Abstract
The newly emerged coronavirus (SARS-CoV-2) continues to infect humans, and no effective treatment has yet been found. Antibody therapy is one way to control infection caused by COVID-19. However, the use of classical antibodies raises complex issues. Heavy chain antibodies (HCAbs) are single-domain antibodies derived from the Camelidae family. The variable part of these antibodies (Nanobodies or VHH) has interesting properties such as small size, cost-effective production, and good tissue permeability, causing VHH to be regarded as an antiviral therapeutics. However, the small size of nanobodies may lead to low antigen binding affinity and rapid renal clearance. In this systematic review, the application of nanobodies in the treatment of COVID-19 infection and other similar infections (MERS and SARS) was reviewed.
Keywords: SARS-CoV-2, COVID-19, Nanobodies, VHH
Highlights
-
•
The clinical use of classical antibodies raises complex issues.
-
•
Nanobodies as the single-chain antibodies are now widely used and tested in the treatment of many diseases.
-
•
Many nanobodies have been produced against various viruses.
-
•
The nanobodies can be considered as prominent agents to treat COVID-19 infectious disease.
1. Introduction
1.1. Description of the new corona crisis
As a mattered pathogen, coronaviruses (CoVs) can infect various organs, including the respiratory tract, liver, gastrointestinal tract, and nervous system [1]. The emergence of Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) diseases in 2002 and 2012, respectively, are associated with the coronavirus [2,3]. In December 2019, a new coronavirus (2019-nCoV) was identified. The new coronavirus, also named SARS-CoV-2, is known as a public health problem worldwide.
1.2. New corona detection and treatments problems
Currently, there is a lack of appropriate medication for COVID-19 infection. Prevention or treatment for COVID-19 includes antibody therapy and the use of drugs such as interferon and other antiviral drugs [4]. The use of methods that can specifically detect viruses and act against them can be highly helpful. Almost all related sciences are concurrently scrambling to come up with applicable approaches to put out the fire of such crisis. For instance nanodrugs as innovative products of nanotechnology show a set of advantages [5]. Likewise, in the fight against COVID-19, antiviral nano-formulated drugs could prevent the replication of viruses, inhibit apoptotic cellular death caused by viruses, or induce an immune reaction against the SARS-CoV-2 infection [6]. Nevertheless, owing to the ability of antibodies to detect and neutralize viruses, they are totally trustworthy to be used as diagnostic and therapeutic agents against viruses.
1.3. Nanobodies potentials
Nanobodies are a new class of the recombinant antibody derived from a particular type of antibodies called heavy-chain antibodies existing in camels and sharks. Unlike traditional antibodies, the variable domain of these types of antibodies is made of a single region [7]. These types of antibodies have specific properties distinguishing them from other types of antibodies such as small size, higher solubility and resistance to denaturation, stability in intolerable conditions (high and low pH, high temperature), recognition of the broad diversity of epitopes, fast tissue permeability, low immunogenicity due to high sequence accommodation with human antibodies caused, and cost-effective production [[7], [8], [9], [10]].
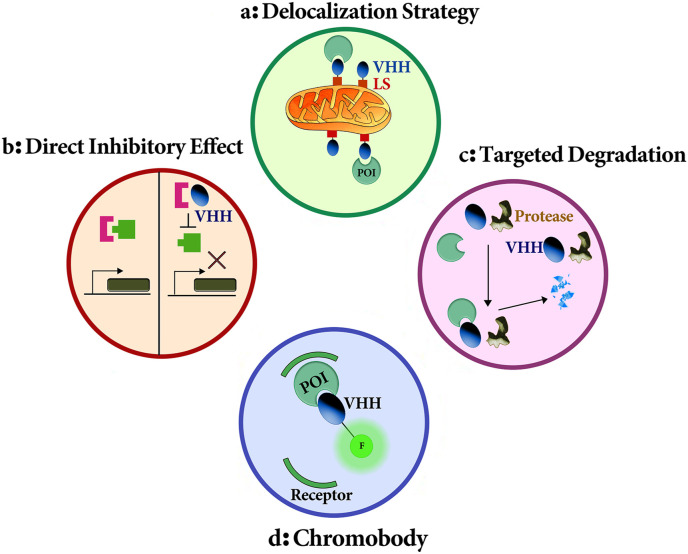
The mechanisms of action of antibodies are include: delocalization strategy, direct inhibitory effect, targeted degradation, chromobody, and detection of protein-protein interactions (Fig. 1 ) [11].
