Abstract
Coronavirus disease 19 (COVID-19) is the latest pandemic resulted from the coronavirus family. Due to the high prevalence of this disease, its high mortality rate, and the lack of effective treatment, the need for affordable and accessible drugs is one of the main challenges in this regard. It has been proved that RdRp, 3CL, Spike, and Nucleocapsid are the most important viral proteins playing vital roles in the processes of proliferation and infection. Therefore, we started studying a wide range of bio-peptides and then conducted molecular docking analyses to investigate their binding affinity for the inhibition of these proteins. After obtaining the best bio-peptides with the highest affinity scores, they were examined for further study and then manipulated to eliminate their side effects. Additionally, the molecular dynamic simulation was performed to validate the structure and interaction stability. The results of this study reveal that glycocin F from Lactococcus lactis and lactococcine G from Lactobacillus plantarum had the high affinities to bind to the viral proteins, and the manipulation of their sequence also led to the side effects’ elimination. In addition, in some cases, their affinities to attach the SARS-CoV-2 proteins have increased. It seems that these two drugs which were discovered and designed, are optimal for treating the COVID-19 infection. However, experimental and pre-clinical studies are necessary to assay their therapeutic effects.
Keywords: COVID-19, Probiotic bacteria, Drug discovery, Bio antimicrobial peptides, Bioinformatics, Drug designing
1. Introduction
The last outbreak of acute respiratory disease has been named COVID-19. Accordingly, it is the third documented disease spreading from other species infected by coronavirus to humans in the last two decades, which has led to a major epidemic [1]. The new virus originated from SARS-CoV with over 95% homology is called SARS-CoV-2 [2,3]. A positive sense RNA from 60 nm to 140 nm in diameter and a Spike on its surface are the main features of this family of viruses. Human coronaviruses generally cause mild respiratory disease. Their genome consists of 6–11 open reading frames (ORFs), which can code non-structural and structural proteins. The proteins transcribed from these sequences are cut by 3-chymotrypsin-like (3CL), which is the main protease of SARS-CoV-2 [4]. Furthermore, M (membrane), E (envelope), N (nucleocapsid), and Spike proteins are its four main structural proteins [5].
Spike is responsible for binding to the host receptor and viral entry. SARS-CoV-2 infects host cells via binding to angiotensin-converting enzyme II (ACE 2) [6], which are dominantly present on the lung epithelial cells [7]. Thus, blocking the spike can be considered as an effective approach to prevent COVID-19 infection.
The N protein of SARS-CoV-2 plays many roles, including setting viral RNA synthesis through replication, forming helical ribonucleoproteins (RNPs) while packaging of the RNA genome, modulating the infected cell metabolism, and transcription. Therefore, this multifunctional RNA-binding protein is crucial for the transcription and replication of viral RNA [8]. The main functions of N protein are attaching to the viral RNA genome and collecting them into a large helical nucleocapsid structure or RNP complex [9]. Many studies have reported that N protein bound to leader RNA to maintain suitable RNA conformation for the replication and transcription of the viral genome [10]. Moreover, several investigations approved that N protein could regulate host-pathogen interactions, including apoptosis, host cell cycle progression, and actin reorganization [[11], [12], [13]]. The N protein is abundantly expressed during infection, so blocking it may modulate the infection [[14], [15], [16]].
3CL is a proven target for the inhibition of SARS-CoV and MERS-CoV. 3CL gene located at the 3’ end with excessive variability [17], is responsible for the replication of virus particles by cleaving the viral polyproteins at 11 distinct site to generate various non-structural proteins that are important for viral replication [18]. Therefore, as there are no host-cell proteases with the specificity to inhibit it, 3CL could be considered as another potential target for SARS-CoV-2 [19]. Moreover, SARS-CoV-2 expresses an RNA-dependent RNA polymerase (RdRp) to generate the daughter RNA genome. RdRp, also known as nsp12 [20], catalyzes the synthesis of a complementary RNA strand by the use of the virus RNA template [21] and helping host transcription factors [22].
Virtual screening policies in antiviral databases are so important to identify potential therapeutic targets [23] and also in the global effort to find the best drug to prevent or control the COVID-19 infection. Correspondingly, we suggested the bio-antimicrobial peptides (bio-AMPs) as therapeutic targets for this fatal disease [24].
In recent years, AMPs have been broadly used as a hopeful solution to conquer dangerous microorganisms. Many microorganisms produce these peptides as their innate immune response component to invade pathogens [25]. The use of AMP can be promising as a therapeutic tool for increasing viral infections, for which no authorized medication or treatment is available [26]. Notably, AMPs are used to treat viral-related infections such as zika virus (ZIKV), dengue virus (DENV) [27], and Influenza A virus (IAV) [28].
Lactoferrin is one of the AMPs playing an inhibitory role in the treatment of some viruses, including herpes simplex virus (HSV) [29], hepatitis C virus (HCV) [30], human immunodeficiency viruses (HIV) [31], rotavirus, and polio [32]. Based on the studies performed on the Lactoferrin antiviral activities and inhibitory effects of this AMP on SARS-CoV-2, this peptide has been recently introduced as a probable therapeutic target for COVID-19 [33].
Therefore, the use of AMPs, which have inhibitory effects on RdRp [21], 3CL [19], S [6], and N [14], as the known proteins of the SARS-CoV-2 virus, is a significant idea and a promising solution for the treatment of COVID-19. One study simulated a computational model of Spike and designed an HR2-based antiviral peptide to inhibit this protein. The affinity of this antiviral peptide was shown to be strong that could completely bind to HR1 of Spike to prevent the formation of the fusion core [34]. Another study has also found that α-helical peptides block ACE2, leading to the inhibition of SARS-CoV-2 [35]. In another in-silico study, the efficacies of lopinavir, ritonavir, hydroxychloroquine, and favipiravir drugs were improved by adding TAT-peptide to them, and their interactions with 3CL have considerably increased [36].
Although previous studies were performed on AMPs and generally on the inhibition of one or two proteins of SARS-CoV-2, the present research was conducted to prevent four basic proteins of SARS-CoV-2 by bio-AMPs from probiotic sources as a new approach. As a result, it was revealed that glycocin F and lactococcine G are the best bio-AMPs to block RdRp, 3CL, S, and N proteins of SARS-CoV-2 with minimal side effects. In addition, it was found that they can be easily consumed in people's daily diet [37].
2. Materials and methods
2.1. Modeling and structure selection of SARS-CoV-2 protein
Initially, the structures of the SARS-CoV-2 Spike, 3CL, RdRp, N proteins, and the ACE2 receptor were modeled by the SWISS-MODEL modeling tools (https://swissmodel.expasy.org/) [38] to obtain the same structures as the mentioned SARS-CoV-2 proteins. Afterward, using the RCSB PDB database (https://www.rcsb.org/) [39], the PDB codes of the similar structures to SARS-CoV-2 were found. Both models and PDB structures were then compared to find the best structure for further studies (Spike PDB ID: 6VW1, 3CL PDB ID: 6m2n, N protein PDB ID: 6m3m, RdRp PDB ID: 7btf).
2.2. Bio-AMPs extraction
StraPep (http://isyslab.info/StraPep/) [40] and PhytAMP (http://phytamp.hammamilab.org/main.php) [41] databases were used to figure out AMPs. For this purpose, approximately 500 bio-peptides were analyzed based on their structures. Bio-AMPs with appropriate structures were selected for molecular docking in terms of having suitable amino acid sequences to prevent dock failure.
2.3. Finding the important amino acids in interaction
The important amino acids of the SARS-CoV-2 Spike, 3CL, RdRp, N proteins, and ACE2 were extracted from several articles to determine the binding site, which are shown in Table 1 .
