Abstract
RNA viruses include many important human and animal pathogens, such as the influenza viruses, respiratory syncytial virus, Ebola virus, measles virus and rabies virus. The genomes of these viruses consist of single or multiple RNA segments that assemble with oligomeric viral nucleoprotein into ribonucleoprotein complexes. Replication and transcription of the viral genome is performed by ~250–450 kDa viral RNA-dependent RNA polymerases that also contain capping or cap-snatching activity. In this Review, we compare recent high-resolution X-ray and cryoelectron microscopy structures of RNA polymerases of negative-sense RNA viruses with segmented and non-segmented genomes, including orthomyxoviruses, peribunyaviruses, phenuiviruses, arenaviruses, rhabdoviruses, pneumoviruses and paramyxoviruses. In addition, we discuss how structural insights into these enzymes contribute to our understanding of the molecular mechanisms of viral transcription and replication, and how we can use these insights to identify targets for antiviral drug design.
Subject terms: Virus structures, Structural biology
In this Review, Fodor and colleagues discuss recent structural insights into the RNA polymerases of negative-sense RNA viruses and their use to identify novel targets for the development of antiviral agents.
Introduction
Negative-sense RNA viruses (NSVs) include many highly prevalent human pathogens that can be responsible for respiratory infections, haemorrhagic fever and encephalitis. NSVs can also cause disease in livestock, arthropods and plants and are considered a substantial economic burden worldwide. In humans, NSVs are responsible for frequent epidemics, such as those caused by human respiratory syncytial virus (HRSV), human parainfluenza viruses, measles virus, mumps virus and the influenza A virus (IAV) and influenza B virus (IBV). NSVs that exist in animal reservoirs can cause occasional zoonotic outbreaks that are often associated with high mortality and morbidity, such as avian IAVs, Ebola virus (EBOV), Lassa mammarenavirus (LASV) and rabies lyssavirus (RABV). A better understanding of the molecular mechanisms underlying NSV replication and the development of new antiviral drugs against NSV infections are therefore essential to combat the impact of these viruses.
The genome of NSVs consists of one or more single-stranded, negative-sense RNA molecules that are assembled with multiple copies of viral nucleoprotein into megadalton-sized viral ribonucleoprotein (vRNP) complexes with a helical configuration1 (Box 1). The nucleoproteins are the main determinants of this helical configuration and oligomerize to form a scaffold for the genomic viral RNA (vRNA). In non-segmented NSVs (nsNSV), these vRNP structures are highly symmetrical and relatively rigid1,2. In segmented NSVs (sNSVs), each individual segment is contained in a distinct vRNP complex that is relatively flexible3,4. In these vRNPs, the vRNA segment forms a pseudocircle through non-covalent RNA–RNA interactions between the 5′ and 3′ termini (Box 1). These 5′ and 3′ termini are typically partially complementary and highly conserved within the same group of viruses.
All NSVs encode an RNA polymerase that contains an RNA-dependent RNA polymerase (RdRP) domain and replicates and transcribes the vRNA genome in the context of vRNPs5,6. Replication of the genome produces a positive-sense antigenome known as a complementary RNA (cRNA) for sNSVs (Box 2), which is encapsidated by nucleoprotein into a complementary RNP (cRNP). The antigenome serves as the template for genome synthesis. The vRNA genome is used as a template for transcription of positive-stranded mRNAs (Box 2). Some NSVs, such as arenaviruses, use an ambisense coding strategy, which involves transcription of the negative-sense genome as well as the cRNA (Box 2). RNA polymerases of NSVs produce mRNAs that contain a 5′ cap structure and, in most virus families, a 3′ polyadenosine (poly(A)) tail (Box 2). In most NSVs, replication and transcription of the genome occurs in the cytoplasm, whereas in orthomyxovirus family NSVs these processes occur in the nucleus.
The RNA polymerases of nsNSVs are capable of de novo synthesis of 5′ cap structures using their capping and methyltransferase (MT) domains7,8. The RNA polymerases of sNSVs lack intrinsic capping and MT activities and instead use a cap-snatching mechanism9. This mechanism involves a cap-binding (CapB) domain and an endonuclease (Endo) domain of the RNA polymerase. The stolen cellular capped RNA fragments are used as primers by the RNA polymerase to initiate viral transcription to generate viral mRNAs with a heterologous host RNA-derived sequence at their 5′ end10. In addition to ensuring efficient translation of viral mRNAs, the host-derived capped RNA primers provide access to alternative open reading frames and thus expand the viral proteome11.
NSVs encode proteins that act as cofactors of the viral RNA polymerase. These include the phosphoprotein (P protein) in paramyxoviruses, pneumoviruses and rhabdoviruses, M2-1 in pneumoviruses and VP24, VP30 and VP35 in filoviruses12–16. These cofactors have been implicated in tethering the polymerase to the nucleoprotein-coated RNA template and assisting in nucleoprotein recruitment to nascent RNA by forming nucleoprotein–P protein complexes to prevent premature oligomerization of nucleoprotein and nonspecific binding of nucleoprotein to RNA17–19.
Structures of NSV RNA polymerases and complexes of these polymerases with their cofactors have been difficult to obtain owing to difficulties in expressing recombinant forms of these proteins and cofactors and in their crystallization. Recent developments in the structural analysis of proteins by cryo-electron microscopy (cryoEM), as well as advances in X-ray crystallography and protein expression technologies, have led to reports of high-resolution structures of RNA polymerases of the orthomyxoviruses IAV, IBV, influenza C virus (ICV) and influenza D virus (IDV)20–24, the peribunyavirus La Crosse virus (LACV)25,26, the phenuivirus severe fever with thrombocytopenia syndrome virus (SFTSF)27,28, the arenaviruses LASV and Machupo virus (MACV)29, the rhabdoviruses vesicular stomatitis virus (VSV) and RABV7,30,31, the pneumoviruses human metapneumovirus (HMPV) and HRSV32–34 and the paramyxovirus human parainfluenza virus type 5 (HPIV)35. Combined with advances in our understanding of the enzymatic activities of RNA polymerases, these structures provide unprecedented structural insights into the replication and transcription machinery of NSVs. An emerging picture is that these multifunctional molecular machines use a modular architecture consisting of a central RdRP domain that is linked to multiple functional domains through flexible linkers. The ability to rearrange these domains enables NSV RNA polymerases to perform and coordinate a set of complex catalytic reactions to produce the different types of RNA that NSVs require to complete an infection cycle.
Box 1 Ribonucleoprotein complexes of negative-sense RNA viruses.
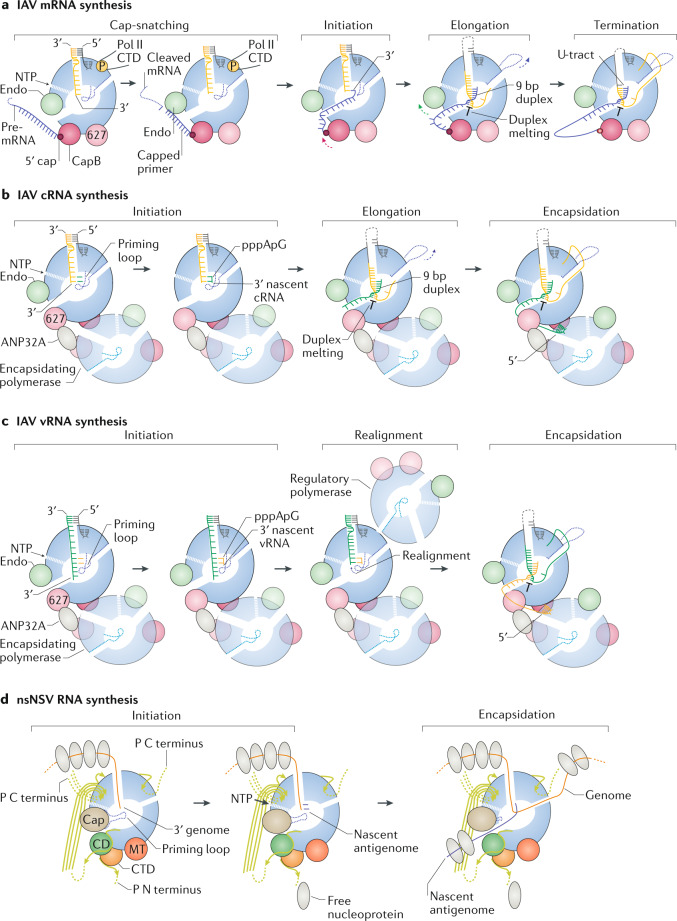
The genome and antigenome of negative-sense RNA viruses (NSVs) are encapsidated by oligomeric nucleoprotein. By coating the genome and antigenome, nucleoprotein protects the RNA from degradation and prevents double-stranded RNA (dsRNA) from forming between RNAs of opposite polarity. All NSV nucleoproteins are composed of two globular domains, a head and a body domain, as in the influenza A virus (IAV) nucleoprotein87, or an N-terminal lobe (NT lobe) and C-terminal lobe (CT lobe), as in the Hantaan virus nucleoprotein88, or an N-terminal domain (NTD) and C-terminal domain (CTD), as in the measles virus nucleoprotein89 (see the figure; schematics of domain organization in part a). The RNA is bound in a positively charged groove located between the domains or lobes. Oligomerization of the nucleoproteins is facilitated by various extensions, such as an internal tail-loop (as in the IAV), flexible N-terminal and/or C-terminal arms (as in the Hantaan and measles virus nucleoproteins), or other mobile subdomains (see the figure; surface and cartoon representations in part b). Ribonucleoproteins (RNPs) of non-segmented NSVs (nsNSVs) form regular helical structures, whereas the RNPs of segmented NSVs (sNSVs) generally form irregular, flexible, supercoiled structures4,88–92 (see the figure; surface representations of nucleoprotein oligomer in part c). These differences can be explained by the intrinsic differences between the nucleoproteins of different virus families. In the RNPs of nsNSVs, the arms that mediate nucleoprotein oligomerization have limited flexibility, such that each nucleoprotein forms identical contacts with each of its neighbours and thus automatically generates a repeated, helical structure (as in the measles virus RNP89). In the RNPs of sNSVs, the arms that mediate formation of nucleoprotein oligomers have flexible hinges that have the ability to form assemblies with very variable geometries. The IAV4,93,94 and Hantaan virus88 RNPs appear as flexible rods comprising two anti-parallel superhelical strands of RNA-bound nucleoproteins with a loop at one end and the polymerase holding together the viral RNA extremities at the other end. The docking of the nucleoprotein atomic structure in cryo-electron microscopy reconstructions of influenza virus viral RNPs (vRNPs) showed that the nucleoproteins in the strands could adopt a wide range of positions4. In addition, nucleoproteins could assume different degrees of rotation relative to the strand axis. Owing to this intrinsic flexibility, only low-resolution structures have been obtained for IAV vRNPs. The RNA-binding groove region generally shows the lowest resolution of the structure, pointing to the RNA as one of the most flexible parts of the complex. This observed flexibility of IAV vRNPs is fully consistent with chemical mappings of the RNA structure within vRNPs, which showed that the RNA can accommodate secondary structures and nucleoprotein is not evenly distributed on the RNA95–97.
