Abstract
SARS-CoV-2 is the etiological agent of COVID-19, which represents a global health emergency that was rapidly declared a pandemic by the World Health Organization. Currently, there is a dearth of effective targeted therapies against viruses. Natural products isolated from traditional herbal plants have had a huge impact on drug development aimed at various diseases. Lapachol is a 1,4- naphthoquinone compound that has been demonstrated to have therapeutic effects against several diseases. SARS-CoV-2 non-structural proteins (nsps) play an important role in the viral replication cycle. Nsp9 seems to play a key role in transcription of the RNA genome of SARS-CoV-2. Virtual screening by docking and molecular dynamics suggests that lapachol derivatives can interact with Nsp9 from SARS-CoV-2. Complexes of lapachol derivatives V, VI, VIII, IX, and XI with the Nsp9 RNA binding site were subjected to molecular dynamics assays, to assess the stability of the complexes via RMSD. All complexes were stable over the course of 100 ns dynamics assays. Analyses of the hydrogen bonds in the complexes showed that lapachol derivatives VI and IX demonstrated strongest binding, with a stable or increasing number of hydrogen bonds over time. Our results demonstrate that Nsp9 from SARS-CoV-2 could be an important target in prospecting for ligands with antiviral potential. In addition, we showed that lapachol derivatives are potential ligands for SARS-CoV-2 Nsp9.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, COVID-19, lapachol derivatives, Nsp9
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged recently as the causative agent of the current widespread coronavirus disease (COVID-19). It was quickly declared a pandemic and represents a global health emergency (Chhikara et al., 2020). Since its identification in December 2019, SARS-CoV-2 has infected more than 84 million people, and led to 1,848,704 deaths (WHO, 2021), posing a worldwide challenge. It is most common modes of human-human transmission include direct transmission via droplets produced during coughing, sneezing, and talking (Doremalen et al., 2020), and indirect transmission via contaminated inanimate objects (Lai et al., 2020). Some infected people can be asymptomatic (Lai et al., 2020). Most symptomatic cases present with symptoms including fever, cough, and acute respiratory disease syndrome (SARS). Severe cases can progress to pneumonia, kidney failure, and death (Zhu et al., 2020).
SARS-CoV-2 is a Coronaviridae family member, and is an enveloped, single-stranded RNA virus (Hsu et al., 2020, p. 2). The viral structure is primarily determined by structural proteins, including spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins (Kim et al., 2020, p. 2). SARS-CoV-2 replication in host cells is not well understood, and the complete mechanism remains to be elucidated. However, knowledge of another coronavirus may suggest how SARS-CoV-2 replicates in host cells.
The SARS-CoV-2 genome has two open reading frames (ORFs), ORF1a and ORF1b, which encode the nonstructural proteins (nsps) (Andersen et al., 2020). ORF1a encodes the polypeptide pp1a, which is cleaved into 11 nsps, and ORF1b encodes the polypeptide pp1ab, which is cleaved into 16 nsps (Kim et al., 2020; Lai & Stohlman, 1981; Yogo et al., 1977). Proteolytic cleavage is performed by viral proteases Nsp3 and Nsp5 (Chan et al., 2020), enabling complex assembly from other nsps to form the replicase-transcriptase complex (RTC) (Gordon et al., 2020). The RTC consists of several enzymes, including RNA-dependent RNA polymerase (Nsp12), the main protease (Nsp5), an endonuclease (Nsp15), and a papain-like protease (Nsp3) (Chan et al., 2020).
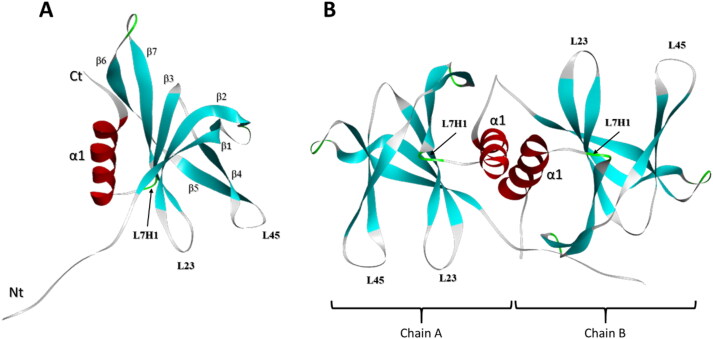
Nsp9, encoded by ORF1a forms a six β strand core that projects extended loops outward, and includes an N-terminal β-strand, and a C-terminal α-helix (Figure 1a) (Littler et al., 2020). The N-and C-terminal regions are essential for Nsp9 dimer’s replicase function (Figure 1b) (Littler et al., 2020) and are important for replication of CoV family viruses, including SARS-CoV (Frieman et al., 2012). SARS-CoV Nsp9 has been demonstrated to bind RNA (Egloff et al., 2004), stabilizing the nascent nucleic acid as it emerges from the nsp7-nsp8 complex during viral replication, and protecting it from nucleases (Bartlam et al., 2005; Zhai et al., 2005). In addition, Nsp9 may interact with a secondary structure element at the 3′ end of the viral genome in several coronaviruses (Robertson et al., 2004). The loops connecting the β2-3 (L23) and β3-4 (L34) strands of Nsp9 are positively charged and glycine rich, suggesting that these regions participate in RNA binding and interaction (Littler et al., 2020). Soluble SARS-CoV Nsp9 dimerizes via interaction of an α-helical GxxxG motif, and any disruption in key conserved residues can impair RNA binding, consequently suppressing SARS-CoV viral replication (Frieman et al., 2012). Mutations in the GxxxG motif of Nsp9 from porcine delta coronaviruses disrupts nucleotide binding capacity (Zeng et al., 2018). Nsp9 replicase is considered an important SARS-CoV virulence factor (Miknis et al., 2009). Since SARS-CoV-2 Nsp9 is 97% homologous to SARS-CoV Nsp9 (Littler et al., 2020; Ponnusamy et al., 2008), this protein may play a crucial role in SARS-CoV-2 replication. It therefore represents a potential target for design and development of antiviral drugs.
Figure 1.
Structural features of Nsp9 monomer and dimer. (a) Structure of Nsp9 monomer. This protein has one C-terminal α-helix associated with seven β strands. Loops L23 (formed between β4 and β5 strands), L45 (between β2 and β3 strands) and L7H1 (between the β7 strand and the α-helix) are indicated. (b) Structure of the Nsp9-dimer composed of A and B chains.
Natural products isolated from plants used in traditional herbal remedies have had a huge impact on drug development against various diseases (Dias et al., 2012). Naphthoquinones are natural metabolites found in plants, bacteria, and fungi that show extraordinary synthetic versatility, and are of great interest in medicinal chemistry (da Silva Júnior et al., 2019). Some naphthoquinones are currently used as medicinal drugs (Aminin & Polonika, 2020). Lapachol is a 1,4- naphthoquinone compound originally extracted from the Bignoniaceae family, first isolated in Tabebuia impetiginosa, but also present in other plants (Hussain & Green, 2017). Previous published studies have demonstrated anticancer, antileishmanial, anti-inflammatory and bactericidal biological properties (Araújo et al., 2019; de Sá et al., 2019; Hussain & Green, 2017; Oliveira et al., 2017). Lapachol has also demonstrated antiviral activity against Epstein-Barr virus and enterovirus in vitro (Pinto et al., 1987; Sacau et al., 2003). In addition, lapachol and its derivatives have been used in pre-clinical and clinical phases of drug development, demonstrating significant promise for cancer treatment (Block et al., 1974).