Table 1.
PDB code, active residue, and important amino acids of SARS-CoV-2 proteins interaction.
| Protein Name | PDB Code | Active Residue and Important Amino Acids in Interaction | Ref. |
|---|---|---|---|
| SARS-CoV-2 Spike | 6VW1 | Asn 501, Ser 494, Gln 493, Leu 455, Phe 486 | [[42], [43], [44]] |
| SARS-CoV-2 RdRp | 7btf | Lys 545, Arg 555, Thr 556, Arg 624, Asp 452, Asp 623, Tyr 619, Val 557, Arg 553, Lys 621, Lys 798, Lys 551, Arg 836, Ser 549, Ala 547 | [45,46] |
| SARS-CoV-2 3CLpro | 6m2n | His 41, Met 49, Glu 166, His 172, His 163, Cys 145 | [[47], [48], [49], [50]] |
| SARS-CoV-2 N protein | 6m3m | Tyr 110, Tyr 112, Ala 56, Thr 55, Arg 89, Tyr88, Phe 111, Gly 148, Ala 51, Ala 91, Thr 50 | [51,52] |
| Human ACE2 Receptor | 1r42 | Gln 42, Lys 353, Tyr 41, Arg 357, His 34, Asp 30, Gln 24, Met 82, Lys 31, Asp 38 | [[42], [43], [44]] |
2.4. Molecular docking analysis
HADDOCK 2.2 (https://alcazar.science.uu.nl/services/HADDOCK2.2/) [53] was used to perform the molecular docking analysis between bio-AMPs and the above-mentioned virus proteins. Furthermore, it was used to study the affinity of the SARS-CoV-2 Spike and human ACE2 receptor. Accordingly, those that had more or close binding affinity to Spike/ACE2 binding energy were kept and those with less binding affinity were ignored in this survey.
2.5. Determination of best score peptides features
The peptides with the best scores were checked for allergenicity, toxicity, anti-angiogenic, interlukine 4 inducing ability, anti-cancer ability, and hemolyticity by AlgPred (https://webs.iiitd.edu.in/raghava/algpred/submission.html) [54], toxinpred (http://crdd.osdd.net/raghava/toxinpred/) [55], target antiangio (http://codes.bio/targetantiangio/) [56], IL-4pred (https://webs.iiitd.edu.in/raghava/il4pred/), ACPred (http://codes.bio/acpred/) [57], and hemopred (http://codes.bio/hemopred/) [58]databases, respectively.
2.6. Mutation and structure modeling
In order to prove the side effects of the common peptides with the best scores, the mutation was performed on their amino acid sequences and they were re-modeled by SWISS-MODEL modeling tools and UCSF Chimera software version 1.14 to find the best-predicted model for the mutated peptides. After the mutation and peptide modeling, all docking analyses were repeated to find out the mutation effect on peptides’ affinity. Additionally, the interaction figures were designed by UCSF Chimera software.
2.7. Molecular dynamics simulation
The molecular dynamics (MD) was carried out for the 2JPK/Spike complex after the mutation, to find the stability of complex structure using version 5 GROningen MAchine for Chemical Simulations (GROMACS) (http://gromacs.org) in the operating system Linux. The complex was then simulated for 45 ns, in order to find out the dynamic behavior and determine the pattern of interaction. The force field was Gromos 54A7, and sodium (Na+) and chloride (Cl-) ions were used to neutralize SPC model for filling water with pressure 1 bar, at the temperature of 300 K, and neutral pH set. Thereafter, Root-Mean-Square Deviation (RMSD) and RootMean-Square Fluctuation (RMSF) diagrams of the simulated complex protein were obtained from studying the physical movements of atoms and molecules.
3. Results
The results of docking analyses are presented in Table 2, Table 3, Table 4, Table 5, Table 6 . The affinity between ACE2 and SARS-CoV-2 Spike protein has been determined as a criterion for comparison. Notably, the Bacteriocin plantaricin ASM1 peptide had the best affinity to the SARS-CoV-2 Spike. Moreover, Bacteriocin lactococcine G had the best affinity for RdRp protein, and Bacteriocin glycocin F had the highest binding affinity for 3CL and N proteins gained from StraPep and PhytAMP databases.
Table 2.
The binding energy between ACE2 and Spike protein of SARS-COV-2.
| Protein interactions | Docking score (kcal/mol) |
|---|---|
| ACE2/SARS-COV-2 Spike | −128.8 ± 3.5 |
Table 3.
The best peptides according to their binding energy with SARS-COV-2 Spike protein.
| peptide code | peptide name | microorganism | sequence | dock score (kcal/mol) |
|---|---|---|---|---|
| 2MVI | Bacteriocin plantaricin ASM1 | Lactobacillus plantarum | KPAWCWYTLAMCGAGYDSGTCDYMYSHCFGVKHSSGGGGSYHC | −149.7 ± 3.2 |
| 2JPK | Bacteriocin lactococcin-G subunit beta | Lactococcus lactis subsp. lactis | KKWGWLAWVDPAYEFIKGFGKGAIKEGNKDKWKNI | −143.7 ± 3.5 |
| 2LJ7 | Defensin Lc-def | Lens culinaris subsp. culinaris | KTCENLSDSFKGPCIPDGNCNKHCKEKEHLLSGRCRDDFRCWCTRNC | −143.0 ± 1.4 |
| 2LG5 | Gallinacin-2 | Gallus gallus | LFCKGGSCHFGGCPSHLIKVGSCFGFRSCCKWPWNA | −140.4 ± 4.8 |
| 2KUY | Bacteriocin glycocin F | Lactobacillus Plantarum | KPAWCWYTLAMCGAGYDSGTCDYMYSHCFGIKHHSSGSSSYHC | −139.0 ± 6.5 |
| 1H5O | Crotamine | Crotalus durissus terrificus | YKQCHKKGGHCFPKEKICLPPSSDFGKMDCRWRWKCCKKGSG | −137.8 ± 6.5 |
| 2RU0 | Actinomycin | Actinomyces sp. oral taxon 171 str. F0337 | GFGCPWNAYECDRHCVSKGYTGGNCRGKIRQTCHCY | −133.1 ± 7.4 |
| 1Z64 | Pleurocidin | Pseudopleuronectes americanus | GWGSFFKKAAHVGKHVGKAALTHYL | −130.9 ± 2.5 |
Table 4.
The best peptides according to their binding energy with SARS-COV-2 RdRp protein.
| peptide code | peptide name | microorganism | sequence | dock score (kcal/mol) |
|---|---|---|---|---|
| 2JPK | Bacteriocin lactococcin-G subunit beta | Lactococcus lactis subsp. lactis | KKWGWLAWVDPAYEFIKGFGKGAIKEGNKDKWKNI | −151.4 ± 9.8 |
| 2MLU | LsbB | Lactococcus lactis subsp. lactis | MKTILRFVAGYDIASHKKKTGGYPWERGKA | −141.5 ± 20.5 |
| 5E5Q | Snakin-1 | Solanum tuberosum | GSNFCDSKCKLRCSKAGLADRCLKYCGICCEECKCVPSGTYGNKHECPCYRDKKNSKGKSKCP | −134.0 ± 8.1 |
Table 5.
