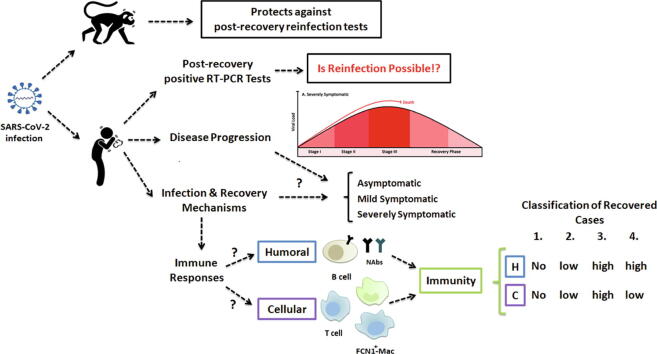
Graphical abstract
Keywords: Coronavirus, COVID-19, Immunity, Recovery, Reinfection, SARS-CoV-2
Abbreviations: ARDS, Acute respiratory distress syndrome; ACE2, Angiotensin-converting enzyme 2; Ang II, Angiotensin II; ADE, Antibody-dependent enhancement; BAL, Bronchoalveolar lavage; COVID-19, Coronavirus disease 2019; ERS, Early recovery stage; FcR, Fc receptor; ISGs, Interferon-stimulated genes; LRS, Late recovery stage; NK, Natural killer; NAb, Neutralizing antibody; N, Nucleocapsid; PBMCs, Peripheral blood mononuclear cells; PSO, Post symptom onset; RT-PCR, Real-time reverse transcriptase–polymerase chain reaction; RBD, Receptor-binding domain; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; sACE2, Soluble ACE2
Abstract
Background
The recent ongoing outbreak of coronavirus disease 2019 (COVID-19), still is an unsolved problem with a growing rate of infected cases and mortality worldwide. The novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is targeting the angiotensin-converting enzyme 2 (ACE2) receptor and mostly causes a respiratory illness. Although acquired and resistance immunity is one of the most important aspects of alleviating the trend of the current pandemic; however, there is still a big gap of knowledge regarding the infection process, immunopathogenesis, recovery, and reinfection.
Aim of Review
To answer the questions regarding “the potential and probability of reinfection in COVID-19 infected cases” or “the efficiency and duration of SARS-CoV-2 infection-induced immunity against reinfection” we critically evaluated the current reports on SARS-CoV-2 immunity and reinfection with special emphasis on comparative studies using animal models that generalize their finding about protection and reinfection. Also, the contribution of humoral immunity in the process of COVID-19 recovery and the role of ACE2 in virus infectivity and pathogenesis has been discussed. Furthermore, innate and cellular immunity and inflammatory responses in the disease and recovery conditions are reviewed and an overall outline of immunologic aspects of COVID-19 progression and recovery in three different stages are presented. Finally, we categorized the infected cases into four different groups based on the acquired immunity and the potential for reinfection.
Key Scientific Concepts of Review
In this review paper, we proposed a new strategy to predict the potential of reinfection in each identified category. This classification may help to distribute resources more meticulously to determine: who needs to be serologically tested for SARS-CoV-2 neutralizing antibodies, what percentage of the population is immune to the virus, and who needs to be vaccinated.
Introduction
The novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the recent ongoing outbreak of coronavirus disease 2019 (COVID-19), initially emerged in the winter of 2019 in China [1]. Presently, by the middle of December 2020, the number of officially confirmed infected cases passed >75 million worldwide, and 1.68 million patients lost their lives by the novel coronavirus [2]. The genome-based analysis revealed a high similarity of SARS-CoV-2 to previously known SARS coronaviruses [1], [3]. Similar to SARS coronavirus, SARS-CoV-2 infects host cells through binding to angiotensin-converting enzyme 2 (ACE2) receptor and causes respiratory illness [4]. Clinical manifestations associated with disease progression typically include fever, dry cough, anosmia, ageusia, mild to severe pneumonia, dyspnea, and coagulopathy [5], [6], [7], [8], [9]. Despite the uninterrupted efforts of scientists around the world, there is still a big gap of knowledge regarding the infection process, clinical symptoms, immunopathogenesis, recovery, and reinfection.
The importance of the studies on the humoral immune responses in COVID-19 patients for vaccine design, antibody-based therapies, and disease management has been emphasized [10]. This review will clarify the role of immune responses in the recovery process of COVID-19 disease, and categorize the infected cases into different groups based on the acquired immunity and possibility of reinfection. Our suggestions will help to improve the health policies for the screening of patients and/or suspected cases and ameliorate diagnostic evaluations. More importantly, this review helps to understand how herd immunity may mitigate future outbreaks of COVID-19. Based on the previous studies, at least 60% of the population needs to acquire protective immunity against COVID-19 through primary recovery or vaccination to achieve herd immunity [11]. Therefore, classification of the infected cases based on the immune responses and possibility of reinfection will help to determine who needs to be evaluated for SARS-CoV-2 neutralizing antibodies (NAb) and what percentage of the population is immune to the SARS-CoV-2, and who needs to be vaccinated. This approach may play a greater role when arguments such as COVID-19 immunity passports and vaccination certificates are brought forward by some governments [12].
Reinfection reports and immunity of macaques
There are several reports of real-time reverse transcriptase-polymerase chain reaction (RT-PCR) based positive tests for COVID-19 patients after recovery and discharge from hospitals in several countries including China and South Korea [13], [14]. Based on reports by the Korea Disease Control and Prevention Agency (KDCA) as early as April 2020, 91 cases of recovered COVID-19 patients were re-identified as positive after being previously discharged from isolation. Since these initial observations, the number has increased to several hundreds of cases [13].
The secondary positive test results were mostly obtained from recovered patients that their full recovery was confirmed by two successive negative RT-PCR test results separated by at least one day [14]. However, it needs to be addressed whether secondary positive SARS-CoV-2 results are due to the reactivation of the previous infection, reinfection, or a false-negative report of the diagnostic tests. Considering the assumption that positive secondary tests are associated with the failure of the immune system for the complete elimination of virus particles or to prevent re-infection with SARS-CoV-2, then a serious concern may arise on the absolute recovery of infected individuals.
Based on a study in rhesus macaques (Macaca mulatta) monkeys, acquired immunity following primary infection with SARS-CoV-2 may protect against subsequent re-infection with the virus [15]. In this study, four monkeys were intratracheally infected with SARS-CoV-2, and clinical symptoms, such as weight loss, body temperature, viral loads in nasal, pharyngeal, and anal swabs, and X-Ray visualized lung pathologies were studied. One of the monkeys was euthanized seven days after infection and various tissues were investigated for virus distribution and histopathological changes. Two of the previously infected monkeys were reinfected with the same dose of virus, 28 days after the primary infection and acquiring recovery. Five days after reinfection, one of the monkeys was euthanized and investigated for viral replication and distribution in tissue samples. Based on the findings, Bao et al. declared that the primary SARS-CoV-2 infection protected monkeys from subsequent exposures [15].
Although the study revealed no indication of COVID-19 disease recurrence and no sign of virus replication after secondary infection, still the findings are insufficient for such a statement. The limited number of experimental animals, lack of preliminary studies to evaluate the minimum infectious doses of the virus for healthy animals, and lack of control test for the infectious potency of the virus are the main constraints of the study. Furthermore, there was no information on the health status of the monkeys before the infection. All monkeys were young which may restrain to extrapolate the results to the human population encountering the COVID-19 pandemic. Finally, in this study, RT-PCR was used to evaluate the shedding status of the monkeys, however, the authors attributed uncertain secondary positive test reports of hundreds of recovered patients to “false negative RT-PCR test results” before the discharging of patients” instead of possible reinfection with the virus after full recovery [15].
