or patients in stable situation with no more symptoms as recovered patients. The symptoms were recorded by healthcare officials at the time of admission to the hospital. A total of 112 features were extracted from the original dataset including symptoms, doctors' medical notes, demographics, and physiological information. We consulted with a medical team to make sure that all of the relevant features are extracted.
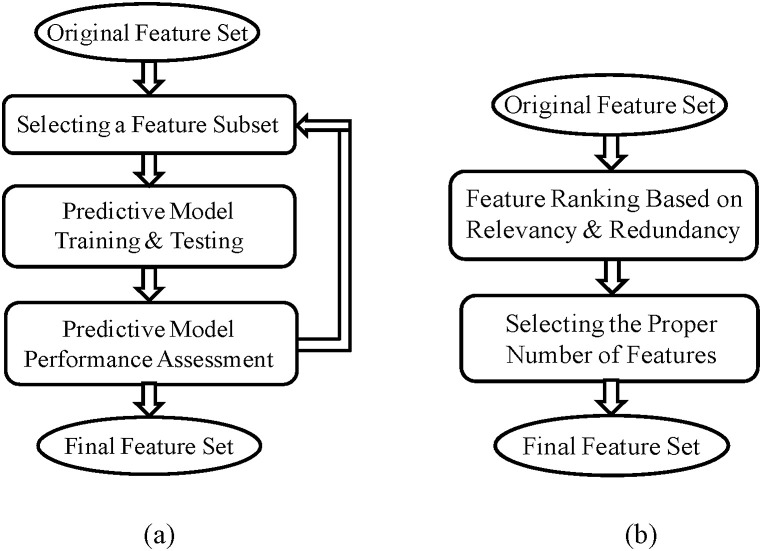
The next step is feature selection. The primary purpose of feature selection is to find the most informative features and eliminate redundant data to reduce the dimensionality and complexity of the model (Pourhomayoun, Nemati, Mortazavi, & Sarrafzadeh, 2015). We used univariate and multivariate filter method and wrapper method to rank the features and select the best feature subset (Pourhomayoun, 2014.). Fig. 2 demonstrates the steps of filter and wrapper method that we used for feature selection.
Fig. 2.
Feature Selection: (a)Wrapper method, (b)Filter method.
Filter methods are very popular (especially for large datasets) since they are usually very fast and much less computationally intensive than wrapper methods. Filter methods use a specific metric to score each individual feature (or a subset of features together). The most popular metrics used in filter methods include correlation coefficient, Fisher score, mutual information, entropy and consistency and chi-square parameters (Pourhomayoun, Nemati, Mortazavi, & Sarrafzadeh, 2015).
After applying different filter and wrapper methods, we chose 57 features out of 112 features. The list of final features is available in Table 1 . The selected features can be categorized into 3 groups: symptoms, pre-existing conditions (or comorbidities), and demographics.
Table 1.
The list of features used in the machine learning algorithm.
| Symptoms | o anorexia | o fever | o shortness of breath |
| o chest pain | o gasp | o somnolence | |
| o chills | o headache | o sore throat | |
| o conjunctivitis | o kidney failure | o sputum | |
| o cough | o lesions on chest radiographs | o septic shock | |
| o diarrhea | o hypertension | o Heart attack | |
| o dizziness | o Myalgia | o cold | |
| o dyspnea | o obnubilation | o cardiac disease | |
| o emesis | o pneumonia | o hypoxia | |
| o expectoration | o myelofibrosis | o fatigue | |
| o eye irritation | o respiratory distress | o rhinorrhea | |
| Pre-existing Conditions | o diabetes | o COPD | o coronary heart disease |
