Abstract
Undoubtedly, the SARS-CoV-2 has become a major concern for all societies due to its catastrophic effects on public health. In addition, mutations and changes in the structure of the virus make it difficult to design effective treatment. Moreover, the amino acid sequence of a protein is a major factor in the formation of the second and tertiary structure in a protein. Amino acid replacement can have noticeable effects on the folding of a protein, especially if an asymmetric change (substitution of polar residue with non-polar, charged with an uncharged, positive charge with a negative charge, or large residue with small residue) occurs. D614G as a spike mutant of SARS-CoV-2 previously identified as an associated risk factor with a high mortality rate of this virus. Using structural bioinformatics, our group determined that D614G mutation could cause extensive changes in SARS-CoV-2 behavior including the secondary structure, receptor binding pattern, 3D conformation, and stability of it.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, Spike, D614G, bioinformatics, molecular dynamics
Introduction
It has been almost eighteen years since the advent of SARS-CoV as a member of the coronavirus in Guangdong, China (Walls et al., 2020). In 2012, Middle-East respiratory syndrome coronavirus (MERS), as another member of coronaviruses, caused the second pandemic of coronaviruses with a prevalence in 27 countries (Chen, 2020). HCoVHKU1, HCoV-229E, HCoV-OC43, and HCoV-NL63 are other coronaviruses that can infect humans, and their pathogenic role has been confirmed (Vinayagam & Sattu, 2020). The main challenge with this viral family is the absence of efficient treatment. Although after the MERS eradication, coronaviruses scenario was forgotten, but the emergence of SARS-CoV-2 has sounded the alarms for human health. The origin of SARS-CoV-2 like MERS and SARS-CoV is bats and the first time was observed in Wuhan (December 2019), China (Cîrstea et al., 2020). Since the outbreak, millions of infections and deaths have been due to SARS-CoV-2. The pandemic SARS-CoV-2 has also affected more than 200 countries and territories. Unlike MERS which its receptor is CD26, SARS-CoV and SARS-CoV-2 interact with human angiotensin-converting enzyme 2 (hACE2) to enter into the cell. So far, many attempts have been undertaken to design and manufacture a drug against COVID-19. Coronavirus could affect various vital organs of human especially who has suffered from underling diseases; however, the many pathophysiological pathways of it remain unclear. In addition, mutation and changes in the structure of coronavirus could also consider as other obstacles to finding proper treatment against it (Cao et al., 2020; Chen, 2020; Cîrstea et al., 2020; Vinayagam & Sattu, 2020; Walls et al., 2020). Among the proposed molecular targets, 3CLpro (Main protease), PLpro (Papin-like protease), RdRp (RNA-dependent RNA polymerase) are main candidates for SARS-CoV-2 inhibition (Morse et al., 2020). Due to the essential role of spike protein in cell entry and pathogenesis of SARS-CoV-2, this protein has attracted the attention of numerous investigators. Spike protein is a homotrimer protein with two functional subunits (S1, S2) in each Mer. S1 binds to the hACE2 and S2 fused viral particles to the cell membrane (Chen & Qiu, 2020). So far, several mutants and spike variants such as V615L, D614G, F817L, and P812S have been identified. Besides, other structural changes such as deletion in cleavage site (PRRAR) and associated flank site (QTQTN) have been reported that alter the molecular behavior of the SARS-CoV-2 (Liu, 2020). According to available evidence, a spike mutant called spikeD614G is more prevalent in some areas, especially in European countries, and has a higher mortality rate (Becerra-Flores & Cardozo, 2020; Eaaswarkhanth et al., 2020; Koyama et al., 2020). Although it was suggested that replacement of Asp614 (native form) with Gly reduces the affinity of the spike to bind to the hACE2 through surface, infectivity of the spikeD614G is higher than native one (Sen, 2020). The present study aims to investigate the structural and molecular properties of mutant D614G by employing bioinformatics and molecular dynamics (MD).
Materials and methods
Spike structural characteristics and immunogenicity evaluation
Important information of spike protein from published papers (Ahmed et al., 2020; Lan et al., 2020; Ou et al., 2020; Rabaan et al., 2020; Yan et al., 2020) and related databases such as UniProt (Garcia et al., 2019) and NCBI were extracted (Yang, 2020). Due to the effect of point mutations on the 3D structure, folding, and stability of a protein, it is expected that the immunogenicity of the spike could also be changed. In this study, the IEDB (Dhanda et al., 2019) server was used to evaluate and compare the immunogenicity of the wild type and D614G mutant spike. The antigen sequence properties (Bepipred Linear Epitope Prediction method) option was used to predict the B-cell linear epitopes. Besides, T-cell epitope prediction was accomplished by MHC I and MHC II binding prediction options.