Further studies with a larger group of animals with different age ranges and controlled conditions are necessary. In this context, another recent study on rhesus macaques suggests primary infection with SARS-CoV-2 may protect against reinfection [16]. In this study, an animal model of SARS-CoV-2 infection was developed with characteristics such as high viral load in the respiratory tract, pathologic lesions in the lungs, and viral pneumonia. Consequently, 35 days post-infection, the previously infected monkeys (following viral clearance) and naive control animals were inoculated with the virus. Immunologic assessments revealed that the induction of humoral and cellular immune responses following primary infection is responsible for protection against re-exposure to the virus. In the infected monkeys, immunity was provided with SARS-CoV-2 specific humoral and cellular immune responses. The anti-spike and NAb responses against multiple subclasses of viral proteins including receptor-binding domain (RBD), the prefusion spike ectodomain, and the nucleocapsid (N) have been developed with diverse effector functions and virus-neutralizing activities such as antibody-dependent complement deposition and antibody-dependent cellular and neutrophil phagocytosis. The study also exhibited infiltration of immune cells including macrophages, neutrophils, and lymphocytes to multifocal regions of inflammation, and induction of anti-spike CD4+ and CD8+ T cell responses [16]. The study revealed protective immunity against re-exposure in non-human primates, however, the period between viral clearance and the second challenge was too short, therefore immune responses were still highly activated in macaques and the titers of NAb were high [16]. It is difficult to extrapolate these findings because of the rapid decline of immune responses in humans after recovery [17].
It would be beneficial to examine COVID-19 positive cases in cohort studies including asymptomatic, mildly symptomatic, and severely symptomatic cases for the development of humoral immunity and virus-specific neutralizing antibodies during disease and after recovery. Also, when the results of RT-PCR tests in recovered cases are positive, other indices of infection and disease, such as clinical symptoms, serological tests, as well as confirmatory tests (virus isolation or alternative quantitative RT-PCR tests) at several points in time also should be considered.
Humoral immunity in COVID-19 recovery
One of the main protective characteristics of humoral immunity is the production of neutralizing antibodies against pathogens, which boosts the defense and recovery process of the infected body. Neutralizing antibodies efficiently block the entry of viruses into the target cells and may lead to the clearance of virus-infected and antigen displaying cells via the involvement of other immune components such as phagocytes and natural killer cells [18]. Preliminary studies revealed the production of IgM and IgG antibodies within week three post-symptom onset (PSO). The study revealed that humoral immune response developed within 3–7 weeks after infection, with a stepwise increase of IgG and decreasing of IgM. However, serum IgM remained detectable for more than one month PSO in some SARS-CoV-2 infected patients because of the prolonged virus replication [19].
Wölfel et al. performed a virological and serological assessment of nine hospitalized COVID-19 patients. Because of the low frequency of neutralizing antibody titers in coronavirus infected cases, a particularly sensitive plaque-reduction neutralization assay was used [20]. Seroconversion started within the second week of disease onset but was not followed by a rapid decline in viral load. Neutralizing antibodies were detectable in all patients; however, titers showed high variation without close correlation with clinical courses [20]. In a cohort study of 208 COVID-19 positive cases, the early antibody response was detected for anti-N IgM and IgA, with a median detection time of five days PSO [21]. In serum of 77.9% of those COVID-19 positive patients, anti-N IgG was detectable 14 days PSO [21]. Also, Jin et al. showed that the positive rate and titer variance of IgG is higher than those of IgM in COVID-19 patients [22].
Wu et al. analyzed neutralizing antibody responses to SARS-CoV-2 in COVID-19 recovered patients [23]. Blood samples of 175 hospitalized patients with mild pneumonia and a history of discharging from the Shanghai Public Health Clinical Centre were analyzed. The titers of NAbs were measured with high variations within 10–15 days PSO. SARS-CoV-2-specific NAbs targeting spike proteins S1, S2, and RBD were measured using a sensitive pseudotyped-lentiviral-vector-based neutralization assay and ELISA test. Monitoring the kinetics of antibody development in selected six patients at different time points revealed that the titers of NAbs were very low before day 10 (ID50 < 200) and then increased sharply and reached a plateau. Among the recovered patients, ten cases had no detectable anti-spike antibody titers (ID50 < 40) and 30% of patients developed low titers of NAbs (ID50 < 500). Also, 17% and 39% of patients had the titers of medium–low (ID50: 500–999) and medium–high (ID50: 1000–2500) for NAbs respectively and only 14% of patients developed high titers (ID50 >2500) despite the similar duration of disease. Assessment of the NAb titers two weeks after discharge revealed no significant differences from the time of discharge, and also patients without detectable NAb levels did not generate NAbs afterward [23]. Moreover, elderly and middle-aged patients had significantly higher plasma neutralization antibody titers and spike-binding antibodies than young patients, and an age-dependent high amount of anti-spike activity positively correlated with plasma C-reactive protein (CRP) levels and lymphopenia, both considered as markers of COVID-19 disease progression and severity [23]. This may be regarded as a controversial report since disease severity and higher mortality rates are mostly associated with aged COVID-19 patients rather than younger patients [23]. Other studies also revealed that anti-viral serological assay and profile of specific antibodies can assist diagnosis and reflects disease course [21], [22].
Although seroconversion, and especially the production of anti-spike IgG antibodies, may neutralize coronaviruses and prevent their binding to ACE2 receptors, the formation of virus-antibody complexes may also lead to pathologic Fc-receptor (FcR) mediated antibody-dependent enhancement (ADE) responses [24], [25]. Antibodies to S and N proteins may also lead to activation of antiviral effector cells by binding to the expressed peptides and Fcγ receptors on the surface of virally-infected key structural cells such as NK cells and may induce antibody-dependent cellular cytotoxicity and opsonophagocytosis of virus particles [26]. Development of neutralizing IgG antibodies targeting different epitopes of SARS-CoV mediates either immune or pathologic responses in virus-infected and immunized animals [27], [28]. Remarkably, anti-N IgG correlates to more severe lung injuries rather than anti-S IgG antibodies, probably through upregulated secretion of pro-inflammatory cytokines and increasing the infiltration rate of neutrophils and eosinophils [27]. Higher affinity, neutralization capacity, and optimal quantity of neutralizing antibodies may promote virus neutralization and protection [29]. Consistent with these findings, ADE response could provide a rational justification for significantly higher anti-S and anti-N antibodies in elderly and middle-aged patients with severe disease and higher mortality rates [23], [30].
Therefore, not only the presence and quantity of neutralizing antibodies against SARS-CoV-2 is highly variable, but also their quality and the functional outcome may differ among recovered patients. Therefore, humoral immunity and plasma neutralizing activity of the COVID-19 patients with a low amount of NAbs is not enough for complete immunization within the recovery process. Consequently, as humoral immunity is not sufficient for recovery of all COVID-19 cases, the contribution of other protective immunologic factors of recovery should be considered. However, precisely which factors aid in recovery are not fully understood. Thus, further investigations are necessary to understand the full scope of immune protective factors elicited during primary infection. Expanded testing for circulating antibodies in COVID-19 positive cases will provide important information regarding the percentage of those with positive RT-PCR results develop antibodies and clearer associations between antibodies and immunity. In this regard, the determination of the quality and immunizing threshold titer of developed neutralizing antibodies mediating immunity against SARS-CoV-2 reinfection also needs to be understood.
Role of ACE2 in SARS-CoV-2 infectivity and recovery
The symptoms and severity of the disease in COVID-19 patients are highly variable and mostly appear as a respiratory illness. However, some people without any clinical symptoms or detectable signs, including people at the presymptomatic stage, may shed and transmit the virus [5], [31], [32], [33]. It should still be clarified whether some COVID-19 patients may only develop a locally limited infection in the mouth and throat, as ACE2 is highly expressed on the epithelial cells of the oral mucosa [34]. However, in COVID-19 patients, similar to other coronavirus infections, such as severe acute respiratory syndrome (SARS) and middle east respiratory syndrome (MERS), the lung is the most sensitive and vulnerable organ [4], [35]. The SARS-CoV-2 pathogenicity with severe respiratory illness, intensive dyspnea, and fatality is due to the targeting of widely distributed ACE2 receptor in alveolar epithelial cells through viral spike protein [4], [36].
