Abstract
2020 will be remembered worldwide for the outbreak of Coronavirus disease (COVID-19), which quickly spread until it was declared as a global pandemic. The main protease (Mpro) of SARS-CoV-2, a key enzyme in coronavirus, represents an attractive pharmacological target for inhibition of SARS-CoV-2 replication. Here, we evaluated whether the anti-inflammatory drug Ibuprofen, may act as a potential SARS-CoV-2 Mpro inhibitor, using an in silico study. From molecular dynamics (MD) simulations, we also evaluated the influence of ionic strength on the affinity and stability of the Ibuprofen–Mpro complexes. The docking analysis shows that R(−)Ibuprofen and S(+)Ibuprofen isomers can interact with multiple key residues of the main protease, through hydrophobic interactions and hydrogen bonds, with favourable binding energies (−6.2 and −5.7 kcal/mol, respectively). MM-GBSA and MM-PBSA calculations confirm the affinity of these complexes, in terms of binding energies. It also demonstrates that the ionic strength modifies significantly their binding affinities. Different structural parameters calculated from the MD simulations (120 ns) reveal that these complexes are conformational stable in the different conditions analysed. In this context, the results suggest that the condition 2 (0.25 NaCl) bind more tightly the Ibuprofen to Mpro than the others conditions. From the frustration analysis, we could characterize two important regions (Cys44-Pro52 and Linker loop) of this protein involved in the interaction with Ibuprofen. In conclusion, our findings allow us to propose that racemic mixtures of the Ibuprofen enantiomers might be a potential treatment option against SARS-CoV-2 Mpro. However, further research is necessary to determinate their possible medicinal use.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, Ibuprofen, Mpro, docking molecular, molecular dynamics
Introduction
In late 2019, a novel coronavirus was identified as the cause of an outbreak of pneumonia cases in Wuhan, the largest city in China’s central region. In February 2020, the World Health Organization (WHO) designated the disease COVID-19, which stands for coronavirus disease 2019 (Rodríguez-Morales et al., 2020; World Health Organization et al., 2020), which quickly spread to several countries until it became a global pandemic. The virus that causes COVID-19 is designated severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2); as the RNA genome is similar to the SARS coronavirus (SARS-CoV). Considering that there is currently only one non-specific antiviral drug (Remdesivir) approved by the FDA for the treatment of certain adult and pediatric patients with COVID-19 (Food and Drug Administration, 2020), one of the biggest challenges in global therapeutic is to develop potent antiviral agents capable of inhibiting the COVID-19 virus, in the shortest possible time. In this direction, one of the best characterized pharmacological targets of this virus is the main protease (Mpro) of SARS-CoV-2, also known as 3-chymotrypsin-like protease (3CLpro) (Jin et al., 2020).
It is characterized by its great similarity with the SARS-CoV Mpro (published in 2003). These proteins are homologous and they have high levels of sequence similarity; however, shown differences in the active site in size and shape, indicating that designing drugs based on SARS-CoV might not be effective (Bzówka et al., 2020). Along with papain-like protease, this enzyme is essential for processing polyproteins, into which the viral RNA is initially translated after it has entered the human cells. The SARS-CoV-2 Mpro protease cleaves at no less than 11 conserved sites on the large polyprotein 1ab (replicase 1ab, 790 kDa) (Boopathi et al., 2020; Gao et al., 2020; Mirza & Froeyen, 2020).
Their structure is a homodimer in which the two subunits are arranged perpendicular to each other. Each subunit has 3 structural domains; between domain I and II lies the substrate binding site, comprising the residues that make up the catalytic dyad (His41 and Cys145). A long loop region (residues 185–200) connects catalytic domains with the domain III. The domain III (residues 201–303) is involved in the dimerization process of the protein, being crucial for the activity of the enzyme (Zhang et al., 2020). Inhibiting their activity would block viral replication. Since its discovery, this protease has been the motivation of several publications intended to understand its mechanism of action and the design of possible inhibitors (Estrada, 2020; Gentile et al., 2020; Macchiagodena et al., 2020).
In silico methods using computational approaches are extremely useful tools for evaluating ligand–protein interactions during the drug discovery process. They facilitate the identification of molecules capable to modulate the biological activity of the protein, providing scientific information on candidate drugs for the treatment of this viral infection. To date, there are numerous publications focused on the virtual screening of potential SARS-CoV-2 Mpro inhibitors on the basis of natural or synthetic compounds from different databases or libraries (Adem et al., 2020; Fischer et al., 2020; Gentile et al., 2020; Omar et al., 2020; Ton et al., 2020; Wu et al., 2020), and also from clinically approved drugs for other diseases (Kandeel & Al-Nazawi, 2020; Kumar et al., 2020).
Particularly, in an emergency like this, repurposing of FDA-approved drugs for other diseases represents a very good option, to speed up clinical trials. Most of these drugs have sufficient experience and dosage, and their safety is well known. Once efficacy is validated, these can be approved by health authorities and the hospital’s ethics committee for the rapid clinical treatment of patients. In recent times, compounds currently used for the treatment of non-infectious diseases as potential antimicrobial alternatives have also been reviewed (Lagadinou et al., 2020; Zimmermann & Curtis, 2017). One example is Ibuprofen, a nonsteroidal anti-inflammatory drug (NSAID) widely used for relief of pain, fever and inflammation, which has demonstrated novel pharmacological actions against bacteria (Dai et al., 2019; Obad et al., 2015; Shah et al., 2018), fungi (Ogundeji et al., 2016; Sanyal et al., 1993) and viruses (Pan et al., 2018; Veljkovic et al., 2015). Interestingly, a short time ago, it was reported a nebulizable formulation of sodium Ibuprofen in high ionic strength with bactericidal properties for the treatment of respiratory infections of fibrotic patients (Muñoz et al., 2018). In addition to exhibiting antiviral properties. The authors of the work highlight that the presence of sodium chloride (NaCl) in the formulation, a condition normally used in a nebulizable solution to fluidize and facilitate the elimination of mucus from the lungs, improves the bactericidal activity of the amphipathic molecule of Ibuprofen soluble, obtaining an effect bactericidal at very low concentrations of Ibuprofen and, in particular, at short periods of time. This phenomenon would be related to the influence of ionic strength on the surface properties of this amphipathic Ibuprofen, since they observe that high concentrations of salts favour the approach and insertion of this compound into the membrane, triggering changes in electrical conductivity and its resulting instability. Besides, this Ibuprofen inhalation does not seem to produce substantial changes in the general state of the animals or significant histopathological alterations in the organs analysed. On the other hand, recently a report suggests that Ibuprofen might impact SARS-CoV-2 induced disease by indirect interaction with actin protein (Veljkovic et al., 2020).
Based on the evidence mentioned, in this work, we proposed to evaluate the binding affinity and identify the ligand–protein interactions of Ibuprofen enantiomers (FDA approved drug with excellent safety record) with the Mpro of SARS-CoV-2 in different simulated environments, using a molecular docking and molecular dynamic (MD) simulations studies. Also, we calculated different structural parameters from trajectories generated by the simulations to explore the dynamic behaviour and the stability of the Ibuprofen–Mpro complexes in the selected environments.
