Abstract
In a study of the fungal diversity on Ulleung Island in Korea, three novel strains of Penicillium were isolated. Different sites on Ulleung Island were selected for collecting endophytic fungi, and three endophytic fungal strains showed unique morphological characteristics. DNA sequence of the internal transcribed spacer, β-tubulin, calmodulin, and RNA polymerase II second largest subunit regions of the strains were analyzed and they showed unique taxonomic position from the other species of Penicillium section Sclerotiora. The new strains were named Penicillium ulleungdoense sp. nov. As the novel endophytic Penicillium taxa were discovered in a unique environment, the data could be meaningful for understanding the geographical distribution of Ascomycetes on Ulleung Island.
Keywords: Penicillium ulleungdoense sp. nov, endophytic Ulleung Island
1. Introduction
The genus Penicillium classified in the order Eurotiales within the family Trichocomaceae [1], was first introduced as P. candidum, P. glaucum, and P. expansum by Johann Heinrich Friedrich Link in 1809 [2]. With association of 26 sections, this genus is subdivided into Aspergilloides, and Penicillium [3,4]. As one of the most common genera in earth, many species belong to this genus could be isolated from various environments including air, soil, food product, and organism [5,6]. Penicillium was originally part of family Trichocomaceae. Since 1990s however, not only morphological characterization but also molecular analysis has been used for identification. Due to the new identification of fungi, Penicillium was redefined as part of family Aspergillaceae [7], and all names of Penicillium were well arranged recently based on polyphasic taxonomy [8].
Based on the color like yellow, and/or orange mycelia, orange, and/or reddish colony reverses, Houbraken and Samson established the Penicillium section Sclerotiora [9]. As the fungi were isolated from soil, plants and insects, the type of host also becomes a standard for identification [3]. With combination of morphological features including colony pattern, conidiophore structure and sclerotia production, the species in section Sclerotiora is identified [10]. The molecular analysis with sequence of internal transcribed spacer (ITS) region and additional DNA marker; β-tubulin (BenA), calmodulin (CaM) and the RNA polymerase II second largest subunit (RPB2) is added to increase the reliability of analysis [8,11]. In worldwide, over 350 species were named as Penicillium [11], and among them, about 100 species of Penicillium were discovered in Korea [12]. Including reporting of P. daejeonium as new kind species by doctor Lim’s team, 12 species were identified as new from Korea [13,14].
In this study, three specific plants (Phedimus takesimensis, Sedum oryzifolium, and Aster spathulifolius) isolated from Ulleung Island were chosen for the collection of novel endophytic fungal strains [15,16]. Ulleung Island is a volcanic island located in the Ulleung Basin in the East Sea (E 131°52′07″, N 37°14′12″). The annual average temperature in the Ulleung Basin is 12 °C with a high humidity; most parts of the island have steep slopes, and strong winds make the inhabitation of plants difficult [17]. Ulleung Island is composed of 89 eastern and western islands, and 65.4% of the land area is made of steep slopes over 40° [18]. Owing to a predominantly dry and salty environment, Ulleung Island has a unique biosystem that includes 48 species of plants [15,19]. Endophytic fungi from Phedimus takesimensis, Sedum oryzifolium, and Aster spathulifolius on Ulleung Island were screened morphologically and molecularly. This study describes a novel strain of P. ulleungdoense sp. nov., isolated from the roots of the three plants on Ulleung Island.
2. Materials and methods
2.1. Isolation of endophytes
The roots of Phedimus takesimensis (E 130°54'32.76", N 37°28'54.97"), Sedum oryzifolium (E 130°47'55.45", N 37°31'07.27"), and Aster spathulifolius (E 130°53'15.88", N 37°32'29.09") were collected from Ulleung Island in 4 April 2017. The root samples were washed with distilled water to remove sand particles and treated with Tween-80 solution for 5 min. The surface was sterilized using 1% perchloric acid solution and subsequently washed with distilled water. After washing, the roots were cut, and the pieces were incubated at 25 °C in Hagem minimal medium containing 80 ppm of streptomycin for pure culture [20,21]. The isolated fungal strains from the roots were subcultured on potato dextrose agar (PDA) and incubated at 25 °C for a week in the dark to allow fungal growth [22]. The isolates KMG401, KMG402, and KMG403 were deposited to the Korean Agricultural Culture Collection (KACC) with allotted no. KACC 48990, KACC 48991, and KACC 48992, respectively.
