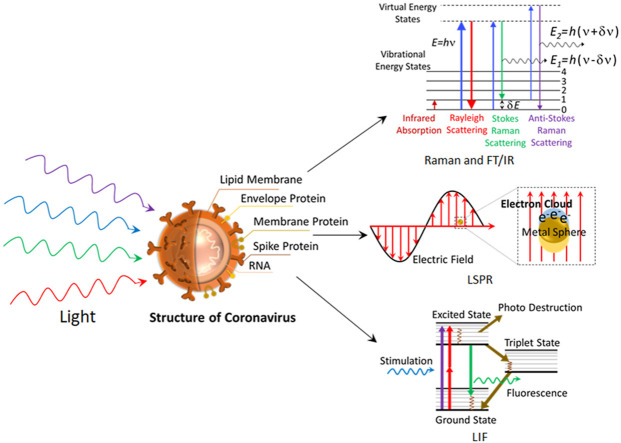
Abstract
The outbreak of life-threatening pandemic like COVID-19 necessitated the development of novel, rapid and cost-effective techniques that facilitate detection of viruses like SARS-CoV-2. The presently popular approach of a collection of samples using the nasopharyngeal swab method and subsequent detection of RNA using the real-time polymerase chain reaction suffers from false-positive results and a longer diagnostic time scale. Alternatively, various optical techniques namely optical sensing, spectroscopy, and imaging shows a great promise in virus detection. Herein, a comprehensive review of the various photonics technologies employed for virus detection, particularly the SARS-CoV family, is discussed. The state-of-art research activities in utilizing the photonics tools such as near-infrared spectroscopy, Fourier transform infrared spectroscopy, Raman spectroscopy, fluorescence-based techniques, super-resolution microscopy, surface plasmon resonance-based detection, for virus detection accounted extensively with an emphasis on coronavirus detection. Further, an account of emerging photonics technologies of SARS-CoV-2 detection and future possibilities is also explained. The progress in the field of optical techniques for virus detection unambiguously show a great promise in the development of rapid photonics-based devices for COVID-19 detection.
Keywords: COVID-19, Virus, Photonics, Optical spectroscopy, Optical imaging, Surface plasmon resonance
Graphical abstract
1. Introduction
The sub-micron sized viruses, described as “organisms at the edge of life” have been responsible for some of the worst human pandemics in history like influenza (1918), smallpox (1978),COVID-19. As of September 17, 2020, COVID-19 caused by the SARS-CoV-2 virus has affected nearly 29 million people and caused 936,521 death causalities (WHO, 2020). Genomic characterization studies on the full-length genome sequences obtained from SARS-CoV-2 infected patients have found a genome similarity of ~80%, ~50%, and ~96% with SARS-CoV, MERS- CoV, and bat coronavirus RaTG13, respectively (Lu et al., 2020). The pathways such as aerosolized droplets assisted transmission, direct, oral-fecal route, etc., are suggested to contribute towards pandemic spread with a viral incubation time as long as 24 days (Guan et al., 2020). In the case of SARS-CoV-2, the spike glycoprotein (S protein) on the virus SARS-CoV-2 normally binds to the receptor, angiotensin-converting enzyme 2 (ACE 2), on the surface of human cells and invade the cell (Yan et al., 2020). Different kinds of clinical samples have been explored for COVID-19 detection. Conventional viral detection techniques usually employ the respiratory specimens collected from the upper and lower respiratory tract. The upper respiratory specimens from nasopharynx or oropharynx swabs have been collected in case of acute infection stage, which usually occurs within 7 days of viral infection. High viral loads have been reported in nasal samples as compared to the specimens obtained from the throat (Zou et al., 2020). The presence of viral RNA could be weakly detected in nasal and throat swabs after two weeks from symptom onset (Walsh et al., 2020). However, sampling of both nasal and oropharynx specimens is recommended to minimize the error in the detection of viruses. Lower respiratory samples are being used to evaluate disease progression by quantitative monitoring of viral load, especially in cases of low viral load (Yu et al., 2020). Higher viral loads and prolonged viral shedding have been observed in lower respiratory specimens compared to upper respiratory samples of critically ill COVID 19 patients (Huang et al., 2020). Bronchoalveolar lavage fluid (BLF) from the lower respiratory tract samples obtained via bronchoscopy or tracheal aspiration has been used for COVID-19 detection in symptomatic subjects with more than a week history. Sputum samples have also been used for viral detection by expectorating deep cough into a sterile container (Mohammadi et al., 2020). Sputum sampling has displayed higher viral RNA detection rates in comparison with nasopharyngeal, oropharyngeal samples. Immunoassay based technologies have been employed for diagnostic purposes using blood serum as the clinical sample. In addition, detection of viral samples has also been performed from saliva and stool specimens, which demands fewer sampling procedures in comparison with respiratory samples (Feng et al., 2020). However, testing of samples from different sites may always improve sensitivity and minimize the false negative test results. This is evident in the clinical study, which has reported RT-PCR test results of samples collected from various sites of COVID-19 infected patients (Wang et al., 2020a). The researchers have demonstrated a detection rate of 93% in bronchoalveolar lavage fluid, 72% in sputum, and 63% in nasopharyngeal swabs, whereas the detection rate was low for pharyngeal swabs (32%) and stool (29%) (Wang et al., 2020a).
The current clinical diagnosis approaches for COVID-19 detection include searching for clinical manifestations, evaluation of epidemiological history, lung imaging, virus response to the antibody in blood serum, and real-time polymerase chain reaction (rT-PCR) of the nasopharyngeal swab sample (Long et al., 2020). The gold standard PCR technique has several pitfalls in the way and the position of nasopharyngeal swab collection, sample transportation to the test center, RNA extraction, enzyme inhibitors, etc. that often lead to false-negative cases (Long et al., 2020). Another commonly used enzyme-linked immunosorbent assay (ELISA) technique offers the advantages of cost-effectiveness, robustness, scalability to large sample size, repeatability, and reproducibility but often requires high-quality antisera (Sakamoto et al., 2018). Moreover, the method is destructive to the samples, and identifying specific viral species/strains can be challenging. As is well known, faster and early diagnosis tools are the key to control pandemic like COVID -19. The high population of asymptotically infected patients and the high reagent requirement for the PCR testing during pandemic necessitate alternative detection/diagnostic tools. With the advancement in the miniaturization of imaging devices and the development of tunable laser sources that allow highly reproducible spectral recording and imaging for sensitive and specific measurements, photonics-based tools are now becoming increasingly popular in clinical science, particularly for virus detection (Wallace, 2020). In this review, we summarize the recent progress made in the field of optical techniques that include spectroscopic as well as imaging techniques for virus detection. Initially, we provide a short account of the various popular methods currently employed for virus detection. Following it, a detailed account of the applicability of techniques such as near-infrared spectroscopy, Fourier-transform infrared spectroscopy, Raman spectroscopy, fluorescence-based techniques, super-resolution microscopy techniques, surface plasmon resonance-based detection, for virus detection, particularly for coronavirus. Further, a brief overview of other light assisted techniques that can potentially detect SARS-CoV-2 is also discussed. Finally, our perspective on the current and future directions in the field of optical technologies for the detection of viruses like SARS-CoV-2 is also presented. Recently, in an interesting review, M. Soler et al. discussed various label-free nanophotonics sensors for the detection of viruses like COVID-19 (Soler et al., 2020a). The researchers accounted mainly for genomic detection and intact virus detection primarily using surface plasmon resonance technique with a little discussion on silicon photonics sensor based detection but not accounted in detail about the spectroscopic signature-based detection of viruses. In another interesting review, A. M Amarni and the colleagues gave a perspective of emerging engineered photonics solutions for COVID-19 (Soler at al. 2020b). In the review, the researchers accounted that beyond the well known SPR biosensors, there are emerging plasmonic nanotechnology and silicon-based photonics and rings and waveguide-based rapid virus screening and diagnosis tools. Additionally, they accounted for various photonics-based disinfection technologies. As compared to these works, the present review article focused on the optical techniques largely using the spectroscopic tools for the virus detection. Several industries have shown interest in exploring the high potential of optical techniques for COVID-19 detection, as given in Supplementary Table 1. A group led by Wilfred Dore from Samsung Strategy and Innovation Center have initiated research on developing SERS based technology, which can enable rapid detection of SARS-COV-2 at an earlier stage from human saliva (Devpost, 2020). Botanisol Analytics, a start-up based at the University of Arizona, has been working on a Raman spectrometer-based COVID-19 detector for faster and cost-effective detection than conventional RT-PCR tests. The team has already received $1.5 million financial support from Air Force Research Laboratory to build the viral detection systems (Optics.org. 2020). Biomark Diagnostics and its affiliated company Bio-Stream have also been trying to develop cost-effective and quick viral detection modalities using optical techniques. Their ultimate goal is to provide the test results from saliva within 30 s by exploring the combination of SERS and machine learning techniques (Newsfile. 2020). Advanced Nanotechnologies (Ant), Ireland, in collaboration with Starna Scientific and Trinity College Schools of Chemistry and Engineering, have been working towards the development of a POIN2T testing device for COVID-19 (Ant. 2020). They have been trying to extend their existing technology ANT Raman Flipper device for viral detection from passengers saliva at various point of interactions such as airport security, cruise ship boarding point, community centers, etc. Similarly, Lambdagen Corporation has also been extending their plasmonic sensing-based point of care Laura digital platform towards COVID-19 testing (LambdaGen. 2020). Creative Biostructure has been using their commercial SPR systems to evaluate the binding affinity between SARS-CoV-2 S protein and human ACE2 receptors (Creative Biostructure. 2020). Xantec has already developed ready to use COVID-19 RBD sensor chip based on SPR technology, which can be utilized in commercial SPR platforms for viral detection (Xantech. 2020). ProMIS Neurosciences, Inc. have also used SPR technology to develop highly accurate antibody test for SARS CoV-2 detection with the collaborative support from BC Neuroimmunology Lab Inc. (BCNI) (ProMIS Neurosciences. 2020). Todos Medical Ltd has been effective in transforming FTIR technology into a successful detection platform for cancer and neurodegenerative diseases. Currently, efforts have been going on to translate the potential of this technology for COVID-19 diagnosis (BioWorld. 2020). LightDeck Diagnostics has raised $11 Million for the development of a rapid, point of care device for COVID 19 detection in 5 min with the aid of laser-based planar waveguide technology (LightDeck Diagnostics. 2020).