MD evaluation
Molecular dynamics simulation is a popular tool for studying the behavior of biomolecules due to the high speed, affordability, and flexibility of this approach (Hildebrand et al., 2019). In this study, the open state crystal structure of the spike with PDB ID 6VYB was employed as the reference structure (Wrapp et al., 2020). The simulation was conducted for the wild and mutant (D614G) type of spike using the GROMACS package (version 2019.1) (Kutzner et al., 2019). During the analysis, desired structures were parameterized with AMBER03 force field (Zhang et al., 2019) and simulation was executed for 100 ns. All structural parameters such as root-mean-square deviation (RMSD) (Park et al., 2019), root-mean-square fluctuation (RMSF) (Narang et al., 2019), solvent accessible surface area (SASA) (Chingin & Barylyuk, 2018), and radius of gyration (Rg) (Lopes et al., 2019) were recorded for target structures. Simulation system components including water, ions, and spike protein were coupled to physiological temperature (37 °C) and pressure (1 bar). Besides, the TIP3P (Liem & Popelier, 2019) water model (triclinic box) was used for solvation. In order to neutralize the net charge of systems 14 Na+ and 12 Cl− ions were added in 0.15 M concentration. Periodic boundary conditions (PBC) were exerted in all three directions of the system. Then, the energy of structures minimized for 1000 steps of steepest descent minimization followed by 1000 steps of conjugate gradient minimization (Li et al., 2019). In the equilibration step an NVT ensemble with a constant temperature of 310 K for 150 ps, and NPT ensemble with a constant pressure of 1 bar for 500 ps were applied. For temperature and pressure coupling, Berendsen (Margraf et al., 2020) and Parrinello-Rahman algorithms (Miller et al., 2016) were employed, respectively. Position restraint process was used in NVT and NPT ensemble steps. In addition, other algorithms such as Lennard-Jones potentials (Bilodeau et al., 2019) and PME (Collier et al., 2009) were employed to handle van der Waals and electrostatic interactions. The structures obtained at this stage were used to perform the desired analyzes. During MD analyzes, 100000 frames (each 1ps) of trajectories were employed.
Tunnel and cavity analysis
Cavities and tunnels in the structure of proteins play an important role in their functions. Any change in these structural attributes due to the impact on the capacity of protein binding to other biomolecules have biological consequences such as inactivation and disruption of protein folding and its activities (Brezovsky et al., 2018). Also, the mutation in the protein sequence is known to be one of the main causes of alterations in molecular behavior (Strokach et al., 2019). In the present investigation, tunnel and binding site analysis of structures were obtained from the accomplished MD using the CAVER Web (v1.0) (Stourac et al., 2019). Due to the importance of choosing the right starting point to implement this method, the mutation position was selected as the starting point. Target point selection is based on known essential residues, identified pockets, or bounded ligands.
Tracking structural and physiochemical changes
Due to the presence of a small and uncharged residue amino acid (Gly) at position 614 of spike, instead of a large and negative charged residue (Asp), significant changes are expected in the domain where the amino acid is located. To get the exact details about the impact of D614G mutation on the spike, our group examined the secondary structure of the desired protein using DSSP (Zhang & Sagui, 2015). DSSP predict secondary structure for each residue by reading atom position and computation of the H-bond energy between all atoms (Zhang & Sagui, 2015). The tertiary structure changes were also investigated by Schrodinger (version 2018.1) (David et al., 2018). In addition, ProtParam (Garg et al., 2016) was used for physiochemical features analysis.
Stability investigation
The structure energy calculation is one of the most common approaches in protein studies to trace the effects of mutations on structural stability. Here, to accurately study energy changes two popular tools in the field of protein engineering were used. The first tool which employed to energy estimation was FoldX (version 5) plugin for YASARA (Buß et al., 2018). Also, the score function module from Rosetta (version 3.12) (Combs et al., 2018), a well-known package in peptide design in parallel was used to calculate and compare the energy of the structure in the wild and mutant spike. Both FoldX and Rosetta tools calculate the energy of the structure by applying linear combination of physics and statistics-based energy terms algorithms (Gerasimavicius et al., 2020). Tools listed in our study provide useful information about the characteristics of protein conformations by calculating the different terms of energy. The unit of the energy calculation for FoldX is kcal/mol, while Rosetta reports this amount in the form of Rosetta Energy Units (REU).
Docking methodology
Docking is one of the most widely used techniques in structural bioinformatics, which studies the biological function of proteins and their pattern of interaction with other biomolecules (Li et al., 2020). To investigate the effect of the D614G mutation on the ability of spike to bind to the hACE2 receptor, the docking process was utilized. For this purpose, the High Ambiguity Driven protein-protein docking (https://bianca.science.uu.nl/haddock2.4/) webserver was employed for protein docking (Kurkcuoglu et al., 2018). In the present study, the docking method was the flexible type and active residues were identified using the analysis of the existing crystal structures with PDB ID 6CS2, 6LZG, and 6M17. During docking, the solvated docking mode was activated and active residues were considered as the fully-flexible segment. Passive residues that were close to the active residues were chosen automatically. The cluster with the best score was used during the docking for analysis. The LigPlot+ v.2.2 was applied to determine schematic diagrams of the protein-protein interactions (Laskowski & Swindells, 2011).