2.2. Morphological analysis
Agar plugs were cultured on PDA and then transferred to malt extract agar (MEA), Czapek yeast autolysate agar (CYA), yeast extract sucrose agar (YESA), czapek yeast autolysate agar with 5% NaCl (CYAS), czapek’s agar (CZ), oatmeal gar (OA), and Creatine sucrose agar (CREA) for morphological analysis [11,23]. Plates were incubated at 25 °C in the dark for 7 days, and plates with CYA were additionally incubated at 30 °C and 37 °C in the dark for 7 days. After incubation, diameters, density of sporulation, obverse and reverse colony colors, and the existence of soluble pigments were recorded. Fungal morphological characterization was identified by using a light microscope (Eclipse 80i; Nikon, Tokyo, Japan) [5,11]. With 85% lactic acid and 99% ethanol, fixed specimen images were acquired.
2.3. DNA extraction, PCR, and sequencing
Strains of the sample were grown on PDA, and DNA was extracted using the Accuprep® Genomic DNA Extraction Kit (Bioneer Corp., Daejeon, Korea). DNA preps were stored at −20 °C until use for PCR. Four regions, internal transcribed spacer (ITS), β-tubulin (BenA), calmodulin (CaM), and RNA polymerase II second largest subunit (RPB2), were amplified with the primer pairs ITS1-ITS4, Bt2a-Bt2b, CMD5-CMD6, and 5Feur-7CReur, respectively [9,11]. Amplifications were performed with different primer sets in a 20 μl reaction mixture [23,24]. The products were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and sequenced using the ABI PRISM BigDye Terminator Cycle Sequencing Kit (PE Biosystems, Foster, CA, USA) with the ABI 310 DNA sequencer (PE Biosystems) [8].
2.4. Phylogenetic analysis
Under manual, the obtained nucleotide sequences were aligned, and submitted for phylogenetic analysis using BioEdit v7.2.5 (Clustal W) [25]. The similarities of fungal sequences were calculated by using Molecular Evolutionary Genetics Analysis (MEGA) 7 software [26]. A maximum likelihood (ML) phylogenetic tree was constructed based on a combination of the markers of ITS, BenA, CaM, and RPB2 for three strains [11]. While constructing phylogenetic tree, RPB2 was not included in the combination of ITS, BenA, and CaM [27]. This is because that some Penicillium species showed no primer sequences of the RPB2. With