Fig. 1.
A schematic picture of nanobody mechanisms. (a) By binding nanobodies to a localization signal (LS), the protein of interest (POI) can be enriched at a specific cellular site, such as mitochondria, (b) Nanobodies can perform a direct inhibitory effect and prevent from a protein activity, (c) Nanobodies can be used to target a for enzymatic degradation, (d) Fusion between the nanobody and the fluorescent protein (FP) results in the formation of a chromobody. POI can be visualized and traced using this chromobody.
Another type of nanobodies called VNAR has been discovered in sharks that is structurally similar to camel nanobodies and has special advantages. First, the advantage of the Shark VNAR is that in the evolutionary relationship, it is far from mammals on the phylogenetic tree. Owing to the large differences between the two species epitopes, it is possible to produce high affinity and selectivity antibodies against most mammalian antigens in the shark's body. Second, longer CDR3 regions in camel and shark nanobodies lead to higher diversity and improved cryptic epitope detection ability of this type of antibody compared to the classical ones. Third, to maintain the osmotic pressure in seawater, shark blood has a high concentration of urea, causing shark antibodies to be highly resistant to this chaotropic detergent [7,8].
Nanobodies derived from animal heavy chain antibodies may cause immunogenic reactions in the human body. Although VHH domains are very similar to their human equivalent, but for therapeutic applications, humanization strategies have been developed to adapt the sequence of amino acid in the framework regions to their human heavy chain variable domain counterparts [12,13].
Also, due to the simple structure of nanobodies and the low complexity of these proteins, nanobodies can be produced in prokaryotic and eukaryotic hosts. Strong production efficiency makes nanobody a powerful tool for defeating infectious disease pandemics [12]. In one study by Gai et al., a specific nanobody against SARS-CoV-2 was produced in Pichia pastoris, and the yield of target protein in fermentation supernatant has even reached 20 g/L [14].
1.4. Neutralizing nanobody in viral infection
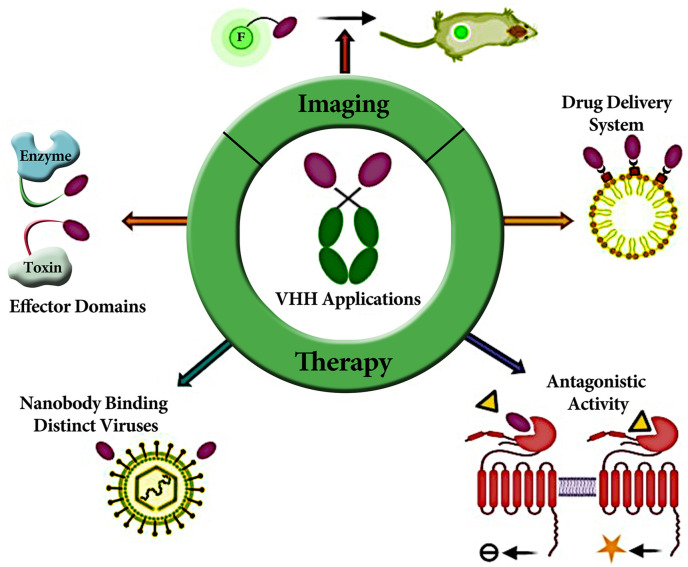
Nanobodies are now widely used and tested in the treatment of many diseases (Fig. 2 ). Currently, many nanobodies have been produced against various viruses such as Hepatitis B virus, influenza virus, poliovirus, rabies virus, HIV, respiratory syncytial virus, FMDV, rotavirus [[15], [16], [17], [18]]. RBD (receptor-binding domain) is the most important domain of the corona virus family to bind with the cell surface receptor and enter the cell. Nanobodies may be able to detect this domain and prevent the virus from entering the cell.
