Abstract
Autophagy acts as a cellular surveillance mechanism to combat invading pathogens. Viruses have evolved various strategies to block autophagy and even subvert it for their replication and release. Here, we demonstrated that ORF3a of the COVID-19 virus SARS-CoV-2 inhibits autophagy activity by blocking fusion of autophagosomes/amphisomes with lysosomes. The late endosome-localized ORF3a directly interacts with and sequestrates the homotypic fusion and protein sorting (HOPS) component VPS39, thereby preventing HOPS complex from interacting with the autophagosomal SNARE protein STX17. This blocks assembly of the STX17-SNAP29-VAMP8 SNARE complex, which mediates autophagosome/amphisome fusion with lysosomes. Expression of ORF3a also damages lysosomes and impairs their function. SARS-CoV-2 virus infection blocks autophagy, resulting in accumulation of autophagosomes/amphisomes, and causes late endosomal sequestration of VPS39. Surprisingly, ORF3a from the SARS virus SARS-CoV fails to interact with HOPS or block autophagy. Our study reveals a mechanism by which SARS-CoV-2 evades lysosomal destruction and provides insights for developing new strategies to treat COVID-19.
Keywords: COVID-19, SARS-CoV-2, ORF3a, SNARE, HOPS, DMV, autophagy
Graphical Abstract
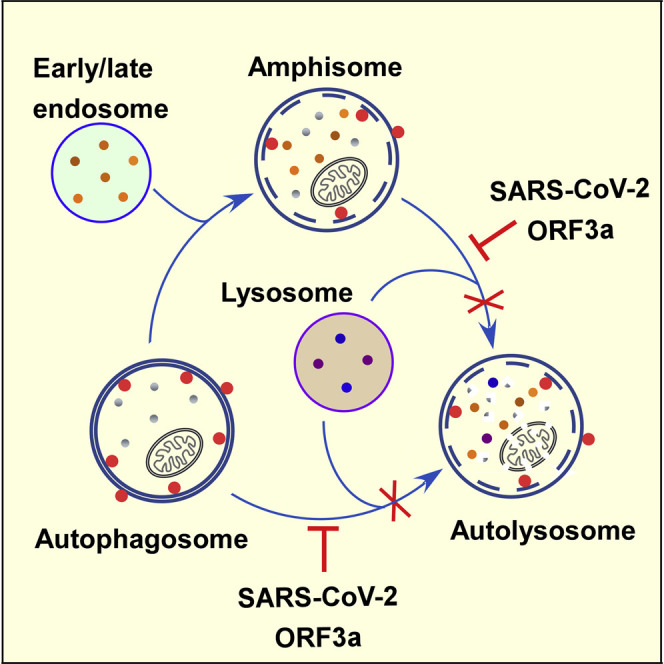
Miao et al. demonstrate that late endosome-localized ORF3a of the COVID-19 virus SARS-CoV-2 sequestrates the HOPS component VPS39. ORF3a blocks autophagosome/amphisome fusion with lysosomes by preventing the assembly of the STX17-SNAP29-VAMP8 SNARE complex. SARS-CoV-2-infected cells also exhibit a defect in autophagosome maturation and sequestration of VPS39 on late endosomes.
Introduction
SARS-CoV-2, the cause of the COVID-19 pandemic, is an enveloped, single-stranded, and positive-sense RNA β-coronavirus (Wang et al., 2020; Wu et al., 2020; Zhou et al., 2020). Very little is known about the mechanism by which SARS-CoV-2 disrupts cellular functions to cause disease pathogenesis. Elucidating how SARS-CoV-2-encoded proteins, especially the accessory proteins not conserved in other coronaviruses, interact with host factors is crucial for us to understand the high infectivity and pathogenicity of this virus and also to develop potential COVID-19 treatments.
Autophagy, a process referring to sequestration of a portion of the cytoplasm in a double-membrane autophagosome and its delivery to lysosomes for degradation, acts as an important cellular mechanism to combat diverse detrimental stresses such as starvation, energy deprivation, and pathogen invasion (Deretic et al., 2013; Feng et al., 2014; Levine et al., 2011; Mizushima et al., 2011). Autophagy involves the initiation and nucleation of a crescent-shaped isolation membrane (IM), which subsequently expands and closes into an autophagosome (Feng et al., 2014; Lamb et al., 2013; Mizushima et al., 2011). Upon closure, autophagosomes fuse with vesicles originated from the endolysomal compartment to form amphisomes before they form degradative autolysosomes (Zhao and Zhang, 2019). A set of autophagy proteins have been identified that act at different steps of the autophagy pathway (Feng et al., 2014; Mizushima et al., 2011; Zhao and Zhang, 2018). Fusion of autophagosomes/amphisomes with late endosomes/lysosomes is mediated by the STX17-SNAP29-VAMP8 SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex (Itakura et al., 2012). The multisubunit homotypic fusion and protein sorting (HOPS) complex, which directly interacts with autophagosome-localized STX17, facilitates the assembly of the SNARE complex for fusion of autophagosomes with late endosomes/lysosomes (Jiang et al., 2014; Takáts et al., 2014).
In response to viral infection, autophagy in host cells is activated by virus-encoded activators of autophagy-inducing signaling, by cellular stresses elicited by infection, and also by Toll-like receptor (TLR)-mediated sensing of viral constituents (Dreux and Chisari, 2010; Jackson, 2015; Viret et al., 2018). Autophagy can act as a defense system by delivering viruses or viral proteins for lysosomal degradation, by trafficking viral nucleic acids and antigens to endolysosomal compartments for activation of innate and adaptive immune responses, and by modulating virus-induced cell death (Deretic et al., 2013; Dreux and Chisari, 2010; Levine et al., 2011). For example, Herpes simplex virus type 1 (HSV-1) and Sindbis virus (SINV) are susceptible to autophagy, and inhibition of autophagy leads to their increased replication and virulence (Choi et al., 2018; Orvedahl et al., 2007). Viruses have evolved various strategies to evade autophagic destruction and even subvert the autophagic machinery for their own benefits (Choi et al., 2018; Jackson, 2015). Positive-stranded RNA viruses such as poliovirus, rhinovirus, coxsackievirus B3 (CVB3), and enterovirus 68 (EVD68) utilize autophagosomes and/or amphisomes to anchor viral RNA replication machineries for replication (Choi et al., 2018; Corona et al., 2018; Jackson et al., 2005; Kemball et al., 2010; Wong et al., 2008). Autophagy also promotes non-lytic release of picornaviruses (e.g., poliovirus and CVB) and parainfluenza virus type 3 (HPIV3) (Bird et al., 2014; Ding et al., 2014; Jackson et al., 2005; Mohamud et al., 2018). The viral proteins exploit different mechanisms to block the fusion of autophagosomes/amphisomes with lysosomes to promote accumulation of autophagic vesicles. The phosphoprotein (P) of HPIV3 binds to SNAP29 to inhibit its interaction with STX17 for fusion of autophagosomes with lysosomes (Ding et al., 2014). CVB3 and EVD68 inhibit fusion of autophagosomes with lysosomes through viral proteinase 3C-mediated cleavage of SNAP29 (Corona et al., 2018; Mohamud et al., 2018). For coronaviruses such as MERS-CoV, murine hepatitis virus (MHV), and SARS-CoV, the replication and transcription complexes (RTCs) are anchored on double-membrane vesicles (DMVs) (Knoops et al., 2008; Snijder et al., 2020). The viral RNA products are localized in the DMVs and transported from the interior to the cytosol for translation or viral assembly via a molecular pore complex spanning the double membrane (Wolff et al., 2020). Autophagic machinery is not required for DMV generation or virus replication in coronavirus-infected cells. The DMVs induced by MHV are linked to the formation of EDEMosomes, which are ER-derived vesicles for export of short-lived ERAD regulators to endosomes/lysosomes (Reggiori et al., 2010). A recent study revealed that β-coronaviruses employ late endosomes/lysosomes for egress (Ghosh et al., 2020). It remains unknown how the endomembrane system is remodeled and the autophagy pathway is modulated in SARS-CoV-2-infected cells.
Here, we investigated the effect of SARS-CoV-2 proteins on autophagy and found that ORF3a, an accessory protein specific to human and animal isolates of the COVID-19 virus SARS-CoV-2 and the SARS virus SARS-CoV, greatly impairs the formation of degradative autolysosomes. ORF3a is localized on late endosomes and directly interacts with and sequestrates the HOPS component VPS39, thereby preventing the interaction of the HOPS complex with autophagosome-localized STX17. Consequently, the assembly of the STX17/SNAP29/VAMP8 complex is blocked. In SARS-CoV-2-infected cells, autophagy is blocked at the step when acidified autolysosomes form, and VPS39 is sequestered on late endosomes. Surprisingly, ORF3a of SARS-CoV fails to interact with HOPS or to block autolysosome formation. Our results reveal a mechanism for viral proteins to block progression of autophagosomes/amphisomes into autolysosomes.
Results
Expression of ORF3a inhibits autophagy activity
We surveyed the effect of proteins encoded by SARS-CoV-2 on autophagy by examining the formation of LC3 puncta and also the accumulation of p62 aggregates. Phosphatidylethanolamine (PE)-conjugated LC3 (known as LC3-II) associates with autophagosomal membranes at different stages, while unlipidated LC3 (LC3-I) is diffusely localized (Klionsky et al., 2016). p62 is an autophagy substrate that accumulates into a large number of aggregates in autophagy-deficient cells (Klionsky et al., 2016). Among the 21 genes encoding SARS-CoV-2 proteins, the number of LC3 puncta and distinct p62 aggregates was significantly increased in HeLa cells expressing ORF3a, ORF7a, M, and NSP6 (Figures 1A–1E, S1A–S1H, and S1M), with ORF3a having the strongest effect. In this study, we focused mainly on SARS-CoV-2 ORF3a, which is simply referred to ORF3a hereafter unless the viral origin is stated. In immunoblotting assays, expression of ORF3a caused an increase in p62 and LC3-II levels as well as the ratio of LC3-II/LC3-I (Figure 1G), indicating that autophagy is impaired by expression of ORF3a. Among 2,782 SARS-CoV-2 strains examined, the two most common nonsynonymous mutations in ORF3a are Q57H (17.4% of strains) and G251V (9.7% of strains) (Issa et al., 2020). Expression of these mutants also caused accumulation of LC3-II and p62 (Figure 1G). Autophagy activity was also impaired in other cells expressing ORF3a such as HEK-293T cells (Figures S1I–S1M).
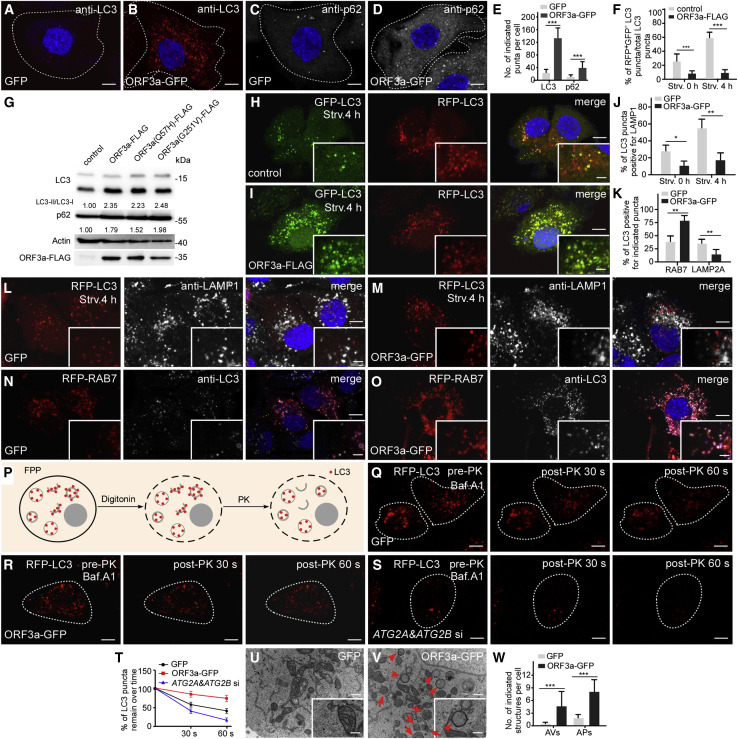
Figure 1.
Expression of ORF3a blocks autophagosome maturation
(A–E) Compared with control cells (A and C), HeLa cells expressing ORF3a-GFP contain more LC3 puncta and p62 puncta (B and D). Cells expressing GFP or ORF3a-GFP are outlined. (E) shows quantification of the number of LC3 puncta in (A and B) and p62 puncta in (C and D) (mean ± SEM., n = 25 cells in each group). ∗∗∗p < 0.001. Scale bars, 5 μm.
(F) Shows the percentage of RFP+GFP−LC3 puncta among total LC3 puncta in control and ORF3a-FLAG-expressing cells (mean ± SEM, n = 27 cells in each group). Strv, starvation. ∗∗∗p < 0.001.