The best peptides according to their binding energy with 3CL of SARS-COV-2.
| peptide code | peptide name | microorganism | sequence | dock score (kcal/mol) |
|---|---|---|---|---|
| 2KUY | Bacteriocin glycocin F | Lactobacillus Plantarum | KPAWCWYTLAMCGAGYDSGTCDYMYSHCFGIKHHSSGSSSYHC | −155.3 ± 7.5 |
| 2KET | Cathelicidin-6 | Bos taurus | GRFKRFRKKFKKLFKKLSPVIPLLHL | −146.3 ± 4.6 |
| 1PXQ | Subtilosin-A | Bacillus subtilis | NKGCATCSIGAACLVDGPIPDFEIAGAXGLXGLWG | −146.0 ± 11.9 |
| 2KEG | Bacteriocin PlnK | Lactobacillus Plantarum | RRSRKNGIGYAIGYAFGAVERAVLGGSRDYNK | −142.9 ± 11.5 |
| 2JOS | Moronecidin | Morone saxatilis | FFHHIFRGIVHVGKTIHRLVTG | −142.5 ± 2.0 |
| 2JPK | Bacteriocin lactococcin-G subunit beta | Lactococcus lactis subsp. lactis | KKWGWLAWVDPAYEFIKGFGKGAIKEGNKDKWKNI | −138.9 ± 10.4 |
| 4GV5 | Crotamine Ile-19 | Crotalus durissus ruruima | YKQCHKKGGHCFPKEKICLPPSSDFGKMDCRWRWKCCKKGSG | −138.6 ± 3.3 |
| 1CW6 | Bacteriocin leucocin-A | Leuconostoc gelidum | KYYGNGVHCTKSGCSVNWGEAFSAGVHRLANGGNGFW | −138.2 ± 3.7 |
| 2G9P | M-zodatoxin-Lt2a | Lachesana tarabaevi | GLFGKLIKKFGRKAISYAVKKARGKH | −132.7 ± 9.7 |
| 1RKK | Polyphemusin-1 | Limulus polyphemus | RRWCFRVCYRGFCYRKCR | −131.2 ± 7.6 |
| 1EWS | Corticostatin-related peptide RK-1 | Oryctolagus cuniculus | MPCSCKKYCDPWEVIDGSCGLFNSKYICCREK | −130.8 ± 4.7 |
Table 6.
The best peptides according to their binding energy with SARS-COV-2 N protein.
| peptide code | peptide name | microorganism | sequence | dock score (kcal/mol) |
|---|---|---|---|---|
| 2KUY | Bacteriocin glycocin F | Lactobacillus Plantarum | KPAWCWYTLAMCGAGYDSGTCDYMYSHCFGIKHHSSGSSSYHC | −143.2 ± 7.1 |
| 1BRZ | Defensin-like protein | Pentadiplandra brazzeana | DKCKKVYENYPVSKCQLANQCNYDCKLDKHARSGECFYDEKRNLQCICDYCEY | −122.4 ± 4.9 |
| 2MVI | Bacteriocin plantaricin ASM1 | Lactobacillus plantarum | KPAWCWYTLAMCGAGYDSGTCDYMYSHCFGVKHSSGGGGSYHC | −120.3 ± 5.5 |
| 2JPK | Bacteriocin lactococcin-G subunit beta | Lactococcus lactis subsp. lactis | KKWGWLAWVDPAYEFIKGFGKGAIKEGNKDKWKNI | −114.4 ± 2.5 |
To investigate the inhibitory effect of bio-AMPs on the Spike, the binding energy of the Spike to the ACE2 was used as a control sample, which was −128.8 ± 3.5 kcal/mol (Table 2). In addition, the binding energy of the Bacteriocin plantaricin ASM1 peptide and the Spike was −149.7 ± 3.2 kcal/mol. All the peptides shown in Table 3 have a higher affinity for Spike compared to ACE2, and as mentioned earlier, Bacteriocin plantaricin ASM1 peptide had the highest affinity for Spike compared to the other peptides.
The ACE2 receptor was used as a control to compare the affinity of peptides with Spike, but unfortunately, because the ligands that normally bind to 3CL, RdRp, and N protein of SARS-CoV-2 are chemical compounds; they cannot be used to compare affinity in studying peptides. Therefore, the obtained highest scores were considered as the best peptides with high affinities for the other three above-mentioned viral proteins. Besides, the obtained scores according to the gained binding energy (For example, scores of −130 or −150) were calculated as high scores in HADDOCK molecular docking scoring system.