Fig. 2.
Application of nanobodies in diagnosis and treatment of diseases.
The present study aims to review research that has used nanobodies to control viral infections similar to what occurs by SARS-CoV-2.
2. Searching strategy and methodology
An evidence-based systematic review of literature was according to the PRISMA guidelines [19]. PubMed, LitCOVID, and Scopus were searched on July 8, 2020, to extract articles published on the use of nanobodies in coronaviruses. The search key words were SARS, Corona, COVID-19, SARS-CoV-2, Corona viruses, MERS-CoV, Nanobody, Camelid antibody, Single chain antibody, Heavy chain anti body and VHH. We used Boolean functions as follows: (SARS OR MERS OR corona OR SARS-CoV OR coronavirus) AND (Nanobody OR Nanobodies OR Camelid antibody OR Single chain antibody OR Heavy chain anti body OR VHH). Two investigators compared the data extraction for consistency, and removed some sources, since they did not have the full text or were in the non-English language.
3. Results
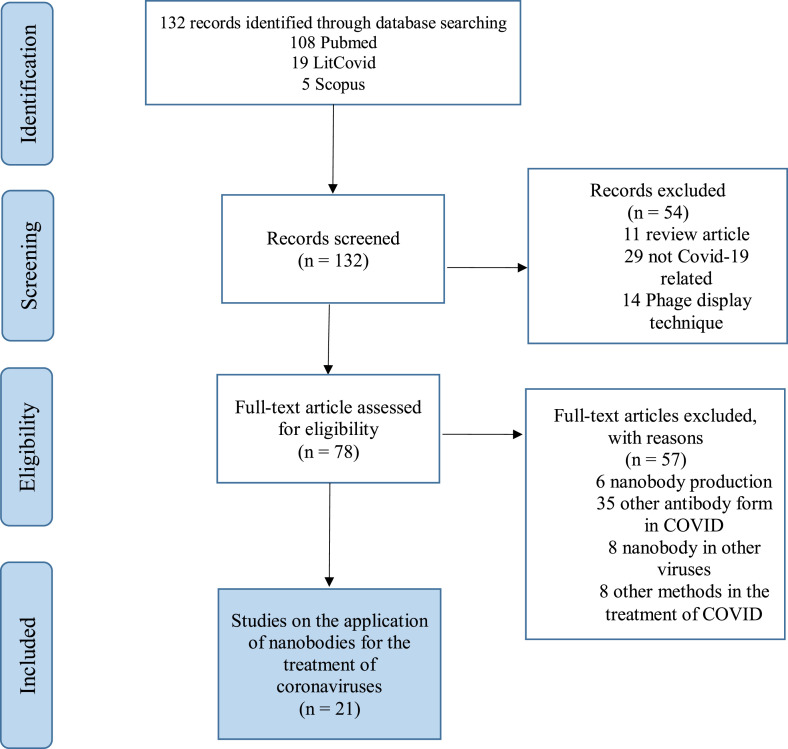
The search yielded 132 studies. After initial screening and reviewing the mined articles, unrelated ones and articles related to the phage display technique were removed. The eligibility of the articles was further reviewed, and irrelevant articles were also removed. Eventually, 21 articles met inclusion criteria (Fig. 3 ). The results related to therapeutic applications of nanobody are mentioned in the first part, and in the second part, brief explanations are given about the role of nanobodies in the detection of coronaviruses infection.
Fig. 3.
Systematic review flow chart of the first literature search according to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines.
3.1. Application of nanobodies in the treatment of coronavirus infection
Raj et al. (2018) identified a number of MERS-CoV–specific VHHs from the bone marrow of MERS-CoV–infected dromedary camels. These VHHs then effectively blocked the entry of viruses to the Huh7 cell (a type of human liver cell line) at picomolar concentrations under experimental conditions. The selected VHHs, with high affinity, bind with the RBD of the viral spike protein. In this study, a camel/human chimeric HCAbs was produced, which consisted of the camel VHH attached to a human Fc domain. This chimeric HCAbs had a long half-life in the serum and protected mice against a deadly MERS-CoV infection [20].