(G) Immunoblotting assays showing that levels of LC3 and p62 are increased in cells expressing ORF3a-FLAG and ORF3a(Q57H or G251V)-FLAG. Levels of LC3-II/LC3-I and p62 levels are normalized by actin and set to 1.00 in control cells.
(H and I) Compared with control cells (H), cells expressing ORF3a-FLAG contain far fewer RFP+GFP−LC3 puncta (I) after 4-h starvation. Scale bars, 5 μm; inserts, 2 μm.
(J) Quantification of the percentage of LC3 puncta co-localizing with LAMP1-labeled lysosomes in control and ORF3a-GFP-expressing cells. Data are shown as mean ± SEM (n = 21 cells in each group). Puncta labeled by LAMP1 with strong fluorescence intensity were counted, while those with very faint fluorescence intensity were not counted. ∗p < 0.05. ∗∗p < 0.01.
(K) Quantification of the percentage of LC3 puncta co-localizing with the indicated puncta in control and ORF3a-GFP-expressing cells. Data are shown as mean ± SEM (n = 20 cells in each group). ∗∗p < 0.01.
(L and M) Compared with control cells (L), LC3 puncta are largely separate from LAMP1-labeled lysosomes in ORF3a-GFP-expressing cells under nutrient-rich conditions and starvation conditions (M). Scale bars, 5 μm; inserts, 2 μm.
(N and O) In ORF3a-GFP-expressing cells, a larger percentage of LC3 puncta co-localize with RAB7-labeled late endosomes (O) than in control cells (N). Scale bars: 5 μm; inserts, 2 μm.
(P) Schematic illustration of the FPP assay. After 5 min treatment with 20 μM digitonin, the plasma membrane of the cell is permeabilized, while the membranes of intracellular organelles are intact. The RFP-LC3 signals inside closed autophagosomes or autolysosomes are protected from addition of PK, while RFP-LC3 localized on isolation membranes and the outer membrane of autophagosomes is digested by PK. RFP-LC3 is shown as red dots. PK, proteinase K.
(Q and R) In FPP assays, time-lapse images show that after 6 h Bafilomycin A1 (Baf.A1) treatment, most of the RFP-LC3 puncta in permeabilized control and ORF3a-GFP-expressing cells are resistant to PK treatment. Scale bars, 5 μm.
(S) In FPP assays, the RFP-LC3 signal disappears in Baf.A1-treated siATG2A/B cells upon PK treatment for 30 and 60 s. Scale bars: 5 μm.
(T) Quantification of the percentage of LC3 puncta that remain over time in FPP assays in control cells, ORF3a-GFP-expressing cells and ATG2A & ATG2B KD cells. Data are shown as mean ± SEM (n = 15 cells in each group).
(U–W) EM analysis indicates that autophagosomes (arrowheads) and amphisomes (arrows) accumulate in ORF3a-GFP-expressing cells (V) but are largely absent in control cells (U) under nutrient-rich conditions. Quantification of the number of autophagosomes and amphisomes per image in (W) is shown as mean ± SEM (n = 66 cells in each group, one image for each cell). ∗∗∗p < 0.001. Scale bars, 0.5 μm; inserts, 0.1 μm.
See also Figures S1 and S2.
We further examined autophagic flux using the RFP-GFP-LC3 assay. Due to quenching of the GFP fluorescence in acidified compartments, yellow puncta (positive for both GFP and RFP signals) represent IMs, autophagosomes or un-acidified amphisomes, while red-only puncta are acidified amphisomes or autolysosomes (Kimura et al., 2007). In ORF3a-expressing cells, yellow LC3 puncta accumulated under nutrient-rich conditions (Figures S1Q and S1R). After 4 h of starvation, numerous red-only puncta were detected in control cells, while LC3 puncta remained yellow in ORF3a-expressing cells (Figures 1F, 1H, and 1I). Yellow LC3 puncta also accumulated in cells expressing M (Figure S1S). Thus, expression of ORF3a or M inhibits autophagy at a step prior to formation of acidified degradative autolysosomes. More red-only LC3 puncta were detected in cells expressing ORF7a or NSP6 compared with control cells (Figures S1P, S1T, and S1U). Accumulation of LC3 and p62 puncta in ORF7a- or NSP6-expressing cells suggests that although the autophagic structures are acidified, their degradative capability is impaired in these cells.
The fusion of autophagosomes/amphisomes with lysosomes is blocked by ORF3a
To determine at which step the autophagy pathway is impaired, we examined the expression of a series of markers that label autophagic structures at different stages. Upon autophagy induction, sequential targeting of the ULK1/FIP200/ATG13 Atg1 complex and the VPS34/Beclin1/ATG14L PI(3)P kinase complex results in generation of PI(3)P-enriched subdomains of the ER, known as omegasomes, followed by the nucleation and expansion of IMs in their vicinity (Axe et al., 2008; Itakura and Mizushima, 2010; Lamb et al., 2013). ATG14L is also associated with nascent autophagosomes and binds to STX17 to promote their fusion with late endosomes/lysosomes (Diao et al., 2015). The PI(3)P effector WIPI2 labels the IM and also nascent autophagosomes with a high PI(3)P level (Cebollero et al., 2012; Wu et al., 2014). Compared with control cells, the number of puncta labeled by ULK1-GFP was not obviously changed in ORF3a-expressing cells under nutrient-rich conditions and also after 1 h of starvation (Figures S1V–S1X). In line with this, mTOR activity, measured by levels of phosphorylated 4EBP-1 and S6K, was not altered in ORF3a-expressing cells (Figure S1C1). The number of structures positive for GFP-DFCP1, which labels omegasomes, was slightly increased (Figure S1V). Levels of endogenous FIP200 and ATG13 co-immunoprecipitated by ULK1-GFP remained largely unaltered, whereas levels of endogenous ATG14 and Beclin1 precipitated by GFP-VPS34 were slightly increased in ORF3a-expressing cells (Figures S1N and S1O). The numbers of puncta formed by GFP-ATG14, WIPI2-GFP, and Flag-STX17 were dramatically increased in cells expressing ORF3a under both nutrient-rich and starvation conditions (Figures S1V, S1Y–S1B1, and S1D1–S1F1). These results suggest that autophagic structures in ORF3a-expressing cells are either at the end stage of IM expansion or autophagosomes/amphisomes.
We further investigated the stage of autophagic structures in ORF3a-expressing cells by examining their colocalization with late endosomes/lysosomes. In control cells, LC3 puncta were largely colocalized with anti-LAMP1-labeled late endosomes/lysosomes after 4 h of starvation (Figures 1J and 1L), while in cells expressing ORF3a, LC3 puncta were largely separate from LAMP1-labeled structures under both nutrient-rich and starvation conditions (Figures 1J, 1M, S2A, and S2B). Structures labeled by LAMP1 are heterogeneous in nature (Cheng et al., 2018). We found that compared with control cells, the LC3 puncta in cells expressing ORF3a were largely colocalized with RFP-RAB7-labeled late endosomes (Figures 1K, 1N, and 1O), and there was much less colocalization of LC3 with LAMP2A, which mainly labels lysosomes (Figures 1K, S2C, and S2D). These results indicate that a portion of autophagosomes in ORF3a-expressing cells have already fused with late endosomes but fail to fuse with lysosomes.
We also performed fluorescence protease protection (FPP) and HaloTag-LC3 (HT-LC3) assays to determine whether autophagic structures are closed. In the FPP assay, the cells were treated with Baf.A1 for 6 h, which blocks autophagosome maturation and/or degradation, to induce accumulation of closed autophagic structures. The treated cells were then permeabilized with digitonin and treated with proteinase K (PK). LC3 on unclosed isolation membranes is digested by PK, while the LC3 signal on inner membranes of closed autophagic structures persists upon PK treatment (Figure 1P). Upon Baf.A1 treatment, RFP-LC3 puncta remained largely fluorescent upon treatment with PK in control and ORF3a-expressing cells (Figures 1Q, 1R, and 1T), while the signal disappeared in Atg2A/B KD cells, in which isolation membranes cannot proceed to autophagosomes (Velikkakath et al., 2012) (Figure 1S). In the HT-LC3 assay, after permeabilization of the plasma membrane to release cytosolic HT-LC3-I, the HT-LC3-labeled IMs and nascent autophagosomes (with LC3 on the outer membrane) are accessible to a membrane-impermeable HaloTag ligand (MIL), while a membrane-permeable HaloTag ligand (MPL) labels HT-LC3 that is located on the inner membranes of closed autophagosomes, amphisomes, and autolysosomes (Takahashi et al., 2018) (Figure S2E). Therefore, MIL+MPL− puncta represent IMs, MIL+MPL+ puncta represent nascent autophagosomes, and MIL−MPL+ puncta represent mature autophagosomes, amphisomes, and autolysosomes (outer membrane-associated LC3-II is largely deconjugated in these structures). After autophagy induction by Torin 1 treatment, more MIL−MPL+ LC3 but fewer MIL+MPL− LC3 puncta accumulated in ORF3a-expressing cells compared with control cells (Figures S2F–S2H). Finally, the autophagic structures were visualized by transmission electron microscopy (TEM) analysis. Compared with control cells, ORF3a-expressing cells accumulated autophagosomes and also amphisomes (Figures 1U–1W), while autolysosomes were rarely detected. EM analysis of cells expressing WIPI2-APEX2 revealed that in control cells, dark APEX2-derived signals were detected in nascent autophagosomes, but not in amphisomes (Figures S2J and S2K). In contrast, dark WIPI2-APEX2 signals were detected on membranes of multiple autophagosomes/amphisomes in ORF3a-expressing cells (Figures S2L and S2M), consistent with the accumulation of WIPI2-GFP puncta. These results indicate that expression of ORF3a results in accumulation of autophagosomes/amphisomes that fail to fuse with lysosomes.
Autophagy is inhibited in SARS-CoV-2 virus-infected cells
We next assessed autophagy activity in cells after SARS-CoV-2 infection. HeLa cells expressing human ACE2 were infected with SARS-CoV-2 virus and harvested at 8, 16, and 24 hpi (hpi, hours post-inoculation). The number of LC3 puncta and p62 aggregates and the protein levels of LC3-II and p62 were dramatically increased in SARS-CoV-2-infected cells (Figures 2A–2D). A large number of WIPI2-GFP puncta also accumulated in virus-infected cells (Figures 2D–2F). In cells transfected with RFP-GFP-LC3, yellow LC3 puncta accumulated 16 h after SARS-CoV-2 infection (Figures 2G–2I). Consistent with this observation, LC3 puncta were largely separate from LAMP1-labeled late endosomes/lysosomes in SARS-CoV-2-infected cells (Figures S2I, S2Q, and S2R). These results indicate that SARS-CoV-2 infection blocks the formation of acidified autolysosomes, resembling the defects in ORF3a-expressing cells.
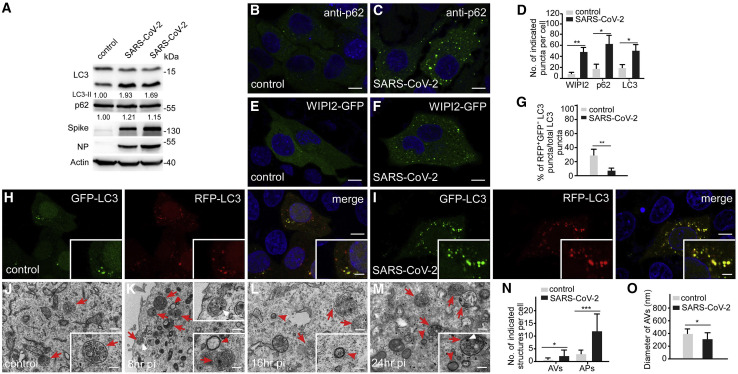
Figure 2.
SARS-CoV-2 infection blocks autophagosome maturation
(A) Immunoblotting assays showing that levels of LC3 and p62 are increased in SARS-CoV-2-infected cells. HeLa cells were transfected with human ACE2, then infected with SARS-CoV-2 virus and harvested at 16 hpi (hpi, hours post-inoculation). Lane 2 and lane 3 show cells infected with 4 × 105 TCID50 (tissue culture infectious dose 50%)/well and 2 × 106 TCID50/well, respectively. Levels of LC3-II and p62 are normalized by actin and set to 1.00 in control cells.
(B–D) Compared with control cells (B), p62 forms more punctate structures in cells infected with SARS-CoV-2 virus (1 × 105 TCID50/well) (C). (D) shows quantification of the indicated puncta in control cells and SARS-CoV-2 virus-infected cells (1 × 105 TCID50/well). Data are shown as mean ± SEM (n = 18 cells in each group). ∗p < 0.05; ∗∗p < 0.01. Scale bars, 5 μm.
(E and F) Compared with control cells (E), WIPI2-GFP forms more punctate structures in cells infected with SARS-CoV-2 virus (1 × 105 TCID50/well) (F). Scale bars, 5 μm.