Box 2 Genome and RNA synthesis products of negative-sense RNA viruses.
The genomes of negative-sense RNA viruses (NSVs) vary in length and segmentation. The non-segmented NSVs (nsNSVs) contain a single negative-sense RNA molecule that encodes several genes, as in the vesicular stomatitis virus (VSV) genome (see the figure). Replication of the genome involves formation of a replication intermediate termed the antigenome, which is used by the viral RNA polymerase to produce more genome copies. Expression of the genes in the genome occurs by a single-entry transcription mechanism, in which the RNA polymerase engages with the genome at the 3′ terminus and initiates or terminates transcription at each gene start or termination site, respectively. All viral mRNAs contain a 5′ cap structure and 3′ polyadenosine (An) tail. In contrast to nsNSVs, the genome of segmented NSVs (sNSVs) contains two to eight segments (see the figure). Furthermore, whereas replication occurs through a replication intermediate, expression of the encoded genes can occur in a number of ways. The La Crosse virus (LACV) segments each encode one gene, and these genes are expressed as viral mRNAs that contain a 5′ cap structure but no polyadenosine tail. The genome segments of Machupo virus (MACV) contain genes in the negative and positive orientation, which are transcribed by the viral RNA polymerase by transcription initiation at the 3′ terminus of the genome or the antigenome, respectively. MACV mRNAs contain a 5′ cap structure but no 3′ polyadenosine tail. Replication of the influenza A virus (IAV) genome segments also involves an antigenome intermediate, referred to as the complementary RNA (cRNA). Transcription starts at the 3′ terminus of the genome and terminates at a stretch of U residues close to the 5′ terminus, resulting in 5′ capped and 3′ polyadenylated products. Transcripts of the NS and M segments are spliced by the host splicing machinery.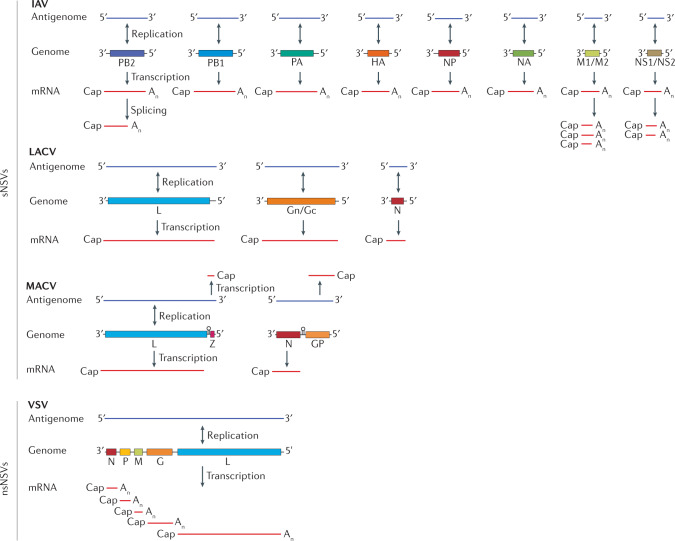
Structure of the RNA polymerase
The RNA polymerases of NSVs have a molecular mass of 250–450 kDa and are expressed as a single polypeptide, the L protein, or as three polypeptides that together form a heterotrimeric RNA polymerase complex. All NSV polymerases comprise an RdRP domain, showing the right hand-like fold with thumb, fingers and palm subdomains that are typical of polymerase domains36, and several other functional domains.
RNA polymerases of sNSVs
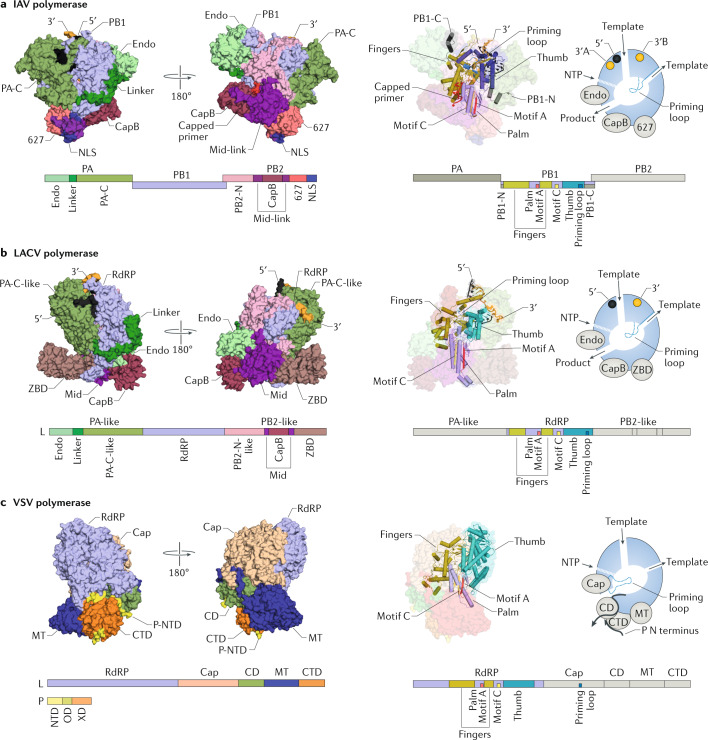
The heterotrimeric RNA polymerase encoded by influenza viruses consists of the polymerase basic 1 (PB1), PB2 and polymerase acidic (PA; known as P3 in ICVs and IDVs) proteins (Fig. 1a). The PB1 subunit is at the centre of the polymerase complex and contains the RdRP domain with the catalytic residues for RNA synthesis20,21,23,37. The PA subunit consists of two domains, the N-terminal Endo domain and the PA C-terminal domain (PA-C), which is deeply integrated into the thumb subdomain of the RdRP domain and connected to the Endo domain by a long linker that wraps around the RdRP domain (Fig. 1a). The PB2 subunit is a modular protein composed of several domains. The N-terminal third of PB2 (PB2-N) is tightly associated with the RdRP domain, while the C-terminal two-thirds of PB2 consists of several discrete domains, including the CapB domain, the middle and linker regions flanking CapB that form a rigid unit denoted as the Mid-link domain, the 627 domain (named for amino acid residue 627 in PB2 that determines host range38) and a C-terminal nuclear localization signal (NLS) domain, all separated by flexible linkers. The Endo domain of PA and the domains in the C-terminal portion of PB2 are linked to the RNA polymerase core by flexible linkers and can be arranged in different configurations relative to the RNA polymerase core, depending on association of the heterotrimeric complex with RNA and with viral and cellular proteins. In the structure of the human IAV RNA polymerase bound to vRNA and capped RNA primer37 (Fig. 1a), the 627 domain packs against the palm and thumb subdomains of the RdRP domain. The CapB domain faces the Endo domain and packs on the Mid-link domain, which is positioned between the 627 domain and the CapB domain. The NLS associates with the 627 domain.
Fig. 1. Domain organization and overall structure of NSV RNA polymerases.
a | Structure of the influenza A virus (IAV) heterotrimeric RNA polymerase bound to viral RNA (vRNA) and a capped RNA primer (Protein Data Bank (PDB) identifier (ID) 6RR7). b | Structure of the La Crosse virus (LACV) L protein with bound vRNA (PDB ID 6Z6B). c | Structure of the vesicular stomatitis virus (VSV) L protein and phosphoprotein (P) monomer complex (PDB ID 6U1X). The structures are rendered in the same orientation after superimposing motif C of the RNA-dependent RNA polymerase (RdRP) domain. CD, connector domain; CTD, C-terminal domain; Endo, endonuclease; Mid-link, middle and linker regions; MT, methyltransferase; NLS, nuclear localization signal; NSV, negative-sense RNA virus; NTD, N-terminal domain; NTP, nucleoside triphosphate; PA, polymerase acidic; PA-C, polymerase acidic C-terminal domain; PB, polymerase basic; P-NTD, P protein N-terminal domain; P-OD, P protein oligomerization domain; P-XD, P protein X domain; ZBD, zinc-binding domain.
The L proteins of sNSVs, such as the LACV L protein (Fig. 1b), are single polypeptide RNA polymerases comprising domains that are similar to those in the trimeric RNA polymerase. All sNSV L proteins consist of a central RdRP domain that is flanked by an N-terminal PA-C-like domain and a C-terminal PB2-N-like domain25–27,39, forming a globular RNA polymerase core. This globular core is linked to an N-terminal Endo domain and a C-terminal extension that, in the LACV L protein, is composed of several distinct domains, including a CapB domain, a middle (Mid) domain (composed of two regions of the L polypeptide flanking CapB) and a zinc-binding domain (ZBD)25 (Fig. 1b). The Endo domain is linked to the RNA polymerase core by a long flexible linker that wraps around the RdRP domain, and is positioned between the RNA polymerase core and the protruding C-terminal extension of the L protein (that is, the PB2-like region), making extensive contacts with multiple regions of the RNA polymerase core on one side and the CapB domain on the other. The C-terminal extension protrudes away from the core and forms an elongated arc-shaped structure that includes the Mid domain, the CapB domain and the ZBD (Fig. 1b). Although the CapB domain is conserved, the other domains of the C-terminal extension vary considerably in sequence and structure between L proteins of different sNSVs. In the SFTSV L protein27, the ZBD is replaced by a lariat domain, whereas the LASV and MACV L proteins29 contain a 627-like domain that is unresolved in current structures (see below). As in the influenza virus polymerases, the Endo domain and domains in the C-terminal extension are linked to the RNA polymerase core by flexible linkers and can be arranged in different configurations relative to the RNA polymerase core25 (see below).