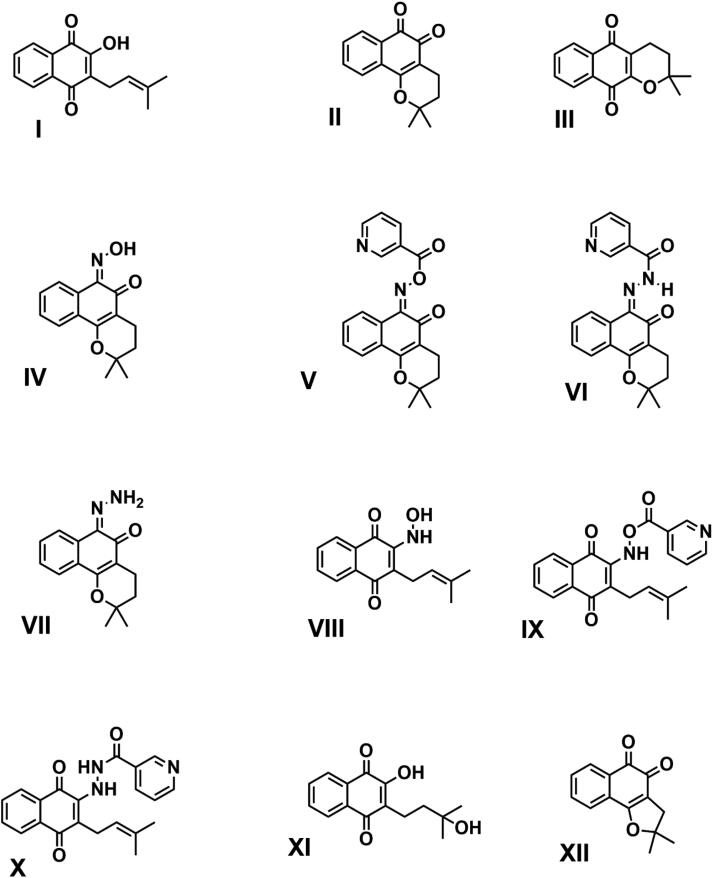
Owing to widespread use of naphthoquinones, especially lapachol (I), β-lapachone (II), and α-lapachone (III) (Figure 2), and their derivatives, there is great interest in studying their biological applications and their use as templates for synthesis of new derivatives. The chemistry of lapachol derivatives has been widely and extensively studied, and derivatives such as nor-β-lapachone (XII), hydroxylapachol (XI), and oxime derived from β-lapachol (IV) have been chemically synthesized (Costa et al., 2016; Santos et al., 2016; Nasiri et al., 2013). Therefore, lapachol derivative chemistry is a source of inspiration for the design and synthesis of new molecules, such as lapachol derivatives V, VI, VIII, IX and X (Figure 2), which to our knowledge are novel, and currently under study in our laboratory. There is a dearth of knowledge on the antiviral activity of lapachol against coronaviruses, especially SARS-CoV-2. Therefore, our in silico data provide evidence that lapachol and its derivatives may serve as SARS-CoV-2 Nsp9 ligands, providing leads for development of candidate drugs against COVID-19.
Figure 2.
Chemical structures of screened compounds. Lapachol (I) and eleven lapachol derivatives were subjected to molecular docking with the RNA binding site of Nsp9, composed of loops L23, L45, and L7H1. Compounds V, VI, VIII, IX, and XI yielded the best scores and stability values.
Methods
Virtual screening and toxicity predictions
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) non-structural protein (nsp) 9 replicase protein structure (PDB:6W4B) (Figure 1) was used as a target to search for protein-ligand interactions with lapachol and its derivatives (Figure 2). Virtual screening comprised prospecting for potential SARS-CoV-2 Nsp9 ligands from a library of 12 lapachol-related compounds. The GOLD 5.8.1 program (Jones et al., 1995) was used to calculate flexible anchoring between proteins and ligands. This program uses specific parameters for its virtual search of compounds in the selected databases. The GOLD program uses a genetic algorithm, propagating multiple copies of flexible ligand models in receptor active sites, randomly recombining segments of these copies until a set of structures converges. The position of the Nsp9 RNA-binding site was obtained from a study of SARS-CoV conducted by Sutton et al. (2004), which defined the RNA binding site as composed of the L23, L45, and L7H1 loops (Figure 1a). These data were corroborated by virtual prediction using the metaPocket 2.0 server (Huang, 2009).
The scoring function used for ranking was ChemPLP, which is defined by its use of a hydrogen bonding term, and multiple linear potentials to model van der Waals and repulsive terms. The parameter for searching an interaction’s degrees of freedom was defined as “Very Flexible,” corresponding to a search efficiency of 200%. All other parameters used options pre-defined by the program. All selected ligands were subjected to 10 iterations using this genetic algorithm. When the best three iterations of each molecule converged to the same docking pose, the five best compounds, according to PLP ranking and convergence, were selected for the next stage. Docking results were also analyzed by generation of 2 D diagrams using LigPlot + (Laskowski & Swindells, 2011). To evaluate potential toxic features, selected molecules were submitted to in silico toxicity prediction using the pkCSM webtool (Pires et al., 2015).
Molecular dynamics
The complexes selected from the results of molecular docking, and the Nsp9 apo protein were subjected to molecular dynamics (MD) simulations using GROMACS 2016 (Abraham et al., 2015). Parameterization of each ligand was performed on the SwissParam server (Zoete et al., 2011), and the MD of each protein-ligand complex was simulated using the CHARMM27 force field with the TIP3P water model. The MD unit cell was triclinic in shape, and water and ions were added to the system. System energy was minimized using the steepest descent algorithm, and minimization was stopped when the maximum force was less than 10.0 kJ/mol. NVT and NPT equilibrium phases were performed at 100 ps each. Solutions were subjected to 100 ns simulations, comprising 10 million simulation steps during the production phase under NTP conditions. Electrostatic interactions were calculated using particle mesh Ewald (PME) for long-range electrostatics (Essmann et al., 1995). Temperature was maintained constant at 300 K using a modified Berendsen thermostat (Berendsen et al., 1984), and pressure was maintained at 1 atm using the Parrinello-Rahman method (Parrinello & Rahman, 1980). Short-range van der Waals and electrostatic forces were set to 1.2.
Post dynamics complex stability analysis
To assess ligand stability, molecular dynamics results were evaluated using tools provided by the GROMACS program. Analyses performed were as follows: root mean square deviation (RMSD), root mean square fluctuation (RMSF), and number of hydrogen bonds between protein and ligand.
Post dynamics binding free energy calculations
The molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method (Massova & Kollman, 2000) with energy decomposition was used to calculate binding free energy between Nsp9 and each ligand. According to (Genheden & Ryde, 2015) crude approximations, it succeeds in rationalizing observed differences between docking studies. The MM-PBSA method was performed using the g_mmpbsa tool (g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations), which calculates three energetic terms: potential energy in vacuum, polar-solvation energy, and non-polar solvation energy.
Principal component analysis
Principal component analysis (PCA) or covariance analysis can be used in molecular dynamics to calculate overall protein molecular motion by analyzing a protein’s dynamic nature and its atomic positional fluctuations (Aarthy et al., 2020). The covar Gromacs tool was used to perform covariance analysis to create and diagonalize a covariance matrix using protein backbone atoms. The two most dominant eigenvectors were used to calculate the overlap between principal components and coordinates of the trajectory.
Results
In silico interactions of lapachol and its derivatives with Nsp9 from SARS-CoV-2
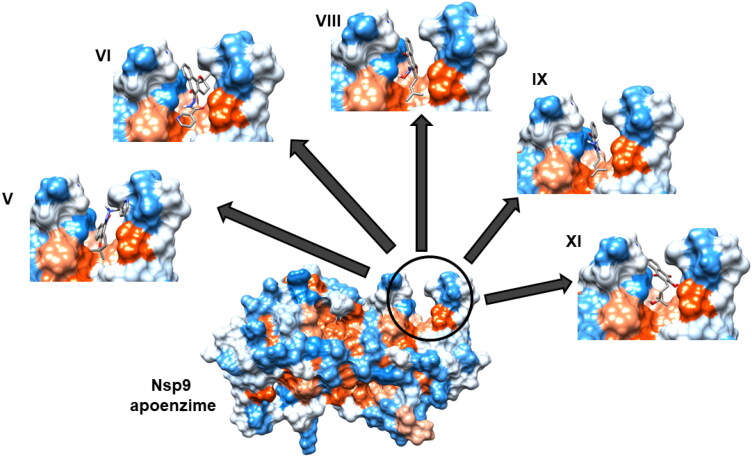
To prospect for ligands to non-structural protein (nsp) 9 of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) (Figure 1), we selected lapachol and 11 lapachol derivatives (Figure 2) for in silico analysis. These compounds were subjected to molecular docking targeting the RNA binding site of Nsp9 composed of loops L23, L45, and L7H1 (Figure 1a) (Sutton et al., 2004). The top five compounds (V, VI, VIII, IX, and XI) were chosen based on best ChemPLP scores, and on their capacity to converge to a single pose within three iterations after docking. Comparison of 3 D docking with an analysis of 2 D diagrammatic docking demonstrated that all five molecules formed a minimum of one hydrogen bond within the RNA binding site from Nsp9, together with hydrophobic interactions (Figures 3 and 4).
Figure 3.
Selected ligands docked in a 3 D model of the Nsp9 RNA binding site. The RNA binding site of Nsp9, formed by loops L23, L45 and L7H1, is indicated within the circle, and interactions with lapachol derivatives V, VI, VIII, IX, and XI are shown. Hydrophobic and hydrophilic regions are presented in orange and blue, respectively.