Results and discussion
Spike structural characteristics and immunogenicity evaluation
To get acquainted with the structure of the spike protein, features including functional domains, components of each subunit, important residues for interaction with hACE2 (Table 1), and cleavage sites (CS) were elicited by reviewing available information about this protein.
Table 1.
Name and position of the SARS-CoV-2 and hACE2 contact residues.
| SARS-CoV-2 | K417-G446-Y449-Y453-L455-F456-A475-F486-N487-Y489-Q493-G496-Q498-T500-N501-G502-Y505 |
|---|---|
| hACE2 | Q24-T27-F28-D30-K31-H34-E35-E37-D38-Y41-Q42-L79-M82-Y83-N330-K353-G354-D355-R357-R393 |
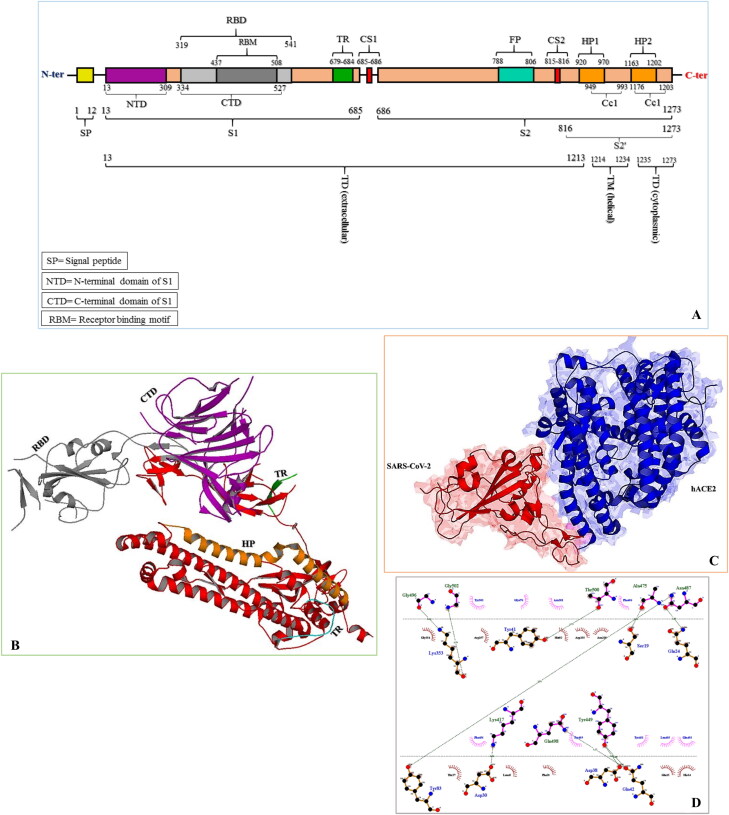
Spike is a large protein with 1273 residues per Mer. This protein is composed of the topological domain (TD) which is divided into cytosolic and extracellular segments, and transmembrane domain (TM). Tropism region (TR), heptad repeats (HP), fusion protein (FP), and coiled coils (Cc) are some important parts of spike. S1 contains sections such as receptor binding domain and region associated with virus tropism, while the S2 consists of heptad repeats, coiled coils, and fusion protein. Comprehensively, the important structural features of the spike are illustrated in Figure 1.
Figure 1.
A, throughout topology of spike functional regions. B, position of important domains in the 3D structure of spike. C, crystallized structure of spike protein and hACE2 complex. D, 2D schematic representation of important residues in spike interaction with its receptor (hydrogen bonds are displayed only).
Examination of the immunogenicity of mutant spike confirmed differences with wild protein. The B-cell linear epitope score was for the wild type (D614) 0.81 and the mutant (G614) 0.091. For T-cell epitopes containing wild and mutant residue, the results are listed in Table 2. Based on the results obtained at this stage, it can be claimed that the wild type has a higher immunogenic capacity than the mutant type for B-cells, MHC-I, and MHC-II. According to the results, it should be noted that mutation at position 614 of spike protein can have a significant effect on its immunogenic ability.
Table 2.
Average score reported for T-cell epitopes. The selected length for MHC-I was 9 and for MHC-II this set on 15.