(G–I) Compared with control cells (H), cells infected with SARS-CoV-2 virus contain many more RFP+GFP+LC3 puncta (I). (G) shows quantification of the percentage of RFP+GFP−LC3 puncta among total LC3 puncta in control and SARS-CoV-2-infected cells (mean ± SEM, n = 20 cells in each group). ∗∗p < 0.01. Scale bars, 5 μm; inserts, 2 μm.
(J–O) EM analysis indicates that many more autophagosomes (red arrowheads) and amphisomes (red arrows) accumulate in SARS-CoV-2 virus-infected cells at 8 hpi (K), 16 hpi (L), and 24 hpi (M) than control cells (J). The white arrowheads indicate virion particles. Quantification of the number of autophagosomes and amphisomes in (N) is shown as mean ± SEM (n = 89 cells in each group, one image for each cell). (O) shows quantification of the diameter of autophagosomes as mean ± SEM (n = 32 autophagosomes in each group). The diameter is about 251 ± 157 nm in SARS-CoV-2 virus-infected cells. ∗p < 0.05, ∗∗∗p < 0.001. Scale bars: 0.5 μm; inserts, 0.1 μm.
See also Figure S2.
We next performed TEM analysis of SARS-CoV-2-infected cells. DMVs dramatically accumulated in the infected cells (Figures S2N and S2O). The content in the DMVs was electron-lucid and distinct from the surrounding cytoplasm. At 24 hpi, fibrous materials, probably corresponding to viral RNAs like those found in SARS-CoV-infected cells (Knoops et al., 2008; Snijder et al., 2020), accumulated in the interior of DMVs. DMVs also merged into large single-membrane vacuoles containing fibrous materials (Figure S2P). Double-membrane autophagosomes, containing cytoplasmic contents, were also formed in the infected cells (Figures 2J–2N), but their size was smaller than control cells (Figure 2O). Amphisome-like structures also accumulated in SARS-CoV-2-infected cells (Figures 2J–2N). Virion particles were detected in amphisome-like structures (Figures 2K and 2M). Taken together, these results show that SARS-CoV-2 infection also inhibits the fusion of autophagosomes/amphisomes with lysosomes.
ORF3a is localized on late endosomes/lysosomes
ORF3a is predicted to be a multi-transmembrane protein. ORF3a protein formed a large number of intracellular punctate structures and also weakly localized on the plasma membrane (Figure 3 A). Coexpression of markers for various endolysosomal compartments showed that ORF3a puncta were separate from EEA-1-labeled early endosomes (Figure S3A), but extensively colocalized with RAB7- and LAMP1-labeled late endosomes/lysosomes (Figures 3A, 3B, and 3E). Structured illumination microscopy (SIM) revealed that ORF3a-GFP overlapped or closely associated with RFP-RAB7 on vesicular membranes (Figure 3C). ORF3a-GFP puncta were largely separate from LAMP2A-labeled or LysoTracker Red-stained lysosomes (Figures 3D, 3E, and S3B).
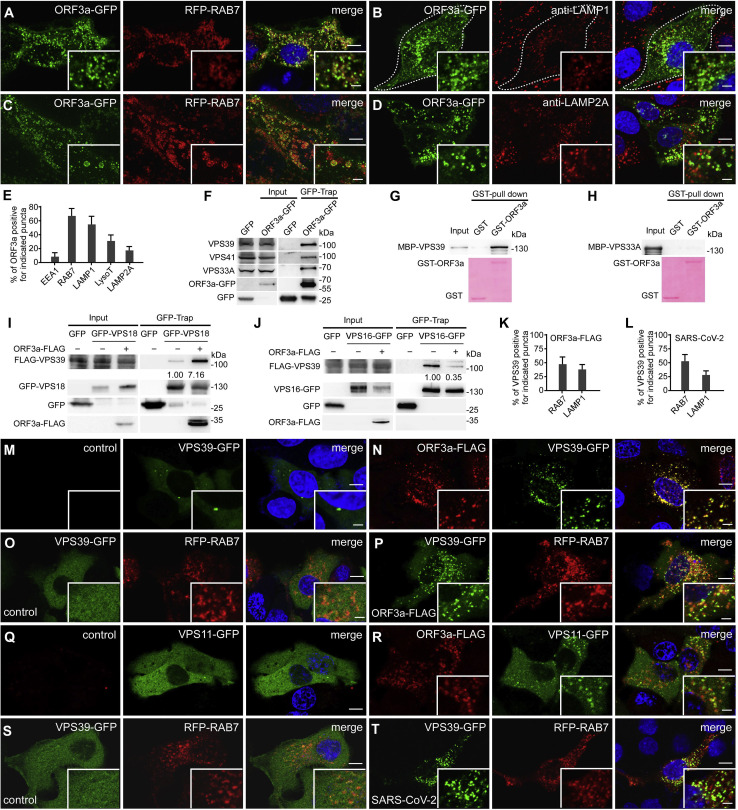
Figure 3.
ORF3a localizes to late endosomes/lysosomes and interacts with components of the HOPS complex
(A) ORF3a-GFP forms a large number of punctate structures that are largely colocalized with late endosomes labeled by RFP-RAB7 in HeLa cells. Weak ORF3a-GFP signal is localized on the plasma membrane. Scale bars, 5 μm; inserts, 2 μm.
(B) ORF3a-GFP puncta partially co-localize with LAMP1-labeled late endosomes/lysosomes, detected by anti-LAMP1. Scale bars, 5 μm; inserts, 2 μm.
(C) 3D-SIM images showing that ORF3a-GFP colocalizes with RFP-RAB7 on vesicles. Images are maximum intensity projections of z stacks (z = 35). Scale bars, 5 μm; inserts, 1 μm.
(D) ORF3a-GFP puncta are separate from LAMP2A-labeled lysosomes, detected by anti-LAMP2A. Scale bars, 5 μm; inserts, 2 μm.
(E) Quantification of the percentage of ORF3a-GFP puncta co-localizing with vesicles labeled by the indicated marker.
(F) Endogenous VPS39, VPS41, and VPS33A are co-precipitated by ORF3a-GFP in GFP-TRAP assays. Extracts of cells expressing GFP vector and ORF3a-GFP were precipitated by GFP-TRAP beads and immunoblotted with anti-VPS39, anti-VPS41, and anti-VPS33A. ~5% of extracts used for GFP-TRAP assays are shown as the input.
(G and H) Human full-length VPS39 (G), but not VPS33A (H), is pulled down by GST-ORF3a in an in vitro GST-pull-down assay. Proteins used for pull-down are shown in the bottom panel.
(I) Levels of FLAG-tagged VPS39 precipitated by GFP-VPS18 are higher in ORF3a-FLAG-expressing cells than control cells in GFP-TRAP assays. Quantification of FLAG-VPS39 level (normalized by GFP-VPS18 level) is also shown.
(J) In GFP-TRAP assays, levels of VPS39 precipitated by VPS16-GFP are lower in ORF3a-FLAG-expressing cells than control cells. Quantification of FLAG-VPS39 level (normalized by VPS16-GFP level) is also shown.
(K) Quantification of the percentage of VPS39-GFP puncta co-localizing with vesicles labeled by the indicated marker in ORF3a-FLAG-expressing cells.
(L) Quantification of the percentage of VPS39-GFP puncta co-localizing with vesicles labeled by the indicated marker in SARS-CoV-2 virus-infected cells.
(M and N) In control cells (M), VPS39-GFP mainly localizes to the cytoplasm and forms very few punctate structures, while in cells expressing ORF3a-FLAG (N), VPS39-GFP forms a larger number of puncta that co-localize with ORF3a-FLAG. There are also some VPS39-GFP puncta showing very weak or no detectable ORF3a signal. Scale bars: 5 μm; inserts, 2 μm.
(O and P) VPS39-GFP is largely diffuse in control cells (O). VPS39-GFP forms punctate structures that partially co-localize or closely associate with RAB7-labeled late endosomes in cells expressing ORF3a-FLAG (P). Scale bars, 5 μm; inserts, 2 μm.
(Q and R) VPS11-GFP is largely diffuse in control cells (Q), while in ORF3a-expressing cells, VPS11-GFP forms some punctate structures that co-localize with ORF3a-FLAG (R). Scale bars: 5 μm; inserts, 2 μm.
(S and T) In SARS-CoV-2 virus-infected cells, VPS39-GFP forms a large number of punctate structures that partially co-localize or closely associate with RAB7-labeled late endosomes (T). VPS39-GFP is diffuse in control cells (S). Scale bars, 5 μm; inserts, 2 μm.
See also Figures S3 and S4.
ORF3a interacts with components of the HOPS complex
We determined whether ORF3a modulates autophagy by interacting with autophagy proteins. ORF3a showed no interaction with components of the ULK1 complex, VPS34, Beclin1, ATG14, WIPI2, STX17, and VAMP8 in GFP-Trap assays (Figure S3F). The HOPS complex consists of the core subunits VPS11, VPS16, VPS18, and VPS33A and the HOPS-specific subunits VPS39 and VPS41. We found that endogenous VPS39, VPS41, and VPS33A or Flag-tagged proteins were strongly co-immunoprecipitated by ORF3a in GFP-Trap assays (Figures 2F, S3C, and S3D). In in vitro pull-down assays, ORF3a directly interacted with VPS39 (Figure 3G) but showed no or very weak binding with other HOPS components (Figure 3H and data not shown). ORF3a showed no interaction with VPS8 (Figure S3E), a specific component of the CORVET (class C core vacuole/endosome tethering) complex. ORF3a also exhibited weak interaction with endogenous RAB7 in coIP assays (Figure S3D).
We next investigated the formation of the HOPS complex in ORF3a-expressing cells. The yeast HOPS complex exhibits an elongated seahorse-like structure with VPS39 and VPS41 located at opposite ends (Bröcker et al., 2012). VPS33 and VPS16 form a subcomplex involved in SNARE protein binding. VPS39 interacts with VPS11 and VPS18, which further connect to VPS33/VPS16, while VPS41 interacts with VPS18 to link VPS33/VPS16 (Bröcker et al., 2012; Plemel et al., 2011; Wartosch et al., 2015). VPS39 also forms a subcomplex with VPS41, whose function remains unknown (Balderhaar and Ungermann, 2013). We found that in ORF3a-expressing cells, higher levels of endogenous VPS39 or FLAG-VPS39 were co-precipitated by GFP-VPS18 or VPS11-GFP (Figures 3I, S3G, and S3H), while their interactions with the SNARE-binding subcomplex VPS33A/VPS16 were reduced (Figures 3J and S3I). The interactions of VPS41 with VPS18, VPS33A with VPS16, or VPS41 with VPS39 remained largely unchanged by ORF3a expression (Figures S3G, S3J, and S3K). Therefore, the formation of the VPS39-containing functional HOPS complex appears to be affected by ORF3a expression.
ORF3a causes accumulation of VPS39 on late endosomes/lysosomes
We next examined whether late endosome/lysosome-localized ORF3a affects the localization of HOPS components. In control cells, VPS39 and the core subunits VPS11, VPS16, and VPS18 were mainly localized in the cytoplasm and only a very few distinct puncta were detected (Figures 3M, 3O, 3Q, and S4A) (Garg et al., 2011; Kim et al., 2001; Peralta et al., 2010). In cells expressing ORF3a-FLAG, VPS39-GFP formed many more punctate structures, whose size was not uniform (Figures 3N, S4C, and S4D). The relatively large VPS39-GFP punctate structures colocalized with ORF3a (Figures 3N and S4C). There were also small VPS39-GFP structures with very weak ORF3a signal (Figure 3N). Even in cells with a very low ORF3a-FLAG level, VPS39-GFP still formed many small punctate structures (Figure S4D). VPS39-GFP puncta in ORF3a-expressing cells partially colocalized or closely associated with RAB7- and LAMP1-labeled late endosomes/lysosomes (Figures 3K, 3O, 3P, S4E, and S4F). Expression of ORF3a also dramatically increased the number of VPS11-GFP- and VPS16-GFP-labeled punctate structures that were colocalized or closely associated with ORF3a (Figures 3Q, 3R, S4A, and S4B). VPS41-GFP formed a large number of puncta that partially colocalized with RAB7-labeled late endosomes/lysosomes (Figure S4G) (Garg et al., 2011; Miao et al., 2020). In ORF3a-expressing cells, there were slightly more VPS41-GFP puncta, and they colocalized with RAB7 (Figure S4H). These results indicate that ORF3a recruits VPS39 and, to a lesser extent, other core components of HOPS to late endosomes/lysosomes.
In SARS-CoV-2-infected cells, VPS39-GFP also accumulated into a large number of punctate structures (Figures 3T and S4J). As in ORF3a-expressing cells, VPS39-GFP puncta were partially colocalized or closely associated with RAB7- and LAMP1-labeled late endosomes/lysosomes (Figures 3L, 3S, 3T, S4I, and S4J).