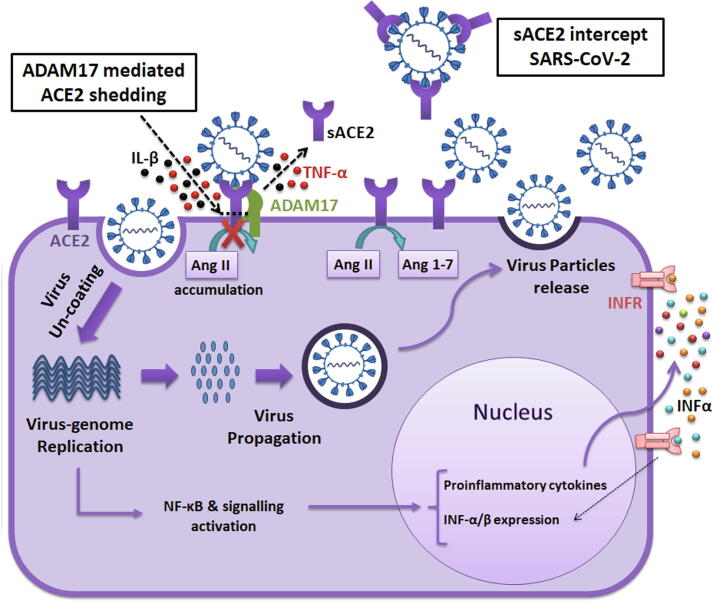
Binding of the virus to the membrane-anchored ACE2 receptors triggers its entry into the cell and subsequently, the virus propagation process will start. ACE2 is a catalytically active protective enzyme in the renin-angiotensin system, which in healthy conditions degrades angiotensin-II peptides [36] (Fig. 1). This receptor is mostly bound to the cell membranes and is barely present in a soluble form in healthy persons [37]. Notably, ADAM17-mediated shedding of sACE2 upon viral spike protein binding to ACE2 receptor and release of some proinflammatory cytokines such as IL-1β and TNFα due to the activity of TNFα-converting enzyme (TACE) upon ACE2 shedding is also described in several studies [38], [39], [40], [41] (Fig. 1). More efficient attachment of SARS-CoV to the membrane-anchored ACE2 is associated with higher induction of ACE2 shedding [42]. Accordingly, similar to SARS coronavirus, the higher affinity of SARS-CoV-2 for the ACE2 receptor might also lead to increased shedding and reduced activity of ACE2 in infected organs [42]. Separation of the ectodomain part of the ACE2 receptor is a byproduct of coronavirus binding and is not necessary for viral entry or spread. However, the ACE2 shedding is associated with down-regulation and reduced expression of ACE2 during infection [42]. It should be taken into account that shedding and subsequent down-regulation of ACE2 receptor upon the attachment of coronavirus, may result in angiotensin-II accumulation (Fig. 1) and development of inflammatory signaling pathway leading to lung injury [36], [42], [43].
Fig. 1.
Target cell infection by SARS-CoV-2 and contribution of ACE2. SARS-CoV-2 uses the ACE2 receptors in target cells, for its entry and infection. ACE2 is a catalytically active protective enzyme, which in normal conditions degrades angiotensin-II peptides. After the attachment of the virus, it enters into the cell and then its genome starts to replication and production of virus proteins, and ACE2 loses its catalytic activity. Upon virus binding to the membrane-anchored ACE2 receptor, ADAM-17 enzyme mediates ACE2 catalytic shedding and release of the sACE2. During this catalytic shedding, the release of some proinflammatory cytokines such as IL-1β and TNFα due to the activity of the TNFα-converting enzyme may also occur. The sACE2 maintains its ability for binding to viral spike protein, so its presence could intercept SARS-CoV-2 and prevents interaction with cell surface ACE2 receptors. Moreover, intracellular virus replication and accumulation of ACE2 substrate (Ang II) activates cell signaling cascades, which may lead to activation of innate immunity receptors by the production of INF-α/β and proinflammatory cytokines. Subsequently, the process of virus propagation and shedding of the infected cells may result in cell damage and apoptosis.
ACE2 deficiency and the presence of high levels of circulating soluble ACE2 (sACE2) are both associated with different disease conditions characterized by increased activity of the renin-angiotensin system, such as hypertension, chronic kidney disease, and heart failure [37], [44], [45]. These conditions have been considered as predisposing factors that promote adverse outcomes and severity of the COVID-19 disease [46]. As the sACE2 maintains its ability for binding to viral spike protein, it has been proposed by some researchers that release of sACE2 from human airway epithelia may limit coronaviruses from interaction with cell surface ACE2 receptors and therefore prevents the spread of the virus inside of the body [40], [47]. Considering the competitive role of sACE2 as an interceptor for viral spike protein (Fig. 1) and the proposed protective effects of recombinant ACE2 against lung injury and SARS infection, sACE2 has been propounded as a novel therapeutic agent to limit the progression of COVID-19 infection [43], [48], [49]. Remarkably, the virus-neutralizing activity associated with the sACE2 shedding into plasma during SARS-CoV-2 infection and with elevated levels of sACE2 in several disease conditions [37], [44], [45] should be studied meticulously. Particularly, the presence of sACE2 in serum needs to be considered because it could influence and enhance the recovery process and intervene in serum-based virus neutralization assays.
Impaired cellular immunity and contribution of inflammatory responses in COVID-19
Convalescence of some COVID-19 patients despite the absence of traceable neutralizing antibodies may suggest the contribution of other possible immune responses, including cell-mediated immunity or release of cytokines [17]. In this context, Qin et al. analyzed and compared peripheral lymphocyte subsets and inflammatory cytokines in the 452 positive cases with severe and non-severe COVID-19 infections [50]. Their study revealed that the severity of the disease is positively correlated with increased levels of inflammatory cytokines and lower numbers of lymphocytes, monocytes, eosinophils, and basophils, and negatively correlated with higher leukocyte counts (including neutrophils). The most affected lymphocytes in COVID-19 patients were T cells including helper (CD4+) T cells, cytotoxic or suppressor (CD8+) T cells, regulatory T cells, and memory T cells [50]. Moreover, in the early stages of SARS-CoV-2 infection, not only the total number of natural killer (NK) and cytotoxic T cells was markedly decreased, also their function was significantly impaired [51].
In several recovered patients, cellular immunity was explored using phenotypical analysis of isolated peripheral blood mononuclear cells (PBMCs) and virus-specific induction of INF-γ among those cells. The higher production of INF-γ in recovered patients was probably associated with a higher number of anti-N and anti-S-RBD specific T cells [17]. However, the high number of anti-SARS-CoV-2 T cells was not maintained two weeks post-discharge [17].
Assessment of PBMSs, during the early recovery stage (ERS), revealed a decreased number of T cells, including CD4+ and CD8+, and an increased level of monocytes including classical CD14++ and CD14++IL1β+. During the late recovery stage (LRS), the monocytes ratio, and the total number of B cells, NK, and T cells were normal again [52]. In another study, specific antiviral CD4+ and CD8+ T cells were detected in 100% and 70% of the recovered patients, respectively. The anti-spike T cell response was robust and positively correlated with the titers of anti-S neutralizing IgG and IgA. Surprisingly, the cross-reactive response may happen against other common circulating coronaviruses, since SARS-CoV-2-reactive CD4+ T cells were detected in 40–60% of non-infected individuals [53].