The purpose of this study is to investigate whether Ibuprofen could be useful in the treatment of COVID-19 by direct interaction with SARS-CoV-2 Mpro.
Materials and methods
Preparation of ligands
Considering that the majority of the approved pharmaceutical forms of Ibuprofen consist of racemic mixtures of the R(−) and S(+) enantiomers, but it is proven that the S(+) enantiomer is the pharmacologically active (Adams et al., 1976; Evans, 2001), both were analysed separately. The 2D structures of R(−)Ibuprofen and S(+)Ibuprofen enantiomers (Figure 1) were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov) in SDF format and then converted to PDB format using the Open Babel (O'Boyle et al., 2011) software. Hydrogen atoms were added to the ligands, the energy was minimised and converted to pdbqt format using the MGL tools of the AutoDockTools4 software (Trott & Olson, 2010) for further analysis.
Figure 1.
2D representation of Ibuprofen enantiomers.
Preparation of protein
Three-dimensional structure of SARS-CoV-2 Mpro (PdbId: 6Y84) with resolution 1.39 Å was collected from Protein Data Bank (PDB, 1971, http://www.rcsb.org/). The crystallized structure of the monomer of SARS-CoV-2 Mpro (Owen et al., 2020) was used for the docking molecular and MD simulations, based on references indicating that only one monomer is active in the SARS-CoV-2 Mpro homodimer (Chen et al., 2006). All the molecules that are not part of the structure of the protein were removed. Hydrogen atoms were added to the proteins and converted to pdbqt format using the MGL tools of the AutoDockTools4 software (Trott & Olson, 2010) for later analysis.
Molecular docking
In order to select the preferred orientation of the enantiomers with the protein to form a stable complex, we docked both enantiomers within the active site of the chain A of the three-dimensional structure of SARS-CoV-2 Mpro (PdbId: 6Y84) using Autodock Vina (Trott & Olson, 2010). The position of the catalytic residues for the Mpro structure was obtained from bibliography (Das et al., 2020; Estrada, 2020; Gentile et al., 2020; Zhang et al., 2020). The box size remained the same for all runs: 30 × 30 × 30 Å. The docked complexes with the most favourable binding modes were selected for molecular dynamics simulations.
The analysis of the 2D interactions of the ligand–protein complexes were analysed with LigPlot + program (Laskowski & Swindells, 2011) and for the 3D visualization PyMol (LLC Schrodinger, 2008) was used.
Molecular dynamic simulations
It is known that the ionic strength determines the protein folding in solution (Gabrielczyk et al., 2017) and influences on the ligand–protein binding affinity (Papaneophytou et al., 2014). As mentioned above, there are evidences of benefits attributed to the inhalation of a hypertonic saline solution (HTS) in the treatment of patients with lung diseases. It has even been described beneficial effects on the bactericidal activity of ibuprofen against agents involved in the development of lung pathologies (Muñoz et al., 2018). Thus, we performed MD simulations to evaluate the influence of ionic strength (NaCl concentration) on the structure of the Ibuprofen–Mpro complexes.
In order to analyse the dynamics of the complexes in different concentrations of NaCl, four different environments were simulated for each complex. Condition 1: only Na + ions to neutralize the complex, condition 2: 0.25 M NaCl, condition 3: 0.50 M NaCl and condition 4: 1 M NaCl. An accurate rule (Machado & Pantano, 2020) was used to define the electrolytic content in simulation boxes and obtain the expected models with NaCl concentrations in the system (0.25 M, 0.5 M and 1 M).
All MD simulations were run using the AMBER20 software package (Case et al., 2020) with the protein being assigned ff99SB (Hornak et al., 2006) parameters and docked ligands being assigned GAFF (Wang et al., 2004) parameters augmented by AM1-BCC (Jakalian et al., 2000, 2002) partial charges. Solvation of the systems were carried out using the TIP3P (Jorgensen et al., 1983) explicit water model and the AMBER pmemd module was used to perform energy minimizations and MD.
A two-stage solvent energy minimization was performed. In the first stage, an energy minimization was performed in order to relax the solvation structure, thus restricting the protein atoms leaving it fixed in order to allow the solvent in the solvent box to move and relax around the protein, this stage consisted of 2.000 steps. In the second step, another minimization of the energy of the solvent was performed but without any restrictions to obtain the optimization of the geometry of the whole system (protein and solvent), this process consisted of 8.000 steps. Then, the thermalization of the system was performed according to the AMBER force field, the system was heated from 0 K to 298 K gradually. A temperature ramp was generated for this purpose, which was increased until the expected temperature was achieved, using the SHAKE (Van Gunsteren & Berendsen, 1977) algorithm in 10.000 steps. After heating, an equilibration for 20 ns was attained at constant temperature and pressure (298 K and 1 Bar) to accommodate the system volume to obtain an adequate density.
Finally, the well-equilibrated complexes were then subject to the production phase without any restrains for a period of 120 ns with a time step of 2fs, and after every 2 ps the structural coordinates were saved. The CPPTRAJ module (Roe & Cheatham, 2013) of Amber tool was used to calculate the root mean square deviation (RMSD), root mean square fluctuation (RMSF) and to analyse the hydrogen bonds from the trajectories generated by the simulations. All the analysis of the 2D interactions of the ligand–protein complexes was using LigPlot + program (Laskowski & Swindells, 2011) and for the 3 D visualization was PyMol (LLC Schrodinger, 2008).
MM-GBSA and MM-PBSA binding free energies estimation
Both Molecular Mechanics-Generalized Born Surface Area (MM-GBSA) and Molecular Mechanics-Poisson Boltzmann surface area (MM-PBSA) analyses were carried out by using the MMPBSA.py (Miller et al., 2012) python script implemented in the AMBER20 package with the aim to estimate the binding free energy of ibuprofen enantiomers with SARS-CoV-2 Mpro.
The binding free energies (reported in kcal/mol) in MM-PBSA method and its complementary MM-GBSA method calculate the difference between bound and unbound state of solvated conformations of a molecule (Equation (1))
| (1) |
The energy terms were extracted every 20 ns of each respective MD trajectory (120 ns) by selecting 1000 uniformly spaced out snapshots. The salt concentration (saltcon) in Generalized Born and the ionic strength (istrng) in Poisson Boltzmann were modified according to the different saline environments of the simulations.
Structural analysis
For localizing frustration in all pdb files extracted from the MD simulations the frustratometer tool (Jenik et al., 2012; Parra et al., 2016) (http://www.frustratometer.tk/) was used. There are three ways to calculate frustration, two of them are at contact level frustration. In this case, the frustration is calculated between residues in contact in the protein structure and are called mutational and configurational. The third way is at single level frustration that calculate frustration per residue. The frustration index (FI) can be classified into three classes, highly frustrated (FI ≤ −1, residues or contacts that are in conflict with the structures), minimally frustrated (FI ≥ 0.78, residues or contacts that are important for protein stability) and neutral (−1 < FI < 0.78, this residues or contact are neutral in structure stability) (Jenik et al., 2012; Parra et al., 2016). The expected values of highly frustrated contacts or residues are 10% for minimally frustrated residues is 50% and for neutral is 40% (Ferreiro et al., 2007). In this study, the configurational FI and the single level frustration were used.