Tamura 3–parameter model, the ML heuristic method was set as the level 3 of Subtree Pruning Regrafting (SPR), and number of bootstrap replicates was 1,000. To set the outgroup for section Sclerotiora, P. levitum CBS 45.48 of section Lanata–Divaricata was used [28]. Through this manual, the initial ML tree was set automatically. The percentage of sequence identity was obtained from a National Center for Biotechnology Information (NCBI) BLASTn search [29,30]. In phylogenetic analysis, the obtained sequences were compared with Penicillium species based on the study of Wang et al. [20].
3. Results
3.1. Phylogenetic analysis
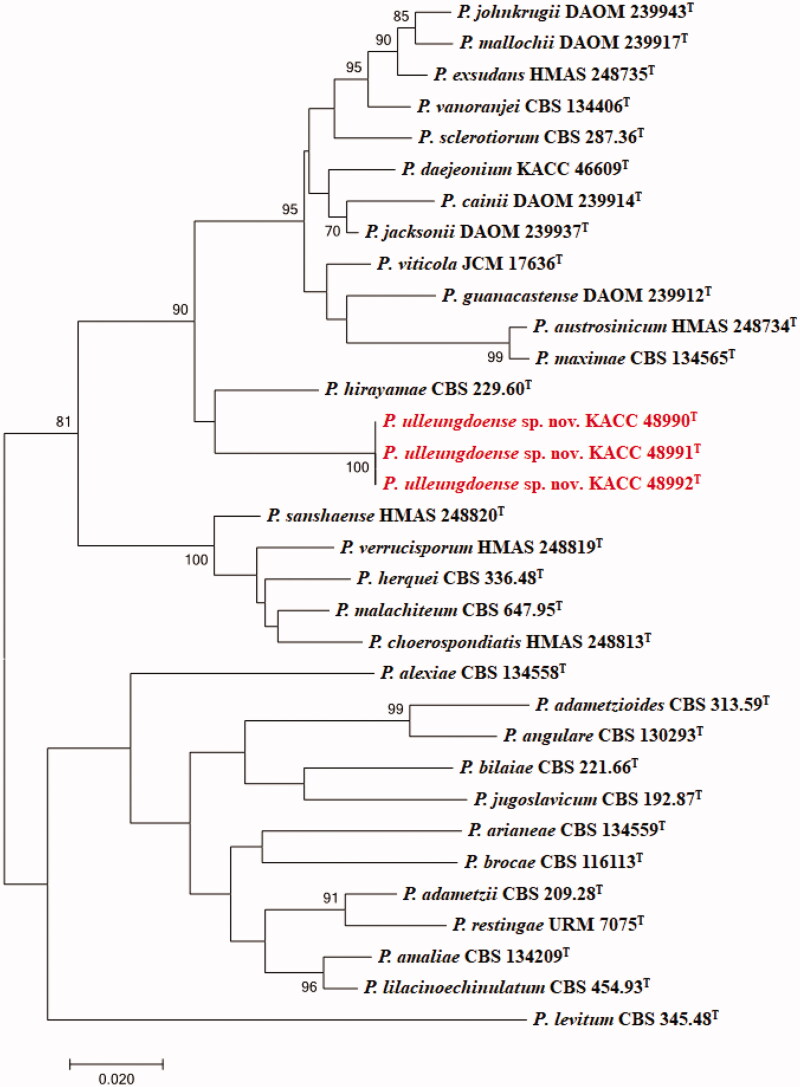
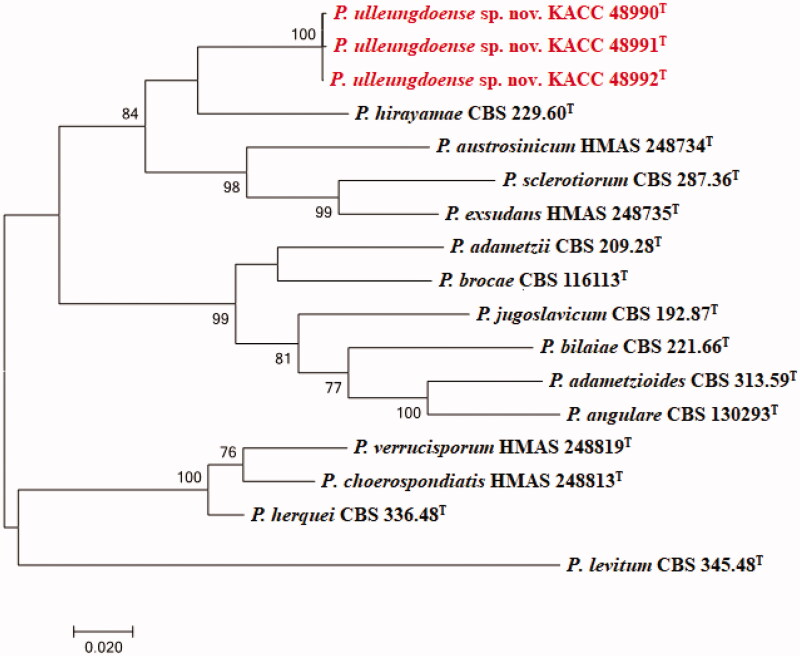
The DNA sequences of the new species were registered in the GenBank database of the NCBI (MN640087–MN640089 for rDNA–ITS, MN737487–MN737489 for BenA, MN745074–MN745076 for CaM, and MN756007–MN756009 for RPB2) (Table 1). The target strains KACC 48990, KACC 48991, and KACC 48992 were positioned using the sequences of the ITS region, BenA region, CaM region, and RPB2 region (Figures 1 and 2). Based on these four genes, the target strains were compared with other strains available in the NCBI database. A phylogenetic tree was constructed following the bootstrap analysis of 1,000 replicates, and all three strains were the most similar to the type strain of Penicillium hirayamae. In addition, BlASTn search was performed against the strains KACC 48990, KACC 48991, and KACC 48992. ITS gene sequence analysis showed that KACC 48990, KACC 48991, and KACC 48992 were the most similar to P. hirayamae with 97% sequence identity. BenA gene sequence analysis showed that KACC 48990, KACC 48991, and KACC 48992 were the most similar to P. hirayamae with 88% sequence identity. CaM gene sequence analysis showed that KACC 48990, KACC 48991, and KACC 48992 were the most similar to P. hirayamae with 91% sequence identity. RPB2 gene sequence analysis showed that KACC 48990, KACC 48991, and KACC 48992 were the most similar to P. hirayamae with 91% sequence identity. Gene sequence analysis with a combination of four primer pairs showed that KACC 48990, KACC 48991, and KACC 48992 were the most similar to P. hirayamae with 93% sequence identity [8,31].
Table 1.