2. Popular techniques for virus detection
Isothermal nucleic acid amplification-based techniques such as reverse transcription loop-mediated isothermal amplification (RT-LAMP) and Rolling Circle Amplification (RCA) have been receiving great attention due to their advantages over conventional RT-PCR test. Unlike RT-PCR, RT-LAMP does not demand the requirement of a thermal cycler for nucleic acid amplification since the isothermal amplification occurs at a constant temperature ~60–65 °C. The amplified DNA in this technique is usually detected by evaluating the turbidimetry or fluorescence/colorimetry. LAMP enables cost-effective, rapid, and specific detection with respect to the PCR technique. However, the complexity in designing the primers and the chances of false-positive results due to non-specific amplification while using pH indicators are the associated limitations (Feng et al., 2020). RCA facilitates sensitive viral detection due to its ability to amplify signals up to 109 fold within 90 min (Wang et al., 2005). This technique performed with minimal reagents can provide high specificity since it minimizes the false-positive results. Researchers have also incorporated isothermal amplification strategies with Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) based assays (SHERLOCK, DETECTR) for realizing cost-effective, sensitive, and specific virus detection (Zhang et al., 2020a; Broughton et al., 2020). This technique does not need complex instrumentation and can be performed in <1 h duration. The separate isothermal amplification and the need for multiple manual operations in these techniques can complicate the detection procedures and elevate the probability of carry-over contaminations (Kumar et al., 2020; Ding et al., 2020). All-In-One Dual CRISPR-Cas12a (AIOD-CRISPR) assay developed by Ding et al. minimize the chances of such contaminations by performing one step and one-pot reaction system, despite its challenging task of experiment optimization (Kumar et al., 2020). Even though mass spectrometry-based tests can facilitate rapid, highly sensitive and specific viral detection, this technique suffers shortcomings in the form of the upfront cost of equipment and the requirement of technical expertise (Nachtigall et al., 2020; Mahmud et al., 2020). Graphene-based Field-effect transistor (FET) biosensors have also shown promising results by detecting spike protein of SARS-Cov-2 in nasopharyngeal swab specimens with a detection limit down to 2.42 × 102 copies/mL (Seo et al., 2020). However, the semiconductor defects are a matter of concern in FET biosensors (Menon et al., 2020). The synthesis of high-quality graphene with identical parameters is difficult, and any variation in the parameters may alter the sensor's response characteristics (Sengupta et al., 2020). The use of electrochemical biosensors provides the benefits of lower sample volume requirement, cost-effective detection, and translational potential to develop miniaturized point of care devices, but also suffers from the lower shelf life and its sensitivity to sample matrix (Menon et al., 2020). The comparison of different methods reported for COVID-19 detection is given in Supplementary Table 2.
3. FTIR spectroscopy
The non-invasive Fourier transform infrared (FTIR) spectroscopy that relies on the absorption of the infrared radiation has been extensively used for molecular structure fingerprinting of protein and nucleic acid. The spectral region of interest for virus studies includes the fundamental vibrations and overtones of most biochemicals, including proteins, nucleic acids, phosphates, and lipids (Vargas et al., 2009). Pizarro et al. have used the FTIR technique in combination with linear discriminant analysis to investigate metabolic profile alterations in the blood plasma of human immunodeficiency virus (HIV)-infected patients and differentiated between the healthy and infected subjects and different stages of HIV (Pizarro et al., 2018). An FTIR study on fibroblast cells infected with Moloney murine sarcoma virus (MSV) showed a high RNA/DNA ratio in the case of malignant cells as compared to healthy cells (Huleihel et al., 2002). The technique further used to differentiate between retrovirus-transformed cells from normal ones and to monitor the impact of an antiviral drug (acyclovir) upon Vero cells infected with herpes simplex virus type 1 (HSV-1) (Erukhimovitch et al., 2006; Huleihel et al., 2001).
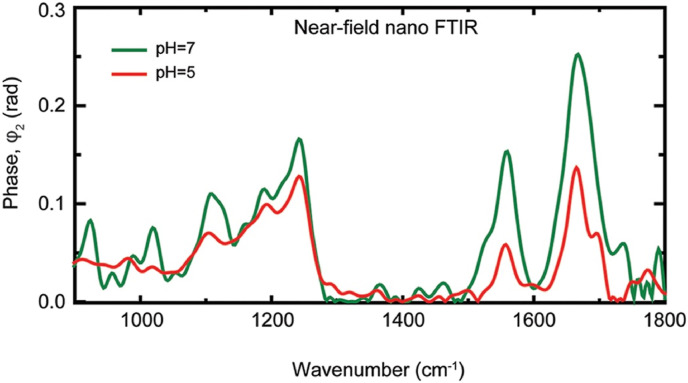
Attenuated total reflection (ATR)-FTIR technique is a variant of IR spectroscopy, where the evanescent wave generated by total internal reflection with the help of a high refractive index crystal was used to probe the sample characteristic signatures in the mid-IR region. Santos et al. have explored the ATR-FTIR technique for the identification and quantification of dengue serotype 3 (DENV-3) virus in blood and serum. In combination with multivariate analytical tools such as principal component analysis-LDA, successive projections algorithm - linear discriminant analysis (SPA-LDA), and genetic algorithm – linear discriminant analysis (GA-LDA), researchers were able to obtain better results for blood and serum samples infected with different levels of viral concentrations (Santos et al., 2017). The ability of ATR-FTIR analysis to monitor disease progression in virally affected cases (e.g., HIV), as well as treatment response, were investigated in serum samples by analyzing the spectral bands corresponding to lipids/fatty acids, carbohydrates, glucose, and proteins (Sitole et al., 2014). A portable ATR-FTIR system was used to diagnose viral hepatitis from blood serum by probing the presence of marker immunoglobulin signal in the case of both hepatitis B (HBV) and hepatitis C (HCV cases) and also found a unique marker band at 1093 cm−1 for the hepatitis B surface antigen (Roy et al., 2019). The potential of ATR- FTIR for viral studies have also tested in the detection of live, attenuated virus in freeze-dried vaccine formulations by investigating the amide III spectral region (1200-1350 cm−1) and proved the efficiency in distinguishing different vaccine doses in the formulation (Hansen et al., 2015). The nanoscale infrared spectroscopic imaging was used to understand the chemical, as well as structural modification, occurs before the membrane fusion in the case of a single archetypal enveloped virus (influenza X31) and at different pH values (Fig. 1 ) (Gamage et al., 2018). A reduction in the amide I and amide II bands observed at acidic pH due to protein unfolding.
Fig. 1.
Nano-FTIR spectra of influenza virus particles at neutral pH (green) and pH 5 (red). Reprinted from (Gamage et al., 2018). (https://creativecommons.org/licenses/by/4.0/). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Raman spectroscopy
The weak inelastically scattered light (shorter energy Stokes and higher energy anti-Stokes scattering) following excitation with monochromatic radiation (Raman scattering) are now routinely used to molecular fingerprint the biological samples as water exhibit low Raman scattering. Sato et al. have used micro-Raman spectroscopy for early-stage viral detection (~12 h) by studying Raman spectral signature (~952 cm−1) during the adenovirus infection onto human embryonic kidney epithelial (HEK293) cell lines (Moor et al., 2014). Rehman et al. have applied the Raman technique for the dengue virus infection detection by analyzing the bands at 1614 cm−1 and 1750 cm−1 that originate from Immunoglobulin antibodies IgG and IgM created in response to dengue infection in blood samples (Rehman et al., 2012). Salman et al. demonstrated the application of Raman spectroscopy for the herpes virus (HSV-1) identification in Vero cell cultures with 100% sensitivity (Salman et al., 2014). The experiments have suggested a lower concentration of proteins and lipids in virus-infected Vero cells. Raman spectroscopy has also been proved beneficial for the in-vitro diagnosis of viral hepatitis from blood serum samples (Saade et al., 2008). Raman bands attributed to lipids and phospholipids at 1170, 1257, and 1344 cm−1 were used as the discriminating factor for identifying infected samples from normal serum samples. The UV-resonance Raman spectroscopy that provides an enhancement of 102-106 was reported to investigate the viruses liked filamentous virus (Wen et al., 1999). Sohail et al. have explored Raman spectroscopy in combination with proximity based machine learning technique for the effective diagnosis of hepatitis C infection with 95% accuracy (Sohail et al., 2018).