Results and discussions
Docking molecular
The enantiomers of Ibuprofen were individually docked into the crystal structure of SARS-CoV-2 Mpro (PdbId: 6Y84). The docking analysis of both enantiomers generated negative values for free energy with the SARS-CoV-2 protease, suggesting a favourable binding affinity for this protein. It stands out that R(−)Ibuprofen (−6.2 kcal/mol) bound to Mpro with a ΔG value lower than S(+)Ibuprofen (−5.7 kcal/mol).
The Ibuprofen–Mpro interactions were analysed and the results contrasted with bibliography in terms of key residues involved in catalytic activity, substrate binding and dimerization process of SARS-CoV-2 Mpro (Estrada, 2020; Gao et al., 2020; Macchiagodena et al., 2020; Zhang et al., 2020).
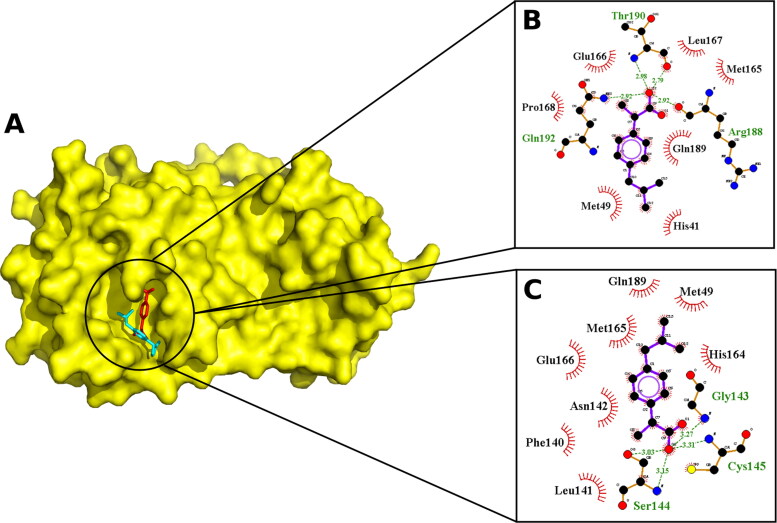
Figure 2 shown the best affinity poses and the interactions of both enantiomers with the SARS-CoV-2 Mpro. According to the results obtained, both enantiomers fully fit within the active site of Mpro (Figure 2(A)) and interact with multiple key residues, establishing hydrophobic contacts with all the molecule and electrostatic bond through the carboxyl group (Figure 2(B,C)).
Figure 2.
Docked poses. A. Molecular surface of SARS-CoV-2 Mpro (PdbId: 6Y84) with R(−)Ibuprofen (red) and S(+)Ibuprofen (cyan) enantiomers. 2D visualization of the interactions visualized by LigPlot + of B. R(−)Ibuprofen and C. S(+)Ibuprofen, H-bond are represented as a dashed line in green and hydrophobic interactions are represented as lines in red.
Upon examining the molecular interactions, it can be seen that both isomers (Figure 2(B,C)) were found to be interacting with catalytic residues (His41-Cys145), the substrate-binding pocket and interacting with residues involved in dimerization process (Phe140 and Glu166). We highlight the fact that the enantiomers interacted differentiated with the residues of the catalytic dyad. This finding is significant considering that most of the approved pharmaceutical forms of this drug consist of racemic mixtures of its enantiomers R(−) and S(+).
In addition, both enantiomers interacted with the Glu166 residue. Key residue that plays a connecting role between the substrate binding site and the dimer interface, necessary for enzymatic activity.
On the other hand, the individual analysis of molecular interactions suggests that the S(+)Ibuprofen may have its own favourable binding with the S1 subsite, a structural feature essential for catalysis. It interacts with Gly143, Ser144 and Cys145 in S1 pocket (Figure 2(C)), an operational element called the “oxyanion hole” (Zhang et al., 2020). This pocket typically consists of backbone amides or positively charged residues, crucial to stabilize the tetrahedral intermediate produced during proteolysis (Su et al., 2020). Moreover, S(+)Ibuprofen interacts with aminoacids implicated in dimerization process (Phe140 and Glu166), positioning it also as a possible candidate to inhibit dimer formation, as mentioned above. Similar results have been described in detail for promising Mpro inhibitors (Ghosh et al., 2020).
It is important to point out that it was found that R(-)Ibuprofen interacts with residues of the Linker loop region (185–200) that links the domain (I + II) with domain III, a region that is being studied for its fundamental role in the control of dimerization and enzymatic activity (Bzówka et al., 2020; Menéndez et al., 2020). Therefore, this interaction with these residues could be significant for the activity.
Considering that the static pose of the ligand–protein complexes obtained by molecular docking do not provide information that represents and characterizes the stability of each complex, both complexes were subjected to molecular dynamics simulations accompanied by calculations of various parameters to examine the real movement and structural modifications that undergo in the different selected conditions.
Molecular Dynamic simulations
As described above, MD simulation studies of 120 ns for each complex, in four different environments were carried out. The trajectories obtained for simulations run were analysed using RMSD, RMSF, ligand–protein binding contacts in terms of hydrogen bonding and binding free energy calculations (MM-GBSA and MM-PBSA).
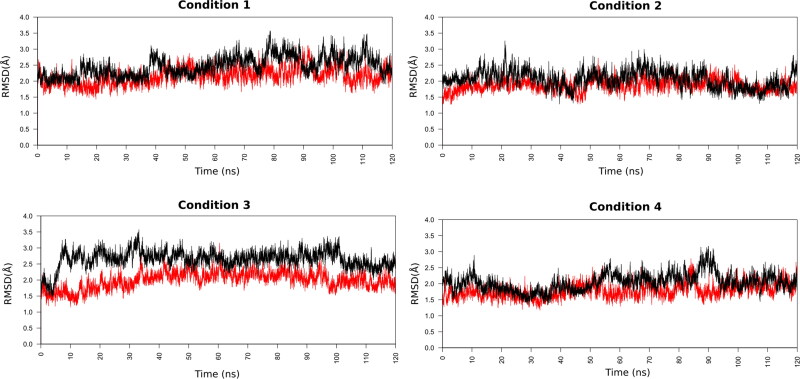
RMSD is a useful parameter to quantify the overall structural stability of a ligand–protein complex after binding of ligand within the active site of protein, in function of a period of time. It can be seen in the RMSD graphs (Figure 3) that the two complexes analysed show deviations within the RMSD range of 1.0 Å to 3.5 Å during the simulations. Values up 3–4 Å are wholly satisfactory for globular proteins, indicating that their bindings are significant stable in all the analysed systems.
Figure 3.
RMSD of the backbone atoms of the docked complexes. In red R(−)Ibuprofen–SARS-CoV-2 complex, in black S(+)Ibuprofen–SARS-CoV-2 complex.