| o hypertensio n |
o Parkinson's disease | o prostate hypertrophy | |
| o chronic kidney disease | o asthma | o Tuberculosis | |
| o hypothyroidism | o cancer | o hepatitis B | |
| o cerebral infarction | o HIV positive | o chronic bronchitis | |
| o cardiac disease | o dyslipidemia | o any chronic disease | |
| Demographics | o age | o country | o province |
| o gender | o city | o travel history |
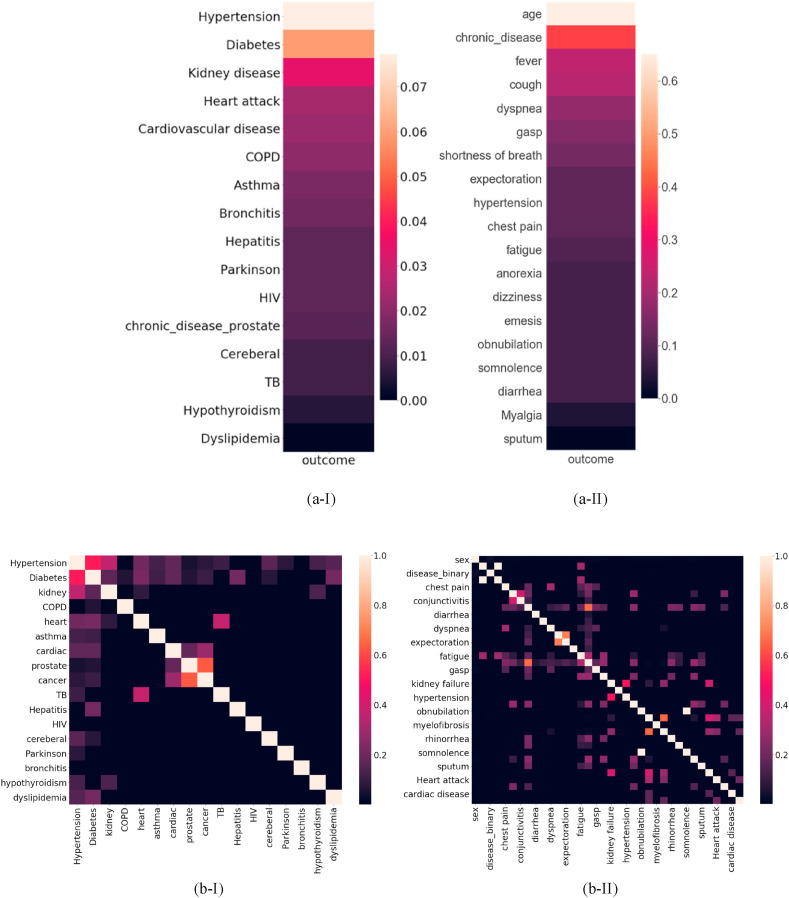
Fig. 3 shows the Correlation Heatmap for dataset features. Fig. 3-(a) shows the correlation between features and the outcome i.e. mortality risk, and Fig. 3-(b) shows the correlation between the features themselves. As Fig. 3-(a) illustrates, some demographic features like age and gender and symptoms like respiratory distress, and pre-existing conditions like diabetes, hypertension and kidney disease are the top features with high correlation to the patient's mortality risk. Hypertension is the major risk factor for heart disease, Chronic Kidney Disease (CKD) and diabetes. On the other hand, CKD is both a common cause of hypertension and also a complication of uncontrolled hypertension (Hamrahian et al., 2016)-(Cheung & Li, 2012). These correlations are clearly demonstrated in Fig. 3-.
Fig. 3.
(a) Correlation heatmap for the most correlated features to the mortality risk: (a-I) Chronic diseases (pre-existing conditions); (a-II) Symptoms. (b) Correlation heatmap for the correlation between features: (b-I) Chronic diseases (pre-existing conditions); (b-II) Symptoms.
2.3. Predictive analytics algorithms
After selecting the best feature subset, we used various machine learning algorithms to build a predictive model. In this research, we used different algorithms including Support Vector Machine (SVM), Neural Networks, Random Forest, Decision Tree, Logistic Regression, and K-Nearest Neighbor (KNN) (Cortes & Vapnik, 1995; Vapnik, 1995; Breiman, 2001).
The Neural Network algorithm achieved the best performance and accuracy. We used grid search to find the best hyperparameters for the neural network. We searched for the following hyperparameters: the number of layers and neurons in each layer (in the range of 3–50), activation functions (‘relu','logistic’), regularization rate, and batch size. The best neural network results were achieved with two hidden layers with 40 neurons in the first layer and 3 neurons in the second layer. We used stochastic gradient optimizer, constant learning rate and the regularization rate of alpha = 0.01.