ORF3a inhibits the interaction of HOPS with autophagosomal STX17
The HOPS complex promotes fusion of autophagosomes/amphisomes with late endosomes/lysosomes (Jiang et al., 2014; Takáts et al., 2014). In control cells after 4 h of starvation, LC3 puncta were colocalized with VPS41-labeled punctate structures (Figure 4 A). The colocalization of LC3 puncta with VPS41-GFP-labeled puncta was greatly reduced in ORF3a-expressing cells (Figures 4B, 4E, S5A, and S5B). VPS39-GFP puncta were also largely separate from LC3 puncta in ORF3a-expressing cells (Figures 4C, 4D, S5C, and S5D).
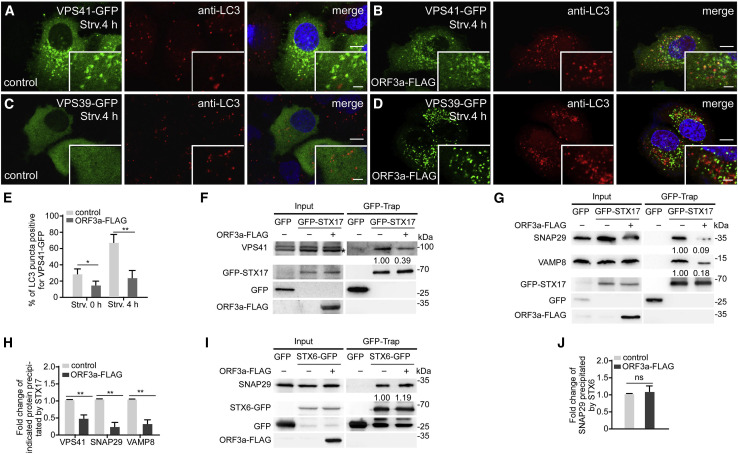
Figure 4.
ORF3a inhibits the formation of the STX17-SNAP29-VAMP8 complex
(A and B) Compared with control cells (A), a smaller percentage of VPS41-GFP-labeled punctate structures co-localize with LC3 puncta in ORF3a-FLAG-expressing cells (B) after 4-h starvation. Scale bars: 5 μm; inserts, 2 μm.
(C and D) LC3-labeled puncta are separate from VPS39-GFP-labeled punctate structures in ORF3a-FLAG-expressing cells (D) after 4-h starvation. VPS39-GFP is diffuse in control cells (C). Scale bars: 5 μm; inserts, 2 μm.
(E) Quantification of the percentage of LC3-labeled puncta co-localizing with VPS41-GFP-labeled punctate structures is shown as mean ± SEM (n = 20 cells in each group). ∗p < 0.05. ∗∗p < 0.01.
(F) In GFP-TRAP assays, levels of endogenous VPS41 precipitated by GFP-STX17 are lower in ORF3a-FLAG-expressing cells than control cells. “∗” indicates the band corresponding to VPS41. Quantification of VPS41 level (normalized by GFP-STX17 level) is also shown.
(G) In GFP-TRAP assays, levels of endogenous SNAP29 and VAMP8 precipitated by GFP-STX17 are much lower in ORF3a-FLAG-expressing cells than control cells. Quantification of SNAP29 and VAMP8 levels (normalized by GFP-STX17 level) is also shown.
(H) Quantification of the fold change of the indicated proteins in control cells and ORF3a-expressing cells is shown as mean ± SEM Results are representative of at least three experiments. ∗∗p < 0.01.
(I) In GFP-TRAP assays, levels of endogenous SNAP29 precipitated by STX6-GFP show no obvious change in ORF3a-FLAG-expressing cells compared with control cells. Quantification of SNAP29 level (normalized by GFP-STX6 level) is also shown.
(J) Quantification of the fold change of SNAP29 precipitated by STX6 in control cells and ORF3a-expressing cells is shown as mean ± SEM Results are representative of at least three experiments. ns: no significant difference.
See also Figure S5.
The HOPS complex directly interacts with autophagosome-localized STX17 to promote autophagosome maturation (Jiang et al., 2014; Takáts et al., 2014). We found that in GFP-Trap assays, levels of VPS41 and VPS39 co-precipitated by GFP-STX17 were dramatically reduced in ORF3a-expressing cells (Figures 4F, 4H, S5E, and S5F). Thus, the interaction of the HOPS complex with STX17 is disrupted by ORF3a.
ORF3a inhibits the formation of STX17-SNAP29-VAMP8 SNARE complex
The HOPS complex promotes the assembly of the STX17-SNAP29-VAMP8 SNARE complex for the fusion of autophagosomes/amphisomes with late endosomes/lysosomes. Expression of ORF3a did not affect levels of endogenous STX17, SNAP29, and VAMP8 (Figure S5G). Levels of endogenous SNAP29 and VAMP8 precipitated by GFP-STX17 were dramatically reduced in ORF3a-expressing cells (Figures 4G and 4H), indicating that the formation of the STX17-SNAP29-VAMP8 complex is greatly impaired by ORF3a. VPS39 and VPS41 failed to interact with the autophagosome-localized R-SNARE YKT6 in GFP-trap assays (Figure S5H). The assembly of YKT6-SNAP29-STX7 SNARE complex, which acts non-redundantly with the STX17-SNAP29-VAMP8 complex for autophagosome maturation (Matsui et al., 2018), was not affected in ORF3a-expressing cells (Figures S5I and S5J). SNAP29 also interacts with the Golgi-localized STX6 (Wong et al., 1999). The interaction of SNAP29 with STX6 was not affected in ORF3a-expressing cells (Figures 4I and 4J). Homotypic and heterotypic fusions of late endosomes and lysosomes are mediated by the Q-SNAREs (STX7, STX8, and VTI1b) and the R-SNARE VAMP8 or VAMP7 (Jahn and Scheller, 2006). HOPS interacts with STX7 (Kim et al., 2001). We found that in ORF3a-expressing cells, levels of endogenous VAMP8 co-precipitated by STX7-GFP were increased (Figures S5K and S5L). Therefore, ORF3a show distinct effects on assembly of different SNARE complexes.
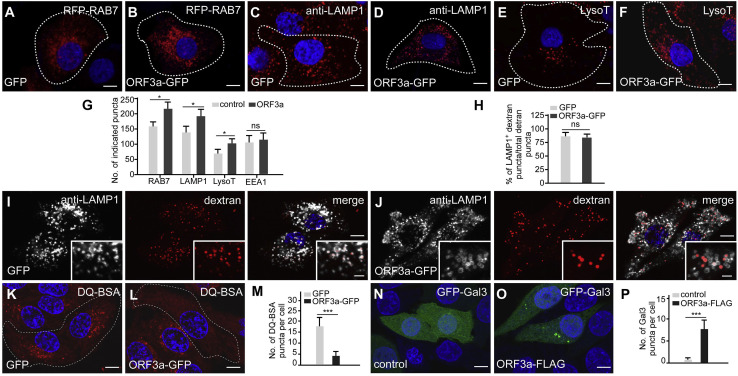
ORF3a affects late endosomal/lysosomal biogenesis and induces lysosomal damage
We noticed that the number of late endosomes/lysosomes labeled by RAB7, LAMP1, and LysoTracker, but not EEA1-labeled early endosomes, was higher in ORF3a-expressing cells (Figures 5A–5G). Compared with control cells, the fluorescence intensities of puncta labeled by anti-LAMP1 and LysoTracker were weaker in ORF3a-expressing cells, especially at vesicles with a strong ORF3a-GFP signal (Figures 3B and S3B). VPS39 KD, but not VPS41 KD, partially reversed the weak LAMP1-staining signals in ORF3a-expressing cells (Figures S6A–S6F). The morphology and distribution of the ER and the Golgi apparatus were not altered (Figures S6G–S6J).
Figure 5.
ORF3a impairs lysosomal function
(A–G) More punctate structures labeled by RFP-RAB7 (B), anti-LAMP1 antibody (D), and LysoTracker (F) are detected in ORF3a-GFP-expressing cells than in control cells (A, C, and E). Cells expressing indicated protein are marked by white dotted lines. Quantification of the number of the indicated punctate structures in GFP control and ORF3a-GFP-expressing cells is shown in (G) as mean ± SEM (n =18 cells in each group). LysoT: LysoTracker. ns: no significant difference; ∗p < 0.05. Scale bars: 5 μm.
(H–J) Similar to control cells (I), endocytosed dextran is largely colocalized with LAMP1-labeled punctate structures in ORF3a-GFP-expressing cells (J). Quantification of the percentage of LAMP1+ dextran puncta among total dextran puncta is shown in (H) as mean ± SEM (n = 20 cells in each group). ns: no significant difference. Scale bars, 5 μm; inserts, 2 μm.
(K–M) Compared with control cells (K), far fewer DQ-BSA-labeled puncta are detected in ORF3a-GFP-expressing cells (L). Cells expressing GFP or ORF3a-GFP are outlined. Quantification of the number of DQ-BSA-labeled punctate structures in GFP control and ORF3a-GFP-expressing cells is shown in (M) as mean ± SEM (n = 25 cells in each group). ∗∗∗p < 0.001. Scale bars, 5 μm.
(N–P) Galectin 3 is diffusely localized in control cells (N) but forms a few punctate structures in ORF3a-FLAG-expressing cells (O). Quantification data of the number of galectin-3-labeled puncta in (P) are shown as mean ± SEM (n = 25 cells in each group). ∗∗∗p < 0.001. Scale bars, 5 μm.
See also Figure S6.
We next investigated whether endocytic trafficking is affected by ORF3a. Dextran-labeled puncta colocalized with LAMP1-labeled late endosomes/lysosomes 2 h after addition in control and also in ORF3a-GFP-expressing cells (Figures 5H–5J). This suggests that the trafficking of dextran-containing endocytic vesicles to late endosomes/lysosomes was not affected by ORF3a. Compared with control cells, levels of mature cathepsin B and cathepsin D and the number of Magic Red Cathepsin B-stained puncta were similar in ORF3a-GFP-expressing cells (Figures S6K–S6O). Degradation of EGFR, triggered by addition of EGF, was also not evidently affected in ORF3a-GFP-expressing cells compared with control cells (Figure S6P). DQ-BSA, which is a bovine serum albumin derivative conjugated to a self-quenched fluorophore, is endocytosed and fluoresces after cleavage by lysosomal proteases (Humphries and Payne, 2012). We found that the number of fluorescent DQ-BSA puncta was much lower in ORF3a-GFP expressing cells than control cells (Figures 5K–5M). Galectin 3, which specifically labels damaged endosomes and lysosomes (Aits et al., 2015), was diffusely localized in control cells, but formed a few puncta in ORF3a-expressing cells (Figures 5N–5P). These results indicate that ORF3a weakly impairs lysosomal function and induces lysosomal damage. TFEB, the master factor regulating autophagy and lysosome biogenesis, undergoes cytoplasmic-to-nuclear transport in response to various cell stressors and lysosomal damage (Nakamura et al., 2020; Raben and Puertollano, 2016). The nuclear localization of TFEB (transcription factor EB) was significantly elevated in ORF3a-expressing cells (Figures S6Q–S6U). However, in ORF3a-expressing cells, only a few genes involved in lysosomal biogenesis such as CTSD (cathepsin D) and CTSA (cathepsin A) showed upregulation, while the mRNA levels of the majority of genes involved in autophagy and lysosomal biogenesis remained unchanged (Figure S6V). Protein levels of LAMP1, LAMP2A, STX17, SNAP29, and VAMP8 showed no obvious changed in ORF3a-expressing cells (Figures S5G and S6W).
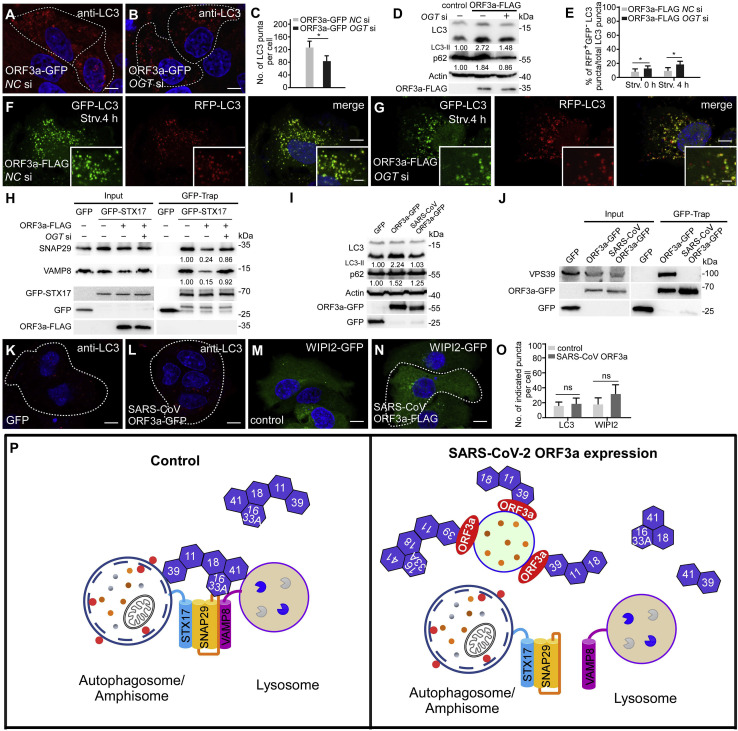
The defect in autophagosome maturation in ORF3a-expressing cells is rescued by OGT knockdown
SNAP29 is post-translationally modified by O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) (Guo et al., 2014). O-GlcNAcylated SNAP29 attenuates the formation of SNAP29-containing trans-SNARE complexes (Guo et al., 2014). In mammalian cells, OGT knockdown or expressing an O-GlcNAcylation-defective SNAP29 mutant promotes the formation of the STX17-SNAP29-VAMP8 complex (Guo et al., 2014). We found that the number of LC3 puncta was reduced in ORF3a-expressing cells with simultaneous depletion of OGT (Figures 6A–6C). Immunoblotting assays showed that levels of LC3-II and p62 were also reduced in ORF3a-expressing cells with depletion of OGT or expression of O-GlcNAcylation-defective mutant SNAP29 (Figures 6D and S7A). The RFP-GFP-LC3 assay also showed that the ratio of red-only puncta was increased in ORF3a-expressing cells with simultaneous depletion of OGT under both nutrient-rich and nutrient-depletion conditions (Figures 6E–6G, S7B, and S7C). Consistent with this, the formation of the STX17-SNAP29-VAMP8 complex was dramatically increased by OGT KD in ORF3a-expressing cells (Figure 6H). Therefore, the autophagy defect caused by ORF3a is ameliorated by enhanced formation of the trans-SNARE complex for autophagosome maturation.