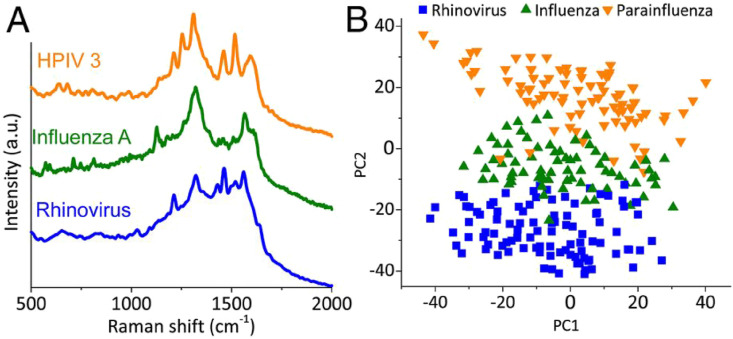
The plasmonic nanostructure enabled improved Raman scattering in surface-enhanced Raman scattering technique (SERS) now finding increased application in virus studies (Yeh et al., 2020). The popular antisense oligonucleotides (ASO) that liberated upon viral destruction have complementary sequences to some unique sites of viral genomes and often form a complex between ASO and viral genome, manifested as different SERS spectra compared to ASO (Ambartsumyan et al., 2020). The production of ASO is straight forward, and it involves the sequencing of the target virus genomes and the selection of a unique sequence for a strain of choice. In another work, upon carrying out PCA and LDA on the recorded Raman spectrum from the serum samples of the patients, the diagnostic sensitivity of 91.4% and a specificity of 83% was obtained for hepatitis B virus detection (Lu et al., 2018). The peak corresponding to L-arginine found to increase whereas spectral lines of saccharides, phenylalanine, tyrosine, as well as valine, and hypoxanthine decreases. In another work, highly ordered Au nanodots deposited electrochemically onto ITO substrates were used as SERS active substrates for the quantitative investigation of (35 fg/mL to 350 pg/mL) HIV (Lee et al., 2015). The usage of the SERS technique for the detection as well as discrimination of influenza, adenovirus, and respiratory syncytial viruses (RSV) was also reported (Shanmukh et al., 2006). SERS also enabled the detection and differentiation of different stages of influenza viruses by exploiting Raman signatures of their unique surface proteins/lipids (Lim et al., 2015). The capability of the SERS technique to detect a nonstructural protein (NS1), an established protein used for the early detection of viral infections, from saliva was also reported (Othman et al., 2017). Similarly, SERS platforms exploiting NS1 protein detection were also reported for the differentiation of the Zika virus and dengue virus (Sánchez-Purrà et al., 2017). Sing et al. have demonstrated SERS based dengue detection from serum samples using a handheld device by the identification of NS1 protein peak (Gahlaut et al., 2020). Recently, a handheld device (Virrion) that capture viruses based on their size with the aid of carbon nanotube forest arrays and subsequent viral recognition using SERS in combination with machine learning algorithms were reported (Yeh et al., 2020). Raman spectra of respiratory viruses obtained using Virrion is shown in Fig. 2 (A) and the PCA results (Fig. 2(B)) demonstrate the effectiveness of the technique in differentiating the viral strains. Besides, oseltamivir-resistant mutant influenza virus in human nasal and saliva specimens has been detected with the aid of SERS-active urchin Au nanoparticles and oseltamivir hexylthiol, which acts as effective receptor for virus capture (Eom et al., 2019). SERS based Magnetic immunoassays has been capable enough to detect influenza virus H3N2 down to 102 TCID50/mL (Sun et al., 2017).
Fig. 2.
VIRRION testing of respiratory viruses. (A) Raman spectra. (B) PCA plot of Raman fingerprint of the different viruses. Each dot represents a collected spectrum. Reproduced from (Yeh et al., 2020) (https://creativecommons.org/licenses/by/4.0/).
The virus titer determination using the label-free SERS technique suffers from nonuniform adsorption, and the high analyte concentration enabled nonlinear signal enhancement. Besides, the irregular ‘hot spots’ on the plasmonic substrate also adversely affect the reproducibility of the SERS signal. The indirect SERS technique, antibodies, molecules attached to the sensor's surface, or aptamers provide the specificity by selective binding with the analyte. Such a binding facilitate the accurate placement of the analyte molecule at desired hotspots. As the SERS signal comes from a reporter molecule, not from the analyte itself, this technique demand reporter molecules must be water-soluble and easily conjugatable to oligonucleotides. In an interesting work, silver-nanorod SERS array functionalized with the receptor ACE2 was employed for the rapid interrogation of SARS-CoV-2 in water without any pre-treatment (Zhang et al., 2020b). The Raman spectroscopy study showed that the recognition binding domain of SARS-CoV-2 spike protein SERS spectral intensity in the assay is significantly quenched for most of the peaks and exhibited a shift from 1189 to 1182 cm−1. The technique offers an opportunity to assess the viral decay in the wastewater treatment plants and the tracking of SARS-CoV-2 in the pipe network. Viral RNA detection from saliva was also reported in a recent study using Raman spectroscopy with a sensitivity of 7.05 × 107 transduction units per ml viral particles (Desai et al., 2020).
Porter et al. have coupled SERS with atomic force microscopy and demonstrated a limit of detection of 1 × 106 FCV/mL in the case of feline calicivirus (FCV) (Porter et al., 2006). Similarly, Popp et al. have performed investigations at a single mosaic virus level using tip-enhanced Raman spectroscopy (TERS), and the Raman bands correspond to coat proteins, and RNA was observed (Cialla et al., 2009). In this technique, the electromagnetic field enhancement and confinement at the sharp scanning probe microscope (SPM) tip due to localized surface plasmon resonance effect and lightning rod effect enable the nanoscale chemical imaging and can be used for imaging of avipoxvirus and adeno-associated virus of diameter 350 nm and 26 nm, respectively (Hermann et al., 2011). The study demonstrated variation in the relative peak intensities and position of most of the spectral features for the different particles from the same virus strain. The potential of this technique for single virus detection and the differentiation of influenza virus and picornavirus was also demonstrated (Deckert et al., 2020). As the SARS-CoV-2 virus diameter fall in a similar range, this technique can be explored for mapping SARS-CoV-2.
5. Fluorescence-based virus detection
The fluorescence emanating from samples following the optical excitation is used for virus studies, particularly for labeled and non-labeled fluorescence microscopy studies. Here, fluorescing transgenic proteins or small chemical express in cells of interest were commonly used as the fluoroprobe. In the labeled detection, fluorescent tags such as quantum dots, covalently attached fluorophores, or other chemicals connected via click chemistry act as labels to investigate the molecule of interest (Moses and Moorhouse 2007; Resch-Genger et al., 2008). In the conventional immunofluorescence (IF), the intercellular structures of the fixed and permeabilized cells were probed via fluorescence of lipids or target proteins whereas oligonucleotides were used in fluorescence in-situ hybridization (FISH) staining (Pernthaler et al., 2001). The viral RNAs of influenza A virus or Hepatitis C virus was visualized by using multiple specific short probes in the single-molecule FISH technique (Chou and Lionnet 2018; Mor et al., 2016; Ramanan et al., 2016). The number of probes bound near the target in an infected cell found to increase drastically by creating a multilayered scaffold. Both these approaches were found to be useful in detecting the Zika virus, HCV, Hepatitis B virus (HBV), or human papillomavirus (HPV) (Lewis et al., 2012; Savidis et al., 2016; Wieland et al., 2014; Zhang et al., 2016). On the other hand, phase-contrast microscopy was used to study viruses such as IAC, adeno-associated virus, HPV, HIV, etc (Bächi 1988). The AdV and HIV were investigated through streptavidin-biotin interactions, functionalization with oligonucleotides, or direct encapsidation in the virion (Herod et al., 2015; Li et al., 2017; Liu et al., 2014; Wan et al., 2014; Zhang et al., 2013). The genetically green fluorescent protein (GFP) and its variants that require no further staining or labeling were used as a generic reporter for infection or an indicator of viral transcription or replication (Baulcombe et al., 1995; Greber and Way 2006). The genome engineered virus-like AdV, HIV, IAV, HSV, etc., provide fluorescence-based spatiotemporal information upon the fusion of GFP with viral proteins (De Martin et al., 1997; Elliott and O'Hare 1999; Manicassamy et al., 2010; Müller et al., 2004). The popularly employing confocal fluorescence microscopy suffers from focused laser beam induced phototoxicity and photobleaching. Spinning disk confocal microscope where the probing of the sample was carried out through multiple pinholes arranged on a Nipkow disk reported to mitigate this issue to a certain extent and thus allow live-cell information but suffer from low spatial resolution (Oreopoulos et al., 2014). The total internal reflection fluorescence microscopy (TIRF-M) reported being more efficient in deciphering the HIV-1 assembly occurring at the cell plasma membrane. With the fluorescein tagged Gag proteins, the researchers were able to visualize the kinetics of HIV-1 Gag assembly in a transfected adherent model cell line (Jouvenet et al., 2008).
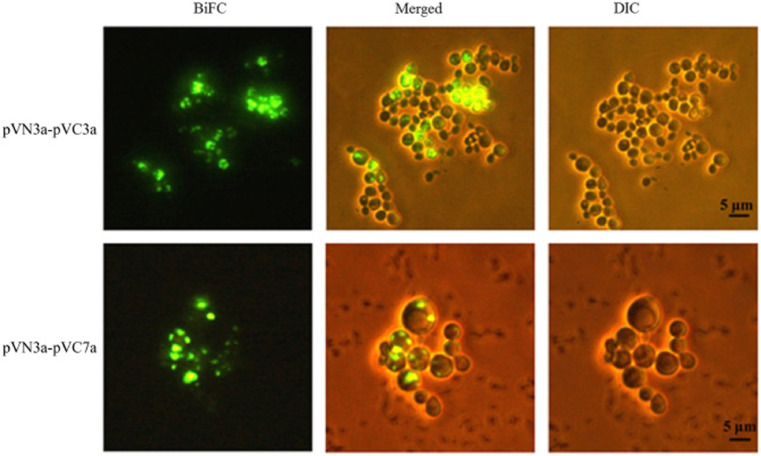
In an important work on SARS-CoV virus, the interaction between the accessory proteins (3a, 3b, 6, 7a, 7b, 8a, 8b, 9b, and ORF14), predicted unknown proteins (PUPs) encoded by the genes, was investigated by an EYFP (enhanced yellow fluorescent protein) bimolecular fluorescence complementation (BiFC) assay (Kong et al., 2015). The authors were able to probe 33 out of 81 interactions which were identified by BiFC, much more than that identified by the yeast two-hybrid (Y2H) system. The strong fluorescence observed from the interactions pVN3a-pVC3a and pVN3a-pVC7a was shown in Fig. 3 .