Zhao et al. (2018) isolated nanobodies against the RBD domain of MERS-CoV, which demonstrated high affinity for this domain and potently inhibited MERS-CoV virus to infected host cells. In this study, the effect of nanobody (NbMS10) and nanobody fused to human-FC antibodies on virus pathogenicity was investigated, and these nanobodies showed high affinity to MERS-CoV and potent cross neutralizing activities against different MERS-CoV strains isolated from various hosts. FC fused nanobody exhibited increased half-life in vivo, and this construct was able to efficiently protect humanized mice from the lethal MERS-CoV test [21].
In the next step, He et al. (2019) built dimeric and trimeric nanobodies to increase the binding efficiency with the RBD of MERS-CoV. These engineered nanobodies had special features, so that they demonstrated higher ability than monomeric nanobodies in different MERS-CoV strains. This trimeric nanobody potently blocked the RBD domain of MERS-CoV from attaching to the cell surface receptor (DPP4) and prevented entering of the virus in the animal's cell. These inventive nanobodies have higher capacity to neutralize MERS-CoV strains archived from different countries and hosts [22]. Furthermore, increasing the size of engineered nanobodies has not negative effects on stability under extreme conditions such as alkaline and acidic pH, high temperature, urea, and protease activity.
Feng et al. (2019) constructed a shark antibodies library with a size of 1.2 × 1010 and high diversity. They used the library to isolate suitable nanobodies against SARS-CoV and MERS-CoV spike proteins. They isolated five shark nanobodies against MERS-CoV and one nanobody against the SARS-CoV spike protein with high binding affinity (Kd = 10.1 nM) [23].
He et al. (2017) used the MERS-CoV RBD domain to immunize alpaca, and after immunization, they constructed a rich diversity phage display library of nanobodies against MERS-CoV. These researchers aimed to build a rich library with high divergence toward MERS-CoV [24]. In addition, Wrapp et al. (2020) described isolation of VHHs (single-domain antibodies) from a llama immunized with coronavirus spikes. In this study, spike proteins were stabilized by prefusion. These VHHs were capable of neutralizing MERS-CoV or SARS-CoV. The results of crystallography of these nanobodies demonstrated two distinct epitopes, and both VHHs inhibited the receptor binding. This study also confirmed a cross-reactivity between SARS-CoV-1 S-VHH and SARS-CoV-2 S. These results showed that this VHH was also able to neutralize pseudotyped SARS-CoV-2 S-like viruses. These findings revealed a molecular principle for beta-coronavirus inhibition by VHHs and indicated that these molecules could be useful for therapy during coronavirus outbreaks [25].
Xiaojing Chi et al. reported five sdAbs against the spike receptor binding domain of SARS-CoV-2. These nanobodies revealed a range of Kd from subnanomolar to 35.5 nM. These antibodies showed no affinity or relatively low affinity to SARS-CoV-2. Neutralization evaluation of these recombinant antibodies was performed using a SARS-CoV-2 spike-pseudotyped particle. The results demonstrated that all five nanobodies had inhibitory effects on the SARS-CoV-2 spike-pseudotyped particle. In addition, these monovalent antibodies showed neutralization activity on the SARS-CoV-2 live virus. The range of EC50 in this experiment was 0.13–0.51 μg/mL. Further investigation showed that two types of these sdABs (named 1E2 and 4D8) had potential to inhibit the binding of SARS-CoV-2 RBD with ACE2. Through fusion with the human IgG1-Fc region, the neutralization potency of constructs was improved, and EC50 reached the sub-nanomolar level [12].
In another study, after screening a VHH library by the phage display method followed by affinity maturation, two nanobodies named H11–H4 and H11-D4 were selected. These nanobodies showed high affinity to RBD (KD = 5 and 10 nM, respectively). These VHHs can block the binding of RBD and spike with ACE2. Additionally, the IgG1- Fc domain was fused to these nanobodies, and the ability of these constructs to block the ACE2 binding with RBD was measured. Using a binding assay, it was revealed that IC50 was 61 and 161 nM for H11–H4-Fc and H11-D4-Fc, respectively. Plaque reduction neutralization test showed that H11–H4-Fc and H11-D4-Fc could neutralize the SARS-CoV-2 virus. (ND = 6 and 18 nM for H11–H4-Fc and H11-D4-Fc, respectively) [13].