Figure 6.
Enhanced autophagosome-lysosome fusion by depletion of OGT partially rescues the autophagy defect in ORF3a-expressing cells
(A–C) Immunostaining with anti-LC3 antibody shows that the accumulation of LC3 puncta in ORF3a-GFP-expressing cells (A) is suppressed by simultaneous depletion of OGT (B). Cells expressing GFP and ORF3a-GFP are outlined. NC, negative control. Numbers of LC3 puncta are quantified and shown as mean ± SEM in (C) (n = 25 cells in each group). NC, negative control. ∗p < 0.05. Scale bars, 5 μm.
(D) Immunoblots of LC3 and p62 show that the increased levels of LC3-II and p62 are inhibited by siOGT in ORF3a-expressing cells. Levels of LC3-II and p62 (normalized by actin levels) are quantified.
(E–G) RFP-GFP-LC3 assays show that the percentage of RFP+GFP−LC3 puncta after 4-h starvation is increased in siOGT-treated ORF3a-expressing cells (G) compared with control siRNA-treated ORF3a-expressing cells (F). Quantification data (n = 27 cells in each group) are shown as mean ± SEM in (E). ∗p < 0.05. Scale bars, 5 μm; inserts, 2 μm.
(H) In GFP-TRAP assays, levels of endogenous SNAP29 and VAMP8 precipitated by GFP-STX17 are increased in siOGT-treated ORF3a-expressing cells compared with control siRNA-treated ORF3a-expressing cells. Quantification of SNAP29 and VAMP8 levels (normalized by GFP-STX17 levels) is shown.
(I) Immunoblotting assays showing that levels of p62 are slightly increased, while levels of LC3-II show no obvious change, in SARS-CoV ORF3a-FLAG-expressing cells compared with control cells. Levels of LC3-II and p62 levels are normalized by actin.
(J) In GFP-TRAP assays, endogenous VPS39 was precipitated by SARS-CoV-2 ORF3a-GFP, but not by SARS-CoV ORF3a-GFP.
(K and L) Compared with control cells (K), expression of SARS-CoV ORF3a-GFP does not evidently increase the number of LC3 puncta (L). Scale bars: 5 μm.
(M and N) Numbers of WIPI2-GFP punctate structures are similar in control cells (M) and SARS-CoV ORF3a-FLAG-expressing cells (N) under nutrient-rich conditions. Scale bars: 5 μm.
(O) Quantification of the numbers of indicated puncta is shown as mean ± SEM (n = 20 cells in each group) ns: no significant difference.
(P) A model showing how ORF3a of SARS-CoV-2 disrupts the HOPS-mediated assembly of the SNARE complex to impair autophagosome maturation. In control cells, the HOPS complex is recruited to autophagosomes by interacting with STX17, which further facilitates the assembly of the STX17-SNAP29-VAMP8 complex for fusion of autophagosomes/amphisomes with late endosomes/lysosomes. In SARS-CoV-2 ORF3a-expressing cells, the HOPS components (such as VPS39, VPS11, and VPS18) are sequestrated on late endosomes/lysosomes by ORF3a, thus, preventing the interaction of HOPS with autophagosomal STX17. 11, VPS11; 16, VPS16; 18, VPA18; 33A, VPS33A; 39, VPS39; 41, VPS41. In control cells, activated RAB7 may be involved in the interaction of HOPS with late endosomes/lysosomes.
See also Figure S7.
SARS-CoV ORF3a fails to interact with the HOPS complex or affect autophagy
ORF3a of SARS-CoV-2 shows ~72% identity and 85% similarity with ORF3a of SARS-CoV (Figure S7M). Consistent with previous reports (Castaño-Rodriguez et al., 2018; Lu et al., 2006), we found that SARS-CoV ORF3a was localized on the plasma membrane and also on a few punctate structures that were largely separate from RAB7- and LAMP1-labeled structures (Figures S7D and S7E). Interestingly, we found that expression of SARS-CoV ORF3a failed to cause accumulation of LC3 and WIPI2 puncta (Figures 6K–6O). In immunoblotting assays, the LC3-II level was not elevated and levels of p62 were only slightly increased by expression of SARS-CoV ORF3a (Figure 6I). The GFP-RFP-LC3 assays showed that a large number of red-only puncta were formed after 4 h of starvation in cells expressing SARS-CoV ORF3a (Figures S7F–S7H). SARS-CoV ORF3a failed to interact with VPS39 in GFP-Trap assays (Figure 6J). The formation of the STX17-SNAP29-VAMP8 complex remained unchanged in SARS-CoV ORF3a-expressing cells (Figure S7I). The DQ-BSA staining assay revealed that lysosomal function was not impaired in SARS-CoV ORF3a-expressing cells (Figures S7J–S7L). SARS-CoV ORF3a has been shown to interact with TRAF3 (Siu et al., 2019). Similar to SARS-CoV ORF3a, SARS-CoV-2 ORF3a also interacted with TRAF3 in a GFP-Trap assay (Figure S7N). TRAF3 KD failed to rescue the autophagy defect in cells expressing SARS-CoV-2 ORF3a (Figures S7O–S7Q). Taken together, these results suggest that expression of the SARS-CoV ORF3a, unlike its counterpart in SARS-CoV-2, causes no autophagy defect.
Discussion
SARS-CoV-2 ORF3a inhibits fusion of autophagosomes/amphisomes with lysosomes via sequestration of HOPS complex components
Here, we showed that expression of SARS-CoV-2 ORF3a inhibits HOPS-facilitated assembly of the STX17-SNAP29-VAMP8 complex, which is required for fusion of autophagosomes/amphisomes with lysosomes. HOPS bridges, stabilizes, and proofreads SNAREs to promote membrane fusion. Targeting of HOPS components to late endosomes/lysosomes is dynamic. Mammalian VPS41 is recruited to lysosomes via binding to the Arf-like GTPase Arl8b (Garg et al., 2011). VPS39 and core subunits of the HOPS complex are primarily localized in the cytosol, and their targeting to lysosomes is promoted by coexpression of Arl8b and VPS41 (Garg et al., 2011). Targeting of the HOPS complex to autophagosomes/amphisomes is mediated by its interaction with STX17 (Jiang et al., 2014; Takáts et al., 2014). SARS-CoV-2 ORF3a is localized on RAB7-labeled late endosomes and also, to a lesser extent, on LysoTracker-stained lysosomes. VPS39 and other subunits of HOPS interact with ORF3a and are sequestrated on late endosomes and also on some unidentified structures with weak or no detectable ORF3a signal. VPS39 and VPS11 have also been identified as SARS-CoV-2 ORF3a interactors in a mass spectrometry-based study (Gordon et al., 2020). In ORF3a-expressing cells, the interaction of VPS39 with VPS11 and VPS18 is enhanced, while its interaction with the VPS16-VPS33A module is attenuated. ORF3a is unlikely to directly block the interaction between VPS39 and the VPS16-33A module, as they are connected via VPS11 and VPS18 (Bröcker et al., 2012). The interactions of VPS11 with multiple components of the HOPS complex, including VPS39, VPS41, VPS16, and VPS18 (Plemel et al., 2011), and also the interaction between VPS41 and VPS39, may prevent the recruitment of VPS16-VPS33A in a stoichiometric manner to assemble functional HOPS complex in ORF3a-expressing cells. Targeting of VPS39 to late endosomes through its direct interaction with ORF3a may impose spatial restrictions on VPS39 and its interacting HOPS components that prevent them from assembling into a functional complex. Sequestration of HOPS components on late endosomes further impairs the targeting of functional HOPS complex to autophagosomes and lysosomes. Consistent with this, the interaction of HOPS with STX17 is greatly inhibited by ORF3a expression. As a consequence, the assembly of the STX17-SNAP29-VAMP8 complex is reduced. Enhanced recruitment of the HOPS complex to late endosomes/lysosomes by the elevated PI(4)P level on these compartments in SUSR2-depleted cells increases the interaction of VPS39 with STX17 and enhances assembly of the STX17-SNAP29-VAMP8 complex for autophagosome/amphisome fusion with lysosomes (Miao et al., 2020). Formation of amphisomes in ORF3a-expressing cells may be mediated by other SNARE complexes such as the YKT6-SNAP29-STX7 complex (Matsui et al., 2018). Enhanced assembly of the STX17-SNAP29-VAMP8 complex by reduced O-GlcNAcylation of SNAP29 partially rescues the autophagy defect in ORF3a-expressing cells, which further argues that the STX17-SNAP29-VAMP8 complex is essential for fusion of autophagosomes/amphisomes with lysosomes.
Our results indicate that SARS-CoV-2 ORF3a sequestrates VPS39 on late endosomes to prevent the assembly of the SNARE complex mediating fusion of autophagosomes/amphisomes with lysosomes (Figure 6P). Inhibiting the assembly of the STX17-SNAP29-VAMP8 complex appears to be a widely employed mechanism by viruses to accumulate autophagosomes/amphisomes for their own benefits. Proteinase 3C of CVB3 and EVD68 mediates cleavage of SNAP29 (Corona et al., 2018; Mohamud et al., 2018), while the HPIV3 P protein binds to SNAP29 to inhibit its interaction with STX17 (Ding et al., 2014). The amount of STX17 is reduced in HCV-replicating cells, preventing the formation of autolysosomes (Ren et al., 2016).
SARS-CoV-2-infected cells also accumulate autophagosomes/amphisomes. The autophagosome size in SARS-CoV-2-infected cells is smaller than in starved cells. NSP6 of various coronaviruses such as IBV, MHV, and SARS-CoV has been shown to limit autophagosome expansion, resulting in the generation of small-sized autophagosomes (Cottam et al., 2011, 2014). SARS-CoV-2-infected cells also show sequestration of VPS39 on late endosomes. The ORF3a protein level has been shown to be high in SARS-CoV-infected cells (Tan et al., 2006). We showed that the viral factors M and ORF7a also affect autophagy. The combinatorial effects caused by different viral factors block the progression of autophagic flux and remodel the endomembrane system for the benefits of virus replication and egress. The exact role of autophagy in SARS-CoV-2 replication and egress has yet to be determined.
The role of autophagosomes/amphisomes and DMVs in virus replication and release
The DMVs formed in cells infected by positive-stranded RNA viruses such as poliovirus, CVB3 and EVD68 exhibit hallmarks of autophagosomes despite being smaller (approximately 200 to 400 nm in diameter) than regular autophagosomes (Choi et al., 2018; Corona et al., 2018; Jackson et al., 2005; Kemball et al., 2010; Suhy et al., 2000; Wong et al., 2008; Wong and Sanyal, 2020). They contain cytosolic contents and are labeled by lipidated LC3. They are able to fuse with late endosomes, but further fusion with lysosomes is blocked (Corona et al., 2018; Jackson et al., 2005; Kemball et al., 2010; Mohamud et al., 2018; Wong et al., 2008). DMVs are also involved in non-lytic extracellular release of virus particles (Bird et al., 2014; Jackson et al., 2005; Mohamud et al., 2018). Thus, these viruses subvert modified autophagosomes and/or amphisomes for replication and extracellular release.