| Sequence | MHC-I | MHC-II |
|---|---|---|
| D614 | 0.01 | 70.28 |
| G614 | 0.00 | 58.14 |
MD Analysis
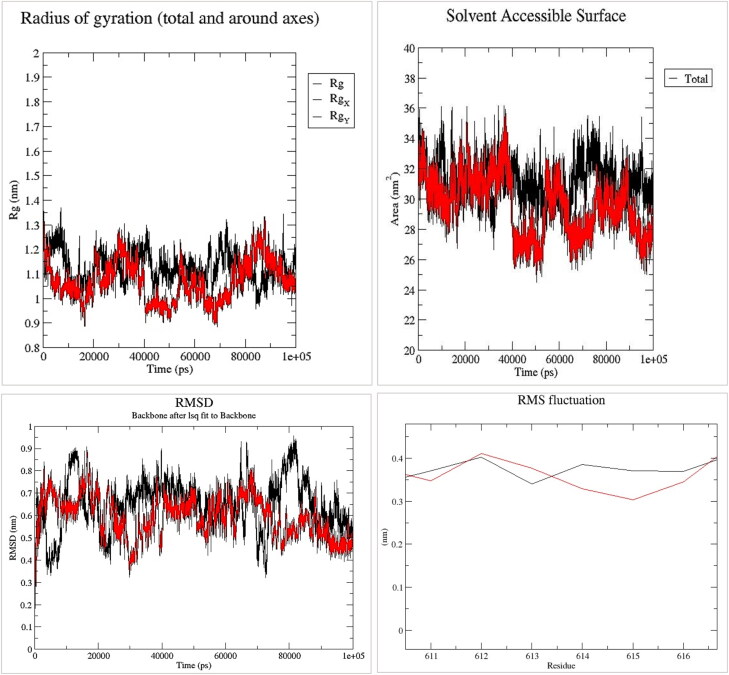
To scrutinize the changes that have occurred, we have focused our calculations on the region (between RBD and TR domain) containing the mutation and spike protein with 6VYB PDB ID has used as the reference structure. During the implementation of the molecular simulation, we find out that the results of this step are consistent with the output of other analyzes, and confirms the existence of a significant structural difference following the mutation. The average of RMSD is 0.6463 nm (σ = 0.116) for the native structure and 0.5932 nm (σ = 0.096) for the mutated protein, which could indicate greater stability of the mutated structure. Also, the obtained values of RMSF for the wild and mutated residue were 0.386 (σ = 0.15) and 0.329 nm (σ = 0.1), respectively, which confirms the reduction of structural fluctuations in the mutation position, and the increase in its stability. An average analysis of the amount of radius of gyration also showed that in the mutated protein, compression in the vicinity of the G614 (1.07 nm) is higher than in the D614 (1.1297 nm), and this increase in compression can decrease the chances of the interaction of mutant spike with proteases and increase its stability. Determining the amount of SASA also indicates a reduction in the available surface area of the mutant spike (Figure 2). Due to the proximity of the mutation position to the cleavage site-1 (CS1), it can be expected that the D614D mutation will affect the spike biological modifications.
Figure 2.
The graphical display of the GROMACS outputs, black refers to the natural spike, and red refers to the mutant spike.
Tunnel and cavity analysis
According to the results of the CAVER Web (v1.0), following of the D614G mutation, remarkable changes occurred in the cavities and tunnels surrounding the target residue (Figure 3).
Figure 3.
Schematic illustration of CAVER Web output. The changes around the mutated residue are easily visible.
We found that due to the difference in size and charge of Gly residue compared to Asp, such changes are expected in the space around this position. Based on the results of the CAVER Web, it is inferred that the tunnel around Gly614 has a greater radius and less length compared to Asp at this position. Given that the G614 is adjacent to the first cutting site of spike protein, this mutation can affect the biological processes associated with this protein. The summary of CAVER Web results is reported in Table 3.
Table 3.
Features of the tunnel adjacent to the mutation position computed by CAVER Web (v1.0).
| Sequence | Bottleneck radius [Å] | Length [Å] | Curvature | Throughput |
|---|---|---|---|---|
| D614 | 1 | 2.5 | 1.1 | 0.88 |
| G614 | 1.1 | 1 | 1 | 0.89 |
Tracking structural and physiochemical changes
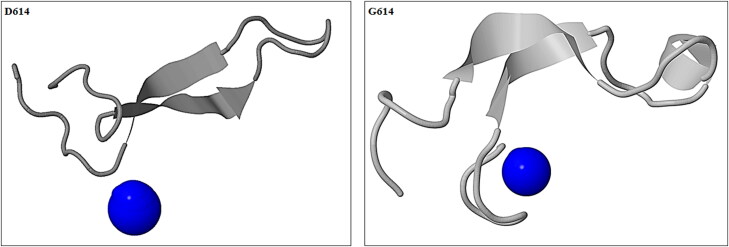
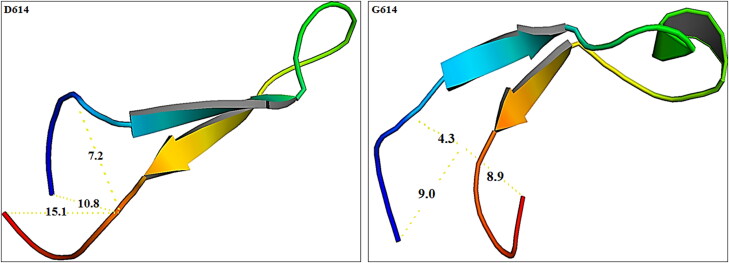
The instability index for target structures was evaluated using the ProtParam tool. Values of The instability index of the native and mutant structure were 17.56 and 11.72, respectively. Increasing the amount of instability index means a reduction in the protein stability. Insight into the physicochemical properties of both wild and mutant revealed that in the G614 protein, the molecular weight decreased compared to the D614, while the isoelectric point increased. Analysis of the changes related to the folding of the area neighbor to position 614 indicates changes in the 3D structure of the target region (Figure 4). Calculating the RMSD of areas close to the mutation between the structure containing D614 and G614 also reflected the existence of broad changes. The amount of RMSD calculated between the wild spike as reference structure and mutant protein was 0.534 nm. As expected, the D614G mutation causes major alterations in the secondary structure of the residues around the mutation position. These changes can undoubtedly affect the biological behavior of the spike, due to its effect on the structural features of this protein. Analysis of DSSP results shows that in the structure containing the D614G there is a great tendency to form helix structures around this position. Moreover, in comparison to the D614 spike protein which extended a more strand elements, what has observed is that in the vicinity of the mutated residue; the tendency to forming of bend and turn secondary structures has increased in the mutant structure of spike in the area near the mutation position.