Importantly, lymphopenia, the decreased count of lymphocytes in peripheral blood samples is one of the common clinical manifestations in COVID-19 patients [5], [50], [54]. Infection of human primary T lymphocytes, lymphocytopenia, and induction of apoptosis pathways by MERS-CoV has been reported [55]. However, compared to SARS-CoV, a significant increase of direct infection of T lymphocytes with SARS-CoV-2 through ACE2 receptor, spike-mediated membrane fusion, upregulation of apoptosis and autophagy also have been found in several studies, which might better explain the lymphopenia [56], [57]. Potential infiltration of T cells into the infected and inflamed tissues, and depletion of T cells due to the direct infection of them, and decimation of the human spleen and lymph nodes are among other justifications for severe lymphopenia [58], [59].
In viral infections, the innate immune system as an important defense barrier develops antiviral activity mediated by the interaction of pathogen-associated molecular patterns with pattern-recognition receptors and stimulating the signaling pathway of INF-α/β production [60]. Although, SARS-CoV-2 infection might induce impaired antigen presentation and interferon responses, production of INF-α/β may cause a local primary inflammatory response in infected tissues and release of proinflammatory cytokines and chemokines leading to both viricidal and tissue damage [5], [61], [62] (Fig. 1). Moreover, the induced expression of interferon-stimulated genes (ISGs) may result in ACE2 upregulation, which is also an ISG and enhance the infection [63].
The assessment of immune cells in lung lavage fluids by single-cell RNA sequencing revealed the existence of highly inflammatory monocyte-derived FCN1+ macrophage cells in severe COVID-19 cases, which may contribute to intense cytokine secretion and storm [64]. Other transcriptome sequencing studies of lung lavage fluid samples from several COVID-19 patients revealed the elevated expression of chemokines (including CXCL1, CXCL2, CCL2, and CCL8) and proinflammatory genes and the chemokine-attracted neutrophils and monocytes [65]. Also, severely infected SARS-CoV-2 patients exhibited a cytokine storm with the intensive release of various pro-inflammatory cytokines and chemokines including IL-6, IL-1β, IL-2, IL-7, IL-8, IL-10, G-CSF, IP-10, MCP-1, MIP1α, and TNFα [7], [50], [66]. This hyper-inflammatory response possibly is associated with dysregulated activation of monocytes and macrophages [67]. Upon seroconversion, the antibodies may bind to the virus, and then via attachment of virus-antibody complex to FcR an FcR mediated ADE response may occur. The ADE response in FcR possessing cells, such as macrophages, monocytes, and myeloid cells, leads to the virus endocytosis and activation of myeloid cells [25], [68], [69], [70]. Also increased ADE response may induce and exacerbate cytokine and chemokine storm, leading to a secondary inflammatory phase, severe lymphopenia, and lung injury [25], [71], [72]. Some studies revealed higher levels of total antibodies in severe COVID-19 patients, which supports the potential role of ADE response in the adverse outcomes of infection [73], [74].
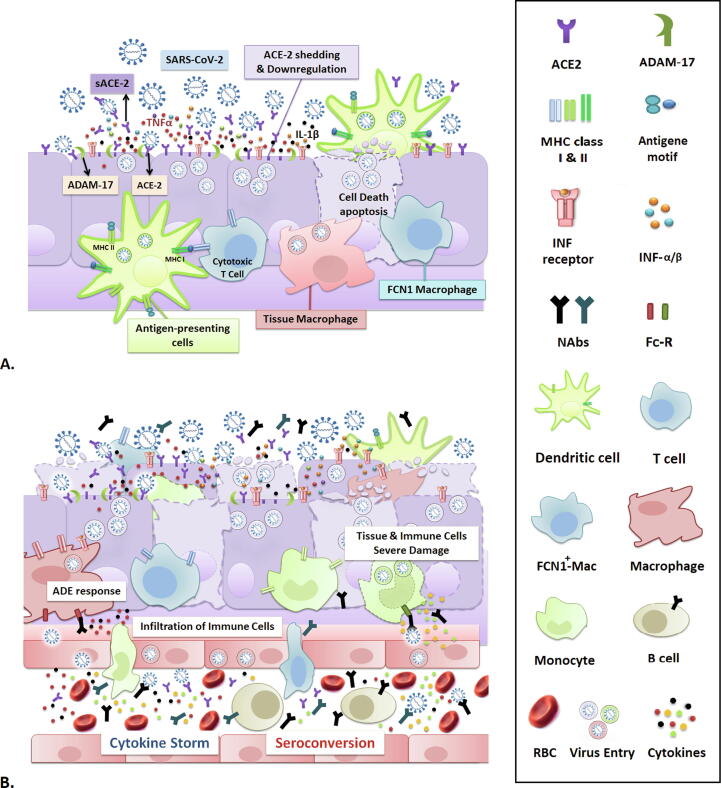
We may postulate that the primary inflammation developed upon viral replication, may lead to ACE2 shedding and release of some proinflammatory cytokines [38], [41]; the resulting tissue damage may recruit immune cells to the infected area, associated with several antiviral cellular immune responses [75] including infiltration and activation of monocyte-derived macrophages [64], neutrophils [65], cytotoxic and suppressor T cells [65], [76], and possibly triggering eosinophil and basophil dependent anti-inflammatory responses [77] (Fig. 2A & B). However, the positive correlation of lymphocytopenia with the severity of the disease, increased CRP level, induction of cytokine storm, seroconversion, and increase of viral replication and inflammation attributable to the FcR-mediated ADE response, support the theory for potential hazardous effects of cellular immune response on final severe lymphocytopenia and dysfunction of the immune system (Fig. 2B). In severe COVID-19 patients, this destructive loop might either synergistically with the application of antiviral and anti-inflammatory drugs influence the decreasing process of viral load and final clearance or may lead to irrecoverable damage and death (Fig. 3). Infection-related destructive effects on the immune and inflammatory responses, such as leukomonocyte counts, functional exhaustion of antiviral lymphocytes, and cytokine release may be normalized after the treatment or during the recovery process [51], [52], [78]. It should still be empirically determined whether the memory T cells may be depleted due to the severe damage of the immune system, as these cells are the main determinant of acquired long-term immunity against reinfection with SARS-CoV-2.
Fig. 2.
Progression of SARS-CoV-2 infection and inflammation. (A) Mild infection and innate immunity responses. SARS-CoV-2 entry and replication may cause primary inflammation through the release of pro-inflammatory cytokines produced by cellular damage and ADAM17-mediated shedding of sACE2, and activation of the interferon pathway. Mild infection possibly occurs before seroconversion and the production of neutralizing IgG. ADE response does not appear in this stage and subsequent events including ACE2 shedding and down-regulation, neutralizing activity of soluble ACE2, low level of inflammation, recruitment of cytotoxic immune cells, activation of innate immune responses, and activity of tissue-resident macrophages may lead to final viral clearance and recovery. (B) Severe infection and cytokine storm. Increased viral replication leads to the upregulation of inflammation and tissue damage, which recruits more immune cells to the infected area. This coincides with seroconversion and development of neutralizing antibodies, resulted in FcR-mediated ADE response upon generation of antibody-virus complexes. ADE response could cause a cytokine storm and may lead to severe lymphopenia via increased infiltration of lymphocytes and finally causes severe damage to immune cells and infected tissue.