β-coronaviruses, such as MERS-CoV, MHV, and SARS-CoV, and the γ-coronavirus IBV also induce formation of DMVs (approximately 200 to 400 nm in diameter) for anchoring the viral RTCs (Choi et al., 2018; Hartenian et al., 2020; Knoops et al., 2008; Maier et al., 2013; Snijder et al., 2020; Wong and Sanyal, 2020). We found that SARS-CoV-2 infection also induces the formation of DMVs in host cells. In cells infected with MHV, SARS-CoV or IBV, autophagy is required neither for the formation of DMVs, nor for viral replication (Zhao et al., 2007). MHV exploits the pathway of EDEMosome formation to generate DMVs (Reggiori et al., 2010). For the release of β-coronaviruses from host cells, new viral particles are assembled in the ER and/or ERGIC and subsequently use a lysosomal exocytic pathway for release (Ghosh et al., 2020; Knoops et al., 2008; Snijder et al., 2020). In SARS-CoV-2-infected cells, LAMP1-labeled lysosomes are filled with viruses during the egress stage (Ghosh et al., 2020). SARS-CoV-2 virions have also been detected in double-membrane structures and smooth-walled secretory vesicles in infected nasal and bronchial human airway epithelial cells (Pizzorno et al., 2020) and in autophagic or endomembrane compartments in lung pneumocytes and kidney endothelial/epithelial cells of COVID-19 patients (Bradley et al., 2020). We also detected virions in amphisomal/endosomal structures at the early stage of infection, which is consistent with the notion that SARS-CoV-2 exploits endocytosis as an entry route in addition to direct fusion with the plasma membrane (Hartenian et al., 2020; Hoffmann et al., 2020; Shang et al., 2020). Autophagy may be differentially modulated at different stages of SARS-CoV-2 infection. At the early stage of infection, activation of the whole autophagy pathway elicits innate and adaptive immune responses. In contrast, at late stages, viral proteins, which have been abundantly translated, inhibit autophagy by reducing the autophagosome size (e.g., NSP6) and blocking autophagosome maturation (e.g., ORF3a) to prevent fusion of virus-containing amphisomal/endosomal compartments (e.g., derived from the TGN or other virion-enclosing structures) with degradative lysosomes.
ORF3a in SARS-CoV-2 and SARS-CoV infection
Expression of ORF3a impairs lysosomal function and also induces lysosomal damage. The fluorescence intensity of structures stained by anti-LAMP1 and LysoTracker is weaker and the number of DQ-BSA-stained puncta is lower in ORF3a-expressing cells than in control cells. Galectin-3-labeled structures are also formed. The level and activity of cathepsin B and degradation of EGFR appears to be largely normal in ORF3a-expressing cells. This could be because soluble hydrolases and lysosomal membrane proteins are sorted to lysosomes via different routes (Braulke and Bonifacino, 2009), and they are differentially affected by ORF3a. HOPS is involved in the maturation of early endosomes and in homotypic and heterotypic fusion of late endosomes and lysosomes and thus is essential for the delivery of endocytic cargo and other materials to lysosomes (Pols et al., 2013a; Solinger and Spang, 2013; Wartosch et al., 2015). The perturbation of lysosomes in ORF3a-expressing cells may result from sequestration of HOPS components on late endosomes, which impairs retrograde transport of transmembrane proteins such as acid-hydrolase receptors from endosomes to the TGN (Bonifacino and Rojas, 2006), and reduces the targeting of HOPS to lysosomes for their fusion with incoming vesicles. Sequestration of VPS41 by VPS39 on late endosomes could also impair its HOPS-independent function in delivering lysosome-associated membrane proteins such as LAMP1 to the lysosomes (Pols et al., 2013b). The impaired lysosomal function caused by ORF3a could facilitate lysosome-mediated extracellular release of SARS-CoV-2 from infected cells (Ghosh et al., 2020). Expression of ORF3a causes a slight expansion of the population of late endosomes and lysosomes. Although nuclear levels of TFEB are increased, upregulation of the majority of genes involved in lysosomal biogenesis is not detected.
SARS-CoV ORF3a is involved in viral pathogenesis. Expression of SARS-CoV ORF3a induces cell death (Chan et al., 2009; Freundt et al., 2010; Yue et al., 2018), elicits ER stress (Minakshi et al., 2009), activates NF-κB and the NLRP3 inflammasome (Siu et al., 2019), and contributes to the release of the virus from infected cells (Castaño-Rodriguez et al., 2018; Lu et al., 2006). SARS-CoV ORF3a is localized on the plasma membrane, TGN, and also on some perinuclear punctate structures (Freundt et al., 2010; Lu et al., 2006; Yue et al., 2018). The few SARS-CoV ORF3a puncta are distinct from late endosomes/lysosomes (Castaño-Rodriguez et al., 2018; Lu et al., 2006). Some studies, however, reported that a small portion of SARS-CoV ORF3a puncta are colocalized with LAMP1 and expression of SARS-CoV ORF3a elicits lysosomal damage and dysfunction (Freundt et al., 2010; Yue et al., 2018). The different localization could be because the examined ORF3a is derived from different virus strains and/or expressed at a different level. We showed here that SARS-CoV ORF3a does not bind to HOPS or block autophagy activity. The unique function of SARS-CoV-2 ORF3a may contribute to the distinct infectivity and pathogenicity of SARS-CoV-2. Our findings imply that disruption of interactions between ORF3a and HOPS may represent a potential therapeutic strategy for COVID-19 treatment.
STAR★methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-LC3 (clone 4E12) | MBL | Cat# M152-3; RRID: AB_1279144 |
| Rabbit polyclonal anti-LC3 | Cell Signaling Technology | Cat# 2775S; RRID:AB_915 950 |
| Rabbit polyclonal anti-p62 | MBL | Cat# PM045; RRID:AB_1279301 |
| Mouse monoclonal anti-WIPI2 | Abcam | Cat# ab105459; RRID :AB_10860881 |
| Mouse monoclonal anti-LAMP1 (clone 1D4B) | BD Biosciences | Cat# 553792; RRID:AB_2134499 |
| Mouse monoclonal anti-Actin (clone 7D2C10) | Proteintech | Cat#60008-1-Ig; RRID:AB_2289225 |
| Mouse monoclonal anti-GM130 (Clone 35) | BD Biosciences | Cat# 610822; RRID:AB_398141 |
| Rabbit polyclonal anti-TOMM20 | Abcam | Cat#ab78547; RRID: AB_2043078 |
| Mouse monoclonal anti-EEA1 (clone 14) | BD Biosciences | Cat# 610457; RRID:AB_397830 |
| Rabbit polyclonal anti-ATG14 | Cell Signaling Technology | Cat# 5504;RRID :AB_10695397 |
| Rabbit monoclonal anti-ULK1 (clone D8H5) | Cell Signaling Technology | Cat# 8054; RRID :AB_11178668 |
| Rabbit monoclonal anti-VPS34 (clone D9A5) | Cell Signaling Technology | Cat# 4263; RRID :AB_2299765 |
| Rabbit polyclonal anti-Beclin1 | MBL | Cat# PD017; RRID :AB_1278767 |
| Mouse monoclonal anti-Flag (clone M2) | Sigma-Aldrich | Cat# F1804; RRID :AB_262044 |
| Rabbit polyclonal anti-STX17 | Sigma-Aldrich | Cat# SAB1304559; RRID:AB_2714021 |
| Rabbit polyclonal anti-VPS41 | Proteintech | Cat# 13869-1-AP, RRID:AB_2215349 |
| Rabbit polyclonal anti-VPS33A | GeneTex | Cat# GTX119416, RRID:AB_10617835 |
| Mouse monoclonal anti-Actin (clone 7D2 C10) | Proteintech | Cat# 60008-1-Ig; RRID :AB_2289225 |
| Mouse monoclonal anti-Myc (clone 9E10) | Sigma-Aldrich | Cat# M5546; RRID :AB_260581 |
| Mouse monoclonal anti-GFP (clone 7.1 and 13.1) | Roche | Cat# 11814460001; RRID :AB_390913 |
| Rabbit polyclonal anti-FIP200 | Proteintech | Cat# 17250-1-AP; RRID:AB_1066642 8 |
| Rabbit monoclonal anti-VAMP8 | Abcam | Cat# ab76021; RRID:AB_1310798 |
| Rabbit monoclonal anti-SNAP29 | Abcam | Cat# ab138500; RRID :AB_2687667 |
| Rabbit polyclonal anti-EGFR | Santa Cruz | Cat# sc-03; RRID: AB_631420 |
| Goat polyclonal anti-CTSD | Santa Cruz | Cat# sc-6486, RRID:AB_637896 |
| Mouse monoclonal anti-SARS-CoV NP | Sino Biological | Cat# 40143-MM05; RRID:AB_2827977 |
| Rabbit polyclonal anti-SARS-CoV-2 Spike | Sino Biological | Cat# 40591-MM42, RRID:AB_2857933 |
| Goat polyclonal anti-CTSB | R and D Systems | Cat# AF953; RRID: AB_ 355738 |
| Bacterial and Virus Strains | ||
| E. coli BL21-CodonPlus (DE3) | Agilent | Cat#280230 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protease inhibitor | Biotool | Cat# B14003 |
| LysoTracker Deep Red | Thermo Fisher Scientific | Cat# L12492 |
| DQ-Red BSA | Thermo Fisher Scientific | Cat# D12051 |
| MAGIC RED CATHEPSIN B Assays | ImmunoChemistry Technologies | Cat# 938 |
| Dextran Texas Red | Thermo Fisher Scientific | Cat#D3328 |
| Cycloheximide | Cell Signaling Technology | Cat# 2112 |
| EGF | Sigma-Aldrich | Cat# E5036 |
| Bafilomycin A1 | Millipore | Cat#19-148 |
| Torin 1 | Cell Signaling Technology | Cat# 14379 |
| GFP-Trap agarose beads | Chromo Tek | Cat#gta-20 |
| Critical Commercial Assays | ||
| RevertAid First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | Cat# K1621 |
| Deposited Data | ||
| Original Data | This paper | |
| Experimental Models: Cell Lines | ||
| HeLa cells | ATCC | CCL-2 |
| HEK293T cells | ATCC | CRL-11268 |
| Oligonucleotides | ||
| Primers used for qRT-PCR experiments, see Table S1 | This paper | N/A |
| siRNA sequence, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| GFP-FIP200 | Laboratory of Dr. Noboru Mizushima (University of Tokyo) | N/A |
| GFP-STX17 | Guo et al., 2014 | N/A |
| ORF3-FLAG | Laboratory of Dr. Peihui Wang (Shandong University) | N/A |
| SARS-ORF3-FLAG | Laboratory of Dr. Peihui Wang (Shandong University) | N/A |
| SARS-ORF3(R11G)-FLAG | This paper | N/A |
| ORF3-GFP | This paper | N/A |
| GFP-YKT6 | This paper | N/A |
| Flag-STX17 | Guo et al., 2014 | N/A |
| STX7-GFP | This paper | N/A |
| Flag-STX7 | This paper | N/A |
| GFP-SNAP29(QM) | Guo et al., 2014 | N/A |
| RFP-Rab5 | This paper | N/A |
| GFP-Rab7 | Wang et al., 2016 | N/A |
| RFP-Rab7 | Wang et al., 2016 | N/A |
| GFP-DFCP1 | Zhao et al., 2017 | N/A |
| ULK1-GFP | Zhao et al., 2017 | N/A |
| WIPI2-GFP | Zhao et al., 2017 | N/A |
| RFP-GFP-LC3 | Kimura et al., 2007 | Addgene Plasmid #21074 |
| VPS41-GFP | Miao et al., 2020 | N/A |
| VPS39-GFP | This paper | N/A |
| VPS11-GFP | This paper | N/A |
| VPS16-GFP | This paper | N/A |
| GFP-VPS18 | This paper | N/A |
| GFP-VPS33A | This paper | N/A |
| GFP-VPS8 | This paper | N/A |
| CNX-GFP | Miao et al., 2020 | N/A |
| RFP-LC3 | Wang et al., 2016 | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.en.softonic.com/ |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/ |
| Clustal Omega | EMBL-EBI | https://www.ebi.ac.uk/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hong Zhang (hongzhang@ibp.ac.cn).
Materials availability
Plasmids and other reagents generated in this study will be available upon request from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
This study did not generate unique datasets or codes.
Experimental model and subject details
Cell lines
HEK293T and HeLa cells obtained from ATCC were cultured in DMEM (SH30022.01B, Hyclone) with 10% FBS (FBS; SH30084.03, Hyclone) supplemented with 50 μg/ml penicillin-streptomycin at 37°C and 5% CO2. None of these cell lines are listed as misidentified or cross-contaminated in the International Cell Line Authentication Committee (ICLAC) database, and all were free of mycoplasma contamination. For serum starvation, cells were washed 3 times with PBS and incubated with DMEM without amino acids (SH4007.01, Hyclone) for the indicated time. To induce autophagy, cells were treated with 1 μM Torin 1 (Cell Signaling Technology, 14379) for 1 h. To inhibit autophagy activity, cells were incubated with 200 nM Bafilomycin A1 (Millipore, 19-148) for 4-6 h.