The average RMSD values of simulated complexes for R(−)Ibuprofen–Mpro and S(+)Ibuprofen–Mpro in condition 1 were 2.15 Å and 2.46 Å, in condition 2 were 1.86 Å and 2.05 Å, in condition 3 were 2.00 Å and 2.65 Å, in condition 4 were 1.78 Å and 2.04 Å, respectively. Based on these average RMSD values, we can clearly infer that the Ibuprofen–Mpro complexes are conformational stable, highlighting that the RMSD value of the R(−)Ibuprofen–Mpro complex was slightly lower than the S(+)Ibuprofen–Mpro complex in all the conditions analysed.
Respect to the impact of the ionic strength (NaCl concentration) on the ligand–protein stability, the RMSD results indicate that the ionic strength did not significantly influence on the structural stability of Ibuprofen–Mpro complexes. However, within this context of overall stability, both complexes in condition 2 (0.25 M NaCl) and condition 4 (1 M NaCl) obtained the lowest average RMSD values after 120 ns of simulation trajectory, suggesting that in these conditions are slightly more stable than in the other conditions.
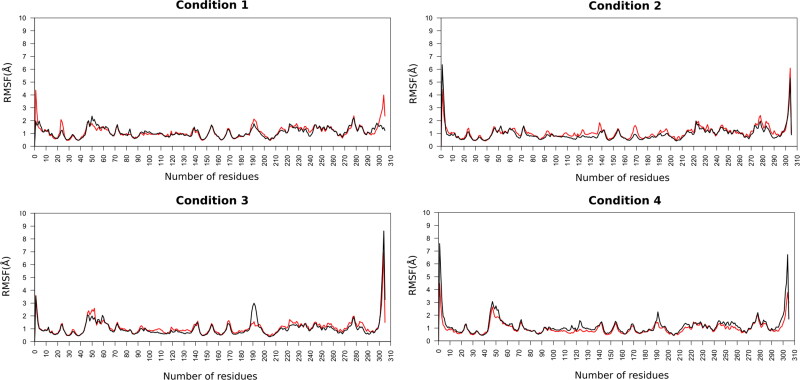
Moreover, the RMSF values were calculated in order to evaluate the fluctuations of each residue of target protein and the ligand in the complex under the different simulated conditions (Figure 4). RMSF is a useful parameter to identify the flexible residues or regions in the protein. The estimated average RMSF values of protein residues (1–304) in complexes for R(−)Ibuprofen–Mpro and S(+)Ibuprofen–Mpro in condition 1 were 1.12 Å and 1.07 Å, in condition 2 were 1.08 Å and 0.95 Å, in condition 3 were 1.09 Å and 1.02 Å, in condition 4 were 0.97 Å and 1.15 Å, respectively.
Figure 4.
RMSF of the backbone atoms of the docked complexes. In red R(−)Ibuprofen–SARS-CoV-2 Mpro complex, in black S(+)Ibuprofen–SARS-CoV-2 Mpro complex.
As a result, we can infer that the protein residues in the different simulated environments had a similar fluctuation profile, with fluctuations within a range of 1–3 Å during whole simulation time. This similarity in the RMSF plots and average values indicates that the ionic strength did not significant effect on the overall flexibility of the protein in all the simulations. However, it is interesting to note a visible difference in two regions of the protein under condition 2. In this condition the residues ranging from 40 to 55 (Loop Cys44-Pro52) and 180–200 (Linker loop 185–200) fluctuated less compared to the other conditions, indicating that these regions have less flexibility. Important fact if we consider that the conformational variations of these loops might modifies the dimerization process and enzymatic activity of the SARS-CoV-2 Mpro.
Additionally, the average RMSF value of R(−)Ibuprofen and S(+)Ibuprofen (305) in complexes with Mpro in condition 1 were 2.35 Å and 1.23 Å, in condition 2 were 1.01 Å and 0.89 Å, in condition 3 were 1.49 Å and 3.20 Å, in condition 4 were 2.95 Å and 1.67 Å, respectively (Figure 3). From these data, it can be inferred that their bindings are stable during the dynamics. These values also indicated that both ligands undergone relatively less fluctuations under the condition 2.
In order to examine the stability of the ligands in the Mpro binding pocket, the conformations of the MD simulation were extracted every 20 ns and the interactions were visualized between the protein and the corresponding enantiomer (Figure S1 [A–H], supplementary material). Both enantiomers remained in the binding pocket throughout the simulation period, forming hydrogen bonds and hydrophobic interactions. It is observed in docked poses that both enantiomers fully fit within the active site of the SARS-CoV-2 Mpro, in all environments analysed. However, it can be appreciated different interactions and postures of isomers within the active site.
Hydrogen bond analysis
Hydrogen bonding is among the most crucial parameters to understand the binding affinity of candidate ligand towards a target protein. Analysing the hydrogen bonds during a simulation clarifies the mode of interaction of the ligand with the different residues of the protein. A large number of H-bonds present in between them evidence a strong binding affinity.
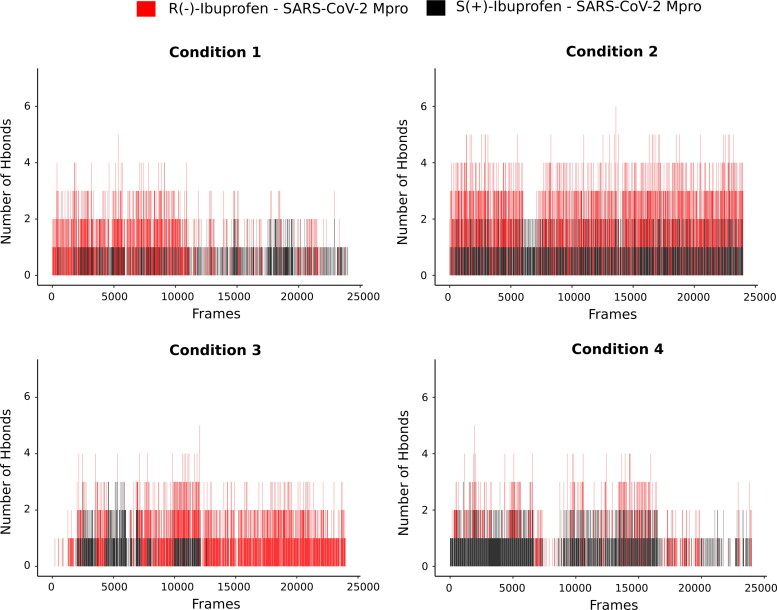
Therefore, we perform an analysis of the number of hydrogen bonds formed during the simulated trajectories (Figure 5), as well as the identity of the protein residues and the atom of the ligand that formed the hydrogen bond.
Figure 5.
Hydrogen bond profiles of the SARS-CoV-2 Mpro complexes having with R(−)Ibuprofen (in red) and S(+)Ibuprofen (in black) in different conditions.
We can observe that the complex with R(−)Ibuprofen, except in condition 4, formed more hydrogen bonds than S(+)Ibuprofen.
Besides, analysing the behaviour of the hydrogen bonds of both enantiomers in the different conditions, it was found that a greater amount of hydrogen bonds was present in condition 2 during 120 ns of MD simulation.