Details of the strains used in phylogenetic analyses.
| Sequence accession numbers |
|||||
|---|---|---|---|---|---|
| Species | Strain numbers |
ITS | BenA | CaM | RPB2 |
| P. adametzii | CBS 209.28T | JN714929 | JN625957 | KC773796 | JN121455 |
| P. adametzioides | CBS 313.59T | JN686433 | JN799642 | JN686387 | JN406578 |
| P. alexiae | CBS 134558T | KC790400 | KC773778 | KC773803 | |
| P. amaliae | CBS 134209T | JX091443 | JX091563 | JX141557 | |
| P. angulare | CBS 130293T | AF125937 | KC773779 | KC773804 | JN406554 |
| P. arianeae | CBS 134559T | KC773833 | KC773784 | KC773811 | |
| P. austrosinicum | HMAS 248734T | KX885061 | KX885041 | KX885051 | KX885032 |
| P. bilaiae | CBS 221.66T | JN714937 | JN625966 | JN626009 | JN406610 |
| P. brocae | CBS 116113T | AF484398 | KC773787 | KC773814 | JN406639 |
| P. cainii | DAOM 239914T | JN686435 | JN686366 | JN686389 | |
| P. citrinum | CBS 139.45T | AF033422 | GU944545 | GU944638 | JF417416 |
| P. coffeae | CBS 119387T | AY742702 | KJ834443 | AY741747 | JN121436 |
| P. guanacastense | DAOM 239912T | JN626098 | JN625967 | JN626010 | |
| P. herquei | CBS 336.48T | JN626101 | JN625970 | JN626013 | JN121494 |
| P. hirayamae | CBS 229.60T | JN626095 | JN625955 | JN626003 | JN121459 |
| P. jacksonii | DAOM 239937T | JN686437 | JN686368 | JN686391 | |
| P. janthinellum | CBS 340.48T | GU981585 | GU981625 | KF296401 | JN121497 |
| P. johnkrugii | DAOM 239943T | JN686447 | JN686378 | JN686401 | |
| P. jugoslavicum | CBS 192.87T | KC773836 | KC773789 | KC773815 | JN406618 |
| P. levitum | CBS 345.48T | GU981607 | GU981654 | KF296394 | KF296432 |
| P. lilacinoechinulatum | CBS 454.93T | AY157489 | KC773790 | KC773816 | |
| P. malachiteum | CBS 647.95T | KC773838 | KC773794 | KC773820 | |
| P. mallochii | DAOM 239917T | JN626104 | JN625973 | JN626016 | |
| P. maximae | CBS 134565T | EU427298 | KC773795 | KC773821 | |
| P. multicolor | CBS 501.73T | JN799647 | JN799645 | JN799646 | EU427262 |
| P. paxilli | CBS 360.48T | GU944577 | JN606844 | JN606566 | JN606610 |
| P. sclerotiorum | CBS 287.36T | JN626132 | JN626001 | JN626044 | JN406585 |
| P. simplicissimum | CBS 372.48T | GU981588 | GU981632 | KF296368 | JN121507 |
| P. ulleungdoense sp. nov. | KACC 48990T | MN640087 | MN737487 | MN745074 | MN756007 |
| KACC 48991 | MN640088 | MN737488 | MN745075 | MN756008 | |
| KACC 48992 | MN640089 | MN737489 | MN745076 | MN756009 | |
| P. vanoranjei | CBS 134406T | KC695696 | KC695686 | KC695691 | |
| P. viticola | JCM 17636T | AB606414 | AB540174 | ||
The accession numbers of the strains KACC 48990, KACC 48991, KACC 48992, and others are available in the NCBI database.
Figure 1.
Phylogenetic tree based on the maximum likelihood analysis of the combined ITS, BenA and CaM dataset for species classified in the Penicillium section. Penicillium levitum was included as an outgroup. Bootstrap analysis was performed with 1,000 replications. Bootstrap support values of ≥ 70% are indicated at the nodes. The bar indicates the number of substitutions per position. T indicates the type strains of the species. Bar, 0.02 substitutions per nucleotide position. The isolates KACC 48990, KACC 48991 and KACC 48992 are marked in red.
Figure 2.
Phylogenetic tree based on the maximum likelihood analysis of RPB2 dataset for species classified in the Penicillium section. Penicillium levitum was included as an outgroup. Bootstrap analysis was performed with 1,000 replications. Bootstrap support values of ≥ 70% are indicated at the nodes. The bar indicates the number of substitutions per position. T indicates the type strains of the species. Bar, 0.02 substitutions per nucleotide position. The isolates KACC 48990, KACC 48991 and KACC 48992 are marked in red.
