Abstract
In the present study, we upgraded Pyrococcus furiosus Argonaute (PfAgo) mediated nucleic acid detection method and established a highly sensitive and accurate molecular diagnosis platform for the large-scale screening of COVID-19 infection. Briefly, RT-PCR was performed with the viral RNA extracted from nasopharyngeal or oropharyngeal swabs as template to amplify conserved regions in the viral genome. Next, PfAgo, guide DNAs and molecular beacons in appropriate buffer were added to the PCR products, followed by incubating at 95 °C for 20–30 min. Subsequently, the fluorescence signal was detected. This method was named as SARS-CoV-2 PAND. The whole procedure is accomplished in approximately an hour with the using time of the Real-time fluorescence quantitative PCR instrument shortened from >1 h to only 3–5 min per batch in comparison with RT-qPCR, hence the shortage of the expensive Real-time PCR instrument is alleviated. Moreover, this platform was also applied to identify SARS-CoV-2 D614G mutant due to its single-nucleotide specificity. The diagnostic results of clinic samples with SARS-CoV-2 PAND displayed 100% consistence with RT-qPCR test.
Keywords: SARS-CoV-2, PfAgo, Molecular diagnosis
Graphical abstract
1. Introduction
The ongoing pandemic of COVID-19 results from a new coronavirus, SARS-CoV-2 (Zhu et al., 2020) has already caused more than 68 million confirmed cases and 1.5 million deaths (https://ncov.dxy.cn/ncovh5/view/pneumonia). To slow down the spread of COVID-19, the infected individuals need to be identified and quarantined efficiently. Molecular diagnosis approaches, including nucleic acid and serological tests offer the possibility for fast pinpoint of infected patients (Shen et al., 2020). Serological test is rapid and requires minimal equipment. But it may take several days for the patient to show a detectable antibody response after onset of initial symptoms (Wölfel et al., 2020). On the other hand, nucleic acid detecting method directly identifies conserved sequences in the SARS-CoV-2 genome and provides the earliest and most sensitive detection of the presence of SARS-CoV-2. Therefore, it is still the primary method for the diagnosis of COVID-19 (Esbin et al., 2020). Currently, the most recommended nucleic acid test is RT-qPCR. The RT-qPCR kits include reverse transcriptase, DNA polymerase, target-specific primers and TaqMan probes. These probes are short oligonucleotides containing 5′-fluorophore and 3′-quencher and anneal to the target DNA. During the extension step of each thermocycle, Taq polymerase moves along the DNA template and degrades the annealed probe through its 5′ to 3′ exonuclease activity. The fluorophore is cleaved off and released from being quenched. The change of fluorescence signal is measured at the end of each thermocycle. Since the outbreak of COVID-19, the worldwide RT-qPCR testing capability increased dramatically and reached millions of tests per day. Even though RT-qPCR test is rapid, specific and economic, it takes hours to perform and requires specialized reagents, expensive equipment and skilled operators (Li et al., 2019). To overcome these limitations, alternative nucleic acid detecting methods, especially programmable endonuclease based methods were innovated (Ding et al., 2020) (Guo et al., 2020) (Hou et al., 2020) (Joung et al., 2020) (Lucia et al., 2020).
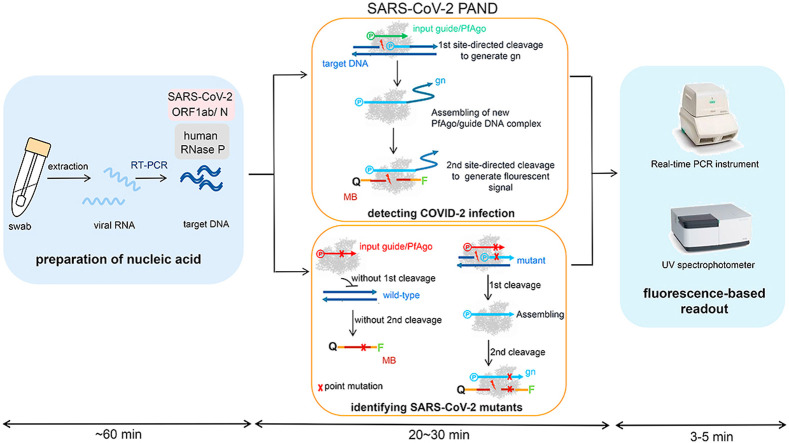
PfAgo is a prokaryotic Argonaute (pAgo) from Pyrococcus furiosus (Swarts DC et al., 2015). As a nucleic-acid-guided endonuclease, PfAgo prefers to cleave DNA substrate under the guidance of short 5′-phosphorylated single-strand DNA without the presence of protospacer-adjacent motif (PAM). In 2019, we developed a PfAgo (Pyrococcus furiosus Argonaute) mediated nucleic acid detection method (PAND) (He et al., 2019). In the present study, we simplified PAND and applied it on the detection of SARS-CoV-2 and its variants (Fig. 1 ). The viral RNA was extracted from nasopharyngeal or oropharyngeal swabs and conserved regions in the viral genome are amplified through RT-PCR. Next, PfAgo, guide DNA and molecular beacon in appropriate buffer were added and mixed with the PCR product, followed by incubating at 95 °C for 20–30 min. During this process, PfAgo cleaved the target DNA