Examining the residues of the protein involved in the formation of the hydrogen bonds with the enantiomers in all simulated trajectories (Figure S2 [A–H], supplementary material), it was observed that for the R(−)Ibuprofen–Mpro complex the residues Gln192 and Thr190 were the residues with the highest hydrogen bonds formed during the entire simulated trajectories in conditions 1, 2 and 3. Whereas, in condition 4 they were Asp187 and Gln189. All these residues comprise the Linker loop (185–200) region.
As for the S(+)Ibuprofen–Mpro complex, it was seen that were Gly143 and Glu166 in conditions 1 and 4. In condition 2, they were Thr190 and Gln192, and in condition 3 they were Gln189 and Thr190.
Binding free energies
Both MM-GBSA and MM-PBSA analyses were carried out all systems to predict their binding affinities (Tables S1–S16, supplementary material). These energies obtained were averaged and standard deviations were determined.
The calculated ΔG binding energy for the complex R(−)Ibuprofen–SARS-CoV-2 Mpro simulated in the different conditions by MM-GBSA method was found to be −21.5825, −27.0388, −24.3249 and −18.5589, respectively. For MM-PBSA: −21.7934, −26.5194, −22.2433 and −17.3998, respectively. By comparing the binding free energy, it was found that the R(−)Ibuprofen simulated in the condition 2 (0.25 M NaCl) bound more tightly to the SARS-CoV-2 Mpro than the others conditions.
With respect to S(+)Ibuprofen, the calculated ΔG binding energy in the different conditions by MM-GBSA was found to be −17.1715, −27.1710, −15.0095 and −20.0240, respectively. For MM-PBSA: −16.9713, −24.6250, −13.7777 and −21.2142, respectively. Again, it was found that the condition 2 bound more tightly the Ibuprofen to the SARS-CoV-2 Mpro than the others conditions.
Both results indicated that the major favourable contributors were van der Waals (VDWAALS) interactions and electrostatic (EEL) and while the polar component of solvation (ΔG polar) contributed unfavourably to the binding of enantiomers. In most conditions, the total binding energy in the MM-GBSA method was lower compared to the MMPBSA.
In addition, by comparation of the estimated ΔG values we can infer that the ionic strength (NaCl concentration) significantly modifies the binding affinity of Ibuprofen–Mpro complexes, being condition 2 the most favourable. A direct correlation can be observed between the number of hydrogen bonds and the binding energies calculations in the different conditions analysed.
On the other hand, we can observe that in most of the analysed conditions the R(−)Ibuprofen–Mpro complex showed better binding free energy than S(+)Ibuprofen–Mpro. Results that are in agreement with the ΔG data obtained from molecular docking studies and higher number of hydrogen bonds.
Structural analysis
In order to perform a structural analysis of R(−)Ibuprofen–SARS-CoV-2 and S(+)Ibuprofen–SARS-CoV-2 complexes, it was decided to analyse the frustration patterns of the structures obtained from the MD simulations. It has been demonstrated that several functional aspects of proteins present enrichment of highly frustrated interactions (Ferreiro et al., 2007, 2014; Freiberger et al., 2019).
Therefore, local frustration was calculated and analysed with the aim of better understanding the functional regions of SARS-CoV-2 Mpro. In addition, due to the differences found in the binding energies analysed in the previous studies it was of interest to detect differences in frustration under the different simulated environments because they would be directly affecting the protein–ligand interactions.
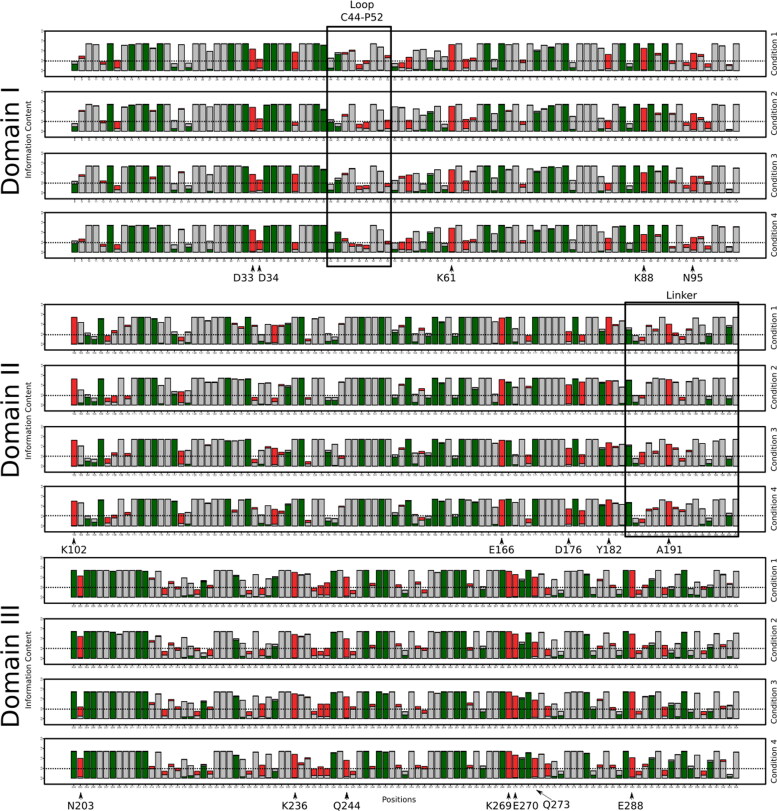
Figure 6 shown the frustration logo for each condition for R(−)Ibuprofen–SARS-CoV-2 complex and S(+)Ibuprofen–SARS-CoV-2 complex in Figure S3, supplementary material. For both complexes, the frustration patterns were very similar and had highly frustrated residues in the three domains.
Figure 6.
Conservation of local frustration for R(−)Ibuprofen–SARS-CoV-2 complex for all conditions. For Domain I (in black square the loop C44-P5), Domain II (in black square Linker Loop) and for Domain III. Marked with narrow residues that are highly frustrated and conserved (Information Content (IC) > 0.5) in all conditions. In green minimally frustrated, in red highly frustrated and in grey neutral residues.
Domain I
In the frustration logos, there are residues that are conserved (Information Content (IC) > 0.5) and highly frustrated in all conditions for both complexes. In Domain I, these residues are Asp33, Lys61 and Asn95. However, some residues are frustrated only in R(−)Ibuprofen–SARS-CoV-2 complex (Figure 6), which are Aps34, Glu55, Asn63 and Lys88; nevertheless, for S(+)Ibuprofen–SARS-CoV-2 these residues are frustrated only in some conditions (Figure S3, supplementary material).
In this domain is located the Loop Cys44-Pro52 which could be regulating the access to the active site because the most important sites for the biological function of SARS-CoV-2 Mpro are located close to this loop (Bzówka et al., 2020). We observed that some residues of this region do not conserve their frustration state.
Domain II
Some residues are highly frustrated and conserved in all conditions for both complexes. These residues are Lys102, Glu166, Asp176 and Tyr182. Whereas, some residues are frustrated only in S(+)Ibuprofen–SARS-CoV-2 complex (Figure S3), which are, Tyr118 and Pro132.