Bacteriocin lactococcin-G subunit beta had the highest affinity to RdRp equal to −151.4 ± 9.8 kcal/mol. Peptides with scores above −130.0 kcal/mol are listed in Table 4.
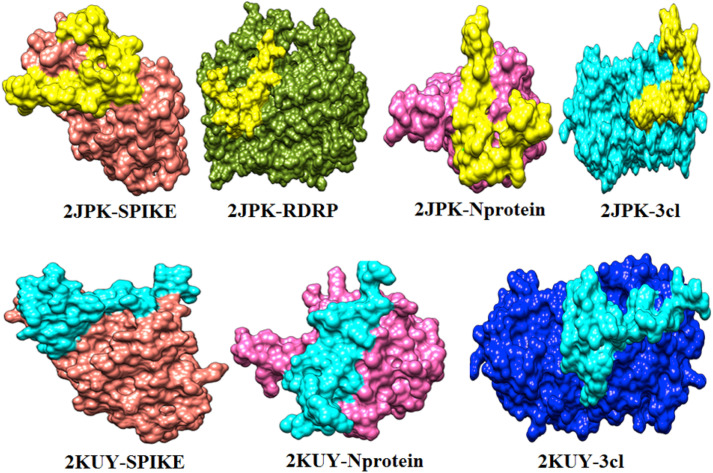
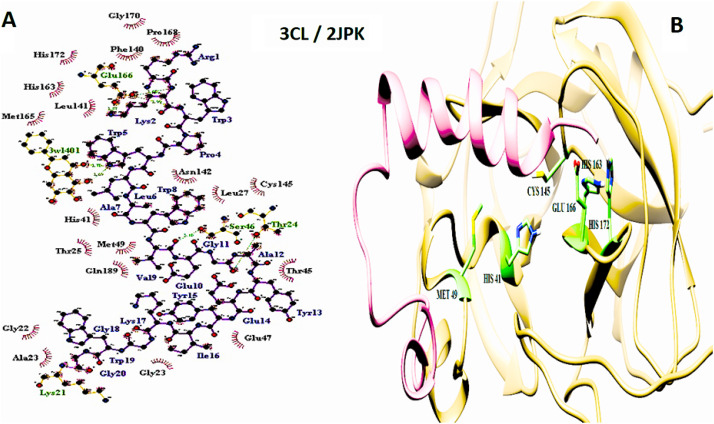
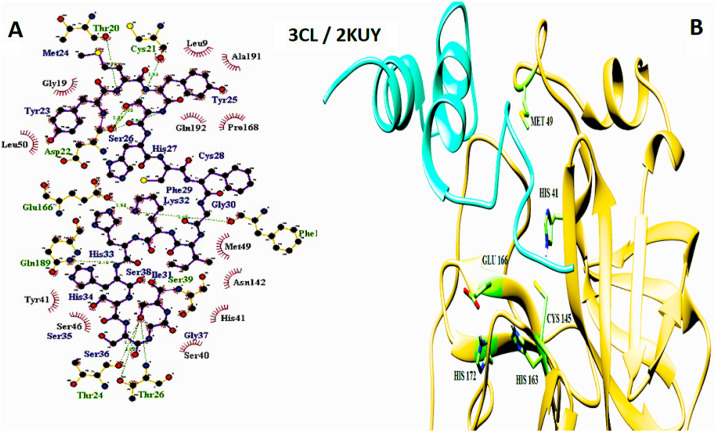
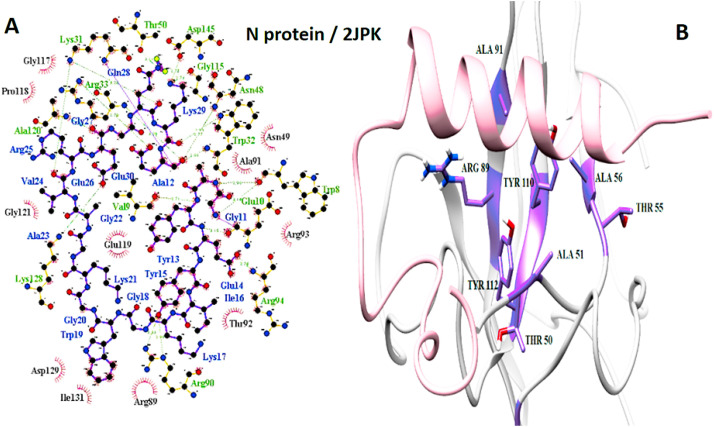
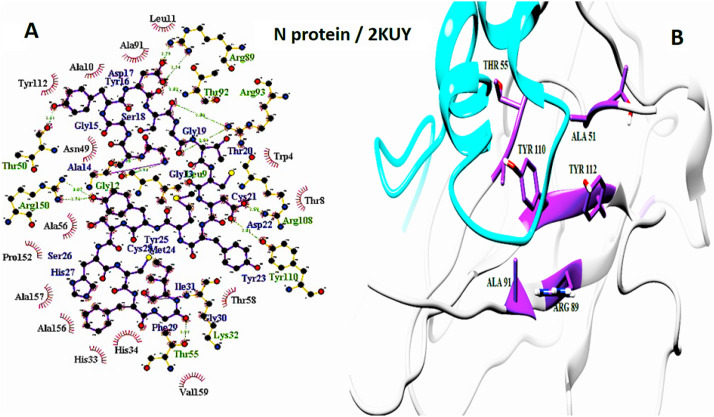
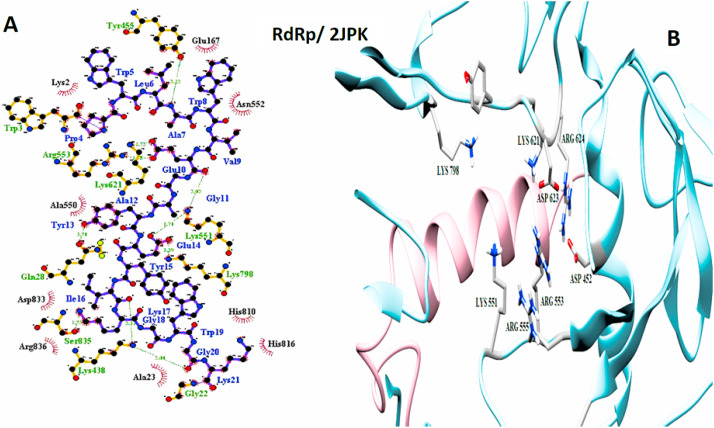
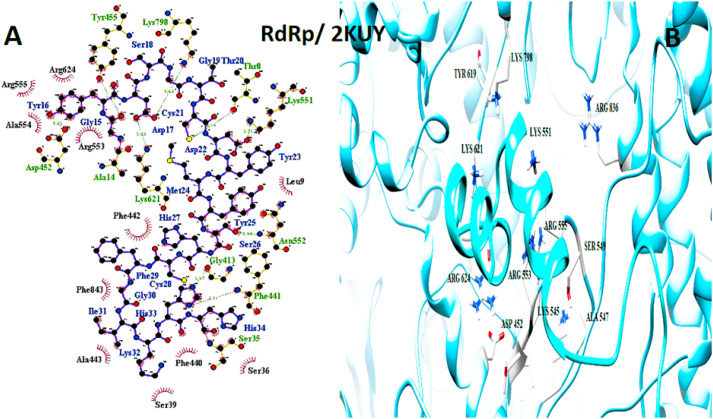
The important interaction between virus proteins and the common top score peptide (PDB code: 2JPK and 2KUY) is shown in Fig. 1 . The virus proteins’ structure was extracted from the RCSB PDB database with the accession number: Spike: 6VW1, RdRp: 7BTF, 3CL: 6M2N, N protein: 6M3M.
Fig. 1.
The interaction of 2KUY (Bacteriocin glycocin F) and 2JPK (Bacteriocin lactococcin-G subunit beta) with SARS-CoV-2 Spike, RdRP, 3CL, and N proteins.
Although 2JPK was common between all four SARS-CoV-2 studied proteins with high affinities, 2KUY was common only among N protein, Spike, and 3CL with high affinities.
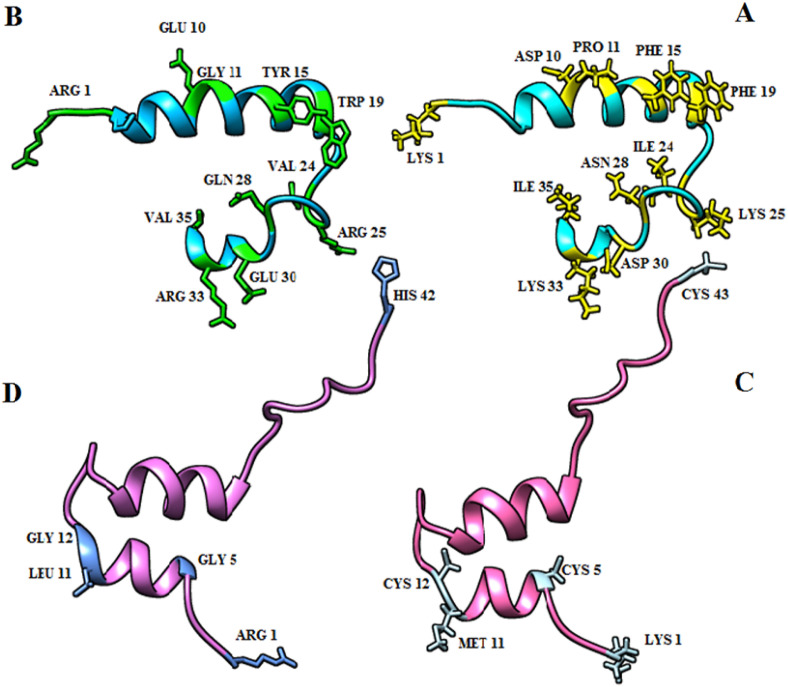
Considering the peptides' side effects, their sequences were mutated to achieve the suitable binding affinity and to resolve the predicted side effects. The primary (without mutation) and secondary (with mutation) structures of the two common peptides (2KUY and 2JPK) were obtained, which are shown in Fig. 2 .
Fig. 2.
The 3-D structure of top common peptides (2KUY and 2JPK) and the final peptide structure for molecular docking analysis as well as the mutated amino acids in the stick model. A: The important amino acids of 2JPK primary structure to change are presented in yellow. B: the structure of 2JPK after the mutation and the mutated amino acids are exhibited in green. C: the important amino acids of 2KUY primary structure to change are shown in sky blue. D: the structure of 2KUY after the mutation and the mutated amino acids are indicated in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
After finding the two common peptides that had a high binding energy, their allergenicity, toxicity, anti-angiogenic, interleukin 4 inducing ability, anti-cancer ability, and hemolyticity were checked. Accordingly, based on the obtained result, some of them had side effects such as allergenicity, toxicity, and hemolyticity. To solve the side effects' problem, a wide range of mutations was done and the binding energy and mentioned features of the mutated peptides were also investigated. A wide random range of mutations was performed to reduce the above-mentioned peptides' side effects. Each mutation was done in the peptide's chain according to its position and each one of the selected amino acids was replaced with the amino acid with similar characteristics such as charge, polarity, and side chain. So, each amino acid was then replaced with an amino acid in its family. The mutated amino acids were positioned in a place where the amino acids caused side effects. It is noteworthy that the important amino acids in the interaction were not mutated as much as possible. The result of each peptide's best mutations and their features are presented in Table 7 .