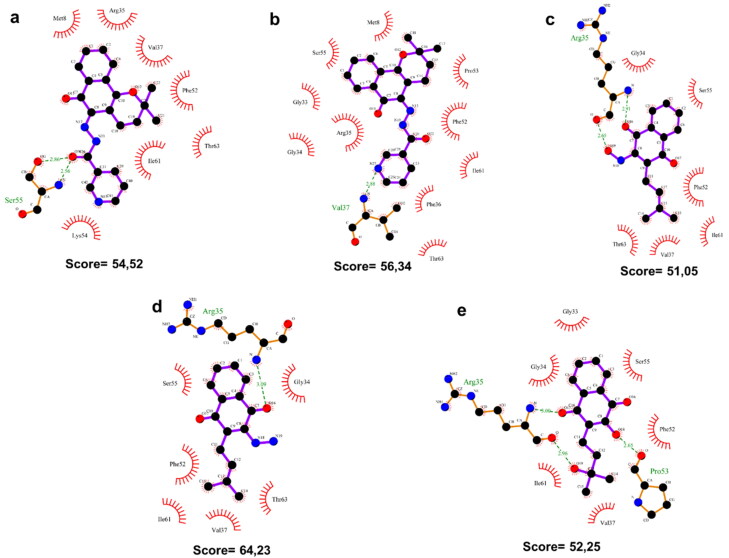
Figure 4.
2 D diagrams of interactions between lapachol derivatives and Nsp9 of SARS-CoV-2. Below each diagram, the respective ChemPLP docking score is shown. Green dashed lines indicate hydrogen bonds. Red arcs indicate hydrophobic interactions with the RNA binding site of Nsp9 of SARS-CoV-2. (a) Lapachol derivative V. (b) Lapachol derivative VI. (c) Lapachol derivative VIII. (d) Lapachol derivative IX. (e) Lapachol derivative XI.
Molecule V formed two hydrogen bonds to Ser35 in the Nsp9 RNA binding site (2,86 and 2,56 Å) and seven hydrophobic interactions (to Met8, Arg35, Val37, Phe52, Thr63, Ile61, and Lys54), producing a docking score of 54.52. Compound VI formed one hydrogen bond to Val37 (2,88 Å), and ten hydrophobic interactions (to Gly33, Gly34, Arg35, Ser55, Met8, Pro53, Phe52, Ile61, Phe36, and Thr63), with a docking score of 56.34. Compounds VIII and IX formed two hydrogen bonds to Arg35 (2.65 and 2.91 Å) or one hydrogen bond (3.09 Å), respectively. Compounds VIII and IX also established six hydrophobic interactions (to Ser55, Phe52, Ile61, Val37, Thr63, and Gly34), producing docking scores of 51.05 and 64.23, respectively. Molecule XI formed two hydrogen bonds to Arg35 (3,00 and 2,96 Å) and one to Pro53 (2,65 Å), and formed six hydrophobic interactions with residues Gly33, Gly34, Ile61, Val37, Phe52 and Ser55, with a docking score of 52.25 (Figure 4).
In silico toxicity predictions for lapachol derivatives
Toxicity predictions evaluated eight features related to potential toxic effects of the selected molecules (Table 1). Except for molecule V, all compounds yielded negative results for AMES toxicity, which assesses potential mutagenicity. The maximum tolerated dose in humans gave the highest values for compounds VIII and IX and the lowest for compound V. Values below or equal to 0.477 log(mg/kg/day) were considered low according to the PKCSM webserver. All molecules showed negative results for hERG inhibition. This feature evaluated the potential for potassium channel inhibition that could lead to fatal ventricular arrhythmia. All molecules except compound VIII displayed negative hepatotoxicity results. All molecules yielded negative skin sensitization results. T. pyriformis is used as a model in toxicity studies. All molecules displayed log(ug/L) values above the limit expected for low toxicity (-0.5 log(µg/L)). Minnow toxicity tests, using fathead fish minnows as a model, gave positive results in silico for all molecules. Expected high acute toxicity occurs at low compound concentrations, with log LC50 scores below 0.3. In general, the molecules displayed little toxicity, except for hepatotoxicity and T. pyriformis toxicity. Molecule V gave the broadest range of toxicities, and molecule VIII displayed the lowest predicted toxicities. It is important to note that as yet, no experimental data corroborate or disagree with these predicted results, as these lapachol derivatives have not been tested in vitro or in vivo.
Table 1.
In silico toxicity predictions for the lapachol derivatives.
| MOLECULES |
|||||
|---|---|---|---|---|---|
| TOXICITY PREDICTIONS | V | VI | VIII | IX | X |
| AMES toxicity | Yes | No | No | No | No |
| Max. tolerated dose log(mg/kg/day) | 0.06 | 0.136 | 0.743 | 0.719 | 0.296 |
| hERG I inhibitor | No | No | No | No | No |
| hERG II inhibitor | No | No | No | No | No |
| Hepatotoxicity | Yes | Yes | No | Yes | Yes |
| Skin Sensitisation | No | No | No | No | No |
| T.pyriformis toxicity log(ug/L) | 0.449 | 0.689 | 0.541 | 0.523 | 0.542 |
| Minnow toxicity log LC50 | 1.713 | 1.455 | 1.278 | 0.718 | 1.334 |
Lapachol derivatives form a highly stable complex with the SARS-CoV-2 Nsp9 RNA binding site
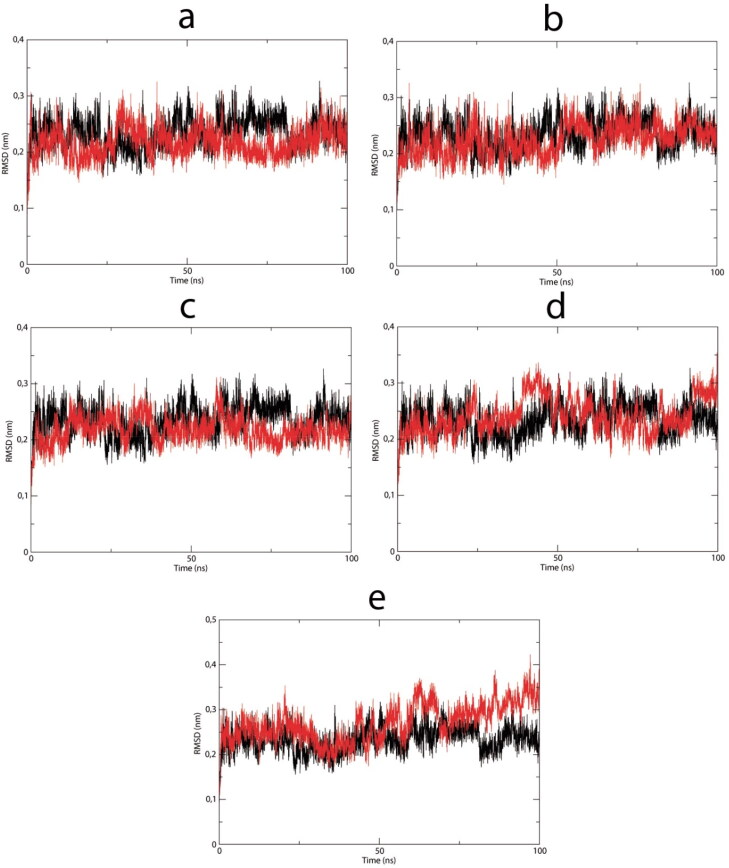
Complexes between the selected lapachol derivatives V, VI, VIII, IX, and XI and the Nsp9 RNA binding site were subjected to molecular dynamics assays to assess the stability of each complex via RMSD. During the 100 ns dynamic simulation, all complexes showed a stable profile (Figure 5), except for the complex with derivative XI. This was confirmed by mean RMSD and low standard deviation values (Table 2). Complexes with lapachol derivatives V, VI, VIII, and IX displayed similar stabilities compared with the apo enzyme RMSD (Figure 5) and by the mean ± standard deviation (Table 2). However, XI showed a slight variation in the last 25 ns compared with the apoenzyme.
Figure 5.
RMSD vs time spanning 100 ns simulations of the selected Nsp9-ligand complexes (red lines): (a) Lapachol derivative V. (b) Lapachol derivative VI. (c) Lapachol derivative VIII. (d) Lapachol derivative IX. (e) Lapachol derivative XI. (black lines): Nsp9 apoenzyme.
Table 2.