In sNSVs, the RdRP, Endo and CapB domains are the structurally most conserved parts of the RNA polymerase9,39. The Endo domain shows a conserved fold across all sNSV polymerases and has the structural characteristics of the PD-(D/E)-XK superfamily of nucleases that use divalent metal ions for nucleic acid cleavage. In the IAV Endo domain, H41, P107, D108, E119 and K134 constitute the active centre, and analogous amino acid residues exist in the Endo domains of other sNSV polymerases. However, in the Endo domain of arenaviruses, the histidine upstream of the conserved PD-(D/E)-XK motif is replaced by an acidic amino acid residue, resulting in a difference in coordination of divalent metal ions in the active site40,41. This difference correlates with the inactivity of the isolated domain, suggesting that a rearrangement of the domain or contribution from the rest of the RNA polymerase is needed for endonuclease activation, potentially providing arenaviruses with a transcription ‘on/off’ switch40. The CapB domains of sNSV polymerases show little homology at the amino acid level but fold into a similar structure, which is composed of a β-sheet formed by five to seven antiparallel β-strands surrounding a single long α-helix or a bundle of several α-helices39. All solved sNSV CapB domains employ the same mechanism of cap binding whereby the methylated guanine of the cap structure is sandwiched between two aromatic amino acid residues by stacking interactions. In the IAV polymerase, H357 and F404 in PB2 form the aromatic sandwich. A histidine performing this role seems to be unique to human and avian IAV whereas phenylalanine, tryptophan or tyrosine perform this role in the cap-binding proteins of other sNSVs described to date.
RNA polymerases of nsNSVs
The L proteins of nsNSVs, such as that of VSV (Fig. 1c), are also single polypeptide RNA polymerases and fold into three catalytic and two structural domains7,31. The core of the nsNSV L protein structure is formed by the N-terminal RdRP domain and the capping (Cap) domain, which possesses polyribonucleotidyl transferase (PRNTase) activity. The Cap domain adopts a kidney-shaped fold and interacts with the RdRP domain over a large interface, forming the RdRP–Cap module7,31 (Fig. 1c). The RdRP–Cap module is linked through the connector domain (CD) to the third catalytic domain of the L protein, the dual-specificity MT domain, possessing both guanine-N7-methyltransferase and nucleoside-2′-O-methyltransferase activities, which is followed by the C-terminal domain (CTD). In the VSV L–P complex, the CD, MT domain and CTD form three globular domains that closely associate with each other as well as with the RdRP–Cap module, forming an overall compact structure.
The CD is only weakly conserved at the amino acid level across different nsNSV families and is believed to have a structural role. The CTD is not conserved at the sequence or structural level, and substantial differences in fold and size have been observed between the CTDs of the pneumovirus and rhabdovirus L proteins7,32. By contrast, the Cap and MT domains are relatively well-conserved and participate in synthesizing a 5′ cap structure. The Cap domain catalyses the formation of GTP-capped pre-mRNA by a mechanism that differs from eukaryotic capping42. The nascent RNA transcript with a 5′ triphosphate is first covalently linked to a catalytic histidine residue (H1241 in the VSV L protein) located in a conserved histidine–arginine (HR) motif, in a reaction that leads to formation of a monophosphate RNA–L protein intermediate. This linkage is attacked by a GTP molecule, resulting in addition of GDP to the monophosphate RNA to form a GTP-capped pre-mRNA. The HR motif and a GxxT motif, which are believed to be involved in GTP binding, are conserved across the Cap domains of nsNSVs. The MT domain is structurally well-conserved across nsNSVs and encodes a dual-function enzyme that methylates the GTP cap of viral mRNAs, first at the 2′-O and then at the N7 position. A GxGxG motif forms the binding site for the methyl donor, S-adenosylmethionine (SAM), and a conserved set of charged residues (K-D-K-E) forms the catalytic tetrad for methyl group addition.
The RNA polymerase cofactor P protein consists of three domains: an N-terminal domain (NTD), a central oligomerization domain (OD) and a C-terminal X domain (XD)18 (Fig. 1c). The NTD is generally unstructured and binds to a nascent nucleoprotein monomer, forming a nucleoprotein–P protein complex to prevent premature oligomerization of nucleoprotein and its nonspecific binding to RNA43. The XD is also intrinsically disordered in solution but associates with nucleoprotein in the context of vRNPs44. The OD mediates the formation of P protein homodimers or homotetramers that interact directly with the L protein, tethering the polymerase to the nucleoprotein-coated RNA template45.
The P protein can associate with the RNA polymerase in several ways. The structure of the VSV L protein has been solved in complex with an N-terminal fragment of P protein in which three motifs have been resolved31 (Fig. 1c). These motifs make clear interactions with the CTD, CD and RdRP domains, locking the CD, MT domain and CTD in a fixed arrangement with respect to the large RdRP–Cap module31. The overall structure of the RABV L–P complex is very similar to that of VSV L–P30, whereas in the HPIV L–P complex (which contains four copies of P protein), the CD, MT domain and CTD have been observed in different positions relative to the RdRP–CAP module35. These differences in L–P interactions most likely reflect different functional states of the L proteins in which they were captured rather than intrinsic differences in L–P interactions between different viruses (see below). In the HRSV and HMPV L–P complexes, which also contain four copies of P protein, the CD, MT domain and CTD remain entirely unresolved, owing to their flexibility32–34 (see below).
Structure of the RdRP domain
The RdRP domain is the only domain that is shared by the RNA polymerases of sNSVs and nsNSVs (Fig. 1). Four channels lead to and from the active site of the RdRP domain and facilitate template RNA entry and exit, nucleoside triphosphate (NTP) entry and product RNA exit26,46 (Fig. 1). In the heterotrimeric RNA polymerase, the three subunits are integrated such that they all contribute to the formation of these channels46.
The RdRP domain folds into a right hand-like shape with thumb, fingers (including fingertips) and palm subdomains that are characteristic of many RdRPs36. The thumb subdomain forms the ‘right-side wall’ of the active site, whereas the palm subdomain forms the ‘floor’ with a central four-stranded β-sheet. The fingers subdomain forms the ‘roof’ and ‘left-side wall’ of the active site. The active site chamber of the RdRP domain is formed by the conserved RNA polymerase motifs A–F, the majority of which are located in the palm subdomain36 (Fig. 1). Specifically, conserved motifs A and C in the palm subdomain contain the aspartate residues that bind to the two metal ions that coordinate nucleotide condensation47. The active site is structurally conserved, whereas a clear divergence at the amino acid sequence level is present in motifs A and C of sNSV and nsNSV RNA polymerases36.
A priming loop protrudes into the active site cavity and is involved in the formation of the first dinucleotide during de novo initiation32,48,49. In sNSV RNA polymerases, the priming loop emerges from the thumb subdomain as a flexible β-hairpin22,25,29 (Fig. 1a,b), whereas in the L protein of nsNSVs, the priming loop is a simple flexible loop that resides in the Cap domain next to the GxxG motif, from which it inserts into the RdRP domain7,32,35 (Fig. 1c).
Binding to promoter RNA
An important feature of all sNSV polymerases is the ability to associate with the conserved, partially complementary termini of the genome and antigenome segments, which represent the vRNA and cRNA promoters, respectively. In influenza virus RNA polymerase complexes, the 5′ end of the vRNA inserts into a pocket formed by the PA and PB1 subunits near the template entry channel21–23,37,50 (Fig. 1a). The same binding pocket is used by the 5′ end of the cRNA. Both the vRNA and cRNA 5′ termini assume a ‘hook’ conformation and bind to the RNA polymerase in a sequence-specific manner. In the LACV RNA polymerase, the vRNA 5′ terminus binds in a similar binding site in an equivalent position close to the template entry channel (Fig. 1b). In influenza virus RNA polymerases, binding of the vRNA 3′ terminus has been observed at two different sites near the polymerase surface, either in a groove near the fingers subdomain23, called the A-site, or at a site formed by residues of the PB1 and PA subunits, called the B-site21,24,37 (Fig. 1a). In contrast to the vRNA 3′ terminus, the cRNA 3′ terminus binds exclusively to the B-site. This B-site is equivalent to the 3′ terminus-binding site observed in the LACV and MACV RNA polymerases26,29 (Fig. 1b).
In influenza viruses, the initiation of replication on the vRNA 3′ terminus or cleavage of capped RNA prior to transcription initiation cannot occur without binding of the vRNA 5′ terminus, consistent with the idea that the vRNA 5′ terminus aids in recruitment of the vRNA 3′ terminus and activates the RNA polymerase. Indeed, the apo form of IAV and ICV polymerases harbours a highly unstable fingertip and priming loop20,37, whereas the same regions in structures of vRNA 5′ terminus-bound polymerases of IAV and IBV are more ordered22,23, suggesting that binding of the vRNA 5′ terminus contributes to the stabilization of the RdRP domain. Similar observations have been made for the LACV, LASV and MACV RNA polymerases25,28,29,51. Although the fingertip and the associated finger extension loops in the RdRP domain of the LASV and MACV RNA polymerases are highly ordered, even in the absence of binding of the RNA 5′ end29, the vRNA 5′ terminus boosts primer-independent and dinucleotide-primed RNA synthesis activity of these polymerases, in line with observations for the influenza virus and LACV polymerases28,29,51. Intriguingly, the vRNA 5′ terminus significantly inhibits capped-RNA-primed transcription for both LASV and MACV polymerases, suggesting that in arenaviruses the vRNA 5′ terminus may serve a regulatory role in the replication and transcription processes29.