3.2. Morphological feature
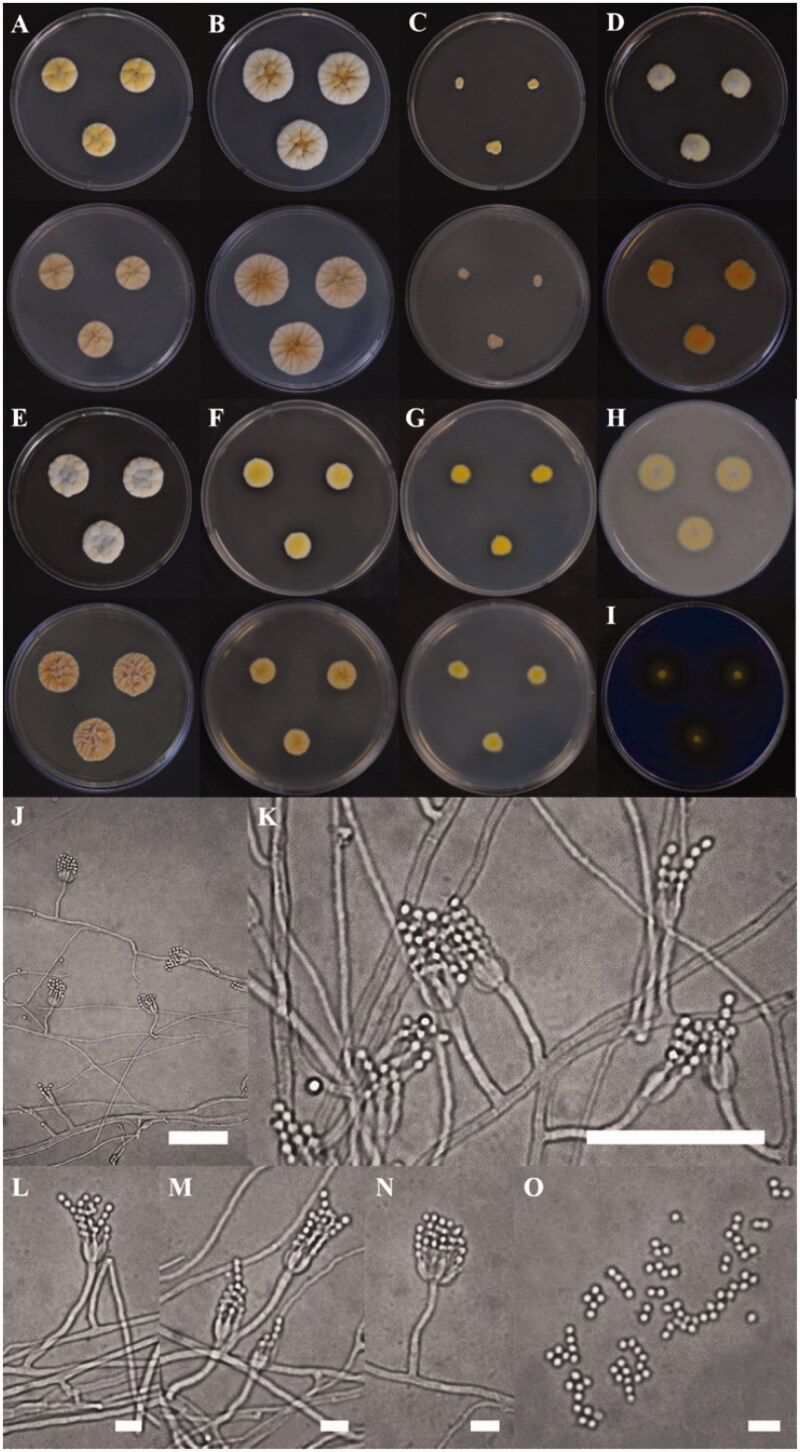
The colony of various plates of the KACC 48990 strain are shown, and the photomicrographs of morphological structures are shown in Figure 3. The detailed fungal morphological descriptions are in the Taxonomy section. Distinct morphological features between P. ulleungdoense, and its related species are summarized in Table 2.
Figure 3.
Morphological characteristics of Penicillium ulleungdoense (KACC 48990) grown on different media. (A) CYA 25 °C, (B) CYA 30 °C, (C) CYA 37 °C, (D) MEA 25 °C, (E) YESA 25 °C, (F) CYAS 25 °C, and (G) CZ 25 °C (top: obverse, bottom: reverse); (H) Colony on OA 25 °C; (I) Colony on CREA 25 °C; (J–N) Conidiophores; (O) Conidia (Scale bar = 50 μm in J–K. Scale bar = 10 μm in L–O).
Table 2.