RMSD and RMSF mean and standard deviation after each dynamic simulation.
| RMSD* | Mol V | Mol VI | Mol VIII | Mol IX | Mol XI | Apo Nsp9 |
|---|---|---|---|---|---|---|
| Mean | 0.22 | 0.22 | 0.22 | 0.24 | 0.27 | 0.23 |
| Standard deviation | 0.03 | 0.03 | 0.02 | 0.03 | 0.04 | 0.03 |
| RMSF ** | ||||||
| Residues (25-65) | ||||||
| Mean | 0.143 | 0.155 | 0.148 | 0.164 | 0.174 | 0.172 |
| Standard deviation | 0.066 | 0.075 | 0.082 | 0.093 | 0.083 | 0.096 |
RMSD mean and standard deviation analyses performed after the first five ns.
RMSF mean and standard deviation analyses performed with binding site residues in the Nsp9 dimer.
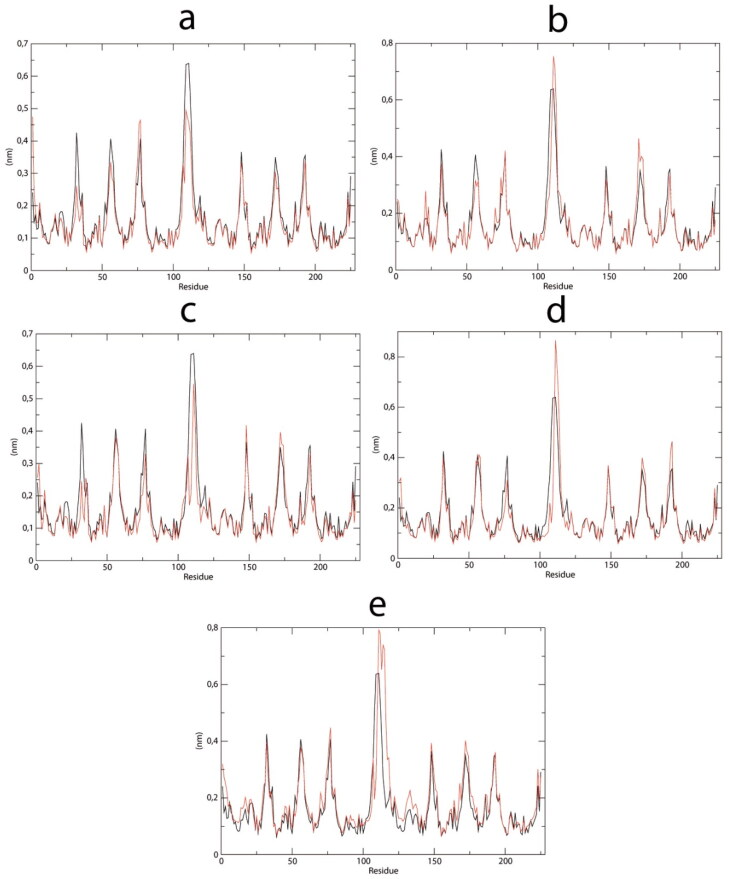
The ligand-binding site is mainly comprised of residues 25 to 65 in Nsp9 chain A (Figure 6). Visual inspection of RMSF analysis (Figure 6) reveals a similar profile of residue fluctuation in all analyzed structures. However, the RMSF mean and standard deviation fluctuate less in residues 25 to 65 in complexes with molecules V, VI, and VIII, compared to apo enzyme (Table 2). These results indicate increased residue restraints, probably due to ligand interactions with these residues.
Figure 6.
RMSF during 100 ns simulations of the selected Nsp9-ligand complexes plotted vs Nsp9 apoenzyme. (a) Lapachol derivative V. (b) Lapachol derivative VI. (c) Lapachol derivative VIII. (d) Lapachol derivative IX. (e) Lapachol derivative XI.
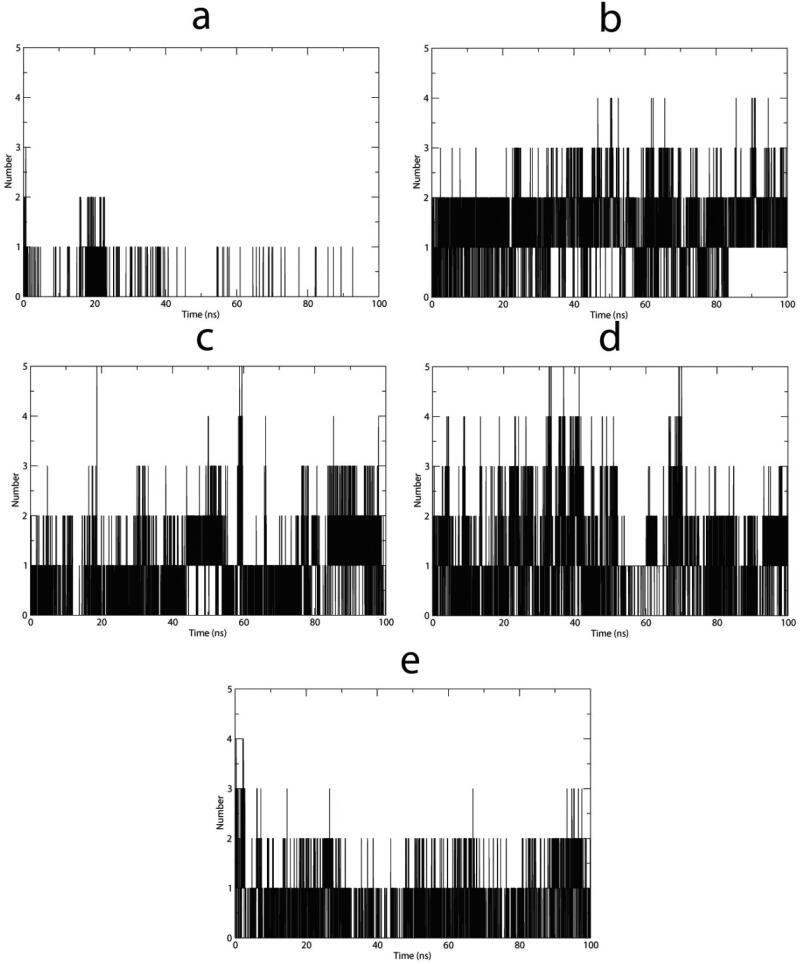
Analysis of hydrogen bonds formed within each complex showed that lapachol derivative VI (Figure 7b) had the most stable hydrogen bond profile with a mean of 1.51 hydrogen bonds plotted along the MD (Table 3). By contrast, complexes with molecules V and XI showed lower means and standard deviations. Similarly, the Nsp9-VIII and IX complexes did not demonstrate a tendency to stabilize, showing high standard deviations (Table 3) and sporadic increased values for hydrogen bonding (Figure 7c and d).
Figure 7.
Hydrogen bonds plotted vs time spanning 100 ns simulations of the selected Nsp9-ligand complexes. (a) Lapachol derivative V. (b) Lapachol derivative VI. (c) Lapachol derivative VIII. (d) Lapachol derivative IX. (e) Lapachol derivative XI.
Table 3.
Mean numbers of hydrogen bonds and standard deviations at the end of each dynamic simulation.
| H BOND | Mol V | Mol VI | Mol VIII | Mol IX | Mol XI |
|---|---|---|---|---|---|
| Mean | 0.07 | 1.51 | 0.82 | 1.06 | 0.62 |
| Standard Deviation | 0.27 | 0.73 | 0.80 | 0.96 | 0.67 |
Binding energy and stability of lapachol derivative/Nsp9 RNA binding site complexes according to MD simulations
The MM/PBSA method is an approach used to estimate free energies of small ligand complexes with macromolecules that requires a small number of computational processes. It has been useful for analyzing binding energies of docked structures and rationalizing observed differences in biological macromolecules’ behaviors (Genheden & Ryde, 2015).