Figure 4.
Displays the change in distances between position 614 and other adjacent positions in both structures. Also, the formation of an alpha helix can be seen in the mutant type.
Stability investigation
The results of this step of investigation are interesting. Based on the observation of FoldX and ROSETTA outputs, our group found out that the D614G mutation increases the stability of spike protein conformation. The energy calculated by FoldX was 32.32 for mutant and 35.27 kcal/mol for the wild spike. In addition, the amount of energy estimated by Rosetta for the mutant and wild spike were −37.06 and −32.47, respectively. In other words, it can be argued that due to the Gly residue which is an uncharged amino acid with a shorter side chain, so, it imposes less entropy and energy on the whole structure than Asp. The lower energy content of the mutant structure indicates that it is more stable compared to the wild type, which can improve the performance of the SARS-CoV-2. In the Table 4 more detail about the calculated energy terms for D614 and G614 spike were listed.
Table 4.
Energy parameters measured by FoldX and ROSETTA for the native and mutant spike (the unit of all energy terms for Rosetta and FoldX were REU and kcal/mol, respectively).
| FoldX |
Torsion energy |
Backbone Hbond | Sidechain Hbond | Energy_ VdW | Electrostatic | Solvation Polar | Solvation Hydrophobic | Van der Waals clashes | Entropy Side Chain | Entropy Main Chain |
|---|---|---|---|---|---|---|---|---|---|---|
| D614 | 1.42 | −8.46 | −0.54 | −17.06 | −0.06 | 28.26 | −20 | 3.65 | 6.79 | 41.27 |
| G614 | 1.20 | −10.83 | −3.83 | −19.80 | −0.65 | 31.39 | −23.32 | 2.30 | 10.93 | 44.92 |
|
ROSETTA |
fa_atr |
fa_rep |
fa_sol |
lk_ball_wtd |
fa_elec |
hbond_sr_bb |
hbond_lr_bb |
hbond_bb_sc |
hbond_sc |
omega |
| D614 | −110.1 | 10.2 | 74.4 | −4.12 | −30.9 | −2.57 | −10.14 | −2.74 | −3.96 | 1.07 |
| G614 | −119.9 | 13.85 | 70.7 | −5.07 | −31.6 | −6.49 | −5.2 | −8.91 | −2.12 | 1.68 |
Note: fa_atr (attractive Lennard-Jones). fa_rep (Repulsive Lennard-Jones). fa_sol (Lazaridis-Karplus solvation energy). lk_ball_wtd (Asymmetric solvation energy). fa_elec (Electrostatic potential). hbond_sr_bb (Backbone-backbone hbonds close in primary sequence). hbond_lr_bb (Backbone-backbone hbonds distant in primary sequence). hbond_bb_sc (Sidechain-backbone hydrogen bond energy). hbond_sc (Sidechain-sidechain hydrogen bond energy). Omega (Omega dihedral in the backbone).