The SVM model was configured with linear kernel, and regularization parameter C = 1.0. The Random Forest algorithm is an ensemble learning method combined of multiple decision tree predictors that are trained based on random data samples and feature subsets (Breiman, 2001). We configured the random forest algorithm with 20 trees in the forest.
2.4. Evaluation
We used 10-fold random cross-validation (with no overlap, with no replacement) to evaluate the developed model. We calculated the Overall Accuracy for all machine learning algorithms to compare. Also, we generated Receiver Operating Characteristic (ROC) curves for every algorithm, and calculated the Area Under Curve (AUC) and Confusion Matrix. Again, we made sure that there is no overlap (no common patient) between training and testing datasets at any level. We have also performed another feature selection during the cross validation and only on the training data to confirm the results, and the selected features match the original feature selection. The next section will provide the results and performance of the developed system.
3. Results
As explained in section II, several metrics such as Accuracy, ROC, AUC, and Confusion Matrix have been used to evaluate the developed model. Table 2 demonstrates the prediction accuracy for predicting mortality in patients with COVID-19 using 10-fold cross-validation for various machine learning algorithms.
Table 2.
The accuracy of mortality prediction in patients with COVID-19 using 10-fold cross-validation.
| Neural Network using 10-fold cross-validation | 89.98% |
|---|---|
| KNN using 10-fold cross-validation | 89.83% |
| SVM using 10-fold cross-validation | 89.02% |
| Random Forest using 10-fold cross-validation | 87.93% |
| Logistic Regression using 10-fold cross-validation | 87.91% |
| Decision Tree using 10-fold cross-validation | 86.87% |
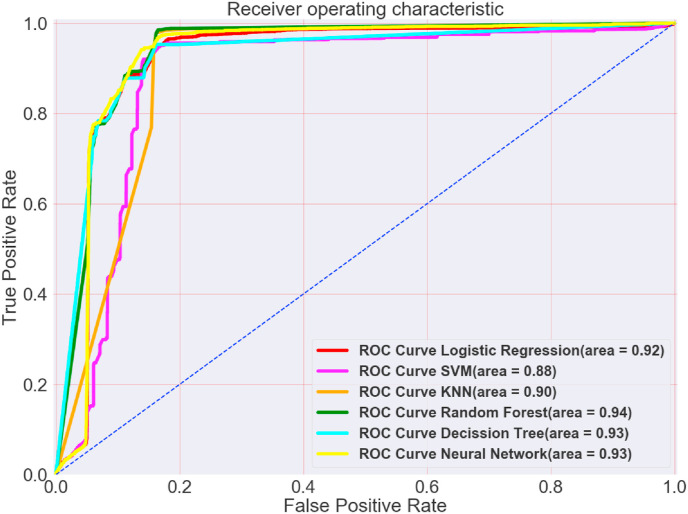
Fig. 4 demonstrates and compares the ROC curves and AUC for every machine learning algorithm that was used in this research.
Fig. 4.
ROC Curve comparison for all algorithms.
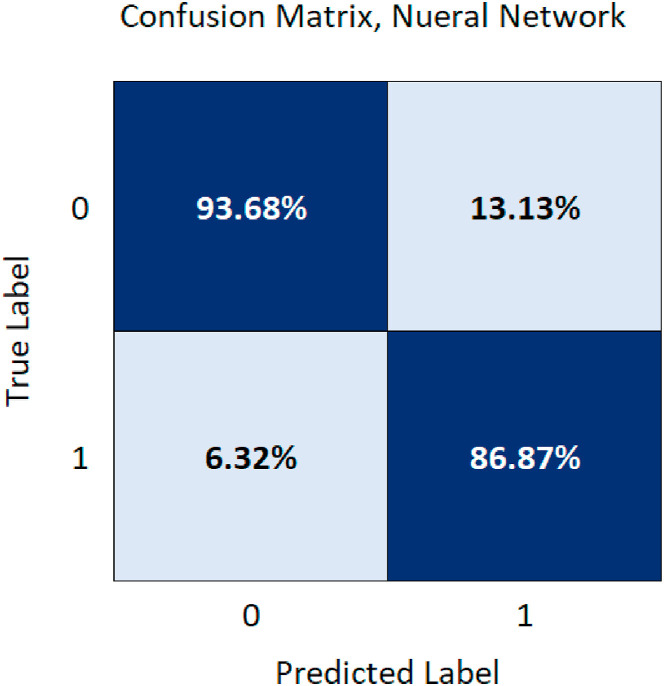
A confusion matrix (Fig. 5 ) is used to describe and visualize the performance of the Neural Network algorithm classifier and also to provide insight on what the model misclassifies. The sensitivity and specificity of the model were calculated using the confusion matrix.
where TP: True positive, TN: True negative, FP: False positive, and FN: False negative.