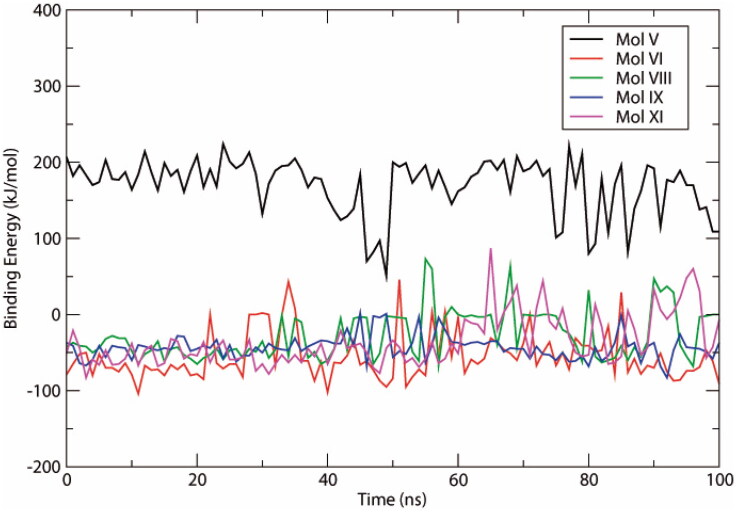
Within each 100 ns simulation, 100 snapshots of the trajectory were obtained at constant intervals of 1 ns for analyses. All complexes showed negative binding energy profiles, with the exception of molecule V (Figure 8). Molecule VI had the most stable profile (Figure 8), and together with molecule IX, showed the lowest mean binding energy among the ligands (Table 4). Molecule IX also showed the lowest standard deviation. These results indicate that van der Waals interactions contribute the most energy to these protein-ligand interactions, and molecules VI and IX display the highest binding affinity.
Figure 8.
Binding energies plotted against time spanning the 100 ns simulations of selected Nsp9-ligand complexes.
Table 4.
Binding energy means ± standard deviations of complexes after molecular dynamics simulation.
| Molecule | Binding Energy | Van der Walls Energy | Eletrostatic Energy | Polar Solvation Energy | SASA energy |
|---|---|---|---|---|---|
| V | 170.138 ± 36.191 | −100.202 ± 42.094 | 229.738 ± 58.107 | 52.777 ± 36.463 | −12.175 ± 5.082 |
| VI | −55.681 ± 31.008 | −116.972 ± 50.833 | −42.593 ± 23.187 | 117.094 ± 49.241 | −13.210 ± 5.794 |
| VIII | −27.810 ± 31.951 | −45.612 ± 42.410 | −8.649 ± 10.312 | 32.182 ± 37.527 | −5.731 ± 5.063 |
| IX | −43.432 ± 15.626 | −91.013 ± 30.072 | −26.228 ± 18.276 | 85.304 ± 31.955 | −11.495 ± 3.493 |
| XI | −35.777 ± 33.400 | −67.041 ± 42.265 | −14.178 ± 12.767 | 53.686 ± 33.573 | −8.243 ± 5.133 |
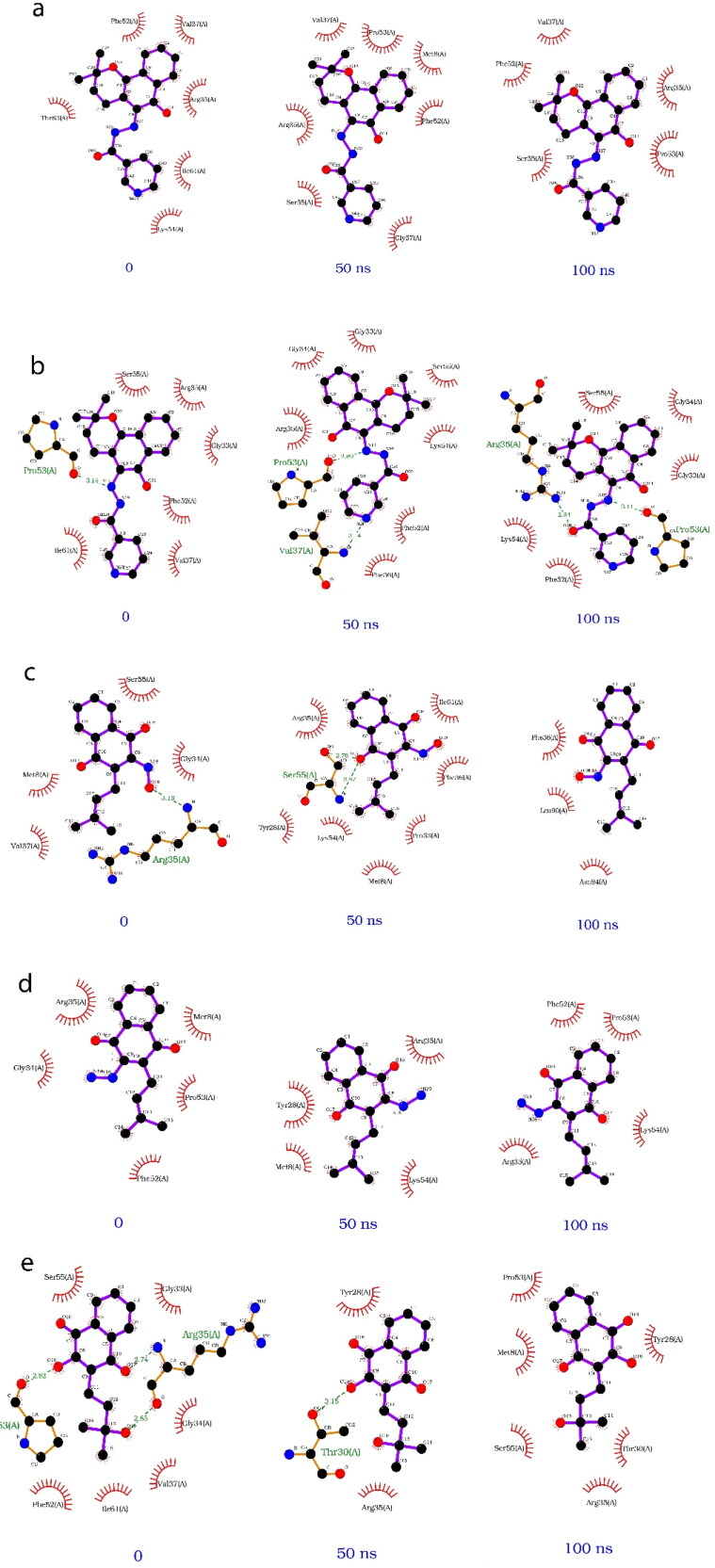
A simple comparison of the dynamics over time of each ligand Nsp9 complex was performed. The protein-ligand interactions are illustrated by the complexes at 0, 50, and 100 ns in each simulation (Figure 9). Lapachol derivative VI displayed the most constant number of hydrophobic interactions and hydrogen bonds over the course of the simulation compared to the other ligands (Figure 9b). This result corroborates prior analysis defining VI as the most stable ligand.
Figure 9.
2 D diagram of interactions at 0, 50, and 100 ns time points in simulations of selected Nsp9-ligand complexes. (a) Lapachol derivative V. (b) Lapachol derivative VI. (c) Lapachol derivative VIII. (d) Lapachol derivative IX. (e) Lapachol derivative XI.
SARS-CoV-2 Nsp9 compactness during MD
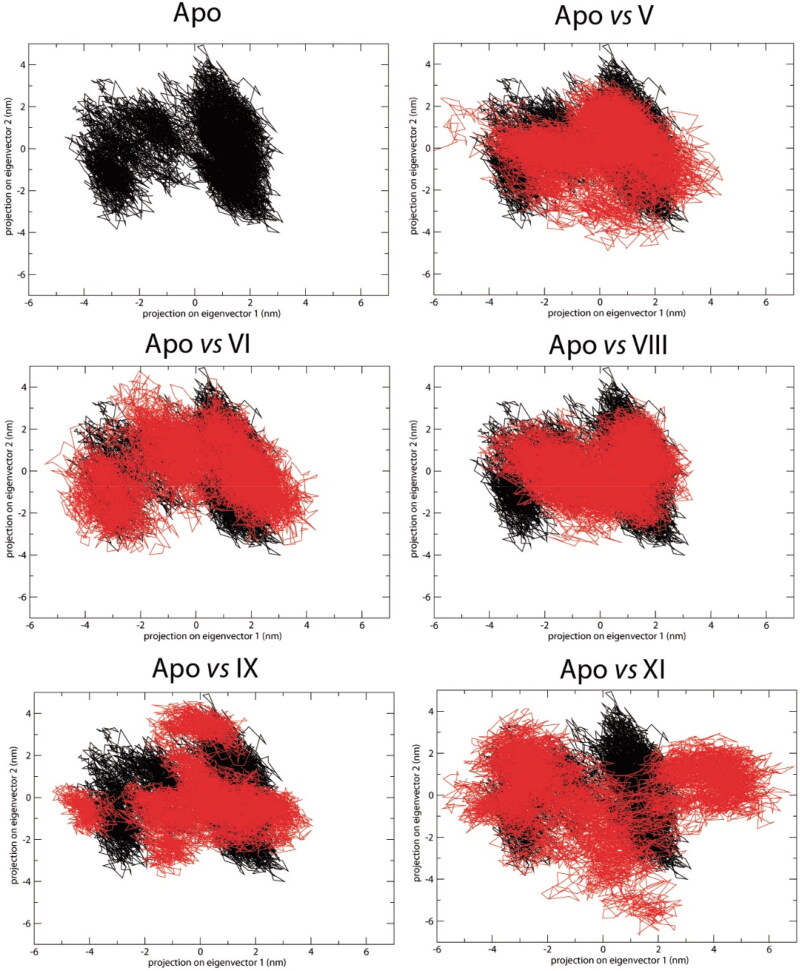
PCA eigenvector projections represent the overlap between the first two principal components and coordinates of each trajectory. They plot the essential subspace occupied by each protein during molecular dynamics. The results show that complexes V, VI, and VIII filled conformational space similar to the apo protein (Figure 10). Thus, complex compactness was maintained during the simulation. By contrast, molecules IX and XI showed exploration of a different conformational space, and a less compact profile compared to the apo structure (Figure 10).