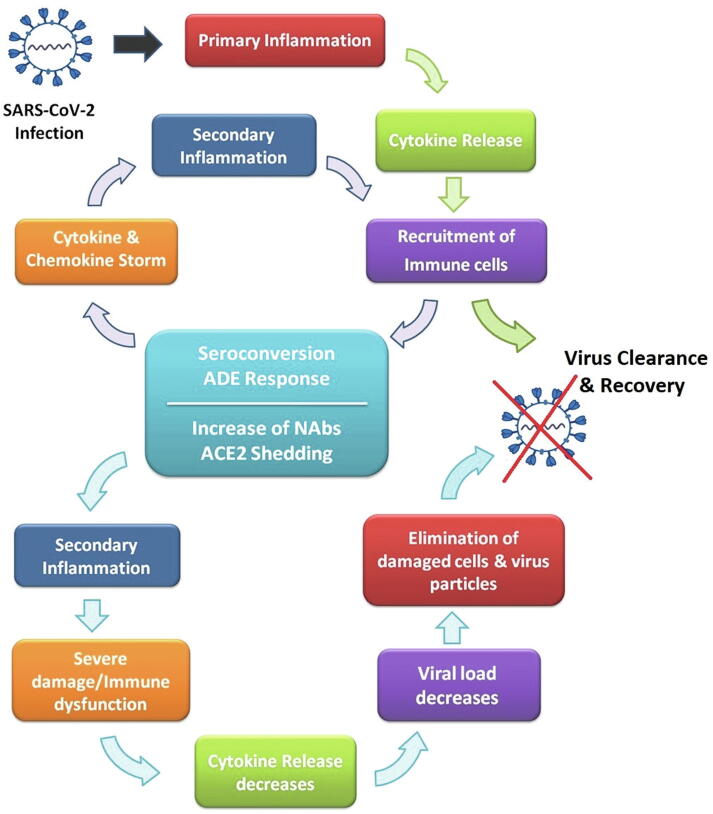
Fig. 3.
The cycle of infection and recovery. Infection loop in mild symptomatic infected cases includes events of primary inflammation, cytokine release, and recruitment of immune cells leading to recovery. In severe symptomatic infected cases, the primary inflammation loop also with the occurrence of seroconversion and ADE response develops cytokine storm and secondary inflammatory responses. In this context, secondary inflammation could cause severe damage and subsequent immune dysfunction. These also lead to loss of ACE2 expressing cells and final ACE2 downregulation, which together with the elevation of neutralizing activity of antibodies and soluble ACE2 resulted in viral clearance and recovery.
An overall view on COVID-19 progression and immunity
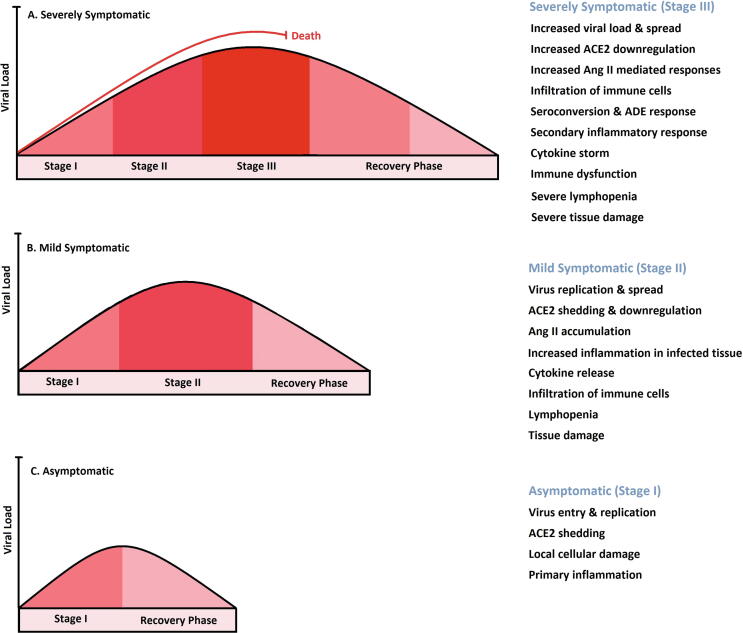
SARS-CoV-2 infection in humans resulted in variable clinical symptoms typically including fever, dry cough, mild pneumonia, anosmia, and less commonly dyspnea, myalgia, headache/dizziness, diarrhea, and nausea [5], [6], [7]. The clinical manifestations are associated with disease progression assorted in asymptomatic, mild to moderately symptomatic, severely symptomatic ending to death or symptoms alleviation, and complete recovery [79]. Approximately, 80% of COVID-19 positive cases exhibit no clinical symptoms or mild to moderate symptoms (with or without mild pneumonia), about 15% progress to severe respiratory disease and 5% develop acute respiratory distress syndrome (ARDS), lung failure, septic shock or multi-organ failure [5], [59], [80].
As depicted in Fig. 4, SARS-CoV-2 infection progress in several stages including (I) asymptomatic incubation period (in average 4–5 days, occasionally more), (II) moderately symptomatic period (10–11.5 days), with various levels and severity of clinical symptoms, (III) severe respiratory symptomatic stage progressing ARDS occurs 8–9 days after symptom appearance and reaches the highest level of viral load [71], [72], [79]. The last stage may lead to death due to respiratory failure and intense hypoxia after day 14 (Fig. 4A). Most severe and hospitalized patients with respiratory failure should be treated with oxygen therapy, mechanical ventilation, and non-specific antiviral and anti-inflammatory drugs [81]. Recovery in COVID-19 patients is characterized by alleviation and disappearing of symptoms, besides viral clearance determined by two negative RT-PCR test results taken at least 24 h apart.
Fig. 4.
COVID-19 disease progression. COVID-19 Infected patients are categorized into groups of severely symptomatic (A), mild symptomatic (B), and asymptomatic (C) based on the clinical manifestations associated with disease progression into different stages. The SARS-CoV-2 infection progresses in several stages including (I) asymptomatic incubation period, (II) moderately symptomatic period, (III) severe respiratory symptomatic stage progressing ARDS. Here disease progress in these categories depicted in graphical curves indicating viral load variations from infection to clearance. Also, the main cellular and molecular mechanisms are addressed for each group.
Based on WHO reports, the median time from symptomatic onset to clinical recovery for mild cases is approximately two weeks and for patients with the severe or critical disease is three to six weeks [82]. Recovery from a severe stage of disease may be slow as symptoms such as cough or pulmonary dysfunction may continue for several weeks due to severe lung damage. Nevertheless, whether the infection may affect the organs such as the brain with non-conventional clinical symptoms after apparent recovery or not, still needs to be clarified. Different possible mechanisms may contribute to virus clearance during the aforementioned stages (I, II, and III) of COVID-19 progression, and innate immune response may play a role as a primary responder at early stages, which activates adaptive immune response [83].
The seroconversion in most COVID-19 patients for total antibody, IgM and IgG, occurred in the second week of disease onset, with delayed seroconversion time for IgG, and was not followed by a rapid decline in viral load [20], [73]. The development of virus-neutralizing antibodies and particularly anti-spike IgG coincides with stage III and ARDS progression due to ADE response (Fig. 2B and Fig. 3). As also seen in SARS-CoV infection, fast and earlier development of antiviral-IgG within the symptom onset with the highest titer in about 14 days, leads to increased mortality rate and severe lung injury [73], [84]. Among most of the recovered patients, virus-neutralizing IgG reaches the highest level within several days after the severity phase [84]. Efficient neutralizing antibody production significantly could intercept viruses and prevents their binding to the target receptors, and leads to lowering viral replication [68]. This approach is applied in most antibody-based vaccination methods too. However, the production of neutralizing antibodies against SARS-CoV-2 may trigger a secondary inflammatory response and cause severe lung damage. Results of several studies using SARS-CoV vaccinated animal models also indicated a higher rate of pulmonary damage associated with increased pulmonary pro-inflammatory response compared to unvaccinated animals [69], [85], [86]. We may assume that adverse outcomes of ADE response due to the interaction of antibodies with the viruses, might also result in pulmonary damages during reinfection with SARS-CoV-2 in previously infected and recovered individuals. However, further studies are needed to clarify this in detail. In this context, a series of prototype DNA vaccines expressing various spike proteins were evaluated for their protective efficacy against intranasal and intratracheal SARS-CoV-2 challenges in rhesus macaques [87]. The vaccine protected monkeys with a substantial reduction in viral loads in bronchoalveolar lavage (BAL) and nasal swabs, and a dramatic reduction of viral replication in upper and lower respiratory tracts. In contrast, the less immunogenic vaccines showed partial protection in BAL but essentially no protection in nasal swabs. This study concluded that protection in both organs is necessary for pandemic control and protection in the upper respiratory tract may be more difficult to achieve [87].