Table 7.
Attributes of common peptides between Spike, RdRp, 3CL, N proteins, before and after mutation.
| Peptide code | Peptide name | Sequence | Spike Docking Score (kcal/mol | RdRp Docking Score (kcal/mol) | 3CL Docking Score (kcal/mol) | NP Docking Score (kcal/mol) | Allergenisity | Anti-cancer | Angiogenicity | Hemolysis | Toxicity | IL4 inducing ability |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2JPK -Wild | Bacteriocin lactococcin-G | KKWGWLAWVDPAYEFIKGFGKGAIKEGNKDKWKNI | −143.7 ± 3.5 | −151.4 ± 9.8 | −138.9 ± 10.4 | −114.4 ± 2.5 | allergen | ACP | Non-Antiangiogenic | hemolytic | non-Toxin | IL4 inducer |
| 2JPK- Mutant | Bacteriocin lactococcin-G | RKWPWLAWVEGAYEYIKGWGKGAVREGQKEKWRNV | −149.2 ± 3.7 | −126.5 ± 26.2 | −159.6 ± 9.8 | −129.6 ± 4.1 | Non-allergen | ACP | Non-Antiangiogenic | Non-hemolytic | Non-toxin | Non IL4 inducer |
| 2KUY-Wild | Bacteriocin glycocin F | KPAWCWYTLAMCGAGYDSGTCDYMYSHCFGIKHHSSGSSSYHC | −139.0 ± 6.5 | −98.0 ± 13.2 | −155.3 ± 7.5 | −143.2 ± 7.1 | allergen | ACP | Antiangiogenic | Non-hemolytic | Toxin | Non IL4 inducer |
| 2KUY-Mutant | Bacteriocin glycocin F | RPAWGWYTLALGGAGYDSGTCDYMYSHCFGIKHHSSGSSSYHG | −133.6 ± 8.6 | −100.8 ± 20.1 | −128.3 ± 10.2 | −137.8 ± 9.5 | Non-allergen | ACP | Antiangiogenic | Non-hemolytic | Non-toxin | Non IL4 inducer |
By mutating the sequence of lactococcin-G, its binding affinity to the SARS-CoV-2 mentioned proteins increased and all side effects of this peptide then removed. Besides, the mutation in the glycocin F sequence solved the peptide side effect, but it led to a little decrease in its binding affinity.
The important amino acids in the interaction of both Bacteriocin lactococcin-G subunit beta and Bacteriocin glycocin F peptides with the mentioned SARS-CoV-2 proteins were plotted by UCSF Chimera, LigPlot, and Discovery studio visualizer software after the mutations.
The interaction of peptides with each one of the mentioned viral protein amino acids are demonstrated in the LigPlot generated figures. Moreover, the important amino acids of each protein are presented in UCSF Chimera made pictures. For each interaction, both UCSF Chimera and LigPlot generated figures are provided in Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10 .
Fig. 3.
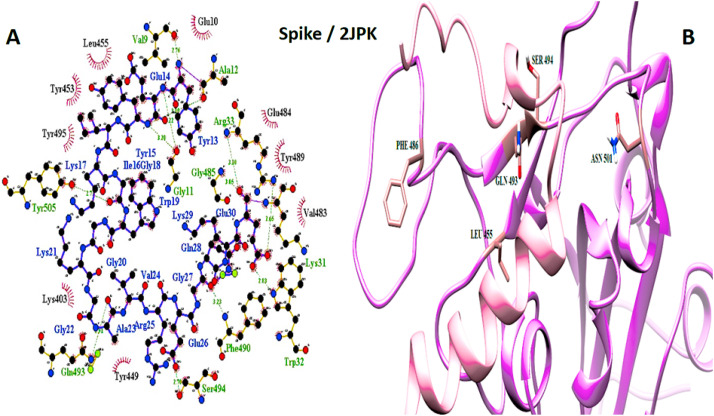
Spike and 2JPK interaction. A: 2-D picture of hydrophobic interactions and H-bonds of Bacteriocin lactococcin-G subunit beta-peptide with Spike protein of SARS-CoV-2 virus using LigPlot. B: 3-D structure of 2JPK/Spike complex; peptide chain is colored in pink, Spike is presented in plum, and the important amino acids of Spike protein in the interaction are exhibited in rosy brown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
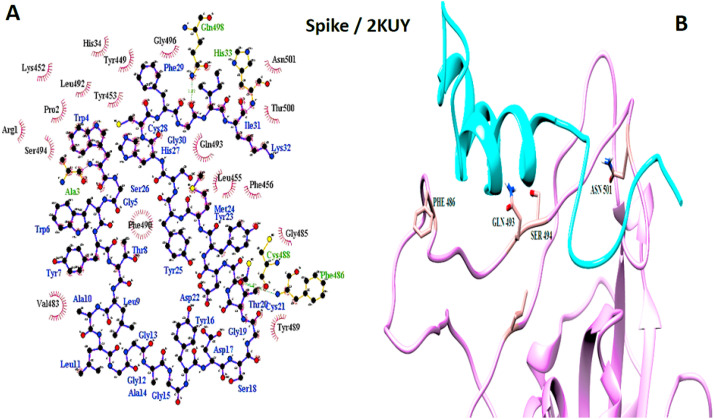
Spike and 2KUY interaction. A: 2-D picture of hydrophobic interactions and H-bonds of Bacteriocin glycocin F peptide with Spike protein of SARS-CoV-2 virus using LigPlot. B: 3-D structure of 2KUY/Spike complex; peptide chain is colored in cyan, Spike is presented in plum, and the important amino acids of Spike protein in the interaction are exhibited in rosy brown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
3CL and 2JPK interaction. A: 2-D picture of hydrophobic interactions and H-bonds of Bacteriocin lactococcin-G subunit beta-peptide with 3CL of SARS-CoV-2 virus using LigPlot. B: 3-D structure of 2JPK/3CL complex; peptide chain is colored in pink, 3CL is presented in khaki, and the important amino acids of 3CL protein in the interaction are exhibited in light green. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
3CL and 2KUY interaction. A: 2-D picture of hydrophobic interactions and H-bonds of Bacteriocin glycocin F peptide with 3CL of SARS-CoV-2 virus using LigPlot. B: 3-D structure of 2KUY/3CL complex; peptide chain is colored in pink, 3CL is presented in khaki, and the important amino acids of 3CL protein in the interaction are exhibited in light green. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 7.
N protein and 2JPK interaction. A: 2-D picture of hydrophobic interactions and H-bonds of Bacteriocin lactococcin-G subunit beta-peptide with N protein of SARS-CoV-2 virus using LigPlot. B: 3-D structure of 2JPK/N protein complex; peptide chain is colored in pink, N protein is presented in light gray, and the important amino acids of N protein in the interaction are exhibited in purple. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 8.
N protein and 2KUY interaction. A: 2-D picture of hydrophobic interactions and H-bonds of Bacteriocin glycocin F peptide with N protein of SARS-CoV-2 virus using LigPlot. B: 3-D structure of 2KUY/N protein complex; peptide chain is colored in cyan, N protein is presented in light gray, and the important amino acids of N protein in the interaction are exhibited in purple. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 9.