Morphological characteristics of KACC 48990 and the reference species Penicillium hirayamae on Czapek yeast autolysate agar (CYA) at 25 °C.
| Characteristics | Study isolate, Penicillium ulleungdoense KACC 48990 | Penicillium hirayamae |
|---|---|---|
| Colony color | bright ivory; reverse silk gray | yellowish brown; reverse orange |
| Colony diameter | 16-18 mm in 7 days | 25-33 mm in 7 days |
| Acid production | moderate | weak |
| Sclerotia | not observed | present; orange |
| Conidiophores | monoverticillate, stipe septate, smooth to rough, 2.1-2.8 × 28.3-50.0 μm | Monoverticillate, single metula rare branching, stipe septate, smooth to rough, 1.8-2.4 × 38.0-44.0 μm |
| Phialides | ampulliform, navicular shape, smooth, 3-8 in number, 2.0-2.5 × 5.5-7.0 μm | ampulliform, pear shape, smooth, 3-7 in number, 2-2.8 × 8.1-8.6 μm |
| Conidia | smooth to rough, globose, 1.9-2.0 × 2.3-2.5 μm | smooth to rough, globose to subglobose, 2.6-2.7 × 2.8-3.0 μm |
aAccording to the description of Samson et al. [9].
3.3. Taxonomy
Penicillium ulleungdoense D.H. Choi & J.G. Kim, sp. nov. (Figure 3).
Fungal Names: KMG 401.
Typus: KACC 48990.
Mycobank: MB835474.
Etymology: The specific epithet refers the colleced space: Korean, Ulleung Island in 4 April 2017, collector D.H. Choi.
DNA barcodes: ITS MN640087, BenA MN737487, CaM MN745074, RPB2 MN756007.
Colony diam, 7d, 25 °C (unless stated otherwise): CYA 16–18 mm; CYA 30 °C 25–28 mm; CYA 37 °C 4–7 mm; MEA 12–17 mm; YES 19–24 mm; CREA 6–10 mm; CZ 8–9 mm; OA 13–16mm.
Colony characteristics: On CYA 25 °C, 7 days: Colonies nearly circular, convex, concentrically sulcate, cobblestone gray mycelia appeared in centers; margins narrow, entire; mycelia white near margin, bright ivory elsewhere; texture floccose; sporulation moderately dense; conidial color light ivory; soluble pigments absent; exudates present; reverse conspicuously, and radially sulcate, generally silk gray (Figure 3(A)). On CYA 30 °C, 7 days: Colonies similar to those on CYA 25 °C, 7 days but larger scale; margins moderate, entire; mycelium white near margin, brown beige elsewhere; reverse generally silk gray or beige red in center but at beige periphery (Figure 3(B)). On CYA 37 °C, 7 days: Colonies irregular, plane; margins low, narrow, undulate; mycelia ivory; texture floccose; sporulation dense; conidial color light ivory; soluble pigments absent; exudates present; reverse generally silk gray (Figure 3(C)). On MEA 25 °C, 7 days: Colonies irregular, convex, agate gray mycelia appeared in centers; margins low, narrow, entire or undulate; mycelia yellow near margin, white elsewhere; texture floccose; sporulation dense; conidial color bright yellow; soluble pigments slight dull yellow; exudates absent; reverse orange in centers but orangish white at periphery (Figure 3(D)). On YES 25 °C, 7 days: Colonies nearly circular, convex, dusty gray and pearl dark gray mycelia appeared in centers; margins undulate; mycelia yellowish white, and papyrus white; texture floccose; sporulation moderately dense; conidial color yellowish white; soluble pigments absent; exudates present; reverse conspicuously, and radially sulcate, beige red in center but at beige periphery (Figure 3(E)). On CYAS 25 °C, 7 days: Colonies nearly circular, highly convex in centers; margins low, narrow, entire; mycelia white near margin, beige elsewhere; texture floccose; sporulation dense; conidial color green beige; soluble pigments absent; exudates present; reverse conspicuously, and radially sulcate, traffic gray in center but at pearl light gray periphery (Figure 3(F)). On CZ 25 °C, 7 days: Colonies nearly circular or irregular, convex; margins low, narrow, entire; mycelia bright beige near margin, yellow elsewhere; texture floccose; sporulation dense; conidial color yellow; soluble pigments absent; exudates present; reverse generally bright ivory (Figure 3(G)). On OA 25 °C, 7 days: Colonies circular, convex; margins low, narrow, entire; mycelia bright ivory near margin, cobblestone gray elsewhere; texture floccose; sporulation dense; conidial color ivory; soluble pigments slight dull yellow; exudates absent (Figure 3(H)). On CREA 25 °C, 7 days: acid production moderately present (Figure 3(F)).
Micromorphology: Conidiophores strictly monoverticillate; stipes septate, smooth–walled, 28.3–50.0 × 2.1–2.8 μm, vesticulate; phialides ampulliform, navicular, smooth, 2–8 per stipe, 5.5–7.0 × 2.0–2.5 μm; conidia subglobose, smooth–walled, 2.3–2.5 × 1.9–2.0 μm; sclerotia not observed.