Figure 10.
Principle component analysis over 100 ns simulations with the selected Nsp9-ligand complexes. Nsp9 apoenzyme is represented in black, and each Nsp9-lapachol derivative complex in red.
Lapachol derivative VI interacts with the Nsp9 replicase protein of SARS-CoV
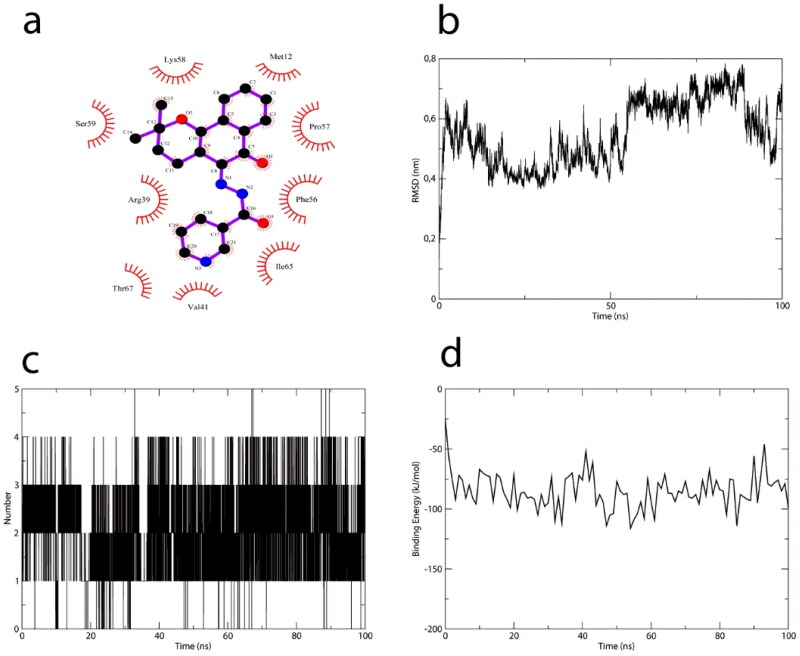
To compare results with another coronavirus target, the severe acute respiratory syndrome-coronavirus (SARS-CoV) Nsp9 replicase protein (PDB:3EE7) was subjected to docking and dynamics with ligand VI, using the same parameters described in Methods. Lapachol derivative VI interacted with Ser59 from the Nsp9 RNA-binding site, and formed nine other hydrophobic interactions (to Met12, Pro57, Ser59, Arg39. Val41, Phe56, Thr67, Ile65, and Lys58), producing a docking score of 57.12 (Figure 11a). Although the RMSD profile of SARS-COV Nsp9 does not seem to reach a stable plateau (Figure 11b), the number of hydrogen bonds seems to increase from one bond to stabilize at four bonds over 100 ns of molecular dynamics (Figure 10c). In addition, binding energy values were under −50 kJ/mol (Figure 11d), similar to SARS-CoV-2 Nsp9-lapachol VI derivative complex values.
Figure 11.
Lapachol derivative VI interacts with the RNA binding site of SARS-CoV Nsp9. (a) 2 D diagram of the lapachol derivative VI/SARS-CoV Nsp9 interaction. Red arcs indicate hydrophobic interactions within the RNA binding site of SARS-CoV Nsp9. (b) RMSD profile of the Nsp9 protein plotted against simulation time. (c) hydrogen bonds in the SARS-CoV Nsp9–lapachol derivative VI complex. (d) binding energy profile of the SARS-CoV Nsp9–lapachol derivative VI complex.
Discussion
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has threatened the global health system (Hsu et al., 2020) and accentuated the urgent need for drugs to combat viral outbreaks. The lack of antivirals and/or vaccines directly impacts our ability to bring pandemics under control, increasing morbidity/mortality, and dissemination of SARS-CoV-2/COVID-19. This situation also impacts populations’ quality of life, overloading healthcare systems worldwide, and resulting in economic hardship. Therefore, drugs that abrogate SARS-CoV-2 replication are needed.
In this study, we docked ligands to the SARS-CoV-2 nonstructural protein (nsp) 9 RNA binding site (a promising drug target) to identify candidate antiviral agents. This methodology has been used to design and identify potential ligands for viruses including Chikungunya virus, dengue virus, and SARS-CoV. For emergency development of antiviral agents, studies have focused on identifying drug targets among essential proteins in virus replication, primarily targeting conserved structures and enzymes such as RNA-dependent-RNA-polymerase (RdRp), main 3-chymotrypsin-like cysteine protease (MPRO), or the receptor-binding domain (RBD) of SARS-CoV-2 spike protein (Bharadwaj et al., 2020; Buonaguro et al., 2020; Rout et al., 2020). However, many other targets are predicted to impact viral activity (Gordon et al., 2020; Wu et al., 2020). Nsp9 is an essential enzyme in the replicase-transcriptase complex (RTC) required for coronavirus replication, that binds to genomic RNA, ensuring production of new RNA genomes, and protecting them from nucleases (Egloff et al., 2004, p. 9). Additionally, it is highly conserved among coronaviruses (Littler et al., 2020; Ponnusamy et al., 2008). Therefore, Nsp9 may represent a good target for direct-acting antivirals, given that inhibiting the RNA binding site of Nsp9 prevents it from binding to the nascent RNA from the nsp7-8 complex during SARS-CoV-2 replication.
Once released from the nsp7-8 complex, the nascent RNA does not form a stable secondary structure (Bartlam et al., 2005; Zhai et al., 2005), and can be degraded by nucleases if Nsp9 action is blocked. Nsp9 has three loops (L23, L45, and L7H1) that have been associated with the RNA binding site and forms an OB-fold that is predicted to interact with oligonucleotides (Flynn & Zou, 2010; Sutton et al., 2004). Moreover, the β-barrel in which the L23, L45, and L7H1 loops are found allows RNA binding capacity through nonspecific interactions (Sutton et al., 2004). In this context, considering that SARS-CoV-2 Nsp9 is a conserved structure among coronaviruses (Littler et al., 2020), we explored the RNA binding site formed by the cited loops as a possible target for lapachol and its derivates to assess their antiviral potential.
Lapachol and its derivatives have been associated with several biological activities in preclinical and clinical trials (Block et al., 1974; Hussain & Green, 2017). Here, we evaluated lapachol, and eleven lapachol derivatives as possible ligands for the RNA binding site of SARS-CoV-2 Nsp9. Among the analyzed compounds, lapachol derivatives V, VI, VIII, IX, and XI demonstrated strong interactions with the target site in molecular docking studies.
Lapachol derivatives have been shown to exhibit improved activity and solubility when associated with new radicals (Oliveira et al., 2002, 2017). Our data corroborate this literature, since chemical modifications of lapachol were able to increase hydrophobic interactions with regions of the RNA binding site, and create new interactions with specific amino acids, as demonstrated by lapachol derivative VI.
To assess the overall stability of each complex (lapachol or a lapachol derivative bound in the SARS-CoV-2 Nsp9 RNA binding site), molecular dynamics analyses were performed to obtain RMSD profiles. Lapachol has previously been described as very stable in molecular dynamics assays against ZIKV nsp3 (Oguntade et al., 2017). However, in our analyses, lapachol did not yield the best docking score. Lapachol derivative VI presented one of the best docking scores, showing stable interaction behavior in RMSD and RMSF. In addition, molecular dynamics assessed hydrogen bond formation in each complex over the course of a 100 ns simulation. Hydrogen bonds are essential for stabilizing ligands within a binding pocket (Kostal, 2016), and are mediators of strong interactions (Maréchal, 2007). Lapachol derivative VI showed a tendency to stabilize existing hydrogen bonds, while the other compounds, especially compound IX, showed an increasing number of hydrogen bonds as the simulation progressed.