In this domain is located the Linker loop, it is a loop that connects Domain II with Domain III (Bzówka et al., 2020). Both complexes present in this loop some residues that change their state of frustration in different conditions and some do not conserve their state of frustration. Ala191 is conserved and frustrated in all conditions in R(−)Ibuprofen–SARS-CoV-2 complex; although, this residue in S(+)Ibuprofen–SARS-CoV-2 complex is conserved and frustrated in all conditions except in condition 4.
Domain I and II
A recent study suggests that the residues Leu32, Asp33, Asp34, Val35, Tyr37, Gln83, Lys88, Tyr101, Lys102, Phe103, Val104, Arg105, Asp108, Phe159, Cys160, Asp176, Leu177 and Glu178 are located in Domain I and II and they could form an allosteric site where the binding to them could affect the dimerization of the protein (Panagiotopoulos et al., 2020). In our study, we observe that some of these residues are highly frustrated in both complexes and in all conditions, so we consider that these residues would be involved in a biological function.
Domain III
In this domain, there are differences in the frustration of the residues for both complexes. Despite this, the residues Lys236, Gln244, Lys269, Glu270 and Glu288 are conserved and frustrated in both complexes and in all conditions. Asn203 is frustrated and conserved only in R(−)Ibuprofen–SARS-CoV-2 complex. We observe more frustrated residues in S(+)Ibuprofen–SARS-CoV-2 complex than in R(−)Ibuprofen–SARS-CoV-2 complex. We suggest that these differences in frustration are given by the chemical entity of the S(+)enantiomer; as it was mentioned, this domain is implicated in dimerization so this enantiomer could be affecting the stability of the dimer.
Contact level frustration
We analysed the frustration at the contact level (configurational index) for Cys44-Pro52 and Linker Loop regions (see Tables S17 and S18, supplementary material) due to the two enantiomers of Ibuprofen not only form hydrophobic and hydrogen bond interactions with these regions (Figure S1) but also, they have differences in the frustration state (Figure 6, S3).
Loop Cys44-Pro52
Table S17 shown the highly frustrated (IC > 0.5) and conserved contacts (IC_Contact: number of structures in which that contact is established > 0.5) of the loop Cys44-Pro52 for R(−)Ibuprofen–SARS-CoV-2 complex (red) and S(+)Ibuprofen–SARS-CoV-2 complex (black).
The R(−)Ibuprofen–SARS-CoV-2 Mpro complex shows more frustrated interactions in condition 2 and 4. It is observed that only in condition 2 for both complexes, the residue of Pro52 is forming a highly frustrated contact with Arg188, this residue is important for substrate binding and it is located in the Linker loop.
Linker loop
Table S18 shown the highly frustrated (IC > 0.5) and conserved contacts (IC_Contact > 0.5) of the Linker loop for R(−)Ibuprofen–SARS-CoV-2 Mpro complex (red) and S(+)Ibuprofen–SARS-CoV-2 complex (black).
Different highly frustrated contacts were observed under different conditions. The R(−)Ibuprofen–SARS-CoV-2 Mpro complex shows more frustrated interactions in condition 1 and 4. In the majority of the conditions, except condition 2, the highly frustrated contact is between the residues that are forming the Linker loop structure. As mentioned above, in condition 2, the contact Pro52-Arg188 is highly frustrated in both complexes and Met165-Gln192 in S(+)Ibuprofen–SARS-CoV-2 complex.
Due to the highly frustrated interaction between Pro52-Arg188 present in both complexes in condition 2, we suggest that this contact could be provided by the concentration of NaCl and this could be contributing favourably to the protein–ligand interaction.
Conclusions
From in silico analysis performed, we could predict the binding affinities and conformational stability of Ibuprofen enantiomers with the main protease of SARS-CoV-2. We also evaluated the influence of ionic strength (NaCl concentrations) on the affinity and stability of the Ibuprofen–Mpro complexes. The aim was to examine if this known and widely used anti-inflammatory drug may be used in the treatment against COVID-19 infection.
Our docking results showed that both ibuprofen enantiomers it successfully docked against the inhibitor region of the main protease of SARS-CoV-2 virus, with bindings energetically favourable. Both interacting with catalytic residues, the substrate-binding pocket and interacting with residues involved in the dimerization process. Binding free energy calculations using the MM-GBSA and MM-PBSA confirm the affinity of these complexes. It also demonstrates that the ionic strength (NaCl concentrations) modifies significantly their binding affinities.
RMSD and RMSF parameters estimated from trajectories generated by the MD simulations (120 ns) reveal that these complexes are conformational stable in all conditions analysed. In this context, the binding energies analysis, the number of hydrogen bonds along with the RMSD-RMSF results suggest that the condition 2 (0.25 NaCl) bound more tightly the Ibuprofen to the SARS-CoV-2 Mpro than the others conditions.
From the frustration analysis, we could characterize two important regions (Cys44-Pro52 and Linker loop) of this protein involved in the interaction with Ibuprofen. Also, we could find that residues involved in the biological activity of the protein are highly frustrated in all the conditions simulated.
We suggest that in condition 2, the NaCl concentration and the ligand are affecting positively to Loop Cys44-Pro52 and Linker loop structures benefiting protein–ligand interactions.
We conclude that besides the known anti-inflammatory effects that help to reduce the harmful effects of inflammation on the host, Ibuprofen can provide an additional beneficial therapeutic result. Considering that viral respiratory infection therapy has three main purposes: (1) to reduce symptoms; (2) to limit viral involvement of ear, sinus, bronchi and lungs; and (3) to decrease viral replication and spread of infection.
We propose that Ibuprofen may represent a potential treatment option of COVID-19 by directly affecting the replication of the virus, further strengthening the concept of drug repurposing. However, these results require further lab experiments and clinical studies to be carried out to validate its effectiveness. The differences in pharmacological activity between the two isoforms need to be understood.