Gai et al. constructed nanobody phage display libraries using four camels immunized with the SARS-CoV-2 RBD domain. Then, 381 nanobodies with the ability to recognize SARS-CoV-2 RBD were isolated. In addition, seven Nbs were able to block the interaction of RBD with the human ACE2 (angiotensin converting enzyme 2). These Nbs bound with RBD with high affinity (Kd = 21.6 nM–106 nM). Among these nanobodies, Nb11-59 had the highest activity against SARS-CoV-2. In this study, Nb11-59 was produced in the Pichia pastoris host with more than 99% purity [14].
In one study, using synthetic nanobody sequences and the yeast surface-displayed library technique, Schoof et al. isolated several nanobodies with high affinity for different epitopes of the SARS-CoV-2 spike protein. These nanobodies were classified into two classes. Class I included nanobodies attached to the RBD domain and prevented the virus from binding with the cell surface receptor (ACE2), and class II encompassed those attached to areas other than the RBD domain and were not fully capable of inhibitory function. In class I nanobodies, Nb6 exhibited the best performance, and the nanobodies could be attached to two RBD domains at the same time. Using the rationally engineered protein, the researchers designed NB6-derived nanobodies in bivalent and trivalent form and examined their functions and affinities. NB6 Nanobody inhibitory effect against SARS-CoV-2 with IC50 values of 2.0–2.4 μM were determined and its trivalent form with an IC50 of 1.2 nM showed a 2000-fold increase of inhibitory activity on SARS-CoV-2 virus. In the next step, the researchers of this project, using saturation mutagenesis, changed the sequences of Nb6-CDRs to increase the affinity of this nanobody to the SARS-CoV-2 spike. The nanobody generated by this method demonstrated 500-fold increased affinity to virus spike and its potency to inhibited SARS-CoV-2 infection increased more than 200-fold compared to Nb6 [26].
In another attempt, Dong et al. used a naive and synthetic llama VHH library to isolate potent nanobodies against the SARS-CoV-2 spike protein in order to neutralize it. The framework of the synthetic library was partially humanized to prevent an immune response in the human body. After panning against the recombinant SARS-CoV-2 spike protein, they finally reached 69 unique nanobodies that had good binding with the spike protein, but 15 of them prevented the spike from binding with the cell surface ACE2 receptor. After conducting experiments, the project researchers concluded that combination of some of these nanobodies had synergistic effects on blocking the interaction of the spike protein with the ACE2 receptor and increased the neutralization of SARS-CoV-2 infection. They used in-silico design via their own signature computer-aided antibody design (CAAD) to design the combination of nanobodies that improve affinity and avidity toward the RBD site of the spike protein and inhibit SARS-CoV-2 infection. Finally, they combined the resulting structure with human IgG Fc domains to make high-strength antibodies against SARS-CoV-2. The obtained antibody showed good results in inhibiting the infection with the following parameters (Kd, 0.25 nM - IC100, 36.7 nM - IC95, 12.2 nM - IC50, 1 nM) [27].
To obtain an effective nanobody against the SARS-CoV-2 virus, Hanke et al. used immunization of the llama against the FC conjugated SARS-CoV-2 spike protein and isolated the nanobodies using the phage display technique. After performing the two rounds of panning, the Ty1 nanobody was demonstrated to have potent binding with the RBD of the spike protein. Virus neutralization test showed that this nanobody neutralized SARS-CoV-2 at an IC50 of 0.77 μg/mL (54 nM). Cryoelectron microscopy structural studies have shown that CDR1 and CDR3 of the Ty1 nanobody play an important role in binding with two points of the RBD spike protein and inhibit the spike protein from binding with the ACE2 receptor. Combination of this nanobody with the FC domain of the classical antibody reduced its IC50 to 12 nm and effectively suppressed the SARS-CoV-2 virus [28]. Table 1 summarizes the nanobodies for the treatment of coronavirus infections.