RdRp and 2JPK interaction. A: 2-D picture of hydrophobic interactions and H-bonds of Bacteriocin lactococcin-G subunit beta-peptide with RdRp of SARS-CoV-2 virus using LigPlot. B: 3-D structure of 2JPK/RdRp complex; peptide chain is colored in pink, RdRp is presented in light blue, and the important amino acids of RdRp in the interaction are exhibited in light gray. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 10.
RdRp and 2KUY interaction. A: 2-D picture of hydrophobic interactions and H-bonds of Bacteriocin glycocin F peptide with RdRp of SARS-CoV-2 virus using LigPlot. B: 3-D structure of 2KUY/RdRp complex; peptide chain is colored in pink, RdRp is presented in light blue, and the important amino acids of RdRp in the interaction are exhibited in light gray. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
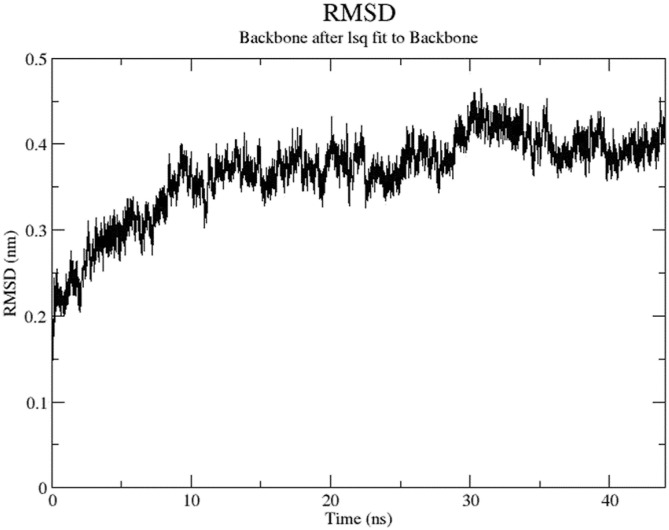
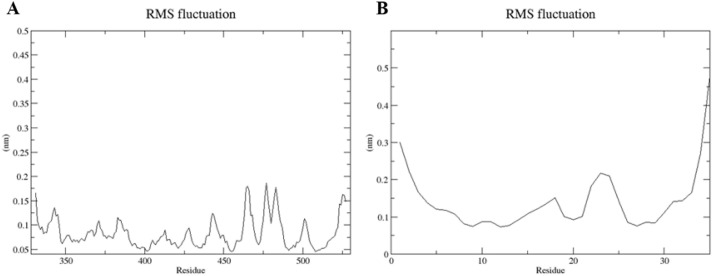
To prove the stability of the peptide/viral protein complexes after the mutation, MD simulation was performed for 2JPK/Spike protein complex for instance. As a result of the RMSD of atomic positions shown in Fig. 11 , it was observed that the complex has reached stability at the end of our simulation. Also, the amino acid fluctuations of 2JPK peptide and Spike protein are illustrated in Fig. 12 .
Fig. 11.
The RMSD graph for the entire MD simulation timescale (45 ns) is illustrated for both 2JPK peptide and Spike protein of SARS-CoV-2 complex.
Fig. 12.
The RMS fluctuation graph. A: the fluctuation of Spike protein amino acids. B: the fluctuation of amino acids of 2JPK peptide.
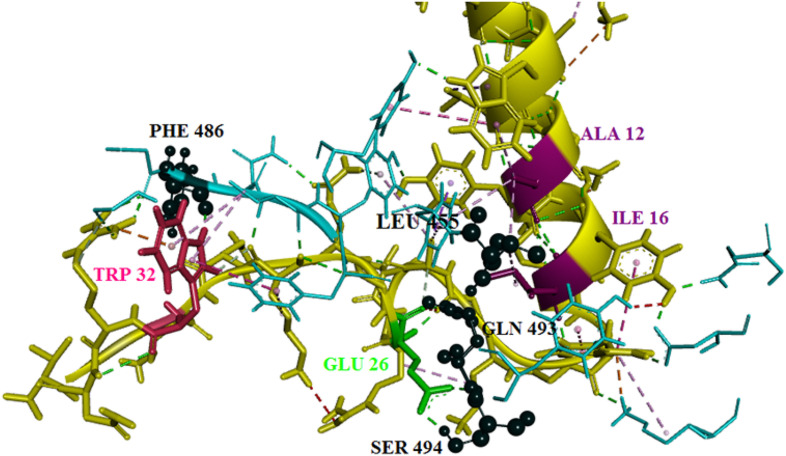
Fig. 13 clearly demonstrates the important amino acids in the interaction between 2JPK peptide and Spike protein after the mutation and MD simulation. The outcomes revealed that PHE 486 from Spike had the highest affinity to TRP 32 of the peptide. Both GLN 493 and SER 494 from Spike had interactions with GLU 26 of the peptide. Additionally, it was identified that LEU 455 had an interaction with both ALA 12 and ILE 16.
Fig. 13.
3-D interaction of 2JPK/Spike protein interaction after the mutation and MD simulation obtained from Discovery Studio. The important amino acids of Spike protein are colored in black and shown in ball and stick styles. The important amino acids of the peptide involved in the interaction are also displayed in three colors as pink, green, and purple. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Since the outbreak of COVID-19, humans have found that few options still exist for the treatment of life-threatening coronavirus infections. The outbreaks of SARS and MERS-CoV have led to performing more studies on the coronaviridae family; however, there is no definite drugs to treat these coronaviruses yet. Over the last 17 years, coronaviruses have shown a transient nature and led to epidemics; this feature prevents prototype coronavirus inhibitors from their development to the preclinical stage. Therefore, it seems that finding broad-spectrum inhibitors to decrease the effects of human coronavirus infection is a challenging research focus.
SARS-CoV-2 enters host cells by binding to ACE2 through the Spike, then viral RNA is transcribed by RdRp, viral RNA is replicated and transcribed, and the N protein is finally synthesized in the cytoplasm. Whereas other viral SPs such as the Spike, M, and E proteins are transcribed and translated in the endoplasmic reticulum (ER). The important proteins of SARS-CoV-2 are processed by 3CL protein, and they are then assembled at the ER–Golgi intermediate compartment (ERGIC) to form a mature virion. Finally, the nascent virion is released from the host cells [59]. Therefore, the inhibition of each of these proteins could be known as a suitable therapeutic target for combating and controlling COVID-19.
In an investigation, nearly 32,297 potential anti-viral phytochemicals were screened to select the top nine hits that can prevent 3CL activity and replication. Accordingly, many of these medicinal plant compounds have been already used to treat various viral diseases successfully [47]. Another in silico study has also presented that ligand-binding is strikingly similar in SARS-CoV and SARS-CoV2 main proteases, and also confirmed that a derivation of chlorophenyl-pyridyl-carboxamide has the strongest affinity to 3CL [48]. Moreover, an investigation identified top compounds among the natural products by virtual screening, which showed that simeprevir and loniflavone had the highest affinities to Spike, and conivaptan and amyrin had the highest ones to the nucleocapsid protein [52]. Several studies by investigating mechanistic studies to clinical trials for COVID-19 presented that remdesivir is a potential drug as a SARS-CoV-2 RNA-chain terminator, which can effectively stop its RdRp [60]. Finally, the Food and Drug Administration (FDA) approved remdesivir as an effective drug on the inhibition of SARS-CoV-2 reproduction [61].
In previous studies, inhibitory agents were usually used for one or two proteins, while in this study, bio-AMPs that had probiotic properties were used to inhibit the viral underlying proteins, including RdRp, 3CL, Spike, and N proteins.