Fig. 3.
Visualization of interactions of pVN3a-pVC3a (top panel) and pVN3a-pVC7a (bottom panel) using a split-EYFP based bimolecular fluorescence complementation assay. The rest of the 31 interactions among accessory proteins were also visualized under the same conditions (data not shown). BiFC, bimolecular fluorescence complementation assay images. DIC, differential interference contrast images. Merged, merged images of DIC and BiFC Reprinted from (Kong et al., 2015), copyright (2015), with permission from Elsevier.
Fluorescence-based virus detection has been employed for some time, and there exists an FDA approved test kit that utilizes fluorescence-based reverse transcript real-time PCR (rRT-PCR) technique for the detection of SARS-CoV-2. Here, the replicated nucleic acid copies from the SARS-CoV-2 were detected by probing the change in fluorescence of the fluorescent probe when it was released from its quencher counterpart upon primer replication. The RNA isolated from the specimen was reverse transcribed into complementary DNA (cDNA) and amplified via PCR processes along with several sets of SARS-CoV-2 primer and probe detection targets. The probe detection sets were a pair of molecules that were bound in tandem as a fluorescence emitter (5(6)-carboxyfluorescein, FAM) and dark quencher (Black Hole Quencher – 1 (BHQ1)). Here, the fluorescence intensity for positive diagnosis increases during the extension phase of each PCR cycle as a consequence of polymerized Taq primer nuclease activity on the 5'end. The nuclease activity ruptures the FAM from the BHQ-1 in the probe. Though FAM has high quantum efficiency at around 520 nm upon 488 nm laser excitation in certain solvents and buffers, the emission falls drastically in an acidic medium (Edinburgh Instruments, 2020).
6. Super-resolution microscopy techniques
The inherent spatial resolution limitation (X, Y ~ 200 nm and Z~600 nm), photobleaching, finite fluorescence rate (saturation), photoblinking, etc. of fluorescence microscopy necessitated the usage of super-resolution microscopy that allow understanding of virus structures and its localization, assembly, replication, and infection within the host cell, etc. Among the various super-resolution microscopes available, techniques like STED (Stimulated Emission Depletion) microscopy or PALM (Photo-Activation Localization Microscopy) and dSTORM (direct Stochastic Optical Reconstruction Microscopy) enable the studies of small viruses like SARS-CoV (<100 nm) (Chojnacki and Eggeling 2018).
According to this position information, the super-resolution microscope technique for virus studies can be categorized into two groups. The first group relies on the targeted shift of the excited fluorophore into the dark state at the fringes of a precisely positioned fluorescence excitation spot, which is the operation principle of STED and related Reversible Saturable Optical (Fluorescence) Transition (RESOLFT) microscopy variant. The second group probe the stochastic switching of the fluorescing molecules in the entire field of view and its localization, which is adopted in PALM, STORM, and its variants. The variants include GSDIM (Ground State Depletion microscopy followed by Individual Molecule return), PAINT (Point Accumulation for Imaging in Nanoscale Topography), and SOFI (Super-resolution Optical Fluctuation Imaging) (Chojnacki and Eggeling 2018). However, most of the super-resolution fluorescence microscopy studies focused on the analysis of fixed virus samples. In addition to high spatial resolution, the study of dynamical processes in live viral samples requires higher imaging speed, accurate labeling, and sample penetration depth. Presently, the popular super-resolution techniques for virological studies are SIM (structured illumination microscopy), SMLM (single-molecule localization microscopy), and STED. A comparison of these techniques adopted from reference (Sydor et al., 2015) is given in supplementary table 3.
One of the earliest viruses that were investigated by a super-resolution fluorescence microscope was HIV-1, and the studies gave insights into the retroviral replication cycle (Chojnacki and Eggeling 2018; Hanne et al., 2016). With the usage of scanning STED-Fluorescence Correlation Spectroscopy, the protein dynamics on the surface of individual HIV-1 particles were investigated (Chojnacki et al., 2017). Analysis of the data confirmed that the virus maturation status played an important role in the mobility of the virus surface envelope glycoprotein Env. The viral protease-mediated cleave of the structural polyprotein, protein Gag, was responsible for the conversion of immature noninfectious budding HIV-1. An increased Env mobility was observed when the disassembly of the immature Gag lattice occurred during the maturation, and the increased mobility of Env found to play an important role in productive virus entry to the cell. The reduction in mobility of the proteins on the immobile environment of the HIV-1 surface was confirmed by comparing the diffusion characteristics study of Env as well as GPI-anchored protein and MHC-I between HIV-1 and cellular surfaces. It was observed that the magnitude of mobility was reduced by two orders of magnitude as compared to the cell plasma membrane. The live-cell studies were done by organic dye compatible tagging approaches wherein the click chemistry along with nonfluorescent tags such as SNAP-/HALO-/CLIP-tag or artificial amino acids were used (Heilemann et al., 2008).
In another interesting work, by using PALM/STORM technique along with TIFR microscopy, Muranyi et al. investigated the fundamental assembly and budding process in HIV-1 and found that the assembly site very close to the plasma membrane and have a diameter around 130 nm (Muranyi et al., 2013). The nanoscopic measurements using the PALM technique of distinct viral particles revealed the conical morphology of the capsid, as well as the localization of the FlAsH, tagged integrase viral protein inside the capsid (Lelek et al., 2012). Also, the study enabled the investigation of the architecture of the envelope glycoproteins. By using a high-resolution dual-color STORM technique, the same group also demonstrated that HIV replication was favored by chromatin organization at the nuclear pore. To investigate the HIV replication, an analysis of histone H3K36me3 and nuclear pore complex was carried out (Lelek et al., 2015). An increase in the cluster size of the HIV capsid (236%) in comparison to the matrix protein was observed during the infection by a detailed study using the dSTORM technique (Pereira et al., 2012). Another interesting study using three-dimensional microscopy showed that the Gag protein recruited the host endosomal sorting complex required for transport to the assembly site of the newly formed virion and facilitate the release of HIV particles from the cell by membrane fission (Van Engelenburg et al., 2014). Advancement in the super-resolution microscopy now allows the investigation of more than a hundred differently labeled Gag clusters at the plasma membrane of the host cell during the virions assembly (Gunzenhä;user et al., 2012). The STED microscopic studies of the receptor and the mutant behavior on the dendritic cell surface for HIV and dengue virus showed the cluster organization of the antigen-presenting cells at the viral entry point. Moreover, the extracellular neck domain was found to be important for the DC-specific ICAM-3 grabbing nonintegrin (DC-SIGN) clustering and virus internalization (Manzo et al., 2012). In a study to understand the relative localization of the core protein and E2 envelope protein of HCV inside an infected cell showed little colocalization of two viral proteins in the individual regions when 0.2 μm diameter lipid droplets distributed very close (1 μm) to the cell membrane (Eggert et al., 2014). In another work, by using the STORM and electron microscopy technique, the role of tetraspanin CD81 in the replication process of influenza A viruses is evaluated (He et al., 2013). The replication and assembly of CD81-positive endosomes at the cell surface and its virus entry was confirmed by using the fluorescently labeled influenza virus particles. Additionally, the study revealed that the fusion of the viral with the cell membrane could be impaired in the absence of CD81. Though little work has been done, all these studies show that the super-resolution microscopic technique provides a great promise in investigating the SARS-CoV-2 and its interaction with cells.
7. Surface plasmon resonance based virus detection
The resonant oscillations within the surface electrons (surface plasmon resonance) at a metal-dielectric interface (with opposite dielectric constants that satisfy relation for the propagation constant) induced by a transverse magnetic, electromagnetic radiation (or p-polarized light) of a specific energy and incident wave vector (angle) has already been successfully exploited to investigate pathogen detection (Homola and Piliarik 2006; Lukose et al., 2018). Here k denotes the free space wavenumber, εm is the dielectric constant of the metal and ns the refractive index of the dielectric material. The energy and momentum matching between the incident light and surface plasmon wave at resonance condition interrogated via angular or wavelength scanning normally manifested as intensity dip. Typically, in the SPR technique, a biomolecule immobilized onto the sensor surface (noble metal) interacts with (binding) bio-particle flowing in the coupling medium and get immobilized to accumulate the biomolecules onto the sensor surface. The accumulation of probing molecules leads to an increase in the refractive index near the sensor surface and change the SPW. Thus, the real-time measurement of refractive index change provides information about the binding efficiency of the biomolecules.