In the context of reinfection, it seems that viral replication in the upper respiratory tract or other entry gates may occur before exposure of the virus to neutralizing antibodies. And high transmissibility of SARS-CoV-2 during respiration and attachment to the ACE2 expressing cells (including airway and alveolar epithelial cells, and macrophages in the lung) may support this process [88]. This may trigger primary pro-inflammatory responses, recruitment of immune cells, and possibly develops secondary inflammatory responses in reinfected tissues, depending on the quality and quantity of the available anti-SARS-CoV-2 antibodies produced from the primary infection. However, generally, this might take a short time for high affinity neutralizing antibodies to increase to sufficient levels or to reach the infected area and act against viruses and prevent further cell entry and replication [29]. In this step, previously acquired specific humoral and cellular immunity against SARS-CoV-2 by and therefore the presence of highly efficient antibodies and virus-specific T cells [53] mediating a fast response could dampen the viral load and lead to rapid viral clearance during reinfection compared with primary infection and non-immunized individuals. This also has been shown in several studies on acquired protection in SARS-CoV-2 infected patients and DNA-vaccinated macaques [15], [16], [87]. In conclusion, the efficacy of antibody-related protection against SARS-CoV-2 reinfection depends on the presence and sufficient amount of protective neutralizing antibodies, and the potency of plasma cells and memory B cells in rapid response against the viral load.
Moreover, non-detectable and low levels of anti-spike antibody titers and plasma neutralizing activity have been reported previously among some COVID-19 recovered patients [23], [73]. This may occur mostly in clinically asymptomatic or mild symptomatic COVID-19 cases and during the pre-symptomatic and incubation stage, before the seroconversion, or in mild symptomatic stage coincide with the beginning of seroconversion and IgG production (Fig. 4B & C). Interestingly, in one study both asymptomatic and mild symptomatic SARS-CoV-2 infected individuals were assessed for clinical manifestations and immunologic responses [89]. Approximately 80% of asymptomatic individuals demonstrated a significantly low level of virus-specific IgG and NAbs, compared to the symptomatic cases. The reduction in IgG and NAb levels has been seen during the early convalescent phase (8 weeks after discharge) in both groups. The average percent of decline for IgG and NAb levels in the asymptomatic group was 71.1% and 8.3%, and for the symptomatic group was 76.6% and 11.7%, respectively. Consequently, in this phase, 40 percent of the asymptomatic group and 12.9 percent of the symptomatic group became seronegative for IgG. Furthermore, the measurement of 32 cytokines and chemokines in plasma revealed no significant differences among healthy individuals and asymptomatic SARS-CoV-2 positive cases, indicating a low level or no induction of inflammatory responses in them [89]. Also, asymptomatic cases revealed a longer duration of virus shedding (median 19 days) in comparison with symptomatic cases (median 14 days) assessed by RT-PCR testing. However, the longer viral shedding confirmed by RT-PCR may be seen as a result of the presence of remnants of the virus genome in the sampling area or false-positive tests, and not because of the presence of culturable infectious virus particles [89], [90].
In these infected individuals, viral clearance may occur before the seroconversion or IgG production reached its peak due to the possible dysfunction of the immune system in antigen presentation, induction of T helper cells, and activation of antibody-producing cells. We may assume that recovery in these COVID-19 positive cases might be due to the contribution of some molecular events including ACE2 shedding and down-regulation [42], neutralizing activity of soluble ACE2 [47], [48], [49], low level of inflammation [89], the successful activity of tissue-resident macrophages and infiltrated monocyte-derived macrophages in depletion of virus-infected cells ending to decreased viral replication and clearance [64], [67]. Interestingly, SARS-CoV-2-specific T cells in unexposed individuals to SARS-CoV-2 also have been reported [53]. The existence of cross-reactive T cell response against the SARS-CoV-2 virus probably may indicate the presence of cellular immunity in a part of the population that were previously infected with other coronaviruses. The possible cross-protective immunity might also result in a higher number of asymptomatic and mild symptomatic COVID-19 cases and influence on the ongoing pandemic [53]. In conclusion, in most patients after recovery, humoral immunity may not be acquired or would be very low level. Also, as reported by several studies, the amount of NAbs is correlated with virus-specific T cell response, and therefore low humoral immune response may demonstrate a low level of acquired cellular immunity too [17], [53]. In these SARS-CoV-2 infected cases, reinfection might be possible, and in case of reinfection, the disease may progress into the severe stages.
The consecutive events occurring in severely symptomatic individuals may probably include: infection with SARS-CoV-2 and replication of the virus, ACE2 downregulation, and angiotensin-II accumulation, triggering of cellular damage and primary inflammation, cytokine release and infiltration of immune cells to the inflamed tissues, seroconversion and production of anti-spike IgG, formation of virus-antibody complex and subsequent ADE response, secondary inflammatory response and cytokine storm, severe immune response feedback and subsequent severe lymphocytopenia, highly severe damage to infected tissues, ARDS and respiratory or multi-organ failure (Fig. 2B).
In the recovery scenario, with starting the treatment of severely symptomatic individuals (those with severe tissue damage and immune dysfunction), the release of cytokines will together with the increase of neutralizing antibodies lead to viral load decline, restoration of peripheral blood lymphocyte count, the activity of cytotoxic T cells and neutrophils in the elimination process of damaged cells and virus-antibody complexes. Finally, complete viral clearance, the disappearance of clinical symptoms, the healing process of damaged tissues, and final recovery may be achieved (Fig. 3). A decrease of IgG and NAb levels and a remarkable reduction of SASR-CoV-2 specific CD4+ and CD8+ T cells after recovery have also been reported [17], [52], [89]. In the recovery process, a short-term immunity will accomplish by the production of efficient neutralizing antibodies; however, a long-term immunity and protection against reinfection mainly depend on the presence and survival of memory T cells and memory B cells, despite severe damage.
Classification of infected cases
As a conclusion, we may classify the recovered and immunized individuals into the following categories:
-
1)
Infected cases with very mild symptoms or asymptomatic without any humoral immune response or elicited memory.
-
2)
Infected cases with mild to moderate symptoms with low humoral immunity and low cellular immunity.
-
3)
Infected cases with moderate or severe symptoms with highly activated humoral immunity and elicited memory.
-
4)
Infected cases with moderate or severe symptoms with highly activated humoral immunity and low cellular immunity.
Among these categories, reinfection may happen in groups 1 and 2, which may also develop the severe disease in the future due to the absence or low levels of acquired immunity. Individuals in group 3 are more protective against further exposures and they may show long-term immunity since they develop increased elicited memory in defense of SARS-CoV-2. Meanwhile, this group also could be further evaluated to investigate the efficacy, half-life, and epitope specificity of antibodies. Although the last group may show rapid response against reinfection; they may not be safe for longer periods because of the non-imprinted memory of immunity. More investigations need to determine the real potency of acquired humoral and cellular immunity and efficacy of vaccines against SARS-CoV-2 further exposures. Also, the pre-existing cross-reactive T cell response [53] against SARS-CoV-2 may also increase the efficacy of acquired cellular immunity. Several studies regarding herd immunity have been shown that at least 60% of the population should be immunized to prevent further outbreaks of COVID-19 in the future [11], [91]. Our study may help to identify which groups are immunized naturally and which groups still need to be vaccinated for developing herd immunity.