Method details
Plasmids
The following plasmids were kind gifts: GalT-YFP from Dr. Wei Liu (Zhejiang University School of Medicine, Hangzhou); FLAG-VPS39, FLAG-VPS41 and MYC-VPS33A from Dr. Chonglin Yang (Yunnan University, Kunming). Other plasmids were constructed as follows. Genes encoding SARS-CoV-2 proteins, including ORF3a, ORF7a, ORF7b, ORF8, ORF9a, ORF14, NSP3, NSP4, NSP5, NSP6, NSP7, NSP8, NSP9, NSP10, NSP13, NSP14, NSP15, E, M, S and N, and SARS-CoV ORF3a were cloned into pcDNA6B-FLAG vector. ORF3a-GFP was cloned using ORF3a-FLAG as a template into pEGFP-N1 vector between the XhoI and HindIII sites. cDNA encoding human ACE2 was cloned into pcDNA6B-FLAG vector. cDNAs encoding human WIPI2, VPS41, VPS11, VPS16, VPS39, ULK1, STX6, STX7 and CNX were cloned into pEGFP-N1 vector. cDNAs encoding VPS33A, VPS18, FIP200, DFCP1, RAB5, RAB7, LC3, YKT6 and STX17 were cloned into pEGFP-C1 vector. cDNAs encoding LC3 and RAB7 were cloned into RFP-N1 vector. STX17 and STX7 cDNAs were cloned into pFlag-CMV4 vector.
Transfection and siRNA in cell lines
Cells were transfected with the indicated plasmids with Lipofectamine 2000 (Life Technologies, 12566014) for 24 h according to the manufacturer’s instructions. Transfection of control or mRNA-specific siRNAs was carried out using Lipofectamine RNAi MAX (Life Technologies, 13778-150) for 72 h according to the manufacturer’s instructions.
Double-stranded siRNAs were purchased from GenePharma (Suzhou, China) and JTSBIO (Wuhan, China). The sequences were as follows:
NC, 5’-UUCUCCGAACGUGUCACGUTT-3’;
human OGT siRNA#1, 5’-GCAGUUCGCUUGUAUCGUATT-3’;
human OGT siRNA#2, 5’-GAUUAAGCCUGUUGAAGUCTT-3’;
human VPS39 siRNA #1, 5’-GGUAAAGAAGCUGAAUGACUCUGAU-3’;
human VPS39 siRNA #2, 5’-CAUUGCAGUGUUGCCUCGAUAUGUU-3;
human VPS41 siRNA#1, 5’-GAGAAUGAAUGUAGAGAUUTT-3’;
human VPS41 siRNA#2, 5’-GGGAUUUGCCAAGUCGAUATT-3’;
human VPS41 siRNA#3, 5’-GGAGAAUGAAUGUAGAGAUTT-3’;
human TRAF3 siRNA, 5’-GAAGGUUUCCUUGUUGCAGAAUGAA-3’.
SARS-CoV-2 virus amplification, titration and infection
All work with live SARS-CoV-2 virus was performed inside biosafety cabinets in the biosafety level 3 facility at the Institute of Microbiology, Chinese Academy of Sciences. Vero E6 cells were used to propagate the SARS-CoV-2 virus (hCoV-19/China/CAS-B001/2020, GISAID No. EPI_ISL_514256-7). The cytopathic efficiency (CPE) assay was performed to determine the virus titer. One day before virus infection, HeLa cells were plated in 24-well format and 6-well format with a concentration of 0.5~1.0×105 per well and 2~4.0×105 per well, respectively. 16~20 hours later, HeLa cells were transfected with human ACE2 and the indicated plasmids with Lipofectamine 2000 according to the manufacturer’s instructions. Then cells in the 6-well format were infected with 2×106 TCID50/ml (tissue culture infectious dose 50%) or 4×105 TCID50/ml virus per well for 1 hour, and cells in the 24-well format were infected with 5×105 TCID50/ml or 1×105 TCID50/ml. After infection for 1 hour, cells were washed 3 times with PBS and then incubated in DMEM medium plus 10% FBS for 8 h, 16 h or 24 h at 37°C in 5% CO2 incubators. Samples for the electron microscopy analysis were fixed with 2.5% glutaraldehyde and 4% PFA in PBS for 2 h at room temperature. This was followed by normal procedures for electron microscopy analysis. Samples for immunostaining assays were fixed with 4% PFA for 2 h at room temperature, and then subjected to normal procedures for immunostaining analysis.
Immunostaining assays
Cells were cultured on coverslips for immunostaining. After washing 3 times with PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), cells were fixed with 4% PFA for 15 min at room temperature, and then permeabilized in cold digitonin (Sigma, D141) for 10 min at 4°C. After blocking with 5% goat serum for 60 min at room temperature, cells were incubated with primary antibodies (diluted in 5% goat serum) overnight at 4°C. Then cells were washed 3 times with PBS and stained with fluorescently-labeled secondary antibodies for 60 min at room temperature. Lysosomes were stained with LysoTracker™ Deep Red (1:1000, L12492, Thermo Fisher Scientific) for 30 min at 37°C and 5% CO2. Coverslips were mounted on microscope slides with DAPI in 50% glycerol. Images were acquired with a confocal microscope (LSM 880 Meta plus Zeiss Axiovert zoom, Zeiss) with a 63×/1.40 oil-immersion objective lens (Plan-Apochromatlan, Zeiss) and a camera (Axiocam HRm, Zeiss).
Co-immunoprecipitation assays
HeLa cells and HEK293T cells were transfected with the indicated plasmids. Cells were harvested 24 h after transfection and lysed on ice for 30 min in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and protease inhibitor cocktail). After centrifugation at 13,000 rpm for 15 min at 4°C, supernatants were collected and immunoprecipitated by GFP-Trap agarose beads (gta-20, ChromoTek) for 1 h at 4°C. The bound proteins were eluted with 2xSDS sample buffer and analyzed by immunoblotting.
Immunoblotting assays
For immunoblotting assays, cells were washed 3 times with cold PBS then lysed on ice for 30 min in lysis buffer supplemented with protease inhibitor cocktail. After centrifuging for 15 min at 4°C at 13,000 rpm, the supernatants were eluted with 5xSDS sample buffer and subjected to SDS-PAGE. The signals were detected using corresponding primary and secondary antibodies.
Protein expression and purification
The full-length human VPS39 gene was PCR-amplified from a human cDNA library and subcloned into the in-house-modified version of pET-32a (Novagen) vector to generate N-terminal MBP-His6-tag-fused recombinant protein, and the SARS-CoV-2 ORF3a gene was inserted into the pGEX-4T-1 vector. Recombinant proteins were expressed in Escherichia coli BL21-CodonPlus (DE3) in LB medium at 18°C, and purified using Ni-NTA agarose beads (for MBP-hVPS39, Qiagen) and glutathione Sepharose 4B beads (for GST-ORF3a, GE Healthcare). The eluates were subjected to gel filtration (Superdex-200, GE Healthcare) with a column buffer containing 20 mM Tris pH 7.5, 300 mM NaCl, 1 mM Dithiothreitol (DTT). For purification of GST-ORF3a, 0.2 mM n-Dodecyl-beta-D-Maltopyranoside (DDM) was additionally added into the final column buffer. All purified proteins were assessed on SDS-PAGE gels.
GST pulldown assay
For capture of purified MBP-hVPS39, 20 μg of GST-ORF3a was incubated with 20 μL glutathione Sepharose beads (GE Healthcare) in 500 μl pulldown buffer, containing 20 mM Tris pH 7.5, 200 mM NaCl, 0.1% NP-40, and 1 mM DTT for 0.5 to 1 h at 4°C. GST protein was used as a control. The beads were washed 3 times with the pulldown buffer and then incubated with 30 μg of MBP-hVPS39A at 4 °C for ~2 h. The reactions were washed 6 times, and the bound proteins were eluted and detected by Western blotting using anti-MBP antibody.
Electron microscopy analysis
For electron microscopy analysis, cells were first transfected with GFP control and ORF3-GFP, then GFP-expressing cells were obtained by fluorescence activated cell sorting (FACS) 24 h later. The cells were then fixed with 2.5% glutaraldehyde in PBS for 2 h at room temperature, followed by post-fixation with 1% OsO4 and 1% potassium ferrocyanide for another 45 min. After washing with ddH2O, cells were treated with thiocarbohydrazide (TCH) solution for 30 min at room temperature. Then cells were washed in ddH2O and placed in 1% OsO4 for 45 min at room temperature. After washing with ddH2O, cells were further treated with a graded series of ethanol solutions for dehydration, then embedded in epoxy resin. Ultrathin sections were stained with uranyl acetate and lead citrate. Images were examined under a 120 kV electron microscope (H-7650B, Hitachi) at 100 kV at room temperature with an AMT CCD camera (XR-41) using DigitalMicrograph software.
For DAB staining, cells were transfected with a construct expressing a WIPI2-APEX2 fusion protein. Monolayer cells were fixed with 2.5% glutaraldehyde in PBS overnight at 4°C and rinsed 5×1 min in PBS on ice. Then cells were treated with 20 mM glycine solution for 5 min and rinsed 5×1 min in PBS. Cells were treated with a freshly diluted solution of 0.5 mg/ml (1.4 mM) DAB tetrahydrochloride for 10 min, then rinsed 5×1 min with chilled buffer. Cells were post-fixed in 1% OsO4 for 10 min on ice. Cells were rinsed 5×1 min in chilled distilled water, and then placed in chilled 2% aqueous uranyl acetate for 30 min. After washing with ddH2O, cells were further dehydrated with a graded series of ethanol solutions and embedded in epoxy resin.
Fluorescence protease protection (FPP)
Cells were plated on glass bottom dishes (801001, NEST) and transfected with the indicated plasmids for 24 h. After washing with KHM buffer (100 mM potassium acetate, 20 mM HEPES, 2 mM MgCl2, pH 7.4), cells were incubated with 20 μM digitonin for 5 min. Then cells were treated with 50 μg/ml proteinase K (P2308, Sigma) and real-time images were immediately recorded by a confocal microscope (LSM 880 Meta plus Zeiss Axiovert zoom, Zeiss).
HaloTag-LC3 assays
HaloTag-LC3 assays were performed as previously described (Takahashi et al., 2018). Briefly, cells were transfected with HaloTag-LC3 for 24 h and treated with 20 μM digitonin in KHM buffer at 37°C for 2 min. Then cells were incubated with membrane-impermeable ligand (MIL) HaloTag® Alexa Fluor® 488 ligand (G1001, Promega) at 37°C for 15 min. Cells were fixed in 4% PFA for 20 min at room temperature, washed 3 times with PBS, and stained with membrane-permeable ligand (MPL) HaloTag® TMR ligand (G8251, Promega) for 30 min at room temperature. Coverslips were mounted on microscope slides with DAPI in 50% glycerol and examined by a confocal microscope (LSM 880 Meta plus Zeiss Axiovert zoom, Zeiss).
Phagocytosis of dextran-Texas Red
Cells were grown on glass coverslips and incubated with dextran-Texas Red (ThermoFisher Scientific, Invitrogen, D2338) for 2 h in culture medium containing 10% FBS. Cells were washed with culture medium and cultured for 30 min before fixation with 4% PFA. The colocalization of dextran and LAMP1 was then analyzed by immunostaining.
Quantitative RT-PCR
Total RNA was extracted from cells transfected with the indicated siRNAs using TRIzol (15596018, ThermoFisher Scientific). cDNAs were synthesized with a RevertAid First Strand cDNA Synthesis Kit (K1622, ThermoFisher Scientific). Quantitative RT-PCR was carried out on a QuantStudio 7 Flex PCR System (Applied Biosystems, CA) with UltraSYBR Mixture (CW2602M, CWBIO). Data were normalized to the Actin level. Results are representative of at least three experiments. The primers are listed in Table S1.
Quantification and statistical analysis
For immunoblotting and co-IP assays, at least 3 independent repeats were performed for each experiment and the density of immunoblot bands was quantified using Image J software (NIH Image). For immunostaining, the cells or images were randomly selected for analysis. Graph plots and P-values were generated using GraphPad Prism 5 software. Statistical comparisons were made using the two-tailed unpaired Student’s t-test and the results are shown as mean ± S.E.M. ns: no significant difference, p>0.05; ∗: p<0.05; ∗∗: p<0.01; ∗∗∗: p<0.001. Statistical parameters including the definitions, exact values of n, what n represents, and statistical significance are reported in the Figures and corresponding Figure Legends.
Acknowledgments
We are grateful to Dr. Isabel Hanson for editing work. This work was supported by the following grants to H. Zhang: National Natural Science Foundation of China (92054301, 31421002, 31561143001, 31630048, and 31790403), Chinese Ministry of Science and Technology (2017YFA0503401), Beijing Municipal Science and Technology Committee (Z181100001318003), Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (grant XDB19000000), and Key Research Program of Frontier Sciences, CAS (grant QYZDY-SSW-SMC006). This work was also supported by a grant from the COVID-19 emergency tackling research project of Shandong University (grant no. 2020XGB03 to P.W.).
Author contributions
H. Zhang designed the experiments. G.M., Y.C., and M.J. performed all mammalian tissue culture experiments. H. Zhao conducted the EM analysis. Y.L. did the SARS-CoV-2 virus infection experiments. Y.S. and Y.B. provided the SARS-CoV-2 virus. P.W. constructed expression plasmids for SARS-CoV-2 proteins. G.M. and H. Zhang wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: December 16, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.devcel.2020.12.010.