In a pioneering work, Wang and the colleagues demonstrated the applicability of surface plasmon resonance imaging for the detection of single influenza A H1N1 viral particles via virus-surface interactions (Wang et al., 2010). The surface functionalization (bare gold, PEG-coated, or anti-influenza A antibody-coated sensor) found to play an important role in the interaction. The interaction between the whole influenza virus and an antibody-based on surface plasmon resonance was investigated by Schofield and Dimmock by immobilizing different antibodies on a CM 5 chip and reported the kinetic parameters of their interactions (Schofield and Dimmock 1996). Later, Hidari and the colleagues reported the whole virus interaction with glycans by immobilizing the glycans on the surface of liposomes and the subsequent immobilization on the L1 sensor chip (Hidari et al., 2007). The potential of this technique for label-free and rapid quantification of small RNA virus belonging to the picornyiridae family (human Enterovirus 71(EV71)) was demonstrated (Prabowo et al., 2017). In the study, the major capsid protein VP1 of EV71 was used as the biomarker, and the signal to noise ratio is enhanced by integrating the area under the reflectivity curve. In another work, the intensity modified-surface plasmon resonance biosensor was developed for the rapid detection (~10 min) of the H7N9 virus and H7N9-RG virus that show similar surface proper properties to avian influenza A H7N9 virus (Chang et al., 2018). The detection limit of the H7N9 virus was estimated to 144 copies/mL, whereas it was measured to be 402 copies/mL for the mimic clinical specimens wherein the H7N9 virus was mixed with nasal mucosa from flu-like syndrome patients. In another interesting work, a gold SPR chip modified with 4-mercaptobenzoid acid (4-MBA) was used for the immobilization of nucleoprotein of Ebola (EBOV-rNPA) and the interaction of different monoclonal antibodies mAb1, mAb2 and mAb3 were investigated and found that affinity constant (KD) for mAb1, mAb2, and mAb3 of Ebola with the immobilized EBOV-rnNP is 809 nM, 350 pM and 52 pM (Sharma et al., 2020). A recent work that utilizes dithiobis (succinimidyl undecanoate) immobilized on NH2 reduced graphene oxide-polyamidoamine nanocomposite as the sensor surface showed the fast detection (<8 min) of ultra-low concentration (0.08 pM) of DENV-2 E-proteins (Omar et al., 2020). Lin et al. have developed a miniaturized optical fibre based SPR sensor for the detection of avian influenza virus from poultry samples (Zhao et al., 2016). Aptamers has been also used as effective receptors for viral detection (avian influenza) via SPR biosensing techniques (Nguyen et al., 2016). Simultaneous detection of nine variants of respiratory viruses has been carried out by attaching virus-specific oligonucleotides onto SPR sensor chip (Shi et al., 2015).
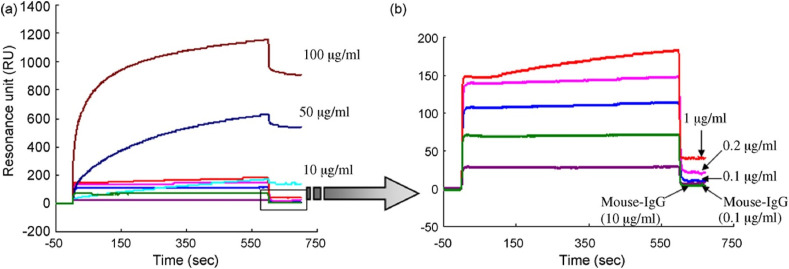
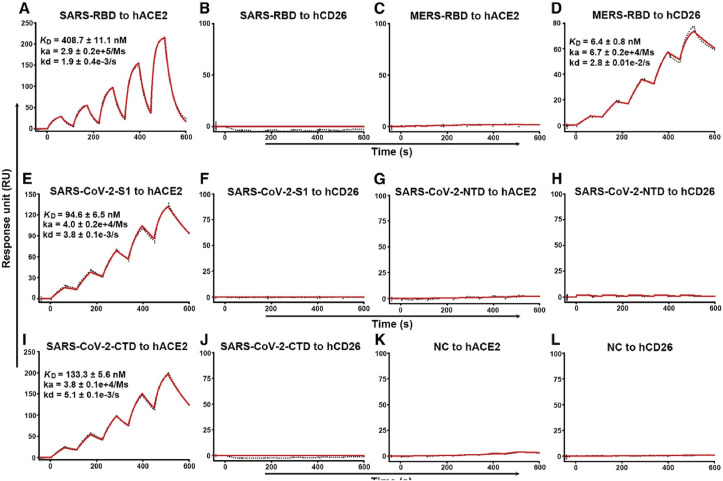
A protein created by the genetic fusion of an anchoring component, gold binding polypeptides (GBPs) and a recognition element, SARS coronaviral surface antigen (SCVme), was employed to diagnose the severe acute respiratory syndrome (SARS) in a surface plasmon resonance (SPR) based biosensor (Park et al., 2009). SPR sensorgrams representing the sensitive as well as specific binding of anti-SCVme to the GBP-E-SCVme were shown in Fig. 4 (a) and (b) respectively. The study showed that the highest detection response for an anti-SCVme (906 RU) was achieved when the packing fraction of the fused protein was 10 g mL−1 and the limit of detection was 200 ng/mL anti-SCVme and the measurement time as less than 10 min. In an important recent study, real-time SPR assays were utilized to investigate the interactions of SARS-CoV-2-CTD (Receptor binding entity of SARS-CoV-2) and SARS-RBD (Receptor binding entity of SARS-CoV) with hACE2 receptor, which is given in Fig. 5 (Wang et al., 2020). A fourfold stronger binding affinity was evident for SARS-CoV-2-CTD towards the entry receptor hACE2 from the SPR results as compared to SARS-RBD. It was found that the C-terminal domain (CTD) of spike protein in two viruses, SARS-CoV and SARS-CoV-2 are antigenically different.
Fig. 4.
SPR sensorgrams for (a) sensitive and (b) selective detection of anti-SCVme using the GBP-E-SCVme immobilized gold sensor chip at various concentrations (0.1, 1, 10, 50, and 100 μg mL−1) of anti-SCVme and (1 and 10 μg mL−1) of mouse IgG as negative controls. Reprinted from (Park et al., 2009), copyright (2009), with permission from Elsevier. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
Specific Interactions between SARS-CoV-2-S1 and SARS-CoV-2-CTD with hACE2, Characterized by SPR. The indicated mFc-tagged proteins in the supernatant were captured by anti-mIgG antibodies that were immobilized on the chip and subsequently tested for binding with gradient concentrations of hACE2 or hCD26, with the following binding profiles shown.
(A) SARS-RBD binding to hACE2. (B) SARS-RBD binding to hCD26. (C) MERS-RBD binding to hACE2. (D) MERS-RBD binding to hCD26. (E) SARS-CoV-2-S1 binding to hACE2. (F) SARS-CoV-2-S1 binding to hCD26. (G) SARS-CoV-2-NTD binding to hACE2. (H) SARS-CoV-2-NTD binding to hCD26. (I) SARS-CoV-2-CTD binding to hACE2. (J) SARS-CoV-2-CTD binding to hCD26. (K) Culture supernatant of HEK293T cells without transfection (NC) binding to hACE2. (L) Culture supernatant of HEK293T cells without transfection (NC) binding to hCD26. The values shown are the mean ± SD of three independent experiments. This article was published in Wang et al. (2020), copyright Elsevier (2020)
In an important work, J. C. Haung and the colleagues developed a localized surface plasmon coupled fluorescence (LSPCF) fiber-optic biosensor to improve the sensitivity of conventional immunoassay detection of SARS-CoV nucleocapsid protein (N protein) (Huang et al., 2009). The authors found a linear relationship between the fluorescence signal and SARS-CoV N(GST-N) protein in buffer solution for the range between 0.1 pg/mL to 1 ng/mL. In another interesting work utilized the surface plasmon resonance to test the biophysical properties of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein, it was found that the SARS-CoV-2 spike glycoprotein bound angiotensin-converting enzyme 2 (ACE2) with much higher affinity than SARS-CoV spike protein (Shang et al., 2020). The binding affinities of the each of the three receptor-binding domains (SARS-CoV-3, chimeric and SARS-CoV) and angiotensin-converting enzyme 2 (ACE2) was measured using surface plasmon resonance and found that chimeric have higher ACE2-binding affinity than SARS-CoV-2 RBD, due to the introduced N–O bridge between receptor binding domain and ACE2 (Shang et al., 2020). The dissociation constant Kd indicates that chimeric and SARS-CoV-2 RBDs have significantly higher ACE2-binding affinities than the SARS-CoV RBD. Amid the pandemic COVID-19, J. Wang and the colleagues developed dual-functional plasmonic biosensors that combine the plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) sensing transduction for the detection of the selected sequence from SARS-CoV-2 (Qiu et al., 2020). In this work, the two-dimensional gold nanoislands (AuNIs) functionalized with complementary DNA receptors was utilized for the detection of SARS-CoV- 2 through nucleic acid hybridization. The thermoplasmonic heating created by the gold nanoislands chip via illumination at plasmonic resonance frequency found to elevate the in-situ hybridization temperature and enable the specific discrimination of two similar gene sequences. The limit of the detection of the dual-functional LSPR sensors for the selected SARS-CoV-2 was estimated to be 0.22 pM. In this work, by utilizing the two different angles of incidence, two different wavelengths were employed for plasmonic resonances of PPT and LSPR, which found to have a pronounced effect on sensing stability, sensitivity, and reliability. The developed technique was found effective to provide real-time, label-free detection of viral sequences, including RdRp-COVID, ORF lab-COVID, and E genes fro SARS-CoV-2. In a very recent work, J-F Masson and the colleagues reported an SPR sensor detecting the nucleocapsid antibodies specific against SARS-CoV-2 in human serum (Djaileb et al., 2020). In the study, an SPR sensing surface coated with a peptide monolayer immobilized with SARS-CoV-2 nucleocapsid recombinant protein provided anti-SARS-CoV-2 detection in the nanomolar range. Recombinant protein functionalization on sensor surface was performed via N-ethyl-N’-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) - N-hydroxysuccinimide (NHS) amine coupling chemistry. The duration of the measurements was about 15 min, which is much faster than conventional PCR based techniques.