Table 1.
List of Nanobodies and type of action.
| Type of nanobody | Type of Virus | Type of action | Binding affinity (Kd) | IC 50 | Ref |
|---|---|---|---|---|---|
| Synthetic VHH from yeast surface-displayed library | SARS-CoV-2 | Block the binding of RBD and spike to ACE2 | Nb6 = 41 nM Nb6 trivalent = N/A Nb6 Redesign = 0.56 nM |
2.0 μM 1.2 nM 1.6 nM |
Schoof et al., 2020 |
| Humanized VHH from llama | SARS-CoV-2 | Block the binding of RBD and spike to ACE2 | 0.25 nM | 1 nM | Dong et al., 2020 |
| VHH from phage display library with Fc domain of classical antibody | SARS-CoV-2 | Block the binding of RBD and spike to ACE2 | Ty1 = 9 nM Ty1-Fc = N/A |
54 nM 12 nM |
Hanke et al., 2020 |
| VHHs from llama | MERS-CoV SARS-CoV-1 SARS-CoV-2 |
Neutralizing SARS-CoV-1 and MERS-CoV RBDs Neutralize SARS-CoV-2 S pseudoviruses. |
VHH 72 = 1.1 nM VHH 55 = 79 pM |
12 nM 1 nM |
Wrapp et al., 2020 |
| Single domain nanobody (sdNb) | SARS-CoV- 2 | Inhibitory effects on SARS-CoV-2 Spike-pseudotyped particle. | 1E2 = 35.5 nM 2F2 = 5.2 nM 3F11 = 3.3 nM 4D8 = 6 nM 5F8 = 0.9 nM |
18.5 nM 22.6 nM 28.6 nM 9.6 nM 39.3 nM |
Chi et al., 2020 |
| VHH form phage display library | SARS-CoV- 2 | Block the binding of RBD and spike to ACE2 | H11–H4 = 12 nM H11–H4-Fc = 5 nM H11-D4 = 39 nM H11-D4-Fc = 18 nM |
N/A 61 nM N/A 161 nM |
Huo et al., 2020 |
| VHH from camel | SARS-CoV-2 | Block the binding of RBD and spike to ACE2 | Nb11-59 = 21 nM Nb16-68 = 36 nM |
35 nM 145 nM |
Gai et al., 2020 |
| VNAR from Shark | MERS-CoV SARS-CoV-1 |
Bind to MERS-CoV and SARS-CoV-1 spike Protein | 10.1 nM | N/A | Feng et al., 2019 |
| VHH from camel | MERS-CoV | Bind to MERS-CoV spike protein (RBD) | Mono-Nb = 0.87 nM Di-Nb = 5.9 pM Tri-Nb = 7 pM |
2.13 nM 0.25 nM 0.38 nM |
He et al., 2019 |
| Chimeric VHH camel with Fc of human antibody | MERS-CoV | Bind to MERS-CoV spike protein | VHH1 = 0.4 nM VHH4 = 0.2 nM VHH83 = 0.1 nM VHH101 = 0.9 nM |
3.63 nM 0.66 nM 0.66 nM 3.63 nM |
Raj et al., 2018 |
| Chimeric VHH camel with Fc of human antibody | MERS-CoV | Bind to MERS-CoV spike protein | NbMS10 = 0.87 nM Fc-NbMS10 = 0.35 nM |
232 nM 153 nM |
Zhao et al., 2018 |
3.2. Application of nanobodies in the detection of coronavirus infection
However, most of the articles obtained are about the therapeutic role of nanobodies; but nanobodies can also be used to diagnose this infection. The development of ELISA-based diagnostic methods will accelerate with the acquisition of specific nanobodies against coronavirus protein components. The rest of the isolated nanobodies with high affinity toward the S protein without blocking function were suitable targets for diagnostic applications to detect SARS-CoV-2 [27].