Supplemental Information
References
- Aits S., Kricker J., Liu B., Ellegaard A.M., Hämälistö S., Tvingsholm S., Corcelle-Termeau E., Høgh S., Farkas T., Holm Jonassen A., et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy. 2015;11:1408–1424. doi: 10.1080/15548627.2015.1063871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar H.J., Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- Bird S.W., Maynard N.D., Covert M.W., Kirkegaard K. Nonlytic viral spread enhanced by autophagy components. Proc. Natl. Acad. Sci. USA. 2014;111:13081–13086. doi: 10.1073/pnas.1401437111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Deutsch G., Lacy J.M., Williams T., et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington state: a case series. Lancet. 2020;396:320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braulke T., Bonifacino J.S. Sorting of lysosomal proteins. Biochim. Biophys. Acta. 2009;1793:605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Bröcker C., Kuhlee A., Gatsogiannis C., Balderhaar H.J., Hönscher C., Engelbrecht-Vandré S., Ungermann C., Raunser S. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. USA. 2012;109:1991–1996. doi: 10.1073/pnas.1117797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Rodriguez C., Honrubia J.M., Gutiérrez-Álvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Báguena C., Queralt-Martín M., et al. Role of severe acute respiratory syndrome coronavirus Viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9 doi: 10.1128/mBio.02325-17. e02325–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero E., van der Vaart A., Zhao M.T., Rieter E., Klionsky D.J., Helms J.B., Reggiori F. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr. Biol. 2012;22:1545–1553. doi: 10.1016/j.cub.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.M., Tsoi H., Chan W.M., Zhai S., Wong C.O., Yao X., Chan W.Y., Tsui S.K., Chan H.Y. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X.T., Xie Y.X., Zhou B., Huang N., Farfel-Becker T., Sheng Z.H. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J. Cell Biol. 2018;217:3127–3139. doi: 10.1083/jcb.201711083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Bowman J.W., Jung J.U. Autophagy during viral infection - a double-edged sword. Nat. Rev. Microbiol. 2018;16:341–354. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona A.K., Saulsbery H.M., Corona Velazquez A.F.C., Jackson W.T. Enteroviruses remodel autophagic trafficking through regulation of host SNARE proteins to promote virus replication and cell exit. Cell Rep. 2018;22:3304–3314. doi: 10.1016/j.celrep.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Saitoh T., Akira S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J., Liu R., Rong Y., Zhao M., Zhang J., Lai Y., Zhou Q., Wilz L.M., Li J., Vivona S., et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Zhang G., Yang X., Zhang S., Chen L., Yan Q., Xu M., Banerjee A.K., Chen M. Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production. Cell Host Microbe. 2014;15:564–577. doi: 10.1016/j.chom.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Dreux M., Chisari F.V. Viruses and the autophagy machinery. Cell Cycle. 2010;9:1295–1307. doi: 10.4161/cc.9.7.11109. [DOI] [PubMed] [Google Scholar]
- Feng Y.C., He D., Yao Z.Y., Klionsky D.J. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt E.C., Yu L., Goldsmith C.S., Welsh S., Cheng A., Yount B., Liu W., Frieman M.B., Buchholz U.J., Screaton G.R., et al. The open reading frame 3a Protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 2010;84:1097–1109. doi: 10.1128/JVI.01662-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S., Sharma M., Ung C., Tuli A., Barral D.C., Hava D.L., Veerapen N., Besra G.S., Hacohen N., Brenner M.B. Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity. 2011;35:182–193. doi: 10.1016/j.immuni.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Dellibovi-Ragheb T.A., Kerviel A., Pak E., Qiu Q., Fisher M., Takvorian P.M., Bleck C., Hsu V.W., Fehr A.R., et al. β-coronaviruses use lysosomes for Egress instead of the biosynthetic secretory pathway. Cell. 2020;183:1520–1535.e14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Liang Q., Li L., Hu Z., Wu F., Zhang P., Ma Y., Zhao B., Kovács A.L., Zhang Z., et al. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 2014;16:1215–1226. doi: 10.1038/ncb3066. [DOI] [PubMed] [Google Scholar]
- Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020;295:12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries W.H., Payne C.K. Imaging lysosomal enzyme activity in live cells using self-quenched substrates. Anal. Biochem. 2012;424:178–183. doi: 10.1016/j.ab.2012.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5 doi: 10.1128/mSystems.00266-20. e00266–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Itakura E., Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W.T. Viruses and the autophagy pathway. Virology. 2015;479–480:450–456. doi: 10.1016/j.virol.2015.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W.T., Giddings T.H., Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., Scheller R.H. SNAREs--engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jiang P., Nishimura T., Sakamaki Y., Itakura E., Hatta T., Natsume T., Mizushima N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemball C.C., Alirezaei M., Flynn C.T., Wood M.R., Harkins S., Kiosses W.B., Whitton J.L. Coxsackievirus infection induces autophagy-like vesicles and Megaphagosomes in pancreatic acinar cells in vivo. J. Virol. 2010;84:12110–12124. doi: 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.Y., Krämer H., Yamamoto A., Kominami E., Kohsaka S., Akazawa C. Molecular characterization of mammalian homologues of class C Vps proteins that interact with syntaxin-7. J. Biol. Chem. 2001;276:29393–29402. doi: 10.1074/jbc.M101778200. [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C.M., Adams P.D., Adeli K., et al. Guidelines for the use and interpretation of assays for monitoring autophagy, (3rd Edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., Tooze S.A. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol.. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H.W. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. USA. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Hawes P.C., Cottam E.M., Mantell J., Verkade P., Monaghan P., Wileman T., Britton P. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio. 2013;4 doi: 10.1128/mBio.00801-13. e00801–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Jiang P.D., Nakano S., Sakamaki Y., Yamamoto H., Mizushima N. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol. 2018;217:2633–2645. doi: 10.1083/jcb.201712058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao G.Y., Zhang Y.J., Chen D., Zhang H. The ER-localized transmembrane protein TMEM39A/SUSR2 regulates autophagy by controlling the trafficking of the PtdIns(4)P phosphatase SAC1. Mol. Cell. 2020;77:618–632.e5. doi: 10.1016/j.molcel.2019.10.035. [DOI] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 2009;4:e8342. doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Mohamud Y., Shi J.Y., Qu J.Y., Poon T., Xue Y.C., Deng H.Y., Zhang J.C., Luo H.L. Enteroviral infection inhibits autophagic flux via disruption of the SNARE complex to enhance viral replication. Cell Rep. 2018;22:3292–3303. doi: 10.1016/j.celrep.2018.02.090. [DOI] [PubMed] [Google Scholar]
- Nakamura S., Shigeyama S., Minami S., Shima T., Akayama S., Matsuda T., Esposito A., Napolitano G., Kuma A., Namba-Hamano T., et al. LC3 lipidation is essential for TFEB activation during the lysosomal damage response to kidney injury. Nat. Cell Biol. 2020;22:1252–1263. doi: 10.1038/s41556-020-00583-9. [DOI] [PubMed] [Google Scholar]
- Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D.A., Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Peralta E.R., Martin B.C., Edinger A.L. Differential effects of TBC1D15 and mammalian Vps39 on Rab7 activation state, lysosomal morphology, and growth factor dependence. J. Biol. Chem. 2010;285:16814–16821. doi: 10.1074/jbc.M110.111633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno A., Padey B., Julien T., Trouillet-Assant S., Traversier A., Errazuriz-Cerda E., Fouret J., Dubois J., Gaymard A., Lescure F.-X., et al. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell. Rep. Med. 2020;1:100059. doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemel R.L., Lobingier B.T., Brett C.L., Angers C.G., Nickerson D.P., Paulsel A., Sprague D., Merz A.J. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol. Biol. Cell. 2011;22:1353–1363. doi: 10.1091/mbc.E10-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols M.S., ten Brink C., Gosavi P., Oorschot V., Klumperman J. The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic. 2013;14:219–232. doi: 10.1111/tra.12027. [DOI] [PubMed] [Google Scholar]
- Pols M.S., van Meel E., Oorschot V., ten Brink C., Fukuda M., Swetha M.G., Mayor S., Klumperman J. hVps41 and VAMP7 function in direct TGN to late endosome transport of lysosomal membrane proteins. Nat. Commun. 2013;4:1361. doi: 10.1038/ncomms2360. [DOI] [PubMed] [Google Scholar]
- Raben N., Puertollano R. TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu. Rev. Cell Dev. Biol. 2016;32:255–278. doi: 10.1146/annurev-cellbio-111315-125407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A.M., Molinari M. Coronaviruses hijack the LC3-I-Positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H.M., Elgner F., Jiang B.F., Himmelsbach K., Medvedev R., Ploen D., Hildt E. The autophagosomal SNARE protein syntaxin 17 is an essential factor for the hepatitis C virus life cycle. J. Virol. 2016;90:5989–6000. doi: 10.1128/JVI.00551-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.L., Yuen K.S., Castaño-Rodriguez C., Ye Z.W., Yeung M.L., Fung S.Y., Yuan S.F., Chan C.P., Yuen K.Y., Enjuanes L., Jin D.-Y. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger J.A., Spang A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS Journal. 2013;280:2743–2757. doi: 10.1111/febs.12151. [DOI] [PubMed] [Google Scholar]
- Suhy D.A., Giddings T.H., Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., He H., Tang Z., Hattori T., Liu Y., Young M.M., Serfass J.M., Chen L., Gebru M., Chen C., et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 2018;9:2855. doi: 10.1038/s41467-018-05254-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Pircs K., Nagy P., Varga Á., Kárpáti M., Hegedűs K., Kramer H., Kovács A.L., Sass M., Juhász G. Interaction of the HOPS complex with syntaxin 17 mediates autophagosome clearance in Drosophila. Mol. Biol. Cell. 2014;25:1338–1354. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Lim S.G., Hong W.J. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antiviral Res. 2006;72:78–88. doi: 10.1016/j.antiviral.2006.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velikkakath A.K., Nishimura T., Oita E., Ishihara N., Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell. 2012;23:896–909. doi: 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viret C., Rozières A., Faure M. Autophagy during early virus-host cell interactions. J. Mol. Biol. 2018;430:1696–1713. doi: 10.1016/j.jmb.2018.04.018. [DOI] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Miao G., Xue X., Guo X., Yuan C., Wang Z., Zhang G., Chen Y., Feng D., Hu J., Zhang H. The vici syndrome protein EPG5 Is a Rab7 effector that determines the fusion specificity of autophagosomes with late endosomes/lysosomes. Mol. Cell. 2016;63:781–795. doi: 10.1016/j.molcel.2016.08.021. [DOI] [PubMed] [Google Scholar]
- Wartosch L., Günesdogan U., Graham S.C., Luzio J.P. Recruitment of VPS33A to HOPS by VPS16 is required for lysosome fusion with endosomes and autophagosomes. Traffic. 2015;16:727–742. doi: 10.1111/tra.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G., Limpens R.W.A.L., Zevenhoven-Dobbe J.C., Laugks U., Zheng S., de Jong A.W.M., Koning R.I., Agard D.A., Grünewald K., Koster A.J., et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.H., Sanyal S. Manipulation of autophagy by (+) RNA viruses. Semin. Cell Dev. Biol. 2020;101:3–11. doi: 10.1016/j.semcdb.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Zhang J.C., Si X.N., Gao G., Mao I., McManus B.M., Luo H.L. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 2008;82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.H., Xu Y., Zhang T., Griffiths G., Lowe S.L., Subramaniam V.N., Seow K.T., Hong W. GS32, a novel Golgi SNARE of 32 kDa, interacts preferentially with syntaxin 6. Mol. Biol. Cell. 1999;10:119–134. doi: 10.1091/mbc.10.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.W., Cheng S.Y., Zhao H.Y., Zou W., Yoshina S., Mitani S., Zhang H., Wang X.C. PI3P phosphatase activity is required for autophagosome maturation and autolysosome formation. EMBO Rep. 2014;15:973–981. doi: 10.15252/embr.201438618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Nabar N.R., Shi C.S., Kamenyeva O., Xiao X., Hwang I.Y., Wang M., Kehrl J.H. SARS-coronavirus open reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., Wang Z., Liu N., Li L., Chen S., et al. The ER-localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell. 2017;67:974–989.e6. doi: 10.1016/j.molcel.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Zhao Y.G., Zhang H. Formation and maturation of autophagosomes in higher eukaryotes: a social network. Curr. Opin. Cell Biol. 2018;53:29–36. doi: 10.1016/j.ceb.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Zhao Y.G., Zhang H. Autophagosome maturation: an epic journey from the ER to lysosomes. J. Cell Biol. 2019;218:757–770. doi: 10.1083/jcb.201810099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.J., Thackray L.B., Miller B.C., Lynn T.M., Becker M.M., Ward E., Mizushima N.N., Denison M.R., Virgin H.W.I.V. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate unique datasets or codes.