Supplementary Material
Acknowledgements
This work used computational resources from CCAD – Universidad Nacional de Córdoba (https://ccad.unc.edu.ar/), which are part of SNCAD – MinCyT, República Argentina. All the authors acknowledge and thank their respective Universities and Research Centers for all supporting this research. CCM, RS and BDM acknowledge CONICET and Dr. Nicolás Wolovick from the Facultad de Matemática, Astronomía y Física (FAMAF), Universidad Nacional de Córdoba, Argentina, for providing access to computing resources.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Adams, S. S., Bresloff, P., & Mason, C. G. (1976). Pharmacological differences between the optical isomers of Ibuprofen: Evidence for metabolic inversion of the (—)-isomer. The Journal of Pharmacy and Pharmacology, 28(3), 256–257. 10.1111/j.2042-7158.1976.tb04144.x [DOI] [PubMed] [Google Scholar]
- Adem, S., Eyupoglu, V., Sarfraz, I., Rasul, A., & Ali, M. (2020). Identification of potent covid-19 main protease (Mpro) inhibitors from natural polyphenols: An in silico strategy unveils a hope against corona . Preprints. 10.20944/preprints202003.0333.v1. [DOI] [PMC free article] [PubMed]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020, Apr 30). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–10. [published online ahead of print]. 2020. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzówka, M., Mitusińska, K., Raczyńska, A., Samol, A., Tuszyński, J. A., & Góra, A. (2020). Structural and evolutionary analysis indicate that the SARS-CoV-2 Mpro is a challenging target for small-molecule inhibitor design. International Journal of Molecular Sciences, 21(9), 3099. 10.3390/ijms21093099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D. A., Darden, T. A., Cheatham, T. E., Simmerling, C. L., Wang, J., Duke, R. E., Luo, R., Walker, R. C., Zhang, W., Merz, K. M., Wang, B., Hayik, S., Roitberg, A., Seabra, G., Kolossvary, I., Wong, K. F., Paesani, F., Vanicek, J., Liu, J. … Roberts, B. P. (2020). Amber 20. University of California. [Google Scholar]
- Chen, H., Wei, P., Huang, C., Tan, L., Liu, Y., & Lai, L. (2006). Only one protomer is active in the dimer of SARS 3c-like proteinase. The Journal of Biological Chemistry, 281(20), 13894–13898. 10.1074/jbc.M510745200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, L., Wu, T-q., Xiong, Y-s., Ni, H-b., Ding, Y., Zhang, W-c., Chu, S-p., Ju, S-q., & Yu, J. (2019). Ibuprofen-mediated potential inhibition of biofilm development and quorum sensing in Pseudomonas aeruginosa. Life Sciences, 237, 116947. 10.1016/j.lfs.2019.116947 [DOI] [PubMed] [Google Scholar]
- Das, S., Sarmah, S., Lyndem, S., & Roy, A. S. (2020). An investigation into the identification of potential inhibitors of SARS CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 1–18. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada, E. (2020). Topological analysis of SARS-CoV-2 main protease. Chaos: An Interdisciplinary Journal of Nonlinear Science, 30(6). 10.1101/2020.04.03.023887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, A. M. (2001). Comparative pharmacology of S (+)-Ibuprofen and (RS)-Ibuprofen. Clinical Rheumatology, 20(S1), 9–14. 10.1007/BF03342662 [DOI] [PubMed] [Google Scholar]
- Ferreiro, D. U., Hegler, J. A., Komives, E. A., & Wolynes, P. G. (2007). Localizing frustration in native proteins and protein assemblies. Proceedings of the National Academy of Sciences of the United States of America, 104(50), 19819–19824. 10.1073/pnas.0709915104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro, D. U., Komives, E. A., & Wolynes, P. G. (2014). Frustration in biomolecules. Quarterly Reviews of Biophysics, 47(4), 285–363. 10.1017/S0033583514000092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, A., Sellner, M., Neranjan, S., Smieško, M., & Lill, M. A. (2020). Potential inhibitors for novel coronavirus protease identified by virtual screening of 606 million compounds. International Journal of Molecular Sciences, 21(10), 3626. 10.3390/ijms21103626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration (FDA). (2020, October 22). Press Announcements. https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19.
- Freiberger, M. I., Guzovsky, A. B., Wolynes, P. G., Parra, R. G., & Ferreiro, D. U. (2019). Local frustration around enzyme active sites. Proceedings of the National Academy of Sciences of the United States of America, 116(10), 4037–4043. 10.1073/pnas.1819859116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielczyk, J., Kluitmann, J., Dammeyer, T., & Jördening, H.-J. (2017). Effects of ionic strength on inclusion body refolding at high concentration. Protein Expression and Purification, 130, 100–106. 10.1016/j.pep.2016.10.004 [DOI] [PubMed] [Google Scholar]
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., … Rao, Z. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (New York, N.Y.), 368(6492), 779–782. 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile, D., Patamia, V., Scala, A., Sciortino, M. T., Piperno, A., & Rescifina, A. (2020). Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: A virtual screening and molecular modelling study. Marine Drugs, 18(4), 225. 10.3390/md18040225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, A. K., Brindisi, M., Shahabi, D., Chapman, M. E., & Mesecar, A. D. (2020). Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem, 15(11), 907–932. 10.1002/cmdc.202000223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak, V., Abel, R., Okur, A., Strockbine, B., Roitberg, A., & Simmerling, C. (2006). Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins: Structure, Function, and Bioinformatics, 65(3), 712–725. 10.1002/prot.21123[Mismatch [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakalian, A., Bush, B. L., Jack, D. B., & Bayly, C. I. (2000). Fast, efficient generation of high-quality atomic charges. AM1‐BCC model: I. Method. Journal of Computational Chemistry, 21(2), 132–146. 13. [DOI] [PubMed] [Google Scholar]
- Jakalian, A., Jack, D. B., & Bayly, C. I. (2002). Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. Journal of Computational Chemistry, 23(16), 1623–1641. 10.1002/jcc.10128 [DOI] [PubMed] [Google Scholar]
- Jenik, M., Parra, R. G., Radusky, L. G., Turjanski, A., Wolynes, P. G., & Ferreiro, D. U. (2012). Protein frustratometer: A tool to localize energetic frustration in protein molecules. Nucleic Acids Research, 40(W1), W348–W351. page gks447. 10.1093/nar/gks447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Kandeel, M., & Al-Nazawi, M. (2020). Virtual screening and repurposing of FDA approved drugs against covid-19 main protease. Life Sciences, 251, 117627. 10.1016/j.lfs.2020.117627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D., Chandel, V., Raj, S., & Rathi, B. (2020). In silico identification of potent FDA approved drugs against coronavirus Covid-19 main protease: A drug repurposing approach. Chemical Biology Letters, 7(3), 166–175. [Google Scholar]
- Lagadinou, M., Onisor, M. O., Rigas, A., Musetescu, D.-V., Gkentzi, D., Assimakopoulos, S. F., Panos, G., & Marangos, M. (2020). Antimicrobial properties on non-antibiotic drugs in the era of increased bacterial resistance. Antibiotics (Basel), 9(3), 107. 10.3390/antibiotics9030107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R. A., & Swindells, M. B. (2011). Ligplot+: multiple ligand–protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling, 51(10), 2778–2786. 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- LLC Schrodinger. (2008). The PyMol molecular graphics system version 1.2 r3pre,