4. Conclusion
As reported in this systematic review study, most researchers use the spike protein, especially RBD domains, to make suitable nanobodies against SARS-CoV and MERS-CoV viruses. It appears that since it is possible to use nanobodies as an inhaler, the resulting nanobodies can be used to prevent the virus from infecting the lungs. Interestingly, owing to the high homology of the SARS-CoV-2 spike protein sequence with its ancestors, the libraries obtained in these studies against SARS-CoV and MERS-CoV can be used to isolate high affinity nanobodies against SARS-CoV-2 in order to prevent entering and infection of this virus.
Acknowledgement
Thanks to guidance and advice from "Clinical Research Development Unit of Baqiyatallah Hospital", Tehran, Iran.
References
- 1.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20(5):269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.M., Lim W., Osterhaus A.D. SARS virus infection of cats and ferrets. Nature. 2003;425(6961) doi: 10.1038/425915a. 915-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Memish Z.A., Cotten M., Meyer B., Watson S.J., Alsahafi A.J., Al Rabeeah A.A., Corman V.M., Sieberg A., Makhdoom H.Q., Assiri A. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia. Emerg. Infect. Dis. 2013;20(6):1012. doi: 10.3201/eid2006.140402. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. Jama. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 5.Campos E.V., Pereira A.E., de Oliveira J.L., Carvalho L.B., Guilger-Casagrande M., de Lima R., Fraceto L.F. How can nanotechnology help to combat COVID-19? Opportunities and urgent need. J. Nanobiotechnol. 2020;18(1):1–23. doi: 10.1186/s12951-020-00685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alphandéry E. The potential of various nanotechnologies for coronavirus diagnosis/treatment highlighted through a literature analysis. Bioconjugate Chem. 2020;31(8):1873–1882. doi: 10.1021/acs.bioconjchem.0c00287. [DOI] [PubMed] [Google Scholar]
- 7.Zare H., Rajabibazl M., Rasooli I., Ebrahimizadeh W., Bakherad H., Ardakani L.S., Gargari S.L.M. Production of nanobodies against prostate-specific membrane antigen (PSMA) recognizing LnCaP cells. Int. J. Biol. Markers. 2014;29(2):169–179. doi: 10.5301/jbm.5000063. [DOI] [PubMed] [Google Scholar]
- 8.Bakherad H., Gargari S.L.M., Rasooli I., RajabiBazl M., Mohammadi M., Ebrahimizadeh W., Ardakani L.S., Zare H. In vivo neutralization of botulinum neurotoxins serotype E with heavy-chain camelid antibodies (VHH) Mol. Biotechnol. 2013;55(2):159–167. doi: 10.1007/s12033-013-9669-1. [DOI] [PubMed] [Google Scholar]
- 9.Ebrahimizadeh W., Gargari S.M., Rajabibazl M., Ardekani L.S., Zare H., Bakherad H. Isolation and characterization of protective anti-LPS nanobody against V. cholerae O1 recognizing Inaba and Ogawa serotypes. Appl. Microbiol. Biotechnol. 2013;97(10):4457–4466. doi: 10.1007/s00253-012-4518-x. [DOI] [PubMed] [Google Scholar]
- 10.Aghamollaei H., Ghanei M., Rasaee M.J., Latifi A.M., Bakherad H., Fasihi‐Ramandi M., Taheri R.A., Gargari S.L.M. Isolation and characterization of a novel nanobody for detection of GRP78 expressing cancer cells. Biotechnol. Appl. Biochem. 2020:1–8. doi: 10.1002/bab.1916. [DOI] [PubMed] [Google Scholar]
- 11.A. Steels, L. Bertier, J. Gettemans, Use, Applications and Mechanisms of Intracellular Actions of Camelid VHHs, Antibody Engineering, IntechOpen2017.