Type strain: KACC 48990, isolated from the root of Phedimus takesimensis in Ulleung Isl, and, Korea, 4 April 2017. The culture is preserved in Korean Agricultural Culture Collection (KACC) in Jeonju, Korea. Molecular markers for the species are MN640087 for rDNA–ITS, MN737487 for β–tubulin, MN745074 for calmodulin, and MN756007 for RNA polymerase II second largest subunit.
Note: In phylogenetic tree, and morphological analysis, KACC 48990 showed the most similarity with P. hirayamae. Though the similarity between KACC 48990, and P. hirayamae, KACC 48990 showed several differences with P. hirayamae; less growth on medium, better acid resistant, and lightly present of soluble pigments.
Additional strains studied: KACC 48991, Republic of Korea. Gyeongsang Province, Ulleung Island, 37°14'33.29"N 131°51'53.43"E, Sedum oryzifolium, 4 April 2017, D.H. Choi, J.G. Kim; KACC 48992, Republic of Korea. Gyeongsang Province, Ulleung Island, 37°14'20.10"N 131°52'08.50"E, Aster spathulifolius, 4 April 2017, D.H. Choi, J.G. Kim.
4. Discussion
With four primer sequences (ITS, BenA, CaM, and RPB2) [20,27], phylogenetic analysis was processed to study the relationship of Penicllium section Sclerotiora. As some of Penicillium species had no primer sequences of the RPB2 region, the construction of phylogenetic tree was done with two kind of versions; combination of ITS, BenA and CaM regions; RPB2 regions [20]. The result for individual markers showed a close relationship with P. hirayamae [8,13] and is confirmed by results of maximum-likelihood phylogenetic trees (Figure 1–2). Similar to many other Eurotiales species, Penicillium can be distinguished based on morphological features and growth temperature [11]. Similar to many other Eurotiales species, Penicillium can be distinguished based on morphological features and growth temperature [11]. Penicillium strains have different growth patterns at different temperatures and in different media. P. hirayamae grew slightly faster on YESA and poorly on CREA. Conidiophores are mostly monoverticillate but rarely have a single metula form [9]. Penicillium species could be distinguished according to the shape of the conidia, phialides, conidiophores, and vesicle. In the result of comparing with Penicillium species based on the study of Zhuang’s team, three isolates KACC 48990, KACC 48991, and KACC 48992 were morphologically similar to P. hirayamae [9,20]. The conidia were globose, joined into chains, smooth to rough, and colorless. Similar to P. hirayamae, ampulliform phialides of conidiophores and yellowish colony color were recognized. Despite these similarities, the isolates could be distinguished from P. hirayamae. Not like P. hirayamae, the colony growth rates of isolates were much smaller and dull yellow soluble pigments were existed slightly on MEA. The reverse color on CYA of P. hirayamae was orange, whereas that of three isolates were silk gray. Another notable feature of three isolates was acid production because P. hirayamae had weak acid production, whereas the isolates had moderate acid production.
Since the initial study of endophytic fungal diversity, various species belonging to Ascomycota have been isolated from Ulleung Island [3,13]. As the novel endophytic Penicillium taxa were discovered in a unique environment, the data could be meaningful for understanding the geographical distribution of Ascomycetes on Ulleung Island. The diversity of endophytes on Ulleung Island is unclear. Further studies including the isolation and analysis of endophytes are important to identify unknown taxa on Ulleung Island. In the present study, a novel strain of Penicillium was identified.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Samson RA, Houbraken J, Thrane U.. Food and indoor fungi. Utrecht: CBS KNAW Biodiversity Center; 2010. [Google Scholar]
- 2.Link JHF. Observationes in ordines plantarum naturales. Dissertatio I. Magazin Der Gesellschaft Naturforschenden Freunde Berlin (in Latin). 1809;3:3–42. [Google Scholar]
- 3.Houbraken J, Samson RA.. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol. 2011;70(1):1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houbraken J, Wang L, Lee HB, et al. . New sections in Penicillium containing novel species producing patulin, pyripyropens or other bioactive compounds. Persoonia. 2016;36:299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitt JI. The genus Penicillium, and its teleomorphic states Eupenicillium, and Talaromyces. London, UK: Academic Press; 1979. [Google Scholar]
- 6.Frisvad JC, Samson RA.. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food, and air–borne Terverticillate Penicillia, and their mycotoxins. Stud Mycol. 2004;49:1–174. [Google Scholar]