The physicochemical and structural properties of bio-AMPs are essential factors in determining their specificity towards the destination cells. Correspondingly, they are efficient agents with different structural and antimicrobial features that can serve as one of the most hopeful future drug candidates to treat COVID-19 infections. For example, Oleg Kit and Yuriy Kit gathered some information on a group of natural peptides with various origins. They proposed that some peptides such as Angiotensins that regulate blood pressure, Bradykinin as an inflammatory mediator, and Beta-casokinin 1 as a bioactive component of milk and dairy products, could be suggested as novel drugs in the treatment of COVID-19 disease [40,41]. Perhaps the best option for choosing the source of peptides is using probiotic bacteria, because they are naturally beneficial to human health, so they can also be used in people's daily diet, which have been stated as a food complement for human health through making useful compounds [62].
In this study, after the investigation of many bio-AMPs, and surveying their effects on Spike, RdRp, 3CL, and N proteins, two common bio-peptides, named glycocin F and lactococcine G, from Lactococcus lactis and Lactobacillus Plantarum were selected with the highest affinity to these proteins, respectively. The other products from these two probiotics with no side effects are used in the food industry.
As claimed by the previous studies, taking vitamin D prevents the proliferation of the SARS-CoV-2 virus, and to the best of our knowledge, dairy products, in turn, contain vitamin D [63]. Notably, using dairy products comprising the Lactococcus lactis and Lactobacillus Plantarum that produce glycocin F and lactococcine G, may Consequently double the inhibitory effect with the consumption of vitamin D.
According to the advantages of the two obtained bio-AMPs, we can produce dairy products based on Lactococcus lactis and Lactobacillus Plantarum probiotic bacteria to control and prevent COVID-19 disease.
Furthermore, the mentioned bio-AMPs could be used as a synthetic medicine with different dosages. If bio-AMPs need to be used as a drug with high concentration, their side effects need to be checked. Thus, glycocin F and lactococcine G were checked in terms of their probable negative effects, and the result reveals that the allergenicity is the most important side effect. Therefore, to solve this problem, peptide manipulation was performed to achieve the proper condition.
Due to the cost of the COVID-19 pandemic for countries, developing a treatment with low cost is very precious. Lactococcus lactis and Lactobacillus Plantarum probiotic bacteria's products, which are abundantly found in dairy products, can be consumed to control this fatal disease. Also, the manipulated glycocin F and lactococcine G can be used as therapeutic targets for the inhibition of SARS-CoV-2 virus entrance, replication, and development in a variety of possible drug delivery mechanisms.
5. Conclusion
Based on this conducted study, it was shown that glycocin F from Lactococcus lactis and lactococcine G from Lactobacillus Plantarum are the common peptides with high-affinity binding to SARS-CoV-2 S, N proteins, and 3CL protease. Moreover, lactococcine G was found to have a high affinity to RdRp protein. The present study revealed that the optimization of glycocin F and lactococcine G can convert these two bio-peptides to suitable therapeutics factors for SARS-CoV-2 protein inhibition with no side effects. Thus, these peptides could be considered as potential drugs to control COVID-19 disease. However, more experimental and pre-clinical studies on peptides’ purification, characterization, and mutation, as well as on the possibility of their anti-viral effects on the SARS-CoV-2 virus are needed to use them as novel medicines for COVID-19.
Funding source
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors wish to thank the National Institute of Genetic Engineering and Biotechnology (NIGEB) for their kind cooperation in this project.
Abbreviation
- Coronavirus disease 19
(COVID-19)
- 3-chymotrypsin-like
(3CL)
- M
(membrane)
- E
(envelope)
- N
(nucleocapsid)
- open reading frames
(ORFs)
- angiotensin-converting enzyme II
(ACE 2)
- ribonucleoproteins
(RNPs)
- RNA-dependent RNA polymerase
(RdRp)
- bio-antimicrobial peptides
(bio-AMPs)
- zika virus
(ZIKV)
- dengue virus
(DENV) (27)
- Influenza A virus
(IAV)
- herpes simplex virus
(HSV) (29)
- hepatitis C virus
(HCV) (30)
- human immunodeficiency viruses
(HIV)
- Root-Mean-Square Deviation
(RMSD) and RootMean-Square
- Fluctuation
(RMSF)
- Food and Drug Administration
(FDA)
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., et al. The species severe acute respiratory syndrome related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 2020;5:536–544. doi: 10.1101/2020.02.07.937862v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soremekun O.S., Omolabi K.F., Soliman M.E.S. Identification and classification of differentially expressed genes reveals potential molecular signatures associated with SARS-CoV-2 infection in lung adenocarcinoma cells. Informatics Medicin Unlocked. 2020:100384. doi: 10.1016/j.imu.2020.100384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S.A., Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry. 2004;43(17):4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 5.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. Jama. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 6.Balmeh N., Mahmoudi S., Mohammadi N., Karabedianhajiabadi A. Predicted therapeutic targets for COVID-19 disease by inhibiting SARS-CoV-2 and its related receptors. Informatics Medicin Unlocked. 2020:100407. doi: 10.1016/j.imu.2020.100407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khodadadi E., Maroufi P., Khodadadi E., Esposito I., Ganbarov K., Espsoito S., et al. Study of combining virtual screening and antiviral treatments of the Sars-CoV-2 (Covid-19) Microb Pathog. 2020:104241. doi: 10.1016/j.micpath.2020.104241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2020;94(4) doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McBride R., Van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang T., Wu M.P., Chen S., Hou M., Hong M., Pan F., et al. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics and system biology. 2005;5(4):925–937. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du L., Zhao G., Lin Y., Chan C., He Y., Jiang S., et al. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine. 2008;26(13):1644–1651. doi: 10.1016/j.vaccine.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surjit M., Liu B., Chow V.T.K., Lal S.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006;281(16):10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh P.-K., Chang S.C., Huang C.-C., Lee T.-T., Hsiao C.-W., Kou Y.-H., et al. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J Virol. 2005;79(22):13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S.-J., Leng C.-H., Lien S., Chi H.-Y., Huang C.-Y., Lin C.-L., et al. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24(16):3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang B., Wang X.-Y., Yuan J.-W., Vabret A., Wu X.-D., Yang R.-F., et al. Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus. Biochem Biophys Biochemical and Biophysical Research Communications. 2005;336(1):110–117. doi: 10.1016/j.bbrc.2005.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Needle D., Lountos G.T., Waugh D.S. Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity. Acta Crystallographica Section D STRUCTURAL BIOLOGY. 2015;71(5):1102–1111. doi: 10.1107/S1399004715003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. 80- [DOI] [PubMed] [Google Scholar]
- 19.H Johansson M. Reversible Michael additions: covalent inhibitors and prodrugs. Mini Rev Med Chem. 2012;12(13):1330–1344. doi: 10.2174/138955712804586693. [DOI] [PubMed] [Google Scholar]
- 20.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020 doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J., Song W., Huang H., Sun Q. Pharmacological therapeutics targeting RNA-dependent RNA polymerase, proteinase and spike protein: from mechanistic studies to clinical trials for COVID-19. J Clin Med. 2020;9(4):1131. doi: 10.3390/jcm9041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zúñiga S., Cruz J.L.G., Sola I., Mateos-Gómez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J Virol. 2010;84(4):2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peele K.A., Chandrasai P., Srihansa T., Krupanidhi S., Sai A.V., Babu D.J., et al. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Informatics Medicin Unlocked. 2020:100345. doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demain A.L. Induction of microbial secondary metabolism. Int Microbiol. 1998;1(4):259–264. doi: 10.2436/IM.V1I4.26. [DOI] [PubMed] [Google Scholar]