No RNA binding has been observed in the currently available structures of nsNSV RNA polymerases, and it remains unclear how these polymerases recognize promoter elements in the genome and antigenome (Fig. 1c). It has been proposed that the N-terminal residues of the VSV and RABV L proteins, which are unresolved in current structures, as well as the CTD of the P protein, might be involved in the binding of template RNA and/or nucleoprotein, owing to their position near the template entry channel30,31,42.
Conformational rearrangements
The NSV RNA polymerases display great conformational flexibility, which is essential for the different modes of initiation that the RNA polymerases use to replicate and transcribe the viral genomic RNA.
RNA polymerases of sNSVs
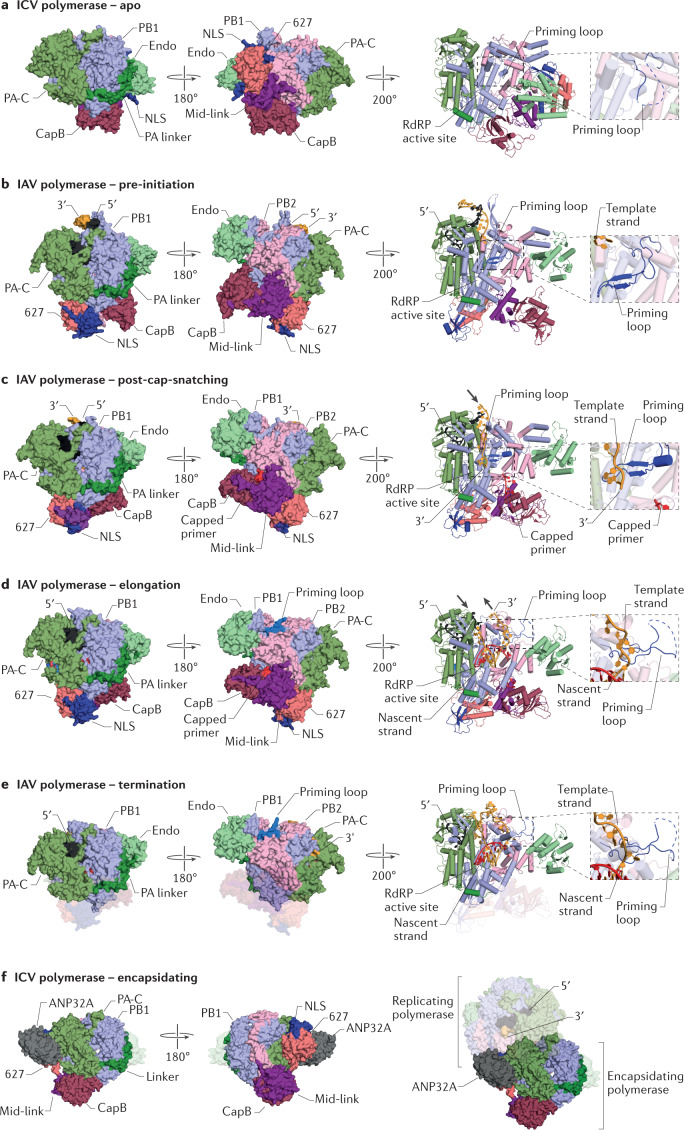
In the heterotrimeric RNA polymerase of influenza viruses, the Endo domain of PA and the C-terminal two-thirds of PB2 are linked to the RNA polymerase core by flexible linkers and can assume different positions relative to the RNA polymerase core (Fig. 2). Several fundamentally distinct conformations of the heterotrimeric RNA polymerase have been reported. In the structure of the influenza virus RNA polymerase apo form, the CapB domain packs against the palm subdomain, whereas the 627 domain is located near the nascent RNA exit channel and the Endo domain20,50 (Fig. 2a). In this conformation, a part of the Mid-link domain inserts into the cap-binding pocket of the CapB domain, preventing cap binding. Furthermore, the Endo domain and CapB domain face away from each other and, consequently, this conformation of the polymerase is unable to perform cap-snatching5. In the active site, the priming loop is disordered. By contrast, where structures have been captured at different stages of transcription, the 627 domain and CapB domain are rearranged through large conformational changes in the Mid-link domain, which repositions the 627 domain next to the palm–thumb subdomain interface and the CapB domain opposite the Endo domain23,24,47 (Fig. 2b–e). In the vRNA-template-bound ‘pre-initiation structure’, the priming loop is ordered, and the cap-binding pocket of the CapB domain is free to bind capped RNA and faces the Endo domain to facilitate cap-snatching (Fig. 2b). In the ‘post-cap-snatching’ structure, the CapB domain is rotated and the cap-binding pocket faces the product exit channel, consistent with a movement that would facilitate insertion of the cleaved capped RNA primer into the RdRP catalytic centre through the product exit channel23 (Fig. 2c). Further conformational rearrangements, including changes in the RdRP domain, take place as the polymerase transits from pre-initiation and initiation to elongation and then to termination24,47 (Fig. 2d,e). Specifically, the priming loop is displaced from the active site into the solvent as a disordered loop47 (Fig. 2b–e). An entirely distinct polymerase conformation has been observed in an asymmetrical dimer of the heterotrimeric polymerase bound by the cellular factor ANP32A52 (Fig. 2f). In this dimer, one of the polymerases has vRNA bound and was proposed to function as a replicase, whereas the second polymerase acts as an encapsidating polymerase that is involved in the assembly of the nascent RNA into an RNP. The conformation of the Endo and CapB domains in the vRNA-bound replicating polymerase is similar to that observed in the apo form of this polymerase (Fig. 2a), and in the active site the priming loop is also disordered. The conformation of the encapsidating polymerase is entirely distinct from that of the replicating polymerase form as well as the cap-snatching-competent vRNA-bound pre-initiation state of the polymerase. Specifically, in the encapsidating polymerase, the PB2 CapB and Mid-link domains pack against the palm and thumb subdomains of the polymerase core and are separated from the 627 and NLS domains, which primarily make contacts with PA-C. The Endo domain of PA and parts of PB2-N and NLS of the encapsidating polymerase are disordered in the structure (Fig. 2f).
Fig. 2. Structures of influenza virus RNA polymerase conformations.
a | Structure of the apo form of the influenza C virus (ICV) RNA polymerase (Protein Data Bank (PDB) identifier (ID) 5D9A). b | Structure of the pre-initiation conformation of the influenza A virus (IAV) RNA polymerase (PDB ID 4WSB). c | Structure of the post-cap-snatching conformation of the IAV RNA polymerase (PDB ID 6RR7). d | Structure of the elongation conformation of the IAV RNA polymerase (PDB ID 6T0V). e | Structure of the termination conformation of the IAV RNA polymerase (PDB ID 6SZU). f | Structure of the encapsidating polymerase of the ICV RNA polymerase bound to ANP32A (PDB ID 6XZR) and the putative replicating–encapsidating polymerase dimer bound to ANP32A. Magnified views show the priming loop. Finger subdomain residues are not shown so that the priming loop is visible. Structures in surface representation in each panel are shown in the same orientation after superimposing motif C of the RNA-dependent RNA polymerase (RdRP) domain. Structures in cartoon representation have been rotated relative to the structures in surface representation to focus on the priming loop. Endo, endonuclease; Mid-link, middle and linker regions; NLS, nuclear localization signal; PA, polymerase acidic; PA-C, polymerase acidic C-terminal domain; PB, polymerase basic.
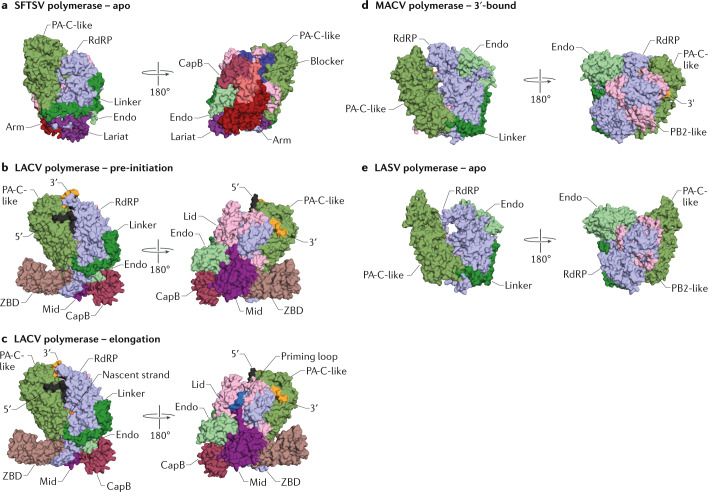
In the L proteins of sNSVs, the positions of the Endo domain, CapB domain and ZBD or 627-like domains also vary greatly and can adopt different configurations relative to the RNA polymerase core25–29 (Fig. 3). In the apo structure of the SFTSV L protein, the configuration of domains is similar to that observed in the apo and replicating polymerase structures of influenza virus. The CapB and Endo domain are packed against each other and are oriented such that the Endo active site faces away from the CapB domain27,28 (Fig. 3a). In addition, the cap-binding pocket of the CapB domain is occupied by the blocker motif, similar to how the Mid-link domain blocks the CapB domain in the apo and replicating polymerase of influenza virus. Consequently, the captured SFTSV L protein structure represents a cap-snatching-incompetent conformation27,28. The LACV L protein has been captured in conformations reflecting pre-initiation and elongation states25 (Fig. 3b,c). In the pre-initiation structure with both the 5′ and 3′ termini of vRNA bound, the Endo domain is rotated by 180° relative to its position in the SFTSV L protein structure and faces the CapB domain to allow cap-snatching25,26 (Fig. 3b). Transition from the pre-initiation to the elongation state with double-stranded RNA (dsRNA) bound in the active site is accompanied by large coordinated rotations of the Endo domain and C-terminal region that are made possible by the conformationally stable Mid domain, which acts as a central hub that mediates contacts between the RNA polymerase core, the CapB domain and the ZBD25. Further movements take place in the RdRP domain, including the extrusion of the priming loop from the active site. In the structures of the apo LASV L protein and the MACV L protein with the vRNA 3′ terminus bound in the B-site29, the Endo domain sits at the top of the fingers subdomain, close to the product exit channel, whereas the CTDs, including the CapB domain, remain unresolved in both structures (Fig. 3d,e).