Fig. 5.
Neural Network confusion matrix for mortality prediction.
The results demonstrate that the developed algorithm is able to accurately predict the mortality risk in patients with COVID-19 based on the patients’ physiological conditions, symptoms, and demographic information. Fig. 6 shows the mortality risk (the probability of death) predicted by the algorithm for sample patients.
Fig. 6.
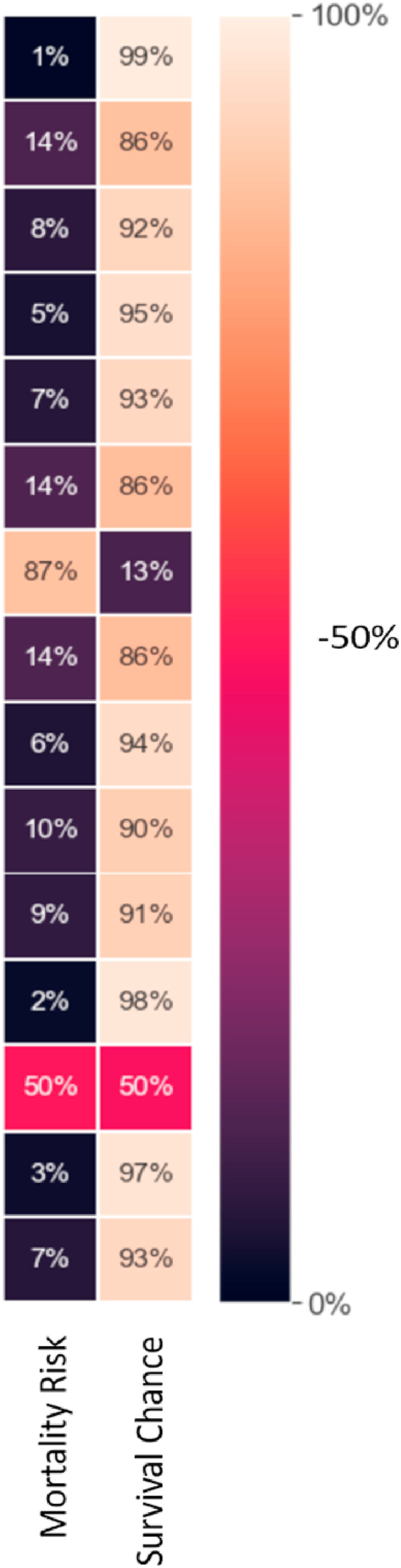
Sample results for predicting the risk of mortality (the probability of death).
4. Discussion
In this study, we processed a large dataset of COVID-19 confirmed cases collected from all around the world, and used state of the art machine learning algorithms to predict the mortality rate for patients with COVID-19. We evaluated the developed algorithms using several different metrics. The evaluation results demonstrate high accuracy and the effectiveness of the developed models.
There are other studies that have shown promising results for predicting mortality rate in COVID-19 patients using blood lab results and clinical data (Yan, 2020). However, in our study, we focused on demographic information, physiological data, patient's symptoms, and pre-existing conditions. We reached an outstanding accuracy of 89.98% using neural network model.
Furthermore, as previous studies mostly focused on data collected from China (Yan, 2020; Du, 2020), we used the hospital data from all around the world to create a more comprehensive model that is applicable to the world population, and is not trained only based on the data of one particular region.