- Macchiagodena, M., Pagliai, M., & Procacci, P. (2020). Identification of potential binders of the Main protease 3clpro of the Covid-19 via structure-based ligand design and molecular modeling. Chemical Physics Letters, 750, 137489. 10.1016/j.cplett.2020.137489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, M. R., & Pantano, S. (2020). Split the charge difference in two! a rule of thumb for adding proper amounts of ions in MD simulations. Journal of Chemical Theory and Computation, 16(3), 1367–1372. 10.1021/acs.jctc.9b00953 [DOI] [PubMed] [Google Scholar]
- Menéndez, C. A., Byléhn, F., Perez-Lemus, G. R., Alvarado, W., & de Pablo, J. J. (2020). Molecular characterization of ebselen binding activity to SARS-CoV-2 main protease. Science Advances, 6(37), eabd0345. 10.1126/sciadv.abd0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, B. R., McGee, T. D., Swails Jr, J. M., Homeyer, N., Gohlke, H., & Roitberg A. E. (2012). MMPBSA.py: An efficient program for end-state free energy calculations. Journal of Chemical Theory and Computation, 8(9), 3314–3321. 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
- Mirza, M. U., & Froeyen, M. (2020). Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. Journal of Pharmaceutical Analysis, 10(4), 320–328. 10.1016/j.jpha.2020.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz, A. J., Alasino, R. V., Garro, A. G., Heredia, V., García, N. H., Cremonezzi, D. C., & Beltramo, D. M. (2018). High concentrations of sodium chloride improve microbicidal activity of Ibuprofen against common cystic fibrosis pathogens. Pharmaceuticals, 11(2), 47. 10.3390/ph11020047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obad, J., Šušković, J., & Kos, B. (2015). Antimicrobial activity of ibuprofen: New perspectives on an “old” non-antibiotic drug. European Journal of Pharmaceutical Sciences: official Journal of the European Federation for Pharmaceutical Sciences, 71, 93–98. 10.1016/j.ejps.2015.02.011 [DOI] [PubMed] [Google Scholar]
- O'Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open babel: An open chemical toolbox. Journal of Cheminformatics, 3(1), 33. 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogundeji, A. O., Pohl, C. H., & Sebolai, O. M. (2016). Repurposing of aspirin and ibuprofen as candidate anti-cryptococcus drugs. Antimicrobial Agents and Chemotherapy, 60(8), 4799–4808. 10.1128/AAC.02810-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar, S., Bouziane, I., Bouslama, Z., & Djemel, A. (2020). In-silico identification of potent inhibitors of covid-19 main protease (Mpro) and angiotensin converting enzyme 2 (Ace2) from natural products: Quercetin, hispidulin, and cirsimaritin exhibited better potential inhibition than hydroxy-chloroquine against covid-19 main protease active site and ACE2. ChemRxiv. Preprint. 10.26434/chemrxiv.12181404 [DOI]
- Owen, C. D., Lukacik, P., Strain-Damerell, C. M., Douangamath, A., Powell, A. J., Fearon, D., Brandao-Neto, J., Crawshaw, A. D., Aragao, D., & Williams, M. (2020). Covid-19 main protease with unliganded active site (2019-ncov, coronavirus disease 2019, SARS-CoV-2). RCSB Protein Data Bank (PDB) ID: 6Y84, 3–7. 10.2210/pdb6YB7/pdb [DOI]
- Pan, T., Peng, Z., Tan, L., Zou, F., Zhou, N., Liu, B., Liang, L., Chen, C., Liu, J., Wu, L., Liu, G., Peng, Z., Liu, W., Ma, X., Zhang, J., Zhu, X., Liu, T., Li, M., Huang, X., … Zhang, H. (2018). Nonsteroidal anti-inflammatory drugs potently inhibit the replication of zika viruses by inducing the degradation of AXL. Journal of Virology, 92(20), e01018–18. 10.1128/JVI.01018-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotopoulos, A. A., Kotzampasi, D.-M., Sourvinos, G., Kampa, M., Pirintsos, S., Castanas, E., & Daskalakis, V. (2020). The natural polyphenol fortunellin is a dimerization inhibitor of the SARS-CoV-2 3c-like proteinase, revealed by molecular simulations. arXiv preprint arXiv:2007.07736.
- Papaneophytou, C. P., Grigoroudis, A. I., McInnes, C., & Kontopidis, G. (2014). Quantification of the effects of ionic strength, viscosity, and hydrophobicity on protein-ligand binding affinity. ACS Medicinal Chemistry Letters, 5(8), 931–936. 10.1021/ml500204e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra, R. G., Schafer, N. P., Radusky, L. G., Tsai, M.-Y., Guzovsky, A. B., Wolynes, P. G., & Ferreiro, D. U. (2016). Protein frustratometer 2: A tool to localize energetic frustration in protein molecules, now with electrostatics. Nucleic Acids Research, 44(W1), W356–W360. page gkw304. 10.1093/nar/gkw304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protein Data Bank . Protein data bank. Nature New Biology, 233, 223. 10.1038/newbio233223b0 [DOI] [Google Scholar]
- Rodríguez-Morales, A. J., MacGregor, K., Kanagarajah, S., Patel, D., & Schlagenhauf, P. (2020). Going global - Travel and the 2019 novel coronavirus. Travel Medicine and Infectious Disease, 33, 101578. 10.1016/j.tmaid.2020.101578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe, D. R., & Cheatham, T. E. III. (2013). PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Sanyal, A. K., Roy, D., Chowdhury, B., & Banerjee, A. B. (1993). Ibuprofen, a unique anti-inflammatory compound with anti-fungal activity against dermatophytes. Letters in Applied Microbiology, 17(3), 109–111. 10.1111/j.1472-765X.1993.tb01436.x [DOI] [Google Scholar]
- Shah, P. N., Marshall-Batty, K. R., Smolen, J. A., Tagaev, J. A., Chen, Q., Rodesney, C. A., Le, H. H., Gordon, V. D., Greenberg, D. E., & Cannon, C. L. (2018). Antimicrobial activity of Ibuprofen against cystic fibrosis-associated gram-negative pathogens. Antimicrobial Agents and Chemotherapy, 62(3), e01574–17. 10.1128/AAC.01574-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H., Yao, S., Zhao, W., Li, M., Liu, J., Shang, W., Xie, H., Ke, C., Gao, M., & Yu, K. (2020). Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3cl protease in vitro, bioRxiv. 10.1101/2020.04.13.038687 [DOI]
- Ton, A.-T., Gentile, F., Hsing, M., Ban, F., & Cherkasov, A. (2020). Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds [Epub ahead of print]. Molecular Informatics, 39(8), 2000028. 10.1002/minf.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gunsteren, W. F., & Berendsen, H. J. C. (1977). Algorithms for macromolecular dynamics and constraint dynamics. Molecular Physics, 34(5), 1311–1327. 10.1080/00268977700102571 [DOI] [Google Scholar]
- Veljkovic, V., Goeijenbier, M., Glisic, S., Veljkovic, N., Perovic, V. R., Sencanski, M., Branch, D-a R., & Paessler, S. (2015). In silico analysis suggests repurposing of Ibuprofen for prevention and treatment of ebola virus disease. F1000Research, 4, 104. 10.12688/f1000research.6436.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljkovic, V., Vergara-Alert, J., Segalés, J., & Paessler, S. (2020). Use of the informational spectrum methodology for rapid biological analysis of the novel coronavirus 2019-ncov: Prediction of potential receptor, natural reservoir, tropism and therapeutic/vaccine target. F1000Research, 9, 52. 10.12688/f1000research.22149.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., & Case, D. A. (2004). Development and testing of a general amber force field. Journal of Computational Chemistry, 25(9), 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2020). Assessment of risk factors for coronavirus disease 2019 (Covid-19) in health workers: Protocol for a case-control study, 26 May 2020. Technical Report. World Health Organization. [Google Scholar]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. [published online ahead of print]. Acta Pharmaceutica Sinica B, 10(5), 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., & Curtis, N. (2017). Antimicrobial effects of antipyretics. Antimicrobial Agents and Chemotherapy, 61(4). 10.1128/AAC.02268-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.