When the lapachol derivative VI/SARS-CoV Nsp9 complex was compared to that of compound VI/SARS-CoV-2 Nsp9, the results showed similar interactive behavior. The main discrepancy in molecular dynamics performance was that the protein RMSD profile of the SARS-CoV Nsp9 protein did not stabilize over time. The hydrogen bonds formed, and the binding energy profiles were highly similar between SARS-CoV and SARS-CoV-2 Nsp9–lapachol derivative VI complexes. The high sequence identity between these two Nsp9 homologs (90%), obtained by the EMBOSS Needle sequence alignment tool (Needleman and Wunsch, 1970), and structural similarity (RMSD = 3.89 Å), obtained by structural alignment using the Chimera program (Pettersen et al., 2004) may explain these functional similarities (Figure S1). Moreover, derivative VI bonding with SARS-CoV Nsp9 displayed similar affinity to that of SARS-CoV-2 Nsp9, suggesting broad-spectrum activity (Littler et al., 2020; Ponnusamy et al., 2008). The affinity of these molecules for other non-structural SARS-CoV-2 proteins is being investigated by our research group. In addition, potential toxic characteristics can be corrected with other rational modifications during in vitro and in vivo tests.
In summary, we have demonstrated that SARS-CoV-2 Nsp9 may be a viable target for antiviral development. We also showed that lapachol derivatives, mainly VI and IX, may function as potential ligands for the SARS-CoV-2 Nsp9 RNA-binding site. Our work shows the importance of studying protein-ligand complex stability via dynamics to corroborate docking studies. Although the results seem promising, it is important to validate this activity. Therefore, these molecules should be further investigated for in vitro and/or in vivo antiviral activity. They may also be used as templates for development of future drugs against SARS-CoV-2 and other coronaviruses. Thus, these data may provide relevant information to advance our ability to combat COVID-19.
Funding Statement
The authors are grateful to FAPEMIG (Minas Gerais Research Foundation - APQ-00587-14 and APQ-03385-18), CAPES – Prevention and Combat of Outbreaks, Endemics, Epidemics and Pandemics (#88881.506794/2020–01), and CNPq (National Council of Technological and Scientific Development - # 142495/2020-4).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Aarthy, H., Panwar, U., & Singh, S. K. (2020). Structural dynamic studies on identification of EGCG analogues for the inhibition of Human Papillomavirus E7. Scientific Reports, 10(1), 1–24. 10.1038/s41598-020-65446-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Aminin, D., & Polonik, S. (2020). 1,4-naphthoquinones: Some biological properties and application. Chemical & Pharmaceutical Bulletin, 68(1), 46–57. 10.1248/cpb.c19-00911 [DOI] [PubMed] [Google Scholar]
- Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., & Garry, R. F. (2020). The proximal origin of SARS-CoV-2. Nature Medicine, 26(4), 450–453. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo, I. A. C., de Paula, R. C., Alves, C. L., Faria, K. F., Oliveira, M. M. D., Mendes, G. G., Dias, E. M. F. A., Ribeiro, R. R., Oliveira, A. B. D., & Silva, S. M. D. (2019). Efficacy of lapachol on treatment of cutaneous and visceral leishmaniasis. Experimental Parasitology, 199, 67–73. 10.1016/j.exppara.2019.02.013 [DOI] [PubMed] [Google Scholar]
- Bartlam, M., Yang, H., & Rao, Z. (2005). Structural insights into SARS coronavirus proteins. Current Opinion in Structural Biology, 15(6), 664–672. 10.1016/j.sbi.2005.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Bharadwaj, S., Azhar, E. I., Kamal, M. A., Bajrai, L. H., Dubey, A., Jha, K., Yadava, U., Kang, S. G. and Dwivedi, V. D. (2020). SARS-CoV-2 Mpro inhibitors: Identification of anti-SARS-CoV-2 Mpro compounds from FDA approved drugs. Journal of Biomolecular Structure and Dynamics, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, J. B., Serpick, A. A., Miller, W., & Wiernik, P. H. (1974). Early clinical studies with lapachol (NSC-11905). Cancer Chemotherapy Reports. Part 2, 4(4), 27–28. [PubMed] [Google Scholar]
- Buonaguro, L., Tagliamonte, M., Tornesello, M. L., & Buonaguro, F. M. (2020). SARS-CoV-2 RNA polymerase as target for antiviral therapy. Journal of Translational Medicine, 18(1), 185. 10.1186/s12967-020-02355-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F.-W., Kok, K.-H., Zhu, Z., Chu, H., To, K. K.-W., Yuan, S., & Yuen, K.-Y. (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes & Infections, 9(1), 221–236. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhikara, B. S., Rathi, B., Singh, J., & Poonam, F. N. U. (2020). Corona virus SARS-CoV-2 disease COVID-19: Infection, prevention and clinical advances of the prospective chemical drug therapeutics. Chemical Biology Letters, 7(1), 63–72. [Google Scholar]
- Costa, M., , Feitosa, A., , Oliveira, F., , Cavalcanti, B., , Da Silva, E., , Dias, G., , Sales, F., , Sousa, B., , Barroso-Neto, I., , Pessoa, C., , Caetano, E., , Di Fiore, S., , Fischer, R., , Ladeira, L., & , Freire, V. (2016). Controlled Release of Nor-β-lapachone by PLGA Microparticles: A Strategy for Improving Cytotoxicity against Prostate Cancer Cells. Molecules, 21(7), 873. https://doi.org/ 10.3390/molecules21070873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Júnior, E. N., Jardim, G. A. M., Jacob, C., Dhawa, U., Ackermann, L., & de Castro, S. L. (2019). Synthesis of quinones with highlighted biological applications: A critical update on the strategies towards bioactive compounds with emphasis on lapachones. European Journal of Medicinal Chemistry, 179, 863–915. 10.1016/j.ejmech.2019.06.056 [DOI] [PubMed] [Google Scholar]
- de Sá, N. P., Cisalpino, P. S., Bertollo, C. M., Santos, P. C., Rosa, C. A., de Souza, D. D. G., Barbeira, P. J. S., Alves, T. M. D. A., Zani, C. L., & Johann, S. (2019). Thiosemicarbazone of lapachol acts on cell membrane in Paracoccidioides brasiliensis. Medical Mycology, 57(3), 332–339. 10.1093/mmy/myy045 [DOI] [PubMed] [Google Scholar]
- Dias, D. A., Urban, S., & Roessner, U. (2012). A historical overview of natural products in drug discovery. Metabolites, 2(2), 303–336. 10.3390/metabo2020303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doremalen, N. V., Bushmaker, T., Morris, D. H., Holbrook, M. G., Gamble, A., Williamson, B. N., Tamin, A., Harcourt, J. L., Thornburg, N. J., Gerber, S. I., & Lloyd-Smith, J. O. (2020). Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. New England Journal of Medicine, 382(16), 1564–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff, M.-P., Ferron, F., Campanacci, V., Longhi, S., Rancurel, C., Dutartre, H., Snijder, E. J., Gorbalenya, A. E., Cambillau, C., & Canard, B. (2004). The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proceedings of the National Academy of Sciences of the United States of America, 101(11), 3792–3796. 10.1073/pnas.0307877101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Flynn, R. L., & Zou, L. (2010). Oligonucleotide/oligosaccharide-binding fold proteins: A growing family of genome guardians. Critical Reviews in Biochemistry and Molecular Biology, 45(4), 266–275. 10.3109/10409238.2010.488216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman, M., Yount, B., Agnihothram, S., Page, C., Donaldson, E., Roberts, A., Vogel, L., Woodruff, B., Scorpio, D., Subbarao, K., & Baric, R. S. (2012). Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. Journal of Virology, 86(2), 884–897. 10.1128/JVI.05957-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden, S., & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery, 10(5), 449–461. 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., O’Meara, M. J., Rezelj, V. V., Guo, J. Z., Swaney, D. L., Tummino, T. A., Hüttenhain, R., Kaake, R. M., Richards, A. L., Tutuncuoglu, B., Foussard, H., Batra, J., Haas, K., Modak, M., … Krogan, N. J. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 583(7816), 459–468. 30 abr. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, L. Y., Chia, P. Y., & Lim, J. F. (2020). The novel coronavirus (SARS-CoV-2) epidemic. Annals of the Academy of Medicine, Singapore, 49(3), 105–103. 10.47102/annals-acadmedsg.202051 [DOI] [PubMed] [Google Scholar]