8. Other light-assisted methods
D. Pan and the colleagues demonstrated nanotechnology-enabled “naked eyed detection of SARS-CoV-2 via a colorimetric assay based on anisotropic gold nanoparticles (Moitra et al., 2020). The viral genome sequencing of the SARS virus revealed that three regions of the virus, NRA-dependent RNA polymerase gene (RdRP gene), enveloped protein gene (E gene) and nucleocapsid phosphoprotein gene (N gene) have conserved sequences. In this work, simultaneous covering of the two specific regions of the viral N-gene sequence was done using a targeting approach mediated by four antisense oligonucleotide sequences. In the presence of the target RNA sequence of SARS-CoV-2, the thiolated ASO capped AuNP start to agglomerate which further amplified upon addition of the RNaseH cleaves and finally leading to the visual precipitation of the plasmonic nanoparticles and the color change. The agglomeration was manifested as a 40 nm redshift in the UV-VIS absorption spectral studies (Moitra et al., 2020). The results are further corroborated with transmission electron microscopy studies and enhanced dark-field hyperspectral imaging studies. The targeting approach of two of the N-gene regions of SARS-CoV-2 ensures the assay can be employed even when one region of the viral gene is mutated. Amos Danielli and his coworkers have recently come up with a fluorescence based technique which provide sensitive detection of Coronavirus-specific RNA sequences from saliva within 15 min (BioOptics World Editors, 2020).
In pursuit of developing a label-free rapid method for COVID-19 detection, researchers working in “CONVAT” project are now exploring the possibility of a laser-based nano-interferometric sensor to detect the virus from saliva (Wallace, 2020; Ruiz-Vega 2020). CONVAT project funded by H2020 European Union Framework Program aims at the development of point of care nano-interferometric biosensor, which can facilitate early diagnosis of COVID-19. The impact and the benefits of such point of care biosensors developed in CONVAT project has been summarized in a recent article (Ruiz-Vega 2020). The CONVAT approach relies on the recognition of the antigens of the virus capsid when the fluid sample passes through the sensor surface with an evanescent field from the propagating laser light in a bimodal waveguide interferometer (Ruiz-Vega 2020). The recognition of the antigen produces a small change in the refractive index, which causes changes in the direction of light travel. A comparison of this change against the precisely measured standard value may enable the instantaneous detection of the coronavirus, even at a very low concentration range (pM–aM) without any additional amplification procedure. Recently, an interferometric scattering (iSCAT) microscopy has developed to observe single nanoparticles, molecules, label-free live-cell imaging, single proteins, and viruses (Taylor et al., 2019; Taylor and Sandoghdar 2019). The same technique is now being explored to investigate the steps involved in the interaction of SARS-CoV-2 with a single cell. A comprehensive table that illustrates the potential of optical techniques for the detection of viruses like COVID-19 is given supplementary Table 4(Cui and Chou, 2020) (Jinglin et al., 2020). (Technology and trends) (Soler et al., 2020).
9. Summary and outlook
The rapid detection and diagnosis of viruses not only help to mitigate the pandemic but also facilitate the discovery of future vaccines. The expensive nature and low throughput of currently employing PCR-based assay demand the development of photonics-based techniques illustrated here for SARS-CoV-2 detection. Despite the great promise, photonics techniques face the biggest challenges from the great diversity of viruses circulating the human organism and the unique microbiome of each individual. However, with the usage of advanced artificial intelligence tools, these techniques expected to provide definite biomarkers for viruses and their interaction with cells. The success of the near-infrared spectroscopy and nanotechnology-enabled Raman spectroscopy in investigating viruses like HIV and influenza ensures the potential of these techniques for studying SARS-CoV-2 with high specificity. With the development of superhydrophobic, slippery, and externally controllable SERS substrates, the rapid detection of coronavirus even at low concentration is possible. While employing magnetic plasmonic nanoparticles for the hot-spot controlling of the SERS substrate, the inner magnetic core provides good magnetic responsivity, whereas the noble metal shell provides the excellent plasmonic activity. The spatial resolution issue of the conventional SERS technique is now mitigated to a certain extent via the TERS technique. The use of labeling strategies (bDNA-FISH) and click chemistry labeling of nucleic acids, lipids, and proteins provide high sensitivity in the fluorescence microscopic imaging-based viral studies. The diffraction limit of conventional fluorescence microscopes to investigate ultra-small viruses like coronavirus can be now mitigated with the usage of a super-resolution microscope and can help the understanding of host-virus interactions and sub-viral structures. The emergence of plasmonic biosensors to detect the viruses like SARS-CoV-2 can be attributed to (i) potential to integrate with complementary analytical tools like colorimetric, SERS, fluorescence imaging (ii) advancement chemical functionalization strategies via top-down or bottom-up approaches (iii) integration of selective receptors to avoid cross-reactivity and specificity. By combining these advantages, SPR based techniques can be employed for the analysis of body fluids such as blood, nasal asperities, urine, etc. With the advancement in technology and miniaturization, photonics-based biosensing platforms will become popular in terms of portability, cost involved, affordability, user-friendliness, and ready-to-use mode. As depicted in this progress, such photonics-based technologies offer a great promise for the rapid detection of deadly dangerous viruses like SARS-CoV-2. Although the COVID-19 may decrease and could disappear or reoccur seasonally in future mutations through the zoonosis from one of the animal reservoirs, the research advancement happening in the field of photonics in detecting viruses may provide novel devices in mitigating new outbreaks from viruses.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
SDG acknowledge (DST project Sanction No. IDP/BDTD/20/2019) and joint Manipal Academy of Higher Education (MAHE) and the FIST program of the Government of India (SR/FST/PSI-174/2012).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ambartsumyan O., Gribanyov D., Kukushkin V., Kopylov A., Zavyalova E. Int. J. Mol. Sci. 2020;21(9):3373. doi: 10.3390/ijms21093373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bächi T. J. Cell Biol. 1988;107(5):1689–1695. doi: 10.1083/jcb.107.5.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D.C., Chapman S., Santa Cruz S. Plant J. 1995;7(6):1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-F., Wang W.-H., Hong Y.-W., Yuan R.-Y., Chen K.-H., Huang Y.-W., Lu P.-L., Chen Y.-H., Chen Y.-M.A., Su L.-C. Anal. Chem. 2018;90(3):1861–1869. doi: 10.1021/acs.analchem.7b03934. [DOI] [PubMed] [Google Scholar]
- Chojnacki J., Eggeling C. Retrovirology. 2018;15(1):41. doi: 10.1186/s12977-018-0424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnacki J., Waithe D., Carravilla P., Huarte N., Galiani S., Enderlein J., Eggeling C. Nat. Commun. 2017;8(1):1–10. doi: 10.1038/s41467-017-00515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.-y., Lionnet T. 2018. Single-Molecule Sensitivity RNA FISH Analysis of Influenza Virus Genome Trafficking. Influenza Virus; pp. 195–211. Springer. [DOI] [PubMed] [Google Scholar]
- Cialla D., Deckert‐ Gaudig T., Budich C., Laue M., Möller R., Naumann D., Deckert V., Popp J. J. Raman Spectrosc. 2009;40(3):240–243. [Google Scholar]
- Cui F., Zhou H.S. Biosens. Bioelectron. 2020:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Martin R., Raidl M., Hofer E., Binder B. Gene Ther. 1997;4(5):493–495. doi: 10.1038/sj.gt.3300408. [DOI] [PubMed] [Google Scholar]
- Deckert V., Deckert-Gaudig T., Cialla D., Popp J., Zell R., Sokolov A., Yi Z., Scully M. arXiv preprint arXiv:2003. 2020 [Google Scholar]
- Desai S., Mishra S.V., Joshi A., Sarkar D., Hole A., Mishra R., Dutt S., Chilakapati M.K., Gupta S., Dutt A. J.Biophotonics. 2020 doi: 10.1002/jbio.202000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devpost 2020. https://devpost.com/software/samsung-research-for-sars-cov-2-detection-with-spectroscopy