Docking analysis
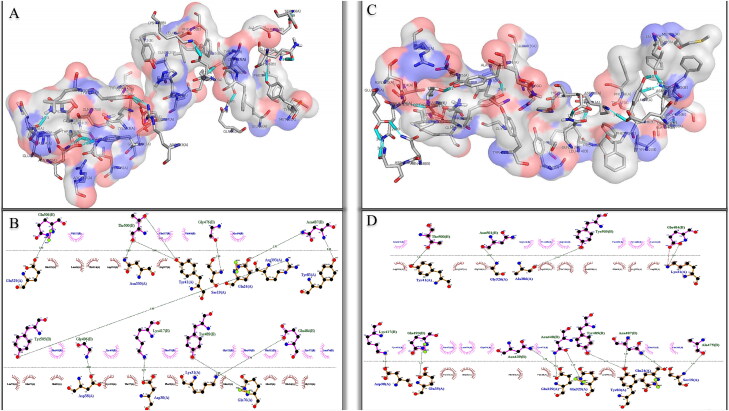
Although laboratory studies have shown that the binding affinity of D614 spike is 5 times higher than the G614 spike at physiological temperatures (37 °C), but changes in the residues binding pattern of spike and hACE2 have not yet reviewed. Undoubtedly, the type and number of amino acids along with the type of formed bonds, which are collectively known as the binding pattern, play an important role in the interaction of one protein with other biomolecules (Perkins et al., 2010). Due to the importance of the subject, the binding pattern of the spike carrying the D614G mutation to its receptor was investigated. Here we obtained valuable information about molecular interactions of spike-hACE2 by using the HADDOCK 2.4 server. In our study, we observed that the pattern of interactions for spikeD614-hACE2 is very different from the mutant spike. In addition to hydrophobic interactions, it was observed that a remarkable difference in hydrogen bonds are excited between the two complexes. For example, in the wild spike, the Thr500 residue form two hydrogen bonds with Tyr41 and one hydrogen bond with Asn330 of hACE2. However, in G614 spike, Thr500 forms hydrogen bonds only with Tyr41. In addition, Asn440 which has not a role in interaction of spikeD614 with hACE2, forms two hydrogen bonds with residues 325 and 329 of receptor in complex of spikeG614-hACE2. The Tyr83 of hACE2 in the complex with spikeD614 bind to the Asn487. However, in the complex with the spikeG614, it forms one hydrogen bond with Tyr489 in addition to Asn487. Moreover, Glu329 of hACE2 forms one hydrogen bond in the complex with the wild spike while this residue in complex of spikeG614-hACE2 forms two hydrogen bonds with Asn439 and Asn440 spike. Variations in the type and number of interactions confirm the widespread effect of the mutation at position 614 on the ability of the spike to bind to its receptor (Figure 5).
Figure 5.
Graphical representation of docking results. A-B and C-D show 3D and 2D interactions of spikeD614 and spikeG614 with hACE2, respectively.
Conclusion
Although it has been a short time since the advent of the COVID-19, due to the importance of the issue, many attempts have been made to identify the molecular biology of this virus. However, many unknown aspects of the virus still exist. In the present study, using bioinformatics tools, we showed that the D614G mutation causes extensive structural changes in the mutation region of spike protein. RMSF, SASA, energy content, immunogenicity values of mutant have decreased in comparison to wild type while Rg value of it has increased and it could be interfered (at least partially) that local and overall structural stability of D614G has increased. In accordance to tertiary structure changes, alterations in the secondary structure and the pattern of protein binding to its receptor for mutant form have observed in MD results. Altogether, higher infectivity of D614G might be related to these structural changes, however, further studies are granted. The experimental stability analysis, binding pattern, and immunogenicity assay studies of D614G might be of great interest to follow to learn in depth about function, feature, and unclear aspects of this more pathogenic mutant of COVID-19 for finding a specific treatment against it in the next future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Ahmed, S. F., Quadeer, A. A., & McKay, M. R. (2020). Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses, 12(3), 254. 10.3390/v12030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra-Flores, M., & Cardozo, T. (2020). SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. International Journal of Clinical Practice, 74(8), e13525. 10.1111/ijcp.13525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau, C. L., Lau, E. Y., Cramer, S. M., & Garde, S. (2019). Conformational Equilibria of Multimodal Chromatography Ligands in Water and Bound to Protein Surfaces. J Phys Chem B, 123(23), 4833–4843. 10.1021/acs.jpcb.9b01218 [DOI] [PubMed] [Google Scholar]
- Brezovsky, J., Kozlikova, B., & Damborsky, J. (2018). Computational analysis of protein tunnels and channels. Methods in Molecular Biology (Clifton, N.J.).), 1685, 25–42. 10.1007/978-1-4939-7366-8_3 [DOI] [PubMed] [Google Scholar]
- Buß, O., Rudat, J., & Ochsenreither, K. (2018). FoldX as protein engineering tool: better than random based approaches? Computational and Structural Biotechnology Journal, 16, 25–33. 10.1016/j.csbj.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., Yin, R., Albrecht, H., Fan, D., & Tan, W. (2020). Cholesterol: A new game player accelerating vasculopathy caused by SARS-CoV-2? American Journal of Physiology. Endocrinology and Metabolism, 319(1), E197–e202. 10.1152/ajpendo.00255.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. Y. (2020). SARS-CoV-2 desensitizes host cells to interferon through inhibition of the JAK-STAT pathway. bioRxiv,