Fig. 3. Structures of sNSV L protein conformations.
a | Structure of the apo form of the severe fever with thrombocytopenia syndrome virus (SFTSV) L protein (Protein Data Bank (PDB) identifier (ID) 6L42). b | Structure of the pre-initiation conformation of the La Crosse virus (LACV) L protein (PDB ID 6Z6B). c | Structure of the elongation conformation of the LACV L protein (PDB ID 6Z8K). d | Structure of the Machupo virus (MACV) L protein bound to the viral RNA 3′ terminus (PDB ID 6KLE). e | Structure of the apo form of the Lassa mammarenavirus (LASV) L protein (PDB ID 6KLC). Structures in each panel are shown in the same orientation after superimposing motif C of the RNA-dependent RNA polymerase (RdRP) domain. Endo, endonuclease; PA, polymerase acidic; PA-C, polymerase acidic C-terminal domain; PB, polymerase basic; sNSV, segmented negative-sense RNA virus; ZBD, zinc-binding domain.
RNA polymerases of nsNSVs
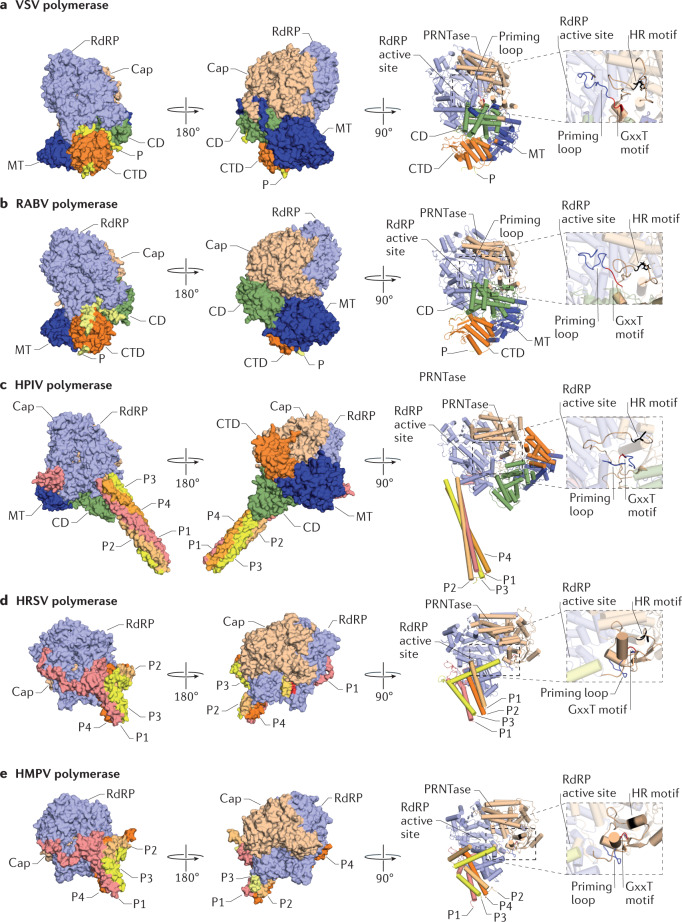
Although no structures of nsNSV L proteins bound to template RNA are available, several conformations of apo L–P complexes have been captured that suggest that conformational rearrangements take place when the polymerase transits from the pre-initiation to the elongation state (Fig. 4). In the VSV and RABV L–P complexes, the Cap domain interacts with a large surface of the RdRP domain and blocks the nascent RNA exit channel7,30,31 (Fig. 4a,b). In the same structures, the CD and CTD pack against the Cap domain, keeping the MT domain away from the nascent RNA exit channel. This conformation is, thus, likely an initiation conformation that must undergo a rearrangement before capping can take place. Such a different state is captured in the apo structure of the HPIV L protein with the CTD moving closer to the Cap domain35 (Fig. 4c). In addition, the priming loop of the CAP domain is organized differently and forms a wider platform in the active site, suggesting that this conformation could reflect a post-initiation structure. Further changes relative to the VSV pre-initiation state can be observed in the apo structures of HRSV and HMPV L–P complexes (Fig. 4d,e). Here, the Cap domain movement exposes the nascent RNA exit channel, whereas the CD, MT domain and CTD are no longer resolved in the EM maps32–34. Furthermore, in these structures, the priming loop of the Cap domain is fully retracted and collapsed onto the Cap domain. The different conformations of the priming loop in the VSV and HRSV L–P complexes suggest that these rearrangements may be coupled to a transition from initiation to elongation32,34.
Fig. 4. Structures of nsNSV L–P complex conformations.
a | Structure of the vesicular stomatitis virus (VSV) L protein–phosphoprotein (P) monomer complex (Protein Data Bank (PDB) identifier (ID) 6U1X). b | Structure of the rabies lyssavirus (RABV) L protein–P monomer complex (PDB ID 6UEB). c | Structure of the human parainfluenza virus type 5 (HPIV) L protein–P tetramer complex (PDB ID 6V86). d | Structure of the human respiratory syncytial virus (HRSV) L protein–P tetramer complex (PDB ID 6UEN). e | Structure of the human metapneumovirus (HMPV) L protein–P tetramer complex (PDB ID 6U5O). Insets show the priming loop and histidine–arginine (HR) and GxxT motifs of the Cap domain. Structures in surface representation in each panel are shown in the same orientation after superimposing motif C of the RNA-dependent RNA polymerase (RdRP) domain. Structures in cartoon representation have been rotated relative to the structures in surface representation to focus on the priming loop. Finger subdomain residues are not shown so that the priming loop and Cap domain active site are visible. CD, connector domain; CTD, C-terminal domain; MT, methyltransferase; nsNSV, non-segmented negative-sense RNA virus; P1–P4, phosphoprotein subunits; PRNTase, polyribonucleotidyl transferase.
The observed conformations are affected by the binding of the cofactor P protein. In the VSV and RABV L–P complexes, which contain an N-terminal fragment of the P protein, the P protein interacts with and locks the CD, MT domain and CTD in a fixed arrangement with respect to the large RdRP–Cap module30,31 (Fig. 4a,b). In the HIPV, HMPV and HRSV L–P complexes, the P protein forms a tetramer through the central OD32–35 (Fig. 4c–e). Strikingly, there is a large degree of variation in the conformation of each of the four P monomers (P1, P2, P3 and P4). These different conformations allow the P protein to wrap around the RdRP domain of the L protein in a tentacular fashion. Although P1 and P4 make extensive contacts with L, P3 makes minimal contacts with L, and P2 interacts almost exclusively with the other P monomers. The L protein of HRSV primarily uses the fingers motif to interact with the OD and XD of P protein, but the palm motif of the RdRP domain is also involved. Interestingly, these L–P interactions do not fix the positions of the CD, the MT domain and CTD, which remain unresolved in the HMPV and HRSV L–P structures.
Transcription and replication
The heterotrimeric RNA polymerase of influenza virus has been captured at different stages of the transcription cycle and is currently the best understood in terms of how the RNA polymerase coordinates the different modes of initiation and termination during transcription and replication. Our current understanding of these processes in the L proteins of sNSVs and nsNSVs is largely based on our knowledge of the influenza virus RNA polymerase.
sNSV RNA polymerases
Transcription by the influenza virus RNA polymerase generates capped and polyadenylated mRNAs that are transcribed from the vRNA segments. Transcription is initiated by binding of the viral polymerase in the context of vRNPs to the serine-5-phosphorylated CTD of host RNA polymerase II (Pol II) via residues in the PA-C and the 627 domain and NLS domain of PB253,54. Pol II CTD binding stabilizes the cap-snatching-competent, pre-initiation conformation of the viral polymerase54, which allows the CapB domain of the viral RNA polymerase to capture the cap structure of nascent host-capped RNA (Fig. 5a). After cleavage of the nascent host transcript by the Endo domain, the 3′ end of the capped primer is transferred into the product exit channel by a 70° rotation of the CapB domain, and the primer is stabilized by interactions with the Mid-link domain and the 3′ end of the vRNA template in the active site23,24,47. The stability of this complex is dependent on both the length of the primer and its 3′ sequence and, consequently, by the number of bases that can pair between the primer and the template55,56. Extension of the primer occurs in a template-dependent manner, although short duplications of nucleotides at the vRNA 3′ terminus have been observed in several sNSV mRNAs, indicative of a prime-and-realign mechanism10,55–57. In the transition from initiation to elongation, the priming loop is expelled from the RdRP active site, opening the template exit channel, and stays extruded during steady-state elongation24,47. During elongation, a 9–10 bp nascent strand–template duplex is maintained in the active site before the template and nascent RNA are separated and directed down their respective exit channels. Evidence from the IAV and IBV RNA polymerases shows that this separation occurs on a conserved aromatic residue of the lid subdomain of the PB2-N domain, at the end of the active site cavity24,47. Other sNSV RNA polymerases also contain a lid subdomain in the PB2-N-like domain, suggesting that this mechanism of strand separation is conserved25. The capped RNA product exits through the product exit channel without being encapsidated. Instead, it is bound by cellular mRNA-binding proteins, such as the nuclear cap-binding complex58. After exiting through the template exit channel, the 3′ terminus of the template is guided along the exterior of the thumb domain to the B-site 3′ binding site24,59, which likely ensures the integrity of the vRNP complex during elongation. During termination, the RNA polymerase stutters on an oligo(U) motif starting at position 17 from the 5′ terminus, involving U17 flipping in and out of the +1 active site position, to generate a poly(A) tail60. The 5′ terminus of the template is not copied and remains bound to its binding site61,62, while the RNA polymerase releases the viral transcript and recycles the vRNA template24.