- Huang, B. (2009). MetaPocket: A meta approach to improve protein ligand binding site prediction. Omics : A Journal of Integrative Biology, 13(4), 325–330. 10.1089/omi.2009.0045 [DOI] [PubMed] [Google Scholar]
- Hussain, H., & Green, I. R. (2017). Lapachol and lapachone analogs: A journey of two decades of patent research(1997-2016). Expert Opinion on Therapeutic Patents, 27(10), 1111–1121. 10.1080/13543776.2017.1339792 [DOI] [PubMed] [Google Scholar]
- Jones, G., Willett, P., & Glen, R. C. (1995). Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. Journal of Molecular Biology, 245(1), 43–53. 10.1016/S0022-2836(95)80037-9 [DOI] [PubMed] [Google Scholar]
- Kim, D., Lee, J. Y., Yang, J. S., Kim, J. W., Kim, V. N., & Chang, H. (2020). The architecture of SARS-CoV-2 transcriptome. Cell, 181(4), 914-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostal, J. (2016). Computational Chemistry in Predictive Toxicology: Status quo et quo vadis? In Fishbein J. C. & Heilman J. M., (Eds.). Advances in molecular toxicology (Vol. 10, pp. 139–186). Elsevier. Chapter 4. [Google Scholar]
- Lai, C.-C., Liu, Y. H., Wang, C. Y., Wang, Y. H., Hsueh, S. C., Yen, M. Y., Ko, W. C., & Hsueh, P. R. (2020). Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. Journal of Microbiology, Immunology, and Infection = Wei Mian Yu Gan Ran Za Zhi, 53(3), 404-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M. M., & Stohlman, S. A. (1981). Comparative analysis of RNA genomes of mouse hepatitis viruses. Journal of Virology, 38(2), 661–670. 10.1128/JVI.38.2.661-670.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R. A., & Swindells, M. B. (2011). LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling, 51(10), 2778–2786. 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- Littler, D. R., Gully, B. S., Colson, R. N., & Rossjohn, J. (2020). Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. Iscience, 23(7), 101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal, Y. (2007). The hydrogen bond: Formation, thermodynamic properties, classification. In Maréchal Y. (Ed.), The hydrogen bond and the water molecule (pp. 3–24). Elsevier. [Google Scholar]
- Massova, I., & Kollman, P. A. (2000). Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspectives in Drug Discovery and Design, 18(1), 113–135. 10.1023/A:1008763014207 [DOI] [Google Scholar]
- Miknis, Z. J., Donaldson, E. F., Umland, T. C., Rimmer, R. A., Baric, R. S., & Schultz, L. W. (2009). Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. Journal of Virology, 83(7), 3007–3018. 10.1128/JVI.01505-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasiri, H. R., , Madej, M. G., , Panisch, R., , Lafontaine, M., , Bats, J. W., , Lancaster, C. R. D., & , Schwalbe, H. (2013). Design, Synthesis, and Biological Testing of Novel Naphthoquinones as Substrate-Based Inhibitors of the Quinol/Fumarate Reductase from Wolinella succinogenes. Journal of Medicinal Chemistry, 56(23), 9530–9541. https://doi.org/ 10.1021/jm400978u [DOI] [PubMed] [Google Scholar]
- Needleman, S. B., & , Wunsch, C. D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology, 48(3), 443–453. https://doi.org/ 10.1016/0022-2836(70)90057-4 [DOI] [PubMed] [Google Scholar]
- Oguntade, S., Ramharack, P., & Soliman, M. E. (2017). Characterizing the ligand-binding landscape of Zika NS3 helicase-promising lead compounds as potential inhibitors. Future Virology, 12(6), 261–273. 10.2217/fvl-2017-0014 [DOI] [Google Scholar]
- Oliveira, K. M., Corrêa, R. S., Barbosa, M. I. F., Ellena, J., Cominetti, M. R., & Batista, A. A. (2017). Ruthenium (II)/triphenylphosphine complexes: An effective way to improve the cytotoxicity of lapachol. Polyhedron, 130, 108–114. 10.1016/j.poly.2017.04.005 [DOI] [Google Scholar]
- Oliveira, M. F., Lemos, T. G., de Mattos, M. C., Segundo, T. A., Santiago, G. M. P., & Braz-Filho, R. (2002). New enamine derivatives of lapachol and biological activity. Anais da Academia Brasileira de Ciencias, 74(2), 211–221. 10.1590/s0001-37652002000200004 [DOI] [PubMed] [Google Scholar]
- Parrinello, M., & Rahman, A. (1980). Crystal structure and pair potentials: A molecular-dynamics study. Physical Review Letters, 45(14), 1196–1199. 10.1103/PhysRevLett.45.1196 [DOI] [Google Scholar]
- Pettersen, E. F., , Goddard, T. D., , Huang, C. C., , Couch, G. S., , Greenblatt, D. M., , Meng, E. C., & , Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. https://doi.org/ 10.1002/jcc.20084 15264254 [DOI] [PubMed] [Google Scholar]
- Pinto, A. V., Pinto, M. d C., Lagrota, M. H., Wigg, M. D., & Aguiar, A. N. (1987). Antiviral activity of naphthoquinones. I. Lapachol derivatives against enteroviruses. Revista Latinoamericana de Microbiologia, 29(1), 15–20. [PubMed] [Google Scholar]
- Pires, D. E. V., Blundell, T. L., & Ascher, D. B. (2015). pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of Medicinal Chemistry, 58(9), 4066–4072. 10.1021/acs.jmedchem.5b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy, R., Moll, R., Weimar, T., Mesters, J. R., & Hilgenfeld, R. (2008). Variable oligomerization modes in coronavirus non-structural protein 9. Journal of Molecular Biology, 383(5), 1081–1096. 10.1016/j.jmb.2008.07.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, M. P., Igel, H., Baertsch, R., Haussler, D., Ares, M., & Scott, W. G. (2004). The Structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biology, 3(1), e5. 10.1371/journal.pbio.0030005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, J., Swain, B. C., & Tripathy, U. (2020). In silico investigation of spice molecules as potent inhibitor of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1819879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacau, E. P., Estévez-Braun, A., Ravelo, A. G., Ferro, E. A., Tokuda, H., Mukainaka, T., & Nishino, H. (2003). Inhibitory effects of lapachol derivatives on epstein-barr virus activation. Bioorganic & Medicinal Chemistry, 11(4), 483–488. 10.1016/S0968-0896(02)00542-4 [DOI] [PubMed] [Google Scholar]
- Santos, V. L. A., de Moraes, M. O., Pessoa, C., da Costa, M. P., Gonsalves, A. A., & Araújo, C. R. M. (2016). Cytotoxicity Activity of Semisynthetic Naphthoquinone-1-oximes against Cancer Cell Lines. Journal of Chemical and Pharmaceutical Research, 8(12), 202–206. [Google Scholar]
- Sutton, G., Fry, E., Carter, L., Sainsbury, S., Walter, T., Nettleship, J., Berrow, N., Owens, R., Gilbert, R., Davidson, A., Siddell, S., Poon, L. L. M., Diprose, J., Alderton, D., Walsh, M., Grimes, J. M., & Stuart, D. I. (2004). The nsp9 replicase protein of SARS-Coronavirus, structure and functional insights. Structure (London, England : 1993)), 12(2), 341–353. 10.1016/j.str.2004.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (2021). Novel Coronavirus (2019-nCoV) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo, Y., Hirano, N., Hino, S., Shibuta, H., & Matumoto, M. (1977). Polyadenylate in the virion RNA of mouse hepatitis virus. Journal of Biochemistry, 82(4), 1103–1108. 10.1093/oxfordjournals.jbchem.a131782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z., Deng, F., Shi, K., Ye, G., Wang, G., Fang, L., Xiao, S., Fu, Z., & Peng, G. (2018). Dimerization of coronavirus nsp9 with diverse modes enhances its nucleic acid binding affinity. Journal of Virology, 92(17), e00692-18. 10.1128/JVI.00692-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, Y., Sun, F., Li, X., Pang, H., Xu, X., Bartlam, M., & Rao, Z. (2005). Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nature Structural & Molecular Biology, 12(11), 980–986. 10.1038/nsmb999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete, V., Cuendet, M. A., Grosdidier, A., & Michielin, O. (2011). SwissParam: A fast force field generation tool for small organic molecules. Journal of Computational Chemistry, 32(11), 2359–2368. 10.1002/jcc.21816 [DOI] [PubMed] [Google Scholar]