- Chen, Y., & Qiu, F. (2020). Spike protein in the detection and treatment of novel coronavirus. Sheng wu yi Xue Gong Cheng Xue za Zhi = Journal of Biomedical Engineering = Shengwu Yixue Gongchengxue Zazhi, 37(2), 246–250. []. 10.7507/1001-5515.202002050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chingin, K., & Barylyuk, K. (2018). Charge-state-dependent variation of signal intensity ratio between unbound protein and protein-ligand complex in electrospray ionization mass spectrometry: the role of solvent-accessible surface area. Analytical Chemistry, 90(9), 5521–5528. 10.1021/acs.analchem.7b05349 [DOI] [PubMed] [Google Scholar]
- Cîrstea, A. E., Buzulică, R. L., Pirici, D., Ceauşu, M. C., Iman, R. V., Gheorghe, O. M., Neamţu, S. D., Stanca, L., Ene, R., Kumar-Singh, S., & Mogoantă, L. (2020). Histopathological findings in the advanced natural evolution of the SARS-CoV-2 infection. Rom J Morphol Embryol, 61(1), 209–218. 10.47162/RJME.61.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier, G., Vellore, N. A., Latour, R. A., & Stuart, S. J. (2009). Development of molecular simulation methods to accurately represent protein-surface interactions: Method assessment for the calculation of electrostatic effects. Biointerphases, 4(4), 57–64. 10.1116/1.3266417 [DOI] [PubMed] [Google Scholar]
- Combs, S. A., Mueller, B. K., & Meiler, J. (2018). Holistic approach to partial covalent interactions in protein structure prediction and design with rosetta. Journal of Chemical Information and Modeling, 58(5), 1021–1036. 10.1021/acs.jcim.7b00398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, T. I., Adelakun, N. S., Omotuyi, O. I., Metibemu, D. S., Ekun, O. E., Eniafe, G. O., Inyang, O. K., Adewumi, B., Enejoh, O. A., Owolabi, R. T., & Oribamise, E. I. (2018). Molecular docking analysis of phyto-constituents from Cannabis sativa with pfDHFR. Bioinformation, 14(9), 574–579. 10.6026/97320630014574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda, S. K., Mahajan, S., Paul, S., Yan, Z., Kim, H., Jespersen, M. C., Jurtz, V., Andreatta, M., Greenbaum, J. A., Marcatili, P., Sette, A., Nielsen, M., & Peters, B. (2019). IEDB-AR: Immune epitope database-analysis resource in 2019. Nucleic Acids Res, 47(W1), W502–w506. 10.1093/nar/gkz452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaaswarkhanth, M., Al Madhoun, A., & Al-Mulla, F. (2020). Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? International Journal of Infectious Diseases : IJID : official Publication of the International Society for Infectious Diseases, 96, 459–460. 10.1016/j.ijid.2020.05.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, L., Bolleman, J., Gehant, S., Redaschi, N., & Martin, M., UniProt Consortium (2019). FAIR adoption, assessment and challenges at UniProt. Scientific Data, 6(1), 175. 10.1038/s41597-019-0180-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg, V. K., Avashthi, H., Tiwari, A., Jain, P. A., Ramkete, P. W., Kayastha, A. M., & Singh, V. K. (2016). MFPPI - Multi FASTA ProtParam Interface. Bioinformation, 12(2), 74–77. 10.6026/97320630012074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimavicius, L., Liu, X., & Marsh, J. A. (2020). Identification of pathogenic missense mutations using protein stability predictors. Scientific Reports, 10(1), 15387. 10.1038/s41598-020-72404-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand, P. W., Rose, A. S., & Tiemann, J. K. S. (2019). Bringing molecular dynamics simulation data into view. Trends in Biochemical Sciences, 44(11), 902–913. 10.1016/j.tibs.2019.06.004 [DOI] [PubMed] [Google Scholar]
- Koyama, T., Weeraratne, D., Snowdon, J. L., & Parida, L. (2020). Emergence of Drift Variants That May Affect COVID-19 Vaccine Development and Antibody Treatment. Pathogens, 9(5), 324. 10.3390/pathogens9050324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkcuoglu, Z., Koukos, P. I., Citro, N., Trellet, M. E., Rodrigues, J. P. G. L. M., Moreira, I. S., Roel-Touris, J., Melquiond, A. S. J., Geng, C., Schaarschmidt, J., Xue, L. C., Vangone, A., & Bonvin, A. M. J. J. (2018). Performance of HADDOCK and a simple contact-based protein-ligand binding affinity predictor in the D3R Grand Challenge 2. J Comput Aided Mol Des , 32(1), 175–185. 10.1007/s10822-017-0049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzner, C., Páll, S., Fechner, M., Esztermann, A., de Groot, B. L., & Grubmüller, H. (2019). More bang for your buck: Improved use of GPU nodes for GROMACS 2018. Journal of Computational Chemistry, 40(27), 2418–2431. 10.1002/jcc.26011 [DOI] [PubMed] [Google Scholar]
- Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., & Wang, X. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 581(7807), 215–220. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., & Swindells, M. B. (2011). LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling, 51(10), 2778–2786. 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- Liem, S. Y., & Popelier, P. L. A. (2019). The influence of water potential in simulation: A catabolite activator protein case study. Journal of Molecular Modeling, 25(8), 216 10.1007/s00894-019-4095-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Li, S., Wei, X., & Zhao, Q. (2019). Scaled Alternating Steepest Descent Algorithm Applied for Protein Structure Determination from Nuclear Magnetic Resonance Data. J Comput Biol, 26(9), 1020–1029. 10.1089/cmb.2019.0013 [DOI] [PubMed] [Google Scholar]