Fig. 5. Mechanisms of RNA synthesis by NSV RNA polymerases.
a | Synthesis of mRNA in influenza virus. b | Synthesis of complementary RNA (cRNA) in influenza virus. c | Synthesis of viral RNA (vRNA) in influenza virus. d | RNA synthesis by a model non-segmented negative-sense RNA virus (nsNSV) RNA polymerase. Arrows indicate direction of C terminus in partial structures of the phosphoprotein (P). CD, connector domain; CTD, C-terminal domain; Endo, endonuclease; MT, methyltransferase; NTP, nucleoside triphosphate; PA, polymerase acidic; Pol II, RNA polymerase II.
Replication by the influenza virus RNA polymerase is a two-step process with the vRNA template first being copied into a cRNA, which next acts as template for vRNA synthesis. cRNA synthesis is initiated de novo, without the assistance of a primer, and starts with the formation of a dinucleotide opposite the terminal 3′ residues of the vRNA segment (Fig. 5b). The replication initiation process is dependent on support by the priming loop and in particular by a conserved proline at the tip of the priming loop48. During synthesis of a new genome from an antigenome template by the influenza virus RNA polymerase, initiation begins at positions 4 and 5 of the cRNA 3′ terminus48,63 (Fig. 5c). The resulting dinucleotide is next realigned to the terminal 3′ residues of the antigenome by backtracking of the template that is mediated by a conformational change in the priming loop37,64. This conformational change has been proposed to be dependent on the binding of a second, regulatory polymerase37,65,66. The regulatory and replicating polymerases form a symmetrical dimer through PA-C, the thumbs subdomain of PB1 and an N-terminal subdomain of PB2 (refs37,67). In the observed dimer, both polymerases are in a conformation similar to that described for the apo form of the polymerase (Fig. 2a). Mutation of residues in the dimer interface or its disruption with a nanobody reduces the efficiency of vRNA synthesis, likely owing to the prevention of template backtracking37. Interestingly, symmetrical dimerization of the polymerase is mutually exclusive with binding of the 3′ termini of the cRNA and vRNA in the B-site, leading to the suggestion that dimer formation might regulate the location of the 3′ end of the template21,37.
During replication, unlike during transcription, the nascent RNA is co-replicatively encapsidated by the binding of viral polymerase to the 5′ end and the recruitment of nucleoprotein to the body of the RNA to form cRNPs and vRNPs. This process is likely mediated by another type of dimerization of the heterotrimeric polymerase, as observed in the complex with the cellular factor ANP32A52 (Fig. 2f). In this asymmetrical polymerase dimer, the encapsidating polymerase is ideally positioned to capture the 5′ end of the nascent RNA product emerging from the active site of the replicating polymerase. ANP32A, an essential factor for the replication of the influenza virus genome, bridges the two polymerase heterotrimers, stabilizing their interaction68,69. The N-terminal leucine-rich repeat (LRR) domain of ANP32A acts as the bridge while most of the C-terminal low complexity acidic region (LCAR) could not be fully resolved in the structures. It has been proposed that the disordered LCAR could act as a molecular whip70 that recruits nucleoprotein in a manner analogous to that proposed for P protein during the replication by nsNSV L proteins.
Currently, it is not known which conformational rearrangements take place during replication and at which point the symmetrical and asymmetrical polymerase dimers form and dissociate. In the proposed replicating polymerase of the asymmetrical polymerase dimer, the 3′ end of the RNA template is unresolved in the active site, and the priming loop is disordered. Further rearrangements must take place upon insertion of the RNA template into the active site before replication can proceed. Furthermore, it is likely that some of the structural rearrangements described for transcription are also relevant for replication. Thus, the priming loop is most likely extruded from the active site of the replicating polymerase, and the already copied 3′ end of the RNA template binds to the B-site as it emerges from the active site through the template exit channel, to maintain RNP integrity during replication.
In comparison with the heterotrimeric polymerase of influenza viruses, much less is known about how L proteins of sNSVs carry out transcription and replication. However, considering that the vRNAs of L-protein-encoding sNSVs are assembled into vRNPs akin to those of influenza virus, and that the L protein associates with the vRNA 5′ and 3′ termini at equivalent binding sites, it is highly likely that transcription and replication also proceed in a manner similar to that of influenza virus.
nsNSV RNA polymerases
To transcribe the genome, the L proteins of nsNSVs initiate transcription at a promoter at the 3′ end of the genome, sequentially transcribing the leader region and five internal genes (N, P, M, G and L) into the leader RNA and five monocistronic mRNAs with a 5′ cap structure and a 3′ poly(A) tail, by using a stop–start transcription mechanism. This mechanism results in the generation of a gradient of mRNA abundance in relation to proximity to the promoter, with the nucleoprotein-encoding mRNA being the most abundant and L mRNA being the least abundant6,42. Initiation and termination are regulated by cis-acting start–stop signals that flank each gene. To replicate the genome, the L protein initiates at the 3′ promoter and produces a full-length copy of the genome, ignoring the internal start–stop signals. The resulting antigenome is neither capped nor polyadenylated and is co-replicatively assembled into an RNP complex by association with nucleoprotein. The replicative intermediate antigenome acts as a template for further rounds of replication to generate genomic RNA for progeny virions.
Although no high-resolution structures have been obtained of the nsNSV RNA polymerases bound to RNA and no pre-initiation or elongation complexes have yet been reported, the currently available apo L–P complexes likely reflect different stages of the RNA synthesis process (Fig. 5d). In the VSV and RABV L–P complexes7,30,31, the priming loop that emerges from the Cap domain protrudes into the active site of the RdRP domain, where it could stabilize the formation of the first dinucleotide using an aromatic residue and/or proline for de novo initiation32,49, suggesting that the captured structures represent the polymerase in a pre-initiation state. This is in contrast to the HPIV, RSV and HMPV L–P structures, in which this loop is retracted from the RdRP active site and is folded into the Cap domain32–35. The retraction of the priming loop is consistent with elongation during RNA synthesis, suggesting that these structures represent the polymerase in elongation mode. Retraction of the priming loop also creates a continuous cavity shared by the RdRP and Cap catalytic sites, which allows the nascent transcript to enter the catalytic cavity of the Cap domain. Capping occurs by an unconventional capping mechanism that involves the covalent attachment of the monophosphorylated 5′ terminus of the nascent transcript to a histidine side chain in the Cap active site, followed by the transfer of the 5′ monophosphate of the nascent transcript onto a GDP acceptor42. The covalent attachment of the 5′ end of the nascent transcript to the Cap active site cavity during capping and the force generated as a result of nascent RNA filling the cavity could result in the release of the CD–MT domain–CTD module from the RdRP–Cap core module. The freeing of the module would expose the MT catalytic site for the subsequent N7 and 2′-O methylation of the 5′ cap after the 5′ end of the nascent transcript has been transferred onto a GDP and released from the Cap domain. During replication, the RNA product likely exits at a site proximal to the N-terminal end of P protein, which is known to associate with monomeric nucleoprotein. Thus, the proximity of this site to the exit for replication products could facilitate the delivery of nucleoprotein to the growing RNA chain, ensuring co-replicative encapsidation of the nascent genome. Interestingly, the phosphorylation status of the N-terminal end of the P protein also has a role in the switch from transcription to replication, although the mechanism by which this occurs is currently unclear42. To reveal the detailed movements of polymerase domains it will be necessary to obtain further snapshots of transcribing and replicating polymerases with bound template RNA and nascent product RNA, as has recently been reported for influenza viruses24,47.
Therapeutic applications
Nucleoside analogues
Several nucleoside analogues are able to inhibit NSV RNA polymerases. Favipiravir, ribavirin and remdesivir are broad-spectrum nucleoside analogues that can inhibit influenza virus, EBOV and HRSV RNA synthesis71–73. These NTPs act by competing with purine incorporation, resulting in the introduction of mutations in the viral genome or leading to polymerase stalling71,74. Resistance to favipiravir can emerge through mutations in thumb and fingers subdomains of RdRP75. The nucleoside analogue N4-hydroxycytidine (NHC) was more recently identified and was found to inhibit IAV, IBV and HRSV infections76 by inducing lethal mutagenesis; no resistance to NHC has been observed to date77. The analogues ALS-8176 and ALS-8112 triphosphate inhibit HMPV and HRSV infections in nonhuman primates, although resistance to these drugs has been observed. The resistance mutations map to homomorph A (a structurally conserved extension of motif A36,78) of the palm subdomain32 and likely alter the binding of the nucleoside analogues in the active site and prevent active site closure when a nucleoside is bound32.
Non-nucleoside inhibitors
Non-nucleoside inhibitors target the auxiliary enzymatic domains, inter-domain interactions or inter-subunit binding of the NSV RNA polymerases. For example, peptides have been designed that can compete with the inter-subunit interactions of the influenza virus heterotrimeric RNA polymerase79,80. The IAV inhibitors baloxavir marboxil (BXA) and pimodivir (VX-787) target the PA Endo and PB2 CapB domains, respectively, of the IAV RNA polymerase and act as transcription inhibitors81–85. BXA can also inhibit the Endo domains of other sNSVs41, including SFTSV and Heartland virus (HRTV), owing to the structural similarity of endonucleases among sNSV polymerases84. However, resistance to BXA and VX-787 rapidly emerges in IAV infections through mutations near the active site of the Endo domain and cap-binding pocket of the CapB domain, respectively86. In an example of using the different capping mechanism of nsNSVs compared with that of the host as a drug target, the HRSV inhibitor BI-compound D (BID) disrupts the capping process of HRSV nascent mRNAs. However, escape mutants resistant to this compound also arise quickly by changes in a pocket near the PRNTase active site34.