- 7.Samson RA, Yilmaz N, Houbraken J, et al. . Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol. 2011;70(1):159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ropars J, Dupont J, Fontanillas E, et al. . Sex in cheese: evidence for sexuality in the fungus Penicillium roqueforti. PLoS One. 2012;7(11):e49665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visagie CM, Houbraken J, Rodriques C, et al. . Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia. 2013;31:42–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera KG, Seifert KA. A taxonomic and phylogenetic revision of the Penicillium sclerotiorum complex. Stud Mycol. 2011;70(1):139–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visagie CM, Houbraken J, Frisvad JC, et al. . Identification and nomenclature of the genus Penicillium. Stud Mycol. 2014;78:343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pangging M, Nguyen TTT, Lee HB.. New records of four species belonging to Eurotiales from soil and freshwater in Korea. Mycobiology. 2019;47(2):154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park MS, Chung D, Baek K, et al. . Three unrecorded species belonging to Penicillium section Sclerotiora from marine environments in Korea. Kor J Mycol. 2019;47:165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen TTT, Pangging M, Bangash NK, et al. . Five new records of the family Aspergillaceae in Korea, Aspergillus europaeus, A. pragensis, A. tennesseensis, Penicillium fluviserpens, and P. scabrosum. Mycobiology. 2020;48(2):81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clay K, Holah J.. Fungal endophyte symbiosis and plant diversity in successional fields. Science. 1999;285(5434):1742–1744. [DOI] [PubMed] [Google Scholar]
- 16.Khan SA, Hamayun M, Kim HY, et al. . Gibberellin production and plant growth promotion by a newly isolated strain of Gliomastix murorum. World J Microbiol Biotechnol. 2009;25:829–833. [PubMed] [Google Scholar]
- 17.Hwang YJ, Ghim SY.. Paenibacillus aceris sp. nov., isolated from the rhizosphere of Acer okamotoanum, a plant native to Ulleungdo Island, Republic of Korea. Int J Syst Evol Microbiol. 2017;67(4):1039–1045. [DOI] [PubMed] [Google Scholar]
- 18.Wang B, Wang L.. Penicillium kongii, a new terverticillate species isolated from plant leaves in China. Mycologia. 2013;105(6):1547–1554. [DOI] [PubMed] [Google Scholar]
- 19.Park SJ, Hwang GJ, Park SJ, et al. . The study of naturalized plants in Ulleungdo. Kor J Env Ecol. Eco. 2007;21:1–12. [Google Scholar]
- 20.Wang XC, Chen K, Zeng ZQ, et al. . Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Sci Rep. 2017;7(1):8233–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan SA, Hamayun M, Kim HY, et al. . A new strain of Arthrinium phaeospermum isolated from Carex kobomugi Ohwi is capable of gibberellin production. Biotechnol Lett. 2009;31(2):283–287. [DOI] [PubMed] [Google Scholar]
- 22.Khan SA, Hamayun M, Kim HY, et al. . Molecular identification and diversity of endophytic fungi isolated from Pinus densiflora in Boeun. Korea. Kor J Mycol. 2009;37:130–133. [Google Scholar]
- 23.Kim HJ, Kim JS, Cheon KH, et al. . Species list of Aspergillus, Penicillium, and Talaromyces in Korea, based on ‘One Fungus One Name’ system. Kor J Mycol. 2016;44:207–219. [Google Scholar]
- 24.Kirk PM, Cannon PF, Minter DW, et al. . Ainsworth and Bisby’s dictionary of the fungi. 10th ed Wallingford: CAB International; 2008. [Google Scholar]
- 25.Thompson JD, Gibson TJ, Plewniak F, et al. . The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Stecher G, Tamura K.. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You YH, Aktaruzzaman M, Heo I, et al. . Talaromyces halophytorum sp. nov. Mycobiology. 2020;48(2):133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markmann M, Tautz D.. Reverse taxonomy: an approach towards determining the diversity of meiobenthic organisms based on ribosomal RNA signature sequences. Philos Trans R Soc Lond B Biol Sci. 2005;360(1462):1917–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visagie CM, Hirooka Y, Tanney JB, et al. . Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol. 2014;78:63–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson SW. Phylogenetic analysis of Penicillium species based on ITS, and LSU–rDNA nucleotide sequences In: Samson RA, Pitt JI (eds) Integration of modern taxonomic methods for Penicillium, and Aspergillus classification. Amsterdam: Harwood Academic Publishers, 2000. pp. 163–178. [Google Scholar]
- 31.Welham KJ, Domin MA, Johnson K, et al. . Characterization of fungal spores by laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2000;14(5):307–310. [DOI] [PubMed] [Google Scholar]