- Li, J., Song, Y., Li, F., Zhang, H., & Liu, W. (2020). FWAVina: A novel optimization algorithm for protein-ligand docking based on the fireworks algorithm. Comput Biol Chem, 88, 107363 10.1016/j.compbiolchem.2020.107363 [DOI] [PubMed] [Google Scholar]
- Liu, Z. (2020). Identification of common deletions in the spike protein of SARS-CoV-2. J Virol, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, J. L. S., Oliveira, D. C. A., Utescher, C. L. A., Quintilio, W., Tenório, E. C. N., Oliveira, C. L. P., Fantini, M. C. A., Rasmussen, M. K., Bordallo, H. N., Sant'Anna, O. A., & Botosso, V. F. (2019). Antigenic and physicochemical characterization of Hepatitis B surface protein under extreme temperature and pH conditions. Vaccine, 37(43), 6415–6425. 10.1016/j.vaccine.2019.09.005 [DOI] [PubMed] [Google Scholar]
- Margraf, J. T., Hennemann, M., & Clark, T. (2020). EMPIRE: A highly parallel semiempirical molecular orbital program: 3: Born-Oppenheimer molecular dynamics. Journal of Molecular Modeling, 26(3), 43. 10.1007/s00894-020-4293-z [DOI] [PubMed] [Google Scholar]
- Miller, R. E., Tadmor, E. B., Gibson, J. S., Bernstein, N., & Pavia, F. (2016). Molecular dynamics at constant Cauchy stress. J Chem Phys, 144(18), 184107. 10.1063/1.4948711 [DOI] [PubMed] [Google Scholar]
- Morse, J. S., Lalonde, T., Xu, S., & Liu, W. R. (2020). Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV. Chembiochem : a European Journal of Chemical Biology, 21(5), 730–738. 10.1002/cbic.202000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang, S. S., Goyal, D., & Goyal, B. (2019). In silico-guided identification of potential inhibitors against β(2)m aggregation in dialysis-related amyloidosis. Journal of Biomolecular Structure and Dynamics. 1–15. 10.1080/07391102.2019.1614093 [DOI] [PubMed] [Google Scholar]
- Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., Guo, L., Guo, R., Chen, T., Hu, J., Xiang, Z., Mu, Z., Chen, X., Chen, J., Hu, K., Jin, Q., Wang, J., & Qian, Z. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications, 11(1), 1620. 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H., Lee, G. R., Kim, D. E., Anishchenko, I., Cong, Q., & Baker, D. (2019). High-accuracy refinement using Rosetta in CASP13. Proteins, 87(12), 1276–1282. 10.1002/prot.25784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, J. R., Diboun, I., Dessailly, B. H., Lees, J. G., & Orengo, C. (2010). Transient protein-protein interactions: Structural, functional, and network properties. Structure (London, England : 1993), 18(10), 1233–1243. 10.1016/j.str.2010.08.007 [DOI] [PubMed] [Google Scholar]
- Rabaan, A. A., Al-Ahmed, S. H., Haque, S., Sah, R., Tiwari, R., Malik, Y. S., Dhama, K., Yatoo, M. I., Bonilla-Aldana, D. K., & Rodriguez-Morales, A. J. (2020). SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Le Infezioni in Medicina, 28(2), 174–184. [PubMed] [Google Scholar]
- Sen, D. (2020). Identification of potential inhibitors of SARS-CoV-2 main protease and spike receptor from 10 important spices through structure-based virtual screening and molecular dynamic study. Journal of Biomolecular Structure and Dynamics, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stourac, J., Vavra, O., Kokkonen, P., Filipovic, J., Pinto, G., Brezovsky, J., Damborsky, J., & Bednar, D. (2019). Caver Web 1.0: Identification of tunnels and channels in proteins and analysis of ligand transport. Nucleic Acids Research, 47(W1), W414–w422. 10.1093/nar/gkz378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strokach, A., Corbi-Verge, C., Teyra, J., & Kim, P. M. (2019). Predicting the effect of mutations on protein folding and protein-protein interactions. Methods in Molecular Biology (Clifton, N.J.).), 1851, 1–17. 10.1007/978-1-4939-8736-8_1 [DOI] [PubMed] [Google Scholar]
- Vinayagam, S., & Sattu, K. (2020). SARS-CoV-2 and coagulation disorders in different organs. Life Sciences, 260, 118431. 10.1016/j.lfs.2020.118431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell, 181(2), 281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.), 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.).), 367(6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. (2020). NCBI's Conserved Domain Database and Tools for Protein Domain Analysis. Curr Protoc Bioinformatics, 69(1), e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., Lazim, R., & Yip, Y. M. (2019). Incorporating polarizability of backbone hydrogen bonds improved folding of short α-helical peptides. Biophysical Journal, 117(11), 2079–2086. 10.1016/j.bpj.2019.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., & Sagui, C. (2015). Secondary structure assignment for conformationally irregular peptides: Comparison between DSSP, STRIDE and KAKSI. Journal of Molecular Graphics & Modelling, 55, 72–84. 10.1016/j.jmgm.2014.10.005 [DOI] [PubMed] [Google Scholar]