Concluding remarks
The past seven years have witnessed unprecedented progress in the structural and functional understanding of the NSV RNA polymerases. High-resolution structures of numerous RNA polymerases of both sNSVs and nsNSVs have been solved. These structures have vastly increased our understanding of the transcription and replication mechanisms used by these viruses. In particular, the work on the influenza virus RNA polymerase has led to a detailed understanding of the molecular mechanisms of viral transcription initiation, elongation and termination, as well as replication initiation. This new mechanistic understanding of RNA synthesis opens avenues to the development of new antiviral agents that target the RNA polymerase of NSVs. This growth in our understanding of the biology of sNSV polymerases has not been replicated for the nsNSV RNA polymerases, and a number of fundamental questions remain. How do nsNSV RNA polymerases bind to the genomic or antigenomic RNAs, and what conformational rearrangements take place when the polymerase proceeds from initiation to elongation to termination? How are the nascent genomic and antigenomic RNAs encapsidated into an RNP? What factors determine whether the RNA polymerase transcribes the genome into mRNA or replicates it into an antigenome? Which host factors have fundamental roles in viral transcription and replication by supporting RNA polymerase function? Given the progress in our understanding of sNSV polymerases, we look forward to further advancements in our knowledge of NSV polymerases in the future.
Acknowledgements
A.J.W.t.V. is supported by joint Wellcome Trust and Royal Society grant 206579/Z/17/Z and the National Institutes of Health grant R21AI147172. J.M.G. is supported by Wellcome Investigator Award 200835/Z/16/Z. E.F. is supported by UK Medical Research Council programme grant MR/R009945/1.
Glossary
- Negative-sense RNA viruses
(NSVs). Viruses with a negative-sense, single-stranded RNA genome.
- Negative-sense RNA
An RNA that must be transcribed by a virally encoded RNA-dependent RNA polymerase into complementary mRNA before viral protein production can occur.
- Nucleoprotein
A protein that binds to the genomic and antigenomic RNA in the context of viral ribonucleoprotein complexes.
- Viral ribonucleoprotein
(vRNP). A complex consisting of viral RNA and nucleoprotein.
- Antigenome
Positive-sense replicative RNA intermediate of viral replication, which is referred to as the complementary RNA for some viruses, such as the influenza viruses.
- Cap-snatching
A process by which the 5′ cap structure is cleaved from cellular capped RNAs by endonuclease activity and subsequently used to prime viral transcription.
- Priming loop
A protein structure that interacts with the first nucleotide of the nascent RNA or terminal base of the template and lowers the activation energy for de novo initiation.
- De novo initiation
The initiation step of nucleic acid synthesis that occurs in the absence of a primer.
- Apo form
A protein in the absence of a ligand.
- Prime-and-realign
A mechanism in which the nascent RNA is separated from the template RNA and reannealed at an identical sequence elsewhere in the template.
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Microbiology thanks R. Fearns, M. Rosenthal and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/2/2021
A Correction to this paper has been published: 10.1038/s41579-021-00524-9
Contributor Information
Aartjan J. W. te Velthuis, Email: ajwt6@cam.ac.uk
Jonathan M. Grimes, Email: jonathan@strubi.ox.ac.uk
Ervin Fodor, Email: ervin.fodor@path.ox.ac.uk.
References
- 1.Luo M, Terrell JR, McManus SA. Nucleocapsid structure of negative strand RNA virus. Viruses. 2020;12:835. doi: 10.3390/v12080835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruigrok RW, Crepin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr. Opin. Microbiol. 2011;14:504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Gallagher JR, Torian U, McCraw DM, Harris AK. Structural studies of influenza virus RNPs by electron microscopy indicate molecular contortions within NP supra-structures. J. Struct. Biol. 2017;197:294–307. doi: 10.1016/j.jsb.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coloma R, et al. Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism. Nat. Microbiol. 2020;5:727–734. doi: 10.1038/s41564-020-0675-3. [DOI] [PubMed] [Google Scholar]
- 5.Te Velthuis AJ, Fodor E. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016;14:479–493. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fearns R, Plemper RK. Polymerases of paramyxoviruses and pneumoviruses. Virus Res. 2017;234:87–102. doi: 10.1016/j.virusres.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang B, et al. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morin B, Rahmeh AA, Whelan SP. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J. 2012;31:1320–1329. doi: 10.1038/emboj.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olschewski S, Cusack S, Rosenthal M. The cap-snatching mechanism of bunyaviruses. Trends Microbiol. 2020;28:293–303. doi: 10.1016/j.tim.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Koppstein D, Ashour J, Bartel DP. Sequencing the cap-snatching repertoire of H1N1 influenza provides insight into the mechanism of viral transcription initiation. Nucleic Acids Res. 2015;43:5052–5064. doi: 10.1093/nar/gkv333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho JSY, et al. Hybrid gene origination creates human-virus chimeric proteins during infection. Cell. 2020;181:1502–1517 e1523. doi: 10.1016/j.cell.2020.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutherland KA, Collins PL, Peeples ME. Synergistic effects of gene-end signal mutations and the M2-1 protein on transcription termination by respiratory syncytial virus. Virology. 2001;288:295–307. doi: 10.1006/viro.2001.1105. [DOI] [PubMed] [Google Scholar]
- 13.Fearns R, Collins PL. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 1999;73:5852–5864. doi: 10.1128/JVI.73.7.5852-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahmeh AA, et al. Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc. Natl Acad. Sci. USA. 2012;109:14628–14633. doi: 10.1073/pnas.1209147109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groseth A, et al. The Ebola virus ribonucleoprotein complex: a novel VP30-L interaction identified. Virus Res. 2009;140:8–14. doi: 10.1016/j.virusres.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu W, et al. Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis. Nat. Commun. 2017;8:15576. doi: 10.1038/ncomms15576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emerson SU, Schubert M. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl Acad. Sci. USA. 1987;84:5655–5659. doi: 10.1073/pnas.84.16.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guseva S, Milles S, Blackledge M, Ruigrok RWH. The nucleoprotein and phosphoprotein of measles virus. Front. Microbiol. 2019;10:1832. doi: 10.3389/fmicb.2019.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishio M, et al. Mapping of domains on the human parainfluenza virus type 2 nucleocapsid protein (NP) required for NP-phosphoprotein or NP-NP interaction. J. Gen. Virol. 1999;80:2017–2022. doi: 10.1099/0022-1317-80-8-2017. [DOI] [PubMed] [Google Scholar]
- 20.Hengrung N, et al. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature. 2015;527:114–117. doi: 10.1038/nature15525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Q, et al. Structural insight into RNA synthesis by influenza D polymerase. Nat. Microbiol. 2019;4:1750–1759. doi: 10.1038/s41564-019-0487-5. [DOI] [PubMed] [Google Scholar]
- 22.Pflug A, Guilligay D, Reich S, Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 23.Reich S, et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature. 2014;516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 24.Wandzik JM, et al. A structure-based model for the complete transcription cycle of influenza polymerase. Cell. 2020;181:877–893 e21. doi: 10.1016/j.cell.2020.03.061. [DOI] [PubMed] [Google Scholar]
- 25.Arragain B, et al. Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes. Nat. Commun. 2020;11:3590. doi: 10.1038/s41467-020-17349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerlach P, Malet H, Cusack S, Reguera J. Structural insights into bunyavirus replication and its regulation by the vRNA promoter. Cell. 2015;161:1267–1279. doi: 10.1016/j.cell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, et al. Structure of severe fever with thrombocytopenia syndrome virus L protein elucidates the mechanisms of viral transcription initiation. Nat. Microbiol. 2020;5:864–871. doi: 10.1038/s41564-020-0712-2. [DOI] [PubMed] [Google Scholar]
- 28.Vogel D, et al. Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein. Nucleic Acids Res. 2020;48:5749–5765. doi: 10.1093/nar/gkaa253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng R, et al. Structural insight into arenavirus replication machinery. Nature. 2020;579:615–619. doi: 10.1038/s41586-020-2114-2. [DOI] [PubMed] [Google Scholar]
- 30.Horwitz JA, Jenni S, Harrison SC, Whelan SPJ. Structure of a rabies virus polymerase complex from electron cryo-microscopy. Proc. Natl Acad. Sci. USA. 2020;117:2099–2107. doi: 10.1073/pnas.1918809117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenni S, et al. Structure of the vesicular stomatitis virus L protein in complex with its phosphoprotein cofactor. Cell Rep. 2020;30:53–60 e55. doi: 10.1016/j.celrep.2019.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan J, et al. Structure of the human metapneumovirus polymerase phosphoprotein complex. Nature. 2020;577:275–279. doi: 10.1038/s41586-019-1759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao D, et al. Cryo-EM structure of the respiratory syncytial virus RNA polymerase. Nat. Commun. 2020;11:368. doi: 10.1038/s41467-019-14246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilman MSA, et al. Structure of the respiratory syncytial virus polymerase complex. Cell. 2019;179:193–204 e114. doi: 10.1016/j.cell.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdella R, Aggarwal M, Okura T, Lamb RA, He Y. Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation. Proc. Natl Acad. Sci. USA. 2020;117:4931–4941. doi: 10.1073/pnas.1919837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.te Velthuis AJ. Common and unique features of viral RNA-dependent polymerases. Cell Mol. Life Sci. 2014;71:4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan H, et al. Structures of influenza A virus RNA polymerase offer insight into viral genome replication. Nature. 2019;573:287–290. doi: 10.1038/s41586-019-1530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subbarao EK, London W, Murphy BR. A single amino-acid in the PB2 gene of influenza-A virus is a determinant of host range. J. Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gogrefe N, Reindl S, Gunther S, Rosenthal M. Structure of a functional cap-binding domain in Rift Valley fever virus L protein. PLoS Pathog. 2019;15:e1007829. doi: 10.1371/journal.ppat.1007829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reguera J, et al. Comparative structural and functional analysis of bunyavirus and arenavirus cap-snatching endonucleases. PLoS Pathog. 2016;12:e1005636. doi: 10.1371/journal.ppat.1005636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reguera J, Weber F, Cusack S. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 2010;6:e1001101. doi: 10.1371/journal.ppat.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogino T, Green TJ. RNA synthesis and capping by non-segmented negative strand RNA viral polymerases: lessons from a prototypic virus. Front. Microbiol. 2019;10:1490. doi: 10.3389/fmicb.2019.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guryanov SG, Liljeroos L, Kasaragod P, Kajander T, Butcher SJ. Crystal structure of the measles virus nucleoprotein core in complex with an N-terminal region of phosphoprotein. J. Virol. 2015;90:2849–2857. doi: 10.1128/JVI.02865-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longhi S, et al. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 2003;278:18638–18648. doi: 10.1074/jbc.M300518200. [DOI] [PubMed] [Google Scholar]