- 25.Wang G., Li X., Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed A., Siman-Tov G., Hall G., Bhalla N., Narayanan A. Human antimicrobial peptides as therapeutics for viral infections. Viruses. 2019;11(8):704. doi: 10.3390/v11080704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boas L.C.P.V., Campos M.L., Berlanda R.L.A., de Carvalho Neves N., Franco O.L. Antiviral peptides as promising therapeutic drugs. Cell Mol Life Sci. 2019;76(18):3525–3542. doi: 10.1007/s00018-019-03138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh I.-N., Hartshorn K.L. The role of antimicrobial peptides in influenza virus infection and their potential as antiviral and immunomodulatory therapy. Pharmaceuticals. 2016;9(3):53. doi: 10.3390/ph9030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenssen H., Sandvik K., Andersen J.H., Hancock R.E.W., Gutteberg T.J. Inhibition of HSV cell-to-cell spread by lactoferrin and lactoferricin. Antivir Res. 2008;79(3):192–198. doi: 10.1016/j.antiviral.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda M., Nozaki A., Sugiyama K., Tanaka T., Naganuma A., Tanaka K., et al. Characterization of antiviral activity of lactoferrin against hepatitis C virus infection in human cultured cells. Virus Res. 2000;66(1):51–63. doi: 10.1016/S0168-1702(99)00121-5. [DOI] [PubMed] [Google Scholar]
- 31.Berkhout B., van Wamel J.L.B., Beljaars L., Meijer D.K.F., Visser S., Floris R. Characterization of the anti-HIV effects of native lactoferrin and other milk proteins and protein-derived peptides. Antivir Res. 2002;55(2):341–355. doi: 10.1016/S0166-3542(02)00069-4. [DOI] [PubMed] [Google Scholar]
- 32.Van der Strate B.W.A., Beljaars L., Molema G., Harmsen M.C., Meijer D.K.F. Antiviral activities of lactoferrin. Antivir Res. 2001;52(3):225–239. doi: 10.1016/s0166-3542(01)00195-4. [DOI] [PubMed] [Google Scholar]
- 33.Elnagdy S., AlKhazindar M. The potential of antimicrobial peptides as an antiviral therapy against COVID-19. ACS Pharmacology & Translational Science. 2020;3(4):780–782. doi: 10.1021/acsptsci.0c00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling R., Dai Y., Huang B., Huang W., Yu J., Lu X., et al. In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. Peptides. 2020;130:170328. doi: 10.1016/j.peptides.2020.170328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14(4):5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansari M.A., Jamal Q.M.S., Rehman S., Almatroudi A., Alzohairy M.A., Alomary M.N., et al. TAT-peptide conjugated repurposing drug against SARS-CoV-2 main protease (3CLpro): potential therapeutic intervention to combat COVID-19. Arabian journal of chemistry. 2020 doi: 10.1016/j.arabjc.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pushpanathan M., Gunasekaran P., Rajendhran J. Antimicrobial peptides: versatile biological properties. Int J Pept Protein Res. 2013 doi: 10.1155/2013/675391. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1) doi: 10.1093/nar/gky427. W296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouranov A., Xie L., de la Cruz J., Chen L., Westbrook J., Bourne P.E., et al. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34(suppl_1):D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Yin T., Xiao X., He D., Xue Z., Jiang X., et al. StraPep: a structure database of bioactive peptides. Database: The Journal of Biological Databases and Curation. 2018;2018 doi: 10.1093/database/bay038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammami R., Ben Hamida J., Vergoten G., Fliss I. PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2009;37(suppl_1):D963–D968. doi: 10.1093/nar/gkn655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choudhary S., Malik Y.S., Tomar S., Tomar S. Identification of SARS-CoV-2 cell entry inhibitors by drug repurposing using in silico structure-based virtual screening approach. Front Immunol. 10 July 2020;11(1664) doi: 10.3389/fimmu.2020.01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu K., Li W., Peng G., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci U S A. 2009;106(47) doi: 10.1073/pnas.0908837106. 19970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L., Zhou R. Structural basis of the potential binding mechanism of remdesivir to SARS-CoV-2 RNA-dependent RNA polymerase. J Phys Chem B. 2020;124(32):6955–6962. doi: 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- 46.Lung J., Lin Y., Yang Y., Chou Y., Shu L., Cheng Y., et al. The potential chemical structure of anti‐SARS‐CoV‐2 RNA‐dependent RNA polymerase. J Med Virol. 2020;92(6):693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharmaceut Anal. 2020;10(4):313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macchiagodena M., Pagliai M., Procacci P. Identification of potential binders of the main protease 3CLpro of the COVID-19 via structure-based ligand design and molecular modeling. Chem Phys Lett. 2020 doi: 10.1016/j.cplett.2020.137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su H., Yao S., Zhao W., Li M., Liu J., Shang W., et al. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. Acta Pharmacol Sin. 2020 doi: 10.1038/s41401-020-0483-6. [DOI] [Google Scholar]
- 50.Chen Y.W., Yiu C.-P.B., Wong K.-Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research. 2020;9 doi: 10.12688/f1000research.22457.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceut Sin B (APSB) 2020;10(7):1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kadioglu O., Saeed M., Johannes Greten H., Efferth T. Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Bull World Health Organ. 2020 doi: 10.2471/BLT.20.255943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Zundert G.C.P., Rodrigues J., Trellet M., Schmitz C., Kastritis P.L., Karaca E., et al. The HADDOCK2. 2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016;428(4):720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 54.Saha S., Raghava G.P.S. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34(suppl_2):W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Raghava G.P.S. Computational peptidology. Springer; 2015. Peptide toxicity prediction; pp. 143–157. [DOI] [PubMed] [Google Scholar]
- 56.Laengsri V., Nantasenamat C., Schaduangrat N., Nuchnoi P., Prachayasittikul V., Shoombuatong W. TargetAntiAngio: a sequence-based tool for the prediction and analysis of anti-angiogenic peptides. Int J Mol Sci. 2019;20(12):2950. doi: 10.3390/ijms20122950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaduangrat N., Nantasenamat C., Prachayasittikul V., Shoombuatong W. ACPred: a computational tool for the prediction and analysis of anticancer peptides. Molecules. 2019;24(10):1973. doi: 10.3390/molecules24101973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Win T.S., Malik A.A., Prachayasittikul V., S Wikberg J.E., Nantasenamat C., Shoombuatong W. HemoPred: a web server for predicting the hemolytic activity of peptides. Future Med Chem. 2017;9(3):275–291. doi: 10.4155/fmc-2016-0188. [DOI] [PubMed] [Google Scholar]
- 59.Liu X., Liu C., Liu G., Luo W., Xia N. COVID-19: Progress in diagnostics, therapy and vaccination. Theranostics. 2020;10(17):7821. doi: 10.7150/thno.47987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hendaus M.A. Remdesivir in the treatment of Coronavirus Disease 2019 (COVID-19): a simplified summary. J Biomol Struct Dyn. 2020:1–6. doi: 10.1080/07391102.2020.1767691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Food and Drug Administration Remdesivir EUA letter of authorization - FDA. https://www.fda.gov/media/137564/download [Internet]. Vol. 364 KB. 2020. p. 6. Available from:
- 62.Parvez S., Malik K.A., Ah Kang S., Kim H. Probiotics and their fermented food products are beneficial for health. J Biomol Struct Dyn. 2006;100(6):1171–1185. doi: 10.1111/j.1365-2672.2006.02963.x. [DOI] [PubMed] [Google Scholar]
- 63.Ilie P.C., Stefanescu S., Smith L. The role of vitamin D in the prevention of coronavirus disease 2019 infection and mortality. Aging Clin Exp Res. 2020;1–4 doi: 10.1007/s40520-020-01570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]