- Ding X., Yin K., Li Z., Lalla R.V., Ballesteros E., Sfeir M.M., Liu C. Nat. Commun. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djaileb A., Charron B., Jodaylami M.H., Thibault V., Coutu J., Stevenson K., Forest S., Live L.S., Boudreau D., Pelletier J.N., Mason J.-F. ChemRxiv. 2020:1–12. doi: 10.26434/chemrxiv.12118914.v1. [DOI] [Google Scholar]
- Eggert D., Rösch K., Reimer R., Herker E. PloS One. 2014;9(7):e102511. doi: 10.1371/journal.pone.0102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G., O'Hare P. J. Virol. 1999;73(5):4110–4119. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom G., Hwang A., Kim H., Yang S., Lee D.K., Song S., Ha K., Jeong J., Jung J., Lim E.-K. ACS Sens. 2019;4(9):2282–2287. doi: 10.1021/acssensors.9b00697. [DOI] [PubMed] [Google Scholar]
- Erukhimovitch V., Talyshinsky M., Souprun Y., Huleihel M. J. Biomed. Optic. 2006;11(6) doi: 10.1117/1.2397554. [DOI] [PubMed] [Google Scholar]
- Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.-B., Cui M., Tao J., Tyrrell D.L., Zhang X.-E., Zhang H., Le X.C. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. 2020. [DOI] [PubMed] [Google Scholar]
- Gahlaut S.K., Savargaonkar D., Sharan C., Yadav S., Mishra P., Singh J.P. Anal. Chem. 2020;92(3):2527–2534. doi: 10.1021/acs.analchem.9b04129. [DOI] [PubMed] [Google Scholar]
- Gamage S., Howard M., Makita H., Cross B., Hastings G., Luo M., Abate Y. PloS One. 2018;13(6) doi: 10.1371/journal.pone.0199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber U.F., Way M. Cell. 2006;124(4):741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier N., Pengo T., Manley S. Nano Lett. 2012;12(9):4705–4710. doi: 10.1021/nl3021076. [DOI] [PubMed] [Google Scholar]
- Hanne J., Zila V., Heilemann M., Müller B., Kräusslich H.G. FEBS Lett. 2016;590(13):1858–1876. doi: 10.1002/1873-3468.12186. [DOI] [PubMed] [Google Scholar]
- Hansen L., De Beer T., Pierre K., Pastoret S., Bonnegarde‐Bernard A., Daoussi R., Vervaet C., Remon J.P. Biotechnol. Prog. 2015;31(4):1107–1118. doi: 10.1002/btpr.2100. [DOI] [PubMed] [Google Scholar]
- He J., Sun E., Bujny M.V., Kim D., Davidson M.W., Zhuang X. PLoS Pathog. 2013;9(10) doi: 10.1371/journal.ppat.1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann M., Van De Linde S., Schüttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., Sauer M. Angew. Chem. Int. Ed. 2008;47(33):6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- Hermann P., Hermelink A., Lausch V., Holland G., Möller L., Bannert N., Naumann D. Analyst. 2011;136(6):1148–1152. doi: 10.1039/c0an00531b. [DOI] [PubMed] [Google Scholar]
- Herod M.R., Pineda R.G., Mautner V., Onion D. Small. 2015;11(7):797–803. doi: 10.1002/smll.201401885. [DOI] [PubMed] [Google Scholar]
- Hidari K.I., Shimada S., Suzuki Y., Suzuki T. Glycoconj. J. 2007;24(9):583–590. doi: 10.1007/s10719-007-9055-y. [DOI] [PubMed] [Google Scholar]
- Homola J., Piliarik M. Springer; 2006. Surface Plasmon Resonance (SPR) Sensors. Surface Plasmon Resonance Based Sensors; pp. 45–67. [DOI] [PubMed] [Google Scholar]
- Huang J.C., Chang Y.-F., Chen K.-H., Su L.-C., Lee C.-W., Chen C.-C., Chen Y.-M.A., Chou C. Biosens. Bioelectron. 2009;25(2):320–325. doi: 10.1016/j.bios.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Chen S., Yang Z., Guan W., Liu D., Lin Z., Zhang Y., Xu Z., Liu X., Li1 Y. Am. J. Respir. Crit. Care Med. 2020;201(11):1435–1438. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huleihel M., Salman A., Erukhimovitch V., Ramesh J., Hammody Z., Mordechai S. J. Biochem. Biophys. 2002;50(2–3):111–121. doi: 10.1016/s0165-022x(01)00177-4. [DOI] [PubMed] [Google Scholar]
- Huleihel M., Talyshinsky M., Erukhimovitch V. J. Spectrosc. 2001;15(2):57–64. doi: 10.1366/00037020360625916. [DOI] [PubMed] [Google Scholar]
- Jinglin H., Minjie Z., Wei L., Guo C., Shuang N., Gao N., Zeyu L., Zongqing Z., Zhibing H., Bo L., Songnan Z., Baohan Z. High Power Laser Part Beams. 2020;32(6) [Google Scholar]
- Jouvenet N., Bieniasz P.D., Simon S.M. Nature. 2008;454(7201):236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J., Shi Y., Wang Z., Pan Y. Acta Pharm. Sin. B. 2015;5(5):487–492. doi: 10.1016/j.apsb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Malik Y.S., Ganesh B., Rahangdale S., Saurabh S., Natesan S., Srivastava A., Sharun K., Yatoo M.I., Tiwari R., Singh R.K., Dhama K. Front. Cell. Infect. Microbiol. 2020;10(1):576875. doi: 10.3389/fcimb.2020.576875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Kim B.-C., Byeung-Keun O., Choi J.-W. J. Biomed. Nanotechnol. 2015;11(12):2223–2230. doi: 10.1166/jbn.2015.2117. [DOI] [PubMed] [Google Scholar]
- Lelek M., Casartelli N., Pellin D., Rizzi E., Souque P., Severgnini M., Di Serio C., Fricke T., Diaz-Griffero F., Zimmer C. Nat. Commun. 2015;6:6483. doi: 10.1038/ncomms7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelek M., Di Nunzio F., Henriques R., Charneau P., Arhel N., Zimmer C. Proc. Natl. Acad. Sci. Unit. States Am. 2012;109(22):8564–8569. doi: 10.1073/pnas.1013267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.S., Jr., Ukpo O.C., Ma X.J., Flanagan J.J., Luo Y., Thorstad W.L., Chernock R.D. Histopathology. 2012;60(6):982–991. doi: 10.1111/j.1365-2559.2011.04169.x. [DOI] [PubMed] [Google Scholar]
- Li Q., Li W., Yin W., Guo J., Zhang Z.-P., Zeng D., Zhang X., Wu Y., Zhang X.-E., Cui Z. ACS Nano. 2017;11(4):3890–3903. doi: 10.1021/acsnano.7b00275. [DOI] [PubMed] [Google Scholar]
- Lim J.-y., Nam J.-s., Yang S.-e., Shin H., Jang Y.-h., Bae G.-U., Kang T., Lim K.-i., Choi Y. Anal. Chem. 2015;87(23):11652–11659. doi: 10.1021/acs.analchem.5b02661. [DOI] [PubMed] [Google Scholar]
- Liu S.-L., Zhang L.-J., Wang Z.-G., Zhang Z.-L., Wu Q.-M., Sun E.-Z., Shi Y.-B., Pang D.-W. Anal. Chem. 2014;86(8):3902–3908. doi: 10.1021/ac500640u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., Zeng B., Li Z., Li X., Li H. Eur. J. Radiol. 2020:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Lin Y., Zheng Z., Tang X., Lin J., Liu X., Liu M., Chen G., Qiu S., Zhou T. Biomed. Optic Express. 2018;9(10):4755–4766. doi: 10.1364/BOE.9.004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukose J., Shetty V., Ballal M., Chidangil S., Sinha R.K. Laser Phys. Lett. 2018;15(7) [Google Scholar]
- Mahmud I., Garret T.J. J. Am. Soc. Mass Spectrom. 2020;31(10):2013–2024. doi: 10.1021/jasms.0c00238. [DOI] [PubMed] [Google Scholar]
- Manicassamy B., Manicassamy S., Belicha-Villanueva A., Pisanelli G., Pulendran B., García-Sastre A. Proc. Natl. Acad. Sci. Unit. States Am. 2010;107(25):11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzo C., Torreno-Pina J.A., Joosten B., Reinieren-Beeren I., Gualda E.J., Loza-Alvarez P., Figdor C.G., Garcia-Parajo M.F., Cambi A. J. Biol. Chem. 2012;287(46):38946–38955. doi: 10.1074/jbc.M112.380121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S., Mathew M.R., Sam S., Keerthi K., Kumar K.G. J. Electroanal. Chem. 2020;878:114596. doi: 10.1016/j.jelechem.2020.114596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi A., Esmaeilzadeh E., Li Y., Bosch R.J., Lia J.Z. EBioMedicine. 2020;59:102903. doi: 10.1016/j.ebiom.2020.102903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra P., Alafeef M., Dighe K., Frieman M., Pan D. ACS Nano. 2020;14(6):7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor K., Ohtani K., Myrzakozha D., Zhanserkenova O., Andriana B.B., Sato H. J. Biomed. Opt. 2014;19(6) doi: 10.1117/1.JBO.19.6.067003. [DOI] [PubMed] [Google Scholar]
- Mor A., White A., Zhang K., Thompson M., Esparza M., Muñoz-Moreno R., Koide K., Lynch K.W., García-Sastre A., Fontoura B.M. Nat. Microbiol. 2016;1(7):1–13. doi: 10.1038/nmicrobiol.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses J.E., Moorhouse A.D. The growing applications of click chemistry. Chem. Soc. Rev. 2007;36:1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- Müller B., Daecke J., Fackler O.T., Dittmar M.T., Zentgraf H., Kräusslich H.-G. J. Virol. 2004;78(19):10803–10813. doi: 10.1128/JVI.78.19.10803-10813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranyi W., Malkusch S., Müller B., Heilemann M., Kräusslich H.-G. PLoS Pathog. 2013;9(2):e1003198. doi: 10.1371/journal.ppat.1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtigall F.M., Pereira A., Trofymchuk O.S., Santos L.S. Nat. Biotechnol. 2020;38:1168–1173. doi: 10.1038/s41587-020-0644-7. [DOI] [PubMed] [Google Scholar]