5. Conclusion
The purpose of this study was to create a predictive algorithm to help hospitals and medical facilities maximize the number of survivors by providing an accurate and reliable tool to help medical decision making and triage COVID-19 patients more effectively and accurately during the pandemic. Our algorithm is able to predict the mortality risk in patients with COVID-19 with high accuracy using the patients’ physiological conditions, symptoms, pre-exiting conditions, and demographic information. This system can help hospitals, medical facilities, and caregivers decide who needs to get attention first before other patients, triage patients when the system is overwhelmed by overcrowding, and also eliminate delays in providing the necessary care. This study could expand to other diseases to help the healthcare system responds more effectively during an outbreak or a pandemic.
Credit author statement
Mohammad Pourhomayoun: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Supervision, Project administration. Mahdi Shakibi: Software, Validation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing, Visualization, Resources, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
References
- Breiman L. Machine Learning; 2001. Random forests. [Google Scholar]
- Cheung B.M.Y., Li C. Diabetes and hypertension: Is there a common metabolic pathway? Current Atherosclerosis Reports. 2012;14:160–166. doi: 10.1007/s11883-012-0227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C., Vapnik V. Machine Learning. Springer; 1995. Support-Vector Networks; pp. 273–297. [Google Scholar]
- Chang Daniel, Chang David, Pourhomayoun Mohammad. IEEE; 2019. “Risk prediction of critical vital signs for ICU patients using recurrent neural network,” the 2019 international conference on computational science and computational intelligence. [Google Scholar]
- Rong-Hui. Du, et al. DOI: https://doi.org/10.1183/13993003.00524-2020.
- Hamrahian S.M., Falkner B. In: Islam M.S., editor. Vol. 956. Springer; Cham: 2016. Hypertension in chronic kidney disease. (Hypertension: From basic research to clinical practice. Advances in experimental medicine and biology). [DOI] [PubMed] [Google Scholar]
- Bogoch I., Watts A., Thomas-Bachli A., Huber C., Kraemer M.U.G., Khan K. Pneumonia of unknown aetiology in wuhan, China: Potential for international spread via commercial air travel. Journal of Travel Medicine. 2020;27(Issue 2) doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalatzis A., Mortazavi B., Pourhomayoun M. Las Vegas; Dec 2018. “Interactive dimensionality reduction for improving patient Adherence in remote health monitoring,” the 2018 international conference on computational science and computational intelligence (CSCI'18) [Google Scholar]
- Li Q., et al. Early transmission dynamics in wuhan, China, of novel coronavirus–infected pneumonia. New England Journal of Medicine. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConghy T., Pon B., Anderson E. 2020. “When does hospital capacity get overwhelmed in USA? Germany? A model of beds needed and available for coronavirus patients” trent.st. [Google Scholar]
- World Health Organization . WHO; Jan 2020. Statement on the second meeting of the International Health Regulations Emergency committee regarding the outbreak of novel coronavirus (2019-nCoV), world health organization (WHO). Archived from the original on 31 january 2020. [Google Scholar]
- Pourhomayoun M. Multiple model analytics for adverse event prediction in remote. Healthcare Innovation & Point-of-Care Technologies; 2014. [Google Scholar]
- Kwon M., et al. Multi-label Classification of Single and Clustered Cervical Cells Using Deep Convolutional Networks. The 14th Int. Conference on Data Science. 2018;(ICDATA’18),:2018. [Google Scholar]
- Pourhomayoun M., Nemati E., Mortazavi B, Sarrafzadeh M., "Context-Aware Data Analytics for Activity Recognition," International Conference on Data Analytics, DATA ANALYTICS 2015, July 19 - 24, 2015.
- WHO Director General . WHO Director-General’s opening remarks at the media briefing on COVID-19, World Health Organization (WHO) World Health Organization (WHO); March 2020. [Google Scholar]
- Xu B., Gutierrez B., Mekaru S., et al. Epidemiological data from the COVID-19 outbreak, real-time case information. Nature Sci Data. 2020;7:106. doi: 10.1038/s41597-020-0448-0. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik V. The nature of statistical learning theory. Springer; 1995. [Google Scholar]
- L. Yan, et al. https://doi.org/10.1101/2020.02.27.20028027.
- Yoo S., Kalatzis A., Amini N., Pourhomayoun M. The 8th ACM MobiHoc2018 Workshop on Pervasive Wireless Healthcare; 2018. Interactive predictive analytics for enhancing patient Adherence in remote health monitoring. [Google Scholar]