Conclusion and perspective
In this review, important molecular immunologic aspects of recovery of COVID-19 patients were described and the importance of ACE2 in immune responses to the virus was addressed. The infected individuals were classified into different categories including recovered patients with higher humoral and cellular immunity, with low humoral and cellular immunity, and also infected cases without any significant immunity. Therefore, individuals with no previous infection history, and also infected cases with low or without humoral or cellular immune responses or low-elicited memory should be considered as special targets of vaccination programs. This article will help policymakers to make the right decisions for screening, lockdown, and issue health certificates ensuring improved diagnostic evaluations, due to clarifying the recovery and immunity process, and estimation of immunized and non-immunized COVID-19 infected individuals. Besides, we suggest further investigation for the development of efficient virus-specific NAbs and generation of memory cells for SARS-CoV-2 infected cases after recovery.
Funding
B.P.L. is supported by grants from the United States National Institutes of Health (R01ES004862, R01ES030300, R01ES023260, and P30ES01247). Other authors received no specific funding for this work.
Declaration of Competing Interest
The authors declare that they have no actual or potential conflict of interests.
Biographies

Mrs. Zahra Khoshkam is a Ph.D. candidate in Molecular and Cell Biology at the University of Tehran under the supervision of Prof. Mehran Habibi-Rezaei. She is working on the neuroinflammatory effects of city-air pollutants and their interaction with neurodegenerative diseases.

Dr. Younes Aftabi is a researcher at the Tuberculosis and Lung Diseases Research Center, Tabriz University of Medical Sciences. He obtained his Ph.D. in Molecular and Cell Biology and currently studies the molecular mechanisms and genetic predispositions of lung diseases.

Prof. Peter Stenvinkel is a senior lecturer at Karolinska University Hospital and Professor of Nephrology at Karolinska Institutet. He conducts translational research with a focus on risk factors for metabolic, cardiovascular, and nutritional complications in chronic kidney disease (CKD). The ultimate goal of his studies is the translation of basic research into novel and improved therapeutic approaches to arrest premature ageing processes in CKD and to decrease the unacceptable high mortality and at the same time improve quality of life for this expanding patient group.

Prof. B. Paige Lawrence is a Professor at the University of Rochester School of Medicine, where she holds appointments in the Departments of Environmental Medicine and Microbiology & Immunology. Dr. Lawrence's expertise is in the disciplines of immunology and toxicology. She has had a long-standing interest in public outreach and science education. Her research lines include Environmental signaling, immune function, and cellular development, the aryl hydrocarbon receptor pathway, neonatal oxygen supplementation, and respiratory viral infection.

Prof. Mehran Habibi-Rezaei is a Professor at the University of Tehran, working on protein science, oxidative stress-related diseases, and neurodegenerative diseases. His lab presently contributes to SARS-CoV-2 studies.

Prof. Gaku Ichihara is a Professor at the Department of Occupational and Environmental Health, Faculty of Pharmaceutical Sciences, Tokyo University of Science, Noda, Japan. He is an expert in Occupational and Environmental Health, Neurotoxicology, and Nanotoxicology.

Dr. Sasan Fereidouni is a Senior Scientist at the Department of Interdisciplinary Life Sciences, University of Veterinary Medicine Vienna. He is an expert in One health, Virology, and Immunopathogenesis with more emphasis on emerging, re-emerging, and zoonotic viruses such as Influenza viruses, Paramyxoviruses, West Nile viruses, and Coronaviruses.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Younes Aftabi, Email: aftabiy@tbzmed.ac.ir.
Sasan Fereidouni, Email: sasan.fereidouni@vetmeduni.ac.at.
References
- 1.Zhou P., Lou Y.X., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Coronavirus Disease (COVID-19) Dashboard | WHO Coronavirus Disease (COVID-19) Dashboard n.d. https://covid19.who.int/ (accessed December 21, 2020).
- 3.Li C, Yang Y, Ren L. Genetic evolution analysis of 2019 novel coronavirus and coronavirus from other species. Infect Genet Evol 2020;82. https://doi.org/10.1016/j.meegid.2020.104285. [DOI] [PMC free article] [PubMed]
- 4.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pacific J Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGonagle D., O’Donnell J.S., Sharif K., Emery P., Bridgewood C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. Lancet Rheumatol. 2020;2:e437–e445. doi: 10.1016/S2665-9913(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaira L.A., Salzano G., Deiana G., De Riu G. Anosmia and Ageusia: Common Findings in COVID-19 Patients. Laryngoscope. 2020;130:1787. doi: 10.1002/lary.28692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siracusano G., Pastori C., Lopalco L. Humoral Immune Responses in COVID-19 Patients: A Window on the State of the Art. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altmann D.M., Douek D.C., Boyton R.J. What policy makers need to know about COVID-19 protective immunity. Lancet. 2020;395:1527–1529. doi: 10.1016/S0140-6736(20)30985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phelan A.L. COVID-19 immunity passports and vaccination certificates: scientific, equitable, and legal challenges. Lancet. 2020;395:1595–1598. doi: 10.1016/S0140-6736(20)31034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahase E. Covid-19: WHO and South Korea investigate reconfirmed cases. BMJ. 2020;369 doi: 10.1136/bmj.m1498. m1498. [DOI] [PubMed] [Google Scholar]
- 14.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y. Positive RT-PCR Test Results in Patients Recovered from COVID-19. JAMA - J Am Med Assoc. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bao L., Deng W., Gao H., Xiao C., Liu J., Xue J. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. BioRxiv. 2020;2020(03) doi: 10.1101/2020.03.13.990226. pp. 13.990226. [DOI] [Google Scholar]
- 16.Chandrashekar A, Liu J, Martinot AJ, McMahan K, Mercado NB, Peter L, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science (80-) 2020:eabc4776. https://doi.org/10.1126/science.abc4776. [DOI] [PMC free article] [PubMed]
- 17.Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity. 2020;52(971–977) doi: 10.1016/j.immuni.2020.04.023. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasui F., Kohara M., Kitabatake M., Nishiwaki T., Fujii H., Tateno C. Phagocytic cells contribute to the antibody-mediated elimination of pulmonary-infected SARS coronavirus. Virology. 2014;454–455:157–168. doi: 10.1016/j.virol.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao A.T., Gao C., Zhang S. Profile of specific antibodies to SARS-CoV-2: The first report. J Infect. 2020;0. doi: 10.1016/j.jinf.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 21.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19) Clin Infect Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin Y., Wang M., Zuo Z., Fan C., Ye F., Cai Z. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49–52. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S. Neutralizing Antibody Responses to SARS-CoV-2 in a COVID-19 Recovered Patient Cohort and Their Implications. SSRN Electron J. 2020 doi: 10.2139/ssrn.3566211. [DOI] [Google Scholar]
- 24.Yang Z.Y., Werner H.C., Kong W.P., Leung K., Traggiai E., Lanzavecchia A. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci U S A. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Y., Cheng Y., Wu Y. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol Sin. 2020;35:266–271. doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.French M.A., Moodley Y. The role of SARS-CoV-2 antibodies in COVID-19: Healing in most, harm at times. Respirology. 2020;25:680–682. doi: 10.1111/resp.13852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasui F., Kai C., Kitabatake M., Inoue S., Yoneda M., Yokochi S. Prior Immunization with Severe Acute Respiratory Syndrome (SARS)-Associated Coronavirus (SARS-CoV) Nucleocapsid Protein Causes Severe Pneumonia in Mice Infected with SARS-CoV. J Immunol. 2008;181:6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H., Li Y., Zhang H., Wang W., Men D., Yang X. Global profiling of SARS-CoV-2 specific IgG/ IgM responses of convalescents using a proteome microarray. MedRxiv. 2020;2020(03) doi: 10.1101/2020.03.20.20039495. pp. 20.20039495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baud D., Qi X., Nielsen-Saines K., Musso D., Pomar L., Favre G. Real estimates of mortality following COVID-19 infection. Lancet Infect Dis. 2020;20:773. doi: 10.1016/S1473-3099(20)30195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Z., Song C., Xu C., Jin G., Chen Y., Xu X. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing. China. Sci China Life Sci. 2020;63:706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:1–5. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert D.W., Yarski M., Warner F.J., Thornhill P., Parkin E.T., Smith A.I. Tumor necrosis factor-α convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J Biol Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel V.B., Clarke N., Wang Z., Fan D., Parajuli N., Basu R. Angiotensin II induced proteolytic cleavage of myocardial ACE2 is mediated by TACE/ADAM-17: A positive feedback mechanism in the RAS. J Mol Cell Cardiol. 2014;66:167–176. doi: 10.1016/j.yjmcc.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 40.Hong P.J., Look D.C., Tan P., Shi L., Hickey M., Gakhar L. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am J Physiol - Lung Cell Mol Physiol. 2009;297. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc Natl Acad Sci U S A. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O. Differential Downregulation of ACE2 by the Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus and Human Coronavirus NL63. J Virol. 2010;84:1198–1205. doi: 10.1128/jvi.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H., Baker A. Recombinant human ACE2: Acing out angiotensin II in ARDS therapy. Crit Care. 2017;21:4–6. doi: 10.1186/s13054-017-1882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wysocki J., Goodling A., Burgaya M., Whitlock K., Ruzinski J., Batlle D. Urine RAS components in mice and people with type 1 diabetes and chronic kidney disease. Am J Physiol - Ren Physiol. 2017;313:F487–F494. doi: 10.1152/ajprenal.00074.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Úri K., Fagyas M., Siket I.M., Kertész A., Csanádi Z., Sándorfi G. New perspectives in the renin-angiotensin-aldosterone system (RAAS) IV: Circulating ACE2 as a biomarker of systolic dysfunction in human hypertension and heart failure. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0087845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan W.J., Liang W.H., Zhao Y., Liang H.R., Chen Z.S., Li Y.M. Comorbidity and its impact on 1,590 patients with Covid-19 in China: A nationwide analysis. Eur Respir J. 2020;55 doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ciaglia E., Vecchione C., Puca A.A. COVID-19 Infection and Circulating ACE2 Levels: Protective Role in Women and Children. Front Pediatr. 2020;8 doi: 10.3389/fped.2020.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batlle D., Wysocki J., Satchell K. Soluble angiotensin-converting enzyme 2: A potential approach for coronavirus infection therapy? Clin Sci. 2020;134:543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 49.Hofmann H., Geier M., Marzi A., Krumbiegel M., Peipp M., Fey G.H. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem Biophys Res Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan. China. Clin Infect Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:1–18. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020;181:1489-1501.e15. https://doi.org/10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed]
- 54.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J. Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med. 2020;2020(02) doi: 10.1101/2020.02.06.20020974. pp. 06.20020974. [DOI] [Google Scholar]
- 55.Chu H., Zhou J., Wong B.H.Y., Li C., Chan J.F.W., Cheng Z.S. Middle East Respiratory Syndrome Coronavirus Efficiently Infects Human Primary T Lymphocytes and Activates the Extrinsic and Intrinsic Apoptosis Pathways. J Infect Dis. 2016;213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X., Xu W., Hu G., Xia S., Sun Z., Liu Z. SARS-CoV-2 infects T lymphocytes through its spike protein-mediated membrane fusion. Cell Mol Immunol. 2020:2–4. doi: 10.1038/s41423-020-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Guo D. Transcriptomic Characteristics of Bronchoalveolar Lavage Fluid and Peripheral Blood Mononuclear Cells in COVID-19 Patients. SSRN Electron J. 2020 doi: 10.2139/ssrn.3549993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.chen yongwen, Feng Z, Diao B, Wang R, Wang G, Wang C, et al. The Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Directly Decimates Human Spleens and Lymph Nodes. MedRxiv 2020;2:2020.03.27.20045427. https://doi.org/10.1101/2020.03.27.20045427.
- 59.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fung T.S., Liu D.X. Human coronavirus: Host-pathogen interaction. Annu Rev Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 61.Yang M. Cell Pyroptosis, a Potential Pathogenic Mechanism of 2019-nCoV Infection. SSRN Electron J. 2020 doi: 10.2139/ssrn.3527420. [DOI] [Google Scholar]
- 62.Taefehshokr N., Taefehshokr S., Hemmat N., Heit B. Covid-19: Perspectives on Innate Immune Evasion. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.580641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181(1016–1035):e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. MedRxiv. 2020;2020(02) doi: 10.1101/2020.02.23.20026690. pp. 23.20026690. [DOI] [Google Scholar]
- 65.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;27(883–890):e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaume M., Yip M.S., Cheung C.Y., Leung H.L., Li P.H., Kien F. Anti-Severe Acute Respiratory Syndrome Coronavirus Spike Antibodies Trigger Infection of Human Immune Cells via a pH- and Cysteine Protease-Independent Fc R Pathway. J Virol. 2011;85:10582–10597. doi: 10.1128/jvi.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang S.F., Tseng S.P., Yen C.H., Yang J.Y., Tsao C.H., Shen C.W. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease. Clin Infect Dis. 2019;2020:1–22. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang B., Zhou X., Zhu C., Feng F., Qiu Y., Feng J. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. MedRxiv. 2020;2020(03) doi: 10.1101/2020.03.12.20035048. pp. 12.20035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKechnie J.L., Blish C.A. The Innate Immune System: Fighting on the Front Lines or Fanning the Flames of COVID-19? Cell Host Microbe. 2020;27:863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Agrati C., Sacchi A., Bordoni V., Cimini E., Notari S., Grassi G. Expansion of myeloid-derived suppressor cells in patients with severe coronavirus disease (COVID-19) Cell Death Differ. 2020:1–12. doi: 10.1038/s41418-020-0572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Isobe Y., Kato T., Arita M. Emerging roles of eosinophils and eosinophil-derived lipid mediators in the resolution of inflammation. Front Immunol. 2012;3:270. doi: 10.3389/fimmu.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen X., Ling J., Mo P., Zhang Y., Jiang Q., Ma Z. Restoration of leukomonocyte counts is associated with viral clearance in COVID-19 hospitalized patients. MedRxiv. 2020;2020(03) doi: 10.1101/2020.03.03.20030437. pp. 03.20030437. [DOI] [Google Scholar]
- 79.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA - J Am Med Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S. Risk Factors Associated with Acute Respiratory Distress Syndrome and Death in Patients with Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aylward B., Liang W. Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19) WHO-China Jt Mission Coronavirus Dis. 2019;2019(2020):16–24. https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf [Google Scholar]
- 83.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M. Immunology of COVID-19: Current State of the Science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang L., Zhang F., Yu W., He T., Yu J., Yi C.E. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol. 2006;78:1–8. doi: 10.1002/jmv.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M. A Double-Inactivated Severe Acute Respiratory Syndrome Coronavirus Vaccine Provides Incomplete Protection in Mice and Induces Increased Eosinophilic Proinflammatory Pulmonary Response upon Challenge. J Virol. 2011;85:12201–12215. doi: 10.1128/jvi.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tseng C Te, Sbrana E, Iwata-Yoshikawa N, Newman PC, Garron T, Atmar RL, et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One 2012;7. https://doi.org/10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed]
- 87.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science (80-) 2020;6284:eabc6284. https://doi.org/10.1126/science.abc6284. [DOI] [PMC free article] [PubMed]
- 88.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020:1–5. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 90.Atkinson B., Petersen E. SARS-CoV-2 shedding and infectivity. Lancet. 2020;395:1339–1340. doi: 10.1016/S0140-6736(20)30868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guerra F.M., Bolotin S., Lim G., Heffernan J., Deeks S.L., Li Y. The basic reproduction number (R0) of measles: a systematic review. Lancet Infect Dis. 2017;17:e420–e428. doi: 10.1016/S1473-3099(17)30307-9. [DOI] [PubMed] [Google Scholar]