- 12.Chi X., Liu X., Wang C., Zhang X., Li X., Hou J., Ren L., Jin Q., Wang J., Yang W. Humanized single domain antibodies neutralize SARS-CoV-2 by targeting the spike receptor binding domain. Nat. Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-18387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27(9):846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 14.Gai J., Ma L., Li G., Zhu M., Qiao P., Li X., Zhang H., Zhang Y., Chen Y., Gong R. 2020. A Potent Neutralizing Nanobody against SARS-CoV-2 with Inhaled Delivery Potential. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarr A.W., Lafaye P., Meredith L., Damier‐Piolle L., Urbanowicz R.A., Meola A., Jestin J.L., Brown R.J., McKeating J.A., Rey F.A. An alpaca nanobody inhibits hepatitis C virus entry and cell‐to‐cell transmission. Hepatology. 2013;58(3):932–939. doi: 10.1002/hep.26430. [DOI] [PubMed] [Google Scholar]
- 16.Bobkov V., Zarca A.M., Van Hout A., Arimont M., Doijen J., Bialkowska M., Toffoli E., Klarenbeek A., van der Woning B., van der Vliet H.J. Nanobody-Fc constructs targeting chemokine receptor CXCR4 potently inhibit signaling and CXCR4-mediated HIV-entry and induce antibody effector functions. Biochem. Pharmacol. 2018;158:413–424. doi: 10.1016/j.bcp.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., Millar A., Power U.F., Stortelers C., Allosery K. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 2016;60(1):6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mu B., Huang X., Bu P., Zhuang J., Cheng Z., Feng J., Yang D., Dong C., Zhang J., Yan X. Influenza virus detection with pentabody-activated nanoparticles. J. Virol Methods. 2010;169(2):282–289. doi: 10.1016/j.jviromet.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int. J. Surg. 2010;8(5):336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Raj V.S., Okba N.M., Gutierrez-Alvarez J., Drabek D., van Dieren B., Widagdo W., Lamers M.M., Widjaja I., Fernandez-Delgado R., Sola I. Chimeric camel/human heavy-chain antibodies protect against MERS-CoV infection. Science advances. 2018;4(8) doi: 10.1126/sciadv.aas9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao G., He L., Sun S., Qiu H., Tai W., Chen J., Li J., Chen Y., Guo Y., Wang Y. A novel nanobody targeting Middle East respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain has potent cross-neutralizing activity and protective efficacy against MERS-CoV. J. Virol. 2018;92(18) doi: 10.1128/JVI.00837-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L., Tai W., Li J., Chen Y., Gao Y., Li J., Sun S., Zhou Y., Du L., Zhao G. Enhanced ability of oligomeric nanobodies targeting MERS coronavirus receptor-binding domain. Viruses. 2019;11(2):166. doi: 10.3390/v11020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng M., Bian H., Wu X., Fu T., Fu Y., Hong J., Fleming B.D., Flajnik M.F., Ho M. Construction and next-generation sequencing analysis of a large phage-displayed VNAR single-domain antibody library from six naive nurse sharks. Antibody therapeutics. 2019;2(1):1–11. doi: 10.1093/abt/tby011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L., Li J., Ren S., Sun S., Guo Y., Qiu H., Liao Y., Ji K., Fan R., Zhao G. Construction and identification of nanobody phage display library targeting Middle East respiratory syndrome coronavirus, Xi bao yu fen zi mian yi xue. za zhi= Chinese journal of cellular and molecular immunology. 2017;33(12):1662–1668. [PubMed] [Google Scholar]
- 25.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., COVID V.-C., Hoffmann M. Cell; 2020. Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Zimanyi M., Deshpande I. 2020. An Ultra-high Affinity Synthetic Nanobody Blocks SARS-CoV-2 Infection by Locking Spike into an Inactive Conformation. bioRxiv. [Google Scholar]
- 27.Dong J., Huang B., Jia Z., Wang B., Kankanamalage S.G., Titong A., Liu Y. Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity. Emerg. Microb. Infect. 2020:1–10. doi: 10.1080/22221751.2020.1768806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanke L., Vidakovics M.L.P., Sheward D., Das H., Schulte T., Morro A.M., Corcoran M., Achour A., Hedestam G.K., Hällberg B.M. 2020. An Alpaca Nanobody Neutralizes SARS-CoV-2 by Blocking Receptor Interaction. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]