- 45.Ivanov I, Crepin T, Jamin M, Ruigrok RW. Structure of the dimerization domain of the rabies virus phosphoprotein. J. Virol. 2010;84:3707–3710. doi: 10.1128/JVI.02557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reguera J, Gerlach P, Cusack S. Towards a structural understanding of RNA synthesis by negative strand RNA viral polymerases. Curr. Opin. Struct. Biol. 2016;36:75–84. doi: 10.1016/j.sbi.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Kouba T, Drncova P, Cusack S. Structural snapshots of actively transcribing influenza polymerase. Nat. Struct. Mol. Biol. 2019;26:460–470. doi: 10.1038/s41594-019-0232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.te Velthuis AJW, Robb NC, Kapanidis AN, Fodor E. The role of the priming loop in influenza A virus RNA synthesis. Nat. Microbiol. 2016;1:16029. doi: 10.1038/nmicrobiol.2016.29. [DOI] [PubMed] [Google Scholar]
- 49.Ogino M, Gupta N, Green TJ, Ogino T. A dual-functional priming-capping loop of rhabdoviral RNA polymerases directs terminal de novo initiation and capping intermediate formation. Nucleic Acids Res. 2019;47:299–309. doi: 10.1093/nar/gky1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thierry E, et al. Influenza polymerase can adopt an alternative configuration involving a radical repacking of PB2 domains. Mol. Cell. 2016;61:125–137. doi: 10.1016/j.molcel.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pyle JD, Whelan SPJ. RNA ligands activate the Machupo virus polymerase and guide promoter usage. Proc. Natl Acad. Sci. USA. 2019;116:10518–10524. doi: 10.1073/pnas.1900790116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carrique L, et al. Host ANP32 mediates the assembly of the influenza virus replicase. Nature. 2020;587:638–643. doi: 10.1038/s41586-020-2927-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lukarska M, et al. Structural basis of an essential interaction between influenza polymerase and Pol II CTD. Nature. 2017;541:117–121. doi: 10.1038/nature20594. [DOI] [PubMed] [Google Scholar]
- 54.Serna Martin I, et al. A mechanism for the activation of the influenza virus transcriptase. Mol. Cell. 2018;70:1101–1110 e1104. doi: 10.1016/j.molcel.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Te Velthuis AJW, Oymans J. Initiation, elongation, and realignment during influenza virus mRNA synthesis. J. Virol. 2018;92:e01775-17. doi: 10.1128/JVI.01775-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Vlugt C, Sikora D, Rocheleau L, Pelchat M. Priming and realignment by the influenza a virus RdRp is dependent on the length of the host primers and the extent of base pairing to viral RNA. Virology. 2019;536:91–100. doi: 10.1016/j.virol.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 57.Yao M, et al. Repetitive prime-and-realign mechanism converts short capped RNA leaders into longer ones that may be more suitable for elongation during rice stripe virus transcription initiation. J. Gen. Virol. 2012;93:194–202. doi: 10.1099/vir.0.033902-0. [DOI] [PubMed] [Google Scholar]
- 58.Bier K, York A, Fodor E. Cellular cap-binding proteins associate with influenza virus mRNAs. J. Gen. Virol. 2011;92:1627–1634. doi: 10.1099/vir.0.029231-0. [DOI] [PubMed] [Google Scholar]
- 59.Walker AP, Sharps J, Fodor E. Mutation of an influenza virus polymerase 3′ RNA promoter binding site inhibits transcription elongation. J. Virol. 2020;94:e00498–00420. doi: 10.1128/JVI.00498-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poon LL, Pritlove DC, Fodor E, Brownlee GG. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 1999;73:3473–3476. doi: 10.1128/JVI.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fodor E, Pritlove DC, Brownlee GG. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tiley LS, Hagen M, Matthews JT, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J. Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng T, Vreede FT, Brownlee GG. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J. Virol. 2006;80:2337–2348. doi: 10.1128/JVI.80.5.2337-2348.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oymans J, Te Velthuis AJW. A mechanism for priming and realignment during influenza A virus replication. J. Virol. 2018;92:e01773-17. doi: 10.1128/JVI.01773-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jorba N, Coloma R, Ortin J. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 2009;5:e1000462. doi: 10.1371/journal.ppat.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jorba N, Area E, Ortin J. Oligomerization of the influenza virus polymerase complex in vivo. J. Gen. Virol. 2008;89:520–524. doi: 10.1099/vir.0.83387-0. [DOI] [PubMed] [Google Scholar]
- 67.Chang S, et al. Cryo-EM structure of influenza virus RNA polymerase complex at 4.3Å resolution. Mol. Cell. 2015;57:925–935. doi: 10.1016/j.molcel.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 68.Long JS, et al. Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature. 2016;529:101–104. doi: 10.1038/nature16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Long JS, et al. Species specific differences in use of ANP32 proteins by influenza A virus. eLife. 2019 doi: 10.7554/eLife.45066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reilly PT, Yu Y, Hamiche A, Wang L. Cracking the ANP32 whips: important functions, unequal requirement, and hints at disease implications. Bioessays. 2014;36:1062–1071. doi: 10.1002/bies.201400058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tchesnokov EP, Feng JY, Porter DP, Gotte M. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delang L, Abdelnabi R, Neyts J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antivir. Res. 2018;153:85–94. doi: 10.1016/j.antiviral.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 73.Graci JD, Cameron CE. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006;16:37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Furuta Y, et al. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goldhill DH, et al. The mechanism of resistance to favipiravir in influenza. Proc. Natl Acad. Sci. USA. 2018;115:11613–11618. doi: 10.1073/pnas.1811345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoon JJ, et al. Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob. Agents Chemother. 2018;62:e00766-18. doi: 10.1128/AAC.00766-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Toots M, et al. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl Med. 2019;11:eaax5866. doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lang DM, Zemla AT, Zhou CL. Highly similar structural frames link the template tunnel and NTP entry tunnel to the exterior surface in RNA-dependent RNA polymerases. Nucleic Acids Res. 2013;41:1464–1482. doi: 10.1093/nar/gks1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li C, et al. A peptide derived from the C-terminus of PB1 inhibits influenza virus replication by interfering with viral polymerase assembly. FEBS J. 2013;280:1139–1149. doi: 10.1111/febs.12107. [DOI] [PubMed] [Google Scholar]
- 80.Ghanem A, et al. Peptide-mediated interference with influenza A virus polymerase. J. Virol. 2007;81:7801–7804. doi: 10.1128/JVI.00724-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamada K, et al. Identification of a novel compound with antiviral activity against influenza A virus depending on PA subunit of viral RNA polymerase. Microbes Infect. 2012;14:740–747. doi: 10.1016/j.micinf.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 82.Clark MP, et al. Discovery of a novel, first-in-class, orally bioavailable azaindole inhibitor (VX-787) of influenza PB2. J. Med. Chem. 2014;57:6668–6678. doi: 10.1021/jm5007275. [DOI] [PubMed] [Google Scholar]
- 83.Pflug A, et al. Capped RNA primer binding to influenza polymerase and implications for the mechanism of cap-binding inhibitors. Nucleic Acids Res. 2018;46:956–971. doi: 10.1093/nar/gkx1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang W, et al. The cap-snatching SFTSV endonuclease domain is an antiviral target. Cell Rep. 2020;30:153–163 e155. doi: 10.1016/j.celrep.2019.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McGowan DC, et al. Design, synthesis, and biological evaluation of novel indoles targeting the influenza PB2 cap binding region. J. Med. Chem. 2019;62:9680–9690. doi: 10.1021/acs.jmedchem.9b01091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Omoto S, et al. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 2018;8:9633. doi: 10.1038/s41598-018-27890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ye Q, Krug RM, Tao YJ. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444:1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 88.Arragain B, et al. High resolution cryo-EM structure of the helical RNA-bound Hantaan virus nucleocapsid reveals its assembly mechanisms. eLife. 2019;8:e43075. doi: 10.7554/eLife.43075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gutsche I, et al. Structural virology. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science. 2015;348:704–707. doi: 10.1126/science.aaa5137. [DOI] [PubMed] [Google Scholar]
- 90.Sugita Y, Matsunami H, Kawaoka Y, Noda T, Wolf M. Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 Å resolution. Nature. 2018;563:137–140. doi: 10.1038/s41586-018-0630-0. [DOI] [PubMed] [Google Scholar]
- 91.Su Z, et al. Electron cryo-microscopy structure of Ebola virus nucleoprotein reveals a mechanism for nucleocapsid-like assembly. Cell. 2018;172:966–978 e912. doi: 10.1016/j.cell.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wan W, et al. Structure and assembly of the Ebola virus nucleocapsid. Nature. 2017;551:394–397. doi: 10.1038/nature24490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arranz R, et al. The structure of native influenza virion ribonucleoproteins. Science. 2012;338:1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- 94.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. Organization of the influenza virus replication machinery. Science. 2012;338:1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dadonaite B, et al. The structure of the influenza A virus genome. Nat. Microbiol. 2019;4:1781–1789. doi: 10.1038/s41564-019-0513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Williams GD, et al. Nucleotide resolution mapping of influenza A virus nucleoprotein-RNA interactions reveals RNA features required for replication. Nat. Commun. 2018;9:465. doi: 10.1038/s41467-018-02886-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee N, et al. Genome-wide analysis of influenza viral RNA and nucleoprotein association. Nucleic Acids Res. 2017;45:8968–8977. doi: 10.1093/nar/gkx584. [DOI] [PMC free article] [PubMed] [Google Scholar]