- Nguyen V.-T., Seo H.B., Kim B.C., Kim S.K., Song C.-S., Gu M.B. Biosens. Bioelectron. 2016;86:293–300. doi: 10.1016/j.bios.2016.06.064. [DOI] [PubMed] [Google Scholar]
- Omar N.A.S., Fen Y.W., Abdullah J., Kamil Y.M., Daniyal W.M.E.M.M., Sadrolhosseini A.R., Mahdi M.A. Sci. Rep. 2020;10(1):1–15. doi: 10.1038/s41598-020-59388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreopoulos J., Berman R., Browne M. Spinning-disk confocal microscopy: present technology and future trends. Methods Cell Biol. 2014:153–175. doi: 10.1016/B978-0-12-420138-5.00009-4. Elsevier. [DOI] [PubMed] [Google Scholar]
- Othman N., Lee K.Y., Radzol A., Mansor W., Ramlan N. IEEE; 2017. Linear Discriminant Analysis for Detection of Salivary NS1 from SERS Spectra. TENCON 2017-2017, IEEE Region 10 Conference; pp. 2876–2879. [Google Scholar]
- Park T.J., Hyun M.S., Lee H.J., Lee S.Y., Ko S. Talanta. 2009;79(2):295–301. doi: 10.1016/j.talanta.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C.F., Rossy J., Owen D.M., Mak J., Gaus K. Virol. J. 2012;9(1):84. doi: 10.1186/1743-422X-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler J., Glöckner F.-O., Schönhuber W., Amann R. Methods Microbiol. 2001;30:207–226. [Google Scholar]
- Pizarro C., Esteban‐ Díez I., Arenzana‐ Rámila I., González‐ Sáiz J.M., tonics J.biopho. 2018;11(3) doi: 10.1002/jbio.201700035. [DOI] [PubMed] [Google Scholar]
- Porter M., Driskell J., Kwarta K., Lipert R., Neill J., Ridpath J. Dev. Biol. 2006;126:31–39. discussion 323. [PubMed] [Google Scholar]
- Prabowo B.A., Wang R.Y., Secario M.K., Ou P.-T., Alom A., Liu J.-J., Liu K.-C. Biosens. Bioelectron. 2017;92:186–191. doi: 10.1016/j.bios.2017.01.043. [DOI] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Ramanan V., Trehan K., Ong M.-L., Luna J.M., Hoffmann H.-H., Espiritu C., Sheahan T.P., Chandrasekar H., Schwartz R.E., Christine K.S. Virology. 2016;494:236–247. doi: 10.1016/j.virol.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A., Anwar S., Firdous S., Ahmed M., Rasheed R., Nawaz M. Laser Phys. 2012;22(6):1085–1089. [Google Scholar]
- Resch-Genger U., Grabolle M., Cavaliere-Jaricot S., Nitschke R., Nann T. Nat. Methods. 2008;5(9):763. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- Roy S., Perez-Guaita D., Bowden S., Heraud P., Wood B.R. Clinical Spectroscopy. 2019;1:100001. [Google Scholar]
- Ruiz-Vega G., Soler M., Lechuga L.M. 2020. JPhys Photonics; pp. 1–8. [Google Scholar]
- Saade J., Pacheco M.T.T., Rodrigues M.R. J. Spectrosc. 2008;22(5):387–395. [Google Scholar]
- Sakamoto S., Putalun W., Vimolmangkang S., Phoolcharoen W., Shoyama Y., Tanaka H., Morimoto S. J. Nat. Med. 2018;72(1):32–42. doi: 10.1007/s11418-017-1144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman A., Shufan E., Zeiri L., Huleihel M. Methods. 2014;68(2):364–370. doi: 10.1016/j.ymeth.2014.02.022. [DOI] [PubMed] [Google Scholar]
- Sánchez-Purr M., Carr -Camps M., de Puig H., Bosch I., Gehrke L., Hamad-Schifferli K. ACS Infect. Dis. 2017;3(10):767–776. doi: 10.1021/acsinfecdis.7b00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M.C., Nascimento Y.M., Araújo J.M., Lima K.M. RSC Adv. 2017;7(41):25640–25649. [Google Scholar]
- Savidis G., Perreira J.M., Portmann J.M., Meraner P., Guo Z., Green S., Brass A.L. Cell Rep. 2016;15(11):2323–2330. doi: 10.1016/j.celrep.2016.05.074. [DOI] [PubMed] [Google Scholar]
- Schofield D., Dimmock N. J. Virol. Methods. 1996;62(1):33–42. doi: 10.1016/0166-0934(96)02086-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Sengupta J., Hussain C.M. Carbon Trends. 2020:100011. doi: 10.1016/j.cartre.2020.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Nature. 2020:1–4. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmukh S., Jones L., Driskell J., Zhao Y., Dluhy R., Tripp R.A. Nano Lett. 2006;6(11):2630–2636. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- Sharma P.K., Kumar J.S., Singh V.V., Biswas U., Sarkar S.S., Alam S.I., Dash P.K., Boopathi M., Ganesan K., Jain R. Anal. Bioanal. Chem. 2020:1–12. doi: 10.1007/s00216-020-02641-5. [DOI] [PubMed] [Google Scholar]
- Shi L., Sun Q., He J.a., Xu H., Liu C., Zhao C., Xu Y., Wu C., Xiang J., Gu D. Bio Med. Mater. Eng. 2015;26(s1):S2207–S2216. doi: 10.3233/BME-151526. [DOI] [PubMed] [Google Scholar]
- Sitole L., Steffens F., Krüger T.P., Meyer D. Omics. 2014;18(8):513–523. doi: 10.1089/omi.2013.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail A., Khan S., Ullah R., Qureshi S.A., Bilal M., Khan A. Biomed. Optic Express. 2018;9(5):2041–2055. doi: 10.1364/BOE.9.002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M., Estevez M.C., Cardenosa-Rubio M., Astua A., Lechuga L.M. ACS Sens. 2020;5(9):2663–2678. doi: 10.1021/acssensors.0c01180. [DOI] [PubMed] [Google Scholar]
- Soler M., Scholtz A., Zeto R., Armani A.M. APL Photonics. 2020;5 doi: 10.1063/5.0021270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Xu L., Zhang F., Song Z., Hu Y., Ji Y., Shen J., Li B., Lu H., Yang H. Biosens. Bioelectron. 2017;89:906–912. doi: 10.1016/j.bios.2016.09.100. [DOI] [PubMed] [Google Scholar]
- Sydor A.M., Czymmek K.J., Puchner E.M., Mennella V. Trends Cell Biol. 2015;25(12):730–748. doi: 10.1016/j.tcb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Taylor R.W., Mahmoodabadi R.G., Rauschenberger V., Giessl A., Schambony A., Sandoghdar V. Nat. Photon. 2019;13(7):480–487. [Google Scholar]
- Taylor R.W., Sandoghdar V. Nano Lett. 2019;19(8):4827–4835. doi: 10.1021/acs.nanolett.9b01822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelenburg S.B., Shtengel G., Sengupta P., Waki K., Jarnik M., Ablan S.D., Freed E.O., Hess H.F., Lippincott-Schwartz J. Science. 2014;343(6171):653–656. doi: 10.1126/science.1247786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas C.A., Wilhelm A.A., Williams J., Lucas P., Reynolds K.A., Riley M.R. Appl. Environ. Microbiol. 2009;75(20):6431–6440. doi: 10.1128/AEM.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X.-Y., Zheng L.-L., Gao P.-F., Yang X.-X., Li C.-M., Li Y.F., Huang C.Z. Sci. Rep. 2014;4:4529. doi: 10.1038/srep04529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P., Ahern S., Carty P.G., O'Brien K.K., O'Murchu E., O'Neill M., Smith S.M., Ryan M., Harringtona P. J. Infect. 2020;81(3):357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Potter S.J., Lin Y., Cunningham A.L., Dwyer D.E., Su Y., Ma X., Hou Y., Saksena N.K. J. Clin. Microbiol. 2005;43(5):2339–2344. doi: 10.1128/JCM.43.5.2339-2344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. J. Am. Med. Assoc. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y.J.C. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Shan X., Patel U., Huang X., Lu J., Li J., Tao N. Proc. Natl. Acad. Sci. Unit. States Am. 2010;107(37):16028–16032. doi: 10.1073/pnas.1005264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z.Q., Armstrong A., Thomas G.J. Biochemistry. 1999;38(10):3148–3156. doi: 10.1021/bi981965m. [DOI] [PubMed] [Google Scholar]
- WHO Coronavirus Disease . 2020. COVID-19) Dashboard.https://covid19.who.int/ [Google Scholar]
- Wieland S., Makowska Z., Campana B., Calabrese D., Dill M.T., Chung J., Chisari F.V., Heim M.H. Hepatology. 2014;59(6):2121–2130. doi: 10.1002/hep.26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.-T., Gulino K., Zhang Y., Sabestien A., Chou T.-W., Zhou B., Lin Z., Albert I., Lu H., Swaminathan V. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117(2):895–901. doi: 10.1073/pnas.1910113117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., Gao G., Wang S., Ma C., Xie R., Wang F., Tan C., Zhu L., Guo Y., Zhang F. Clin. Infect. Dis. 2020;71(15):793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Abudayyeh O.O., Gootenberg J.S. vol. 20200321. 2020. pp. 1–8. (A Protocol for Detection of COVID-19 Using CRISPR Diagnostics). [Google Scholar]
- Zhang D., Zhang X., Ma R., Deng S., Wang X., Zhang X., Huang X., Liu Y., Li G., Qu J. medRxiv. 2020 doi: 10.1101/2020.05.02.20086876. [DOI] [Google Scholar]
- Zhang X., Lu W., Zheng Y., Wang W., Bai L., Chen L., Feng Y., Zhang Z., Yuan Z. J. Clin. Invest. 2016;126(3):1079–1092. doi: 10.1172/JCI83339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ke X., Zheng Z., Zhang C., Zhang Z., Zhang F., Hu Q., He Z., Wang H. ACS Nano. 2013;7(5):3896–3904. doi: 10.1021/nn305189n. [DOI] [PubMed] [Google Scholar]
- Zhao X., Tsao Y.-C., Lee F.-J., Tsai W.-H., Wang C.-H., Chuang T.-L., Wu M.-S., Lin C.-W. J. Virol. Methods. 2016;233:15–22. doi: 10.1016/j.jviromet.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiri M., Wu J. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.