under the guidance of the input guide and generated a short 5′-phosphorylated single-stranded DNA, which in turn, bonded to apo form of PfAgo molecules and served as the guide for the second round of site-directed cleavage to complementary molecular beacons, leading to the split of the quencher from the fluorophore. Subsequently, the fluorescence signal was detected with Real-time fluorescence quantitative PCR instrument or fluorescence spectrometer. This method was named as SARS-CoV-2 PAND. The sensitivity of this method reached 1 copy per reaction. SARS-CoV-2 PAND is compatible with RT-PCR testing reagent easily available, but the working time of Real-time fluorescence quantitative PCR instruments is shortened from >1 h to only 3–5 min per batch in comparison with RT-qPCR test, hence, solves the problem of the shortage of the expensive Real-time PCR instruments. Meanwhile, the targets are amplified with easy-handled regular RT-PCR, followed by a simple enzymatic digestion and no skilled operator is required. Moreover, one shortcoming of RT-qPCR test is the high false-negative rate (Younes et al., 2020). SARS-CoV-2 PAND is more sensitive and reproducible because amplification of the targets is performed in a RT-PCR system without the interference of TaqMan probes. In addition, SARS-CoV-2 PAND is capable of identifying a single-base mutation in the genome of SARS-CoV-2, which cannot be accomplished with RT-qPCR assay.
Fig. 1.
Schematic of recommended SARS-CoV-2 PAND workflow. MB: molecular beacon; gn: newly generated guide; Q: quencher; F: fluorophore.
2. Materials and method
2.1. Primers, enzymes and reagents
All the DNA guides and molecular beacons used in this study were synthesized by Sangon (China). One-step RT-qPCR kits were purchased from ThermoFisher Scientific (USA), Szybio (China) and Yeasen (China). SYBR Gold for resolving urea-denaturing polyacrylamide gel electrophoresis (PAGE) was purchased from ThermoFisher Scientific, USA.
2.2. Construction of plasmids bearing N gene and ORF1ab gene
Nucleocapsid protein (N) and ORF1ab gene fragments were assembled as previously described (Chen et al., 1994). Six primers were synthesized for each target (Fig. S1). Two oligonucleotides (5 μmol) encoding 5′-ends of each target were mixed with lower amount of the other two oligonucleotides (0.5 μmol) and PCR was carried out under the following conditions (25 cycles): 98 °C, 30 s for denaturing; 58 °C, 30 s for annealing; 72 °C, 10 s for elongation. The PCR products were then amplified with primers bearing homologous region with pUC57 and cloned into pUC57 plasmid through T5 exonuclease mediated cloning method (She et al., 2018). The recombinant plasmids were named as pUC57-N and pUC57-O, respectively.
2.3. TBE-denaturing PAGE
The nucleic acid polyacrylamide gel (20%) was prepared by mixing 4.2 g urea, 5 ml of 40% acrylamide, 1 ml of 10 × TBE (Tris-borate-EDTA) buffer, 2 ml formamide, 9 μl of 10% APS and 1 μl TEMED, followed by adding sterile dH2O to a total volume of 10 ml. Next, 10 μl of the sample was mixed with 10 μl of 2 × loading buffer (95% formamide, 0.25% SDS, 0.25% bromophenol blue and 0.25% xylene cyanol FF), and incubated at 95 °C for 5 min. Gel electrophoresis was performed with 0.5 × TBE buffer at 150 V for 90 min, then the gels were stained with SYBR Gold (Invitrogen, USA).
2.4. The recommended workflow of SARS-CoV-2 PAND
Expression and purification of the recombinant PfAgo was performed as previously described (He et al., 2019). RT-PCR mixture in a final volume of 25 μl was prepared as previously described. In brief, 5 μl of the extracted viral RNA, RT-PCR mix, three primer pairs for ORF1ab/N/Human RNase P gene (400 nM for each primer, Table S1) were mixed. Human RNase P gene was used as the reference gene. Reaction was performed as follows: 1 cycle of reverse transcription at 55 °C for 10 min; 94 °C for 3 min, 45 thermo-cycles of denaturing at 94 °C for 15 s, extending at 60 °C for 20 s. Subsequently, 1 μl of purified PfAgo (60 pmol), 0.5 μl of 5′-phosphorylated input guide DNA (3 pmol each) and 0.5 μl of molecular beacons (10 pmol each) (Table S2) and 3 μl of 10 × reaction buffer containing 200 mM HEPES (pH 7.5), 2.5 M NaCl, and 5 mM MnCl2 was added to the PCR products to a final volume of 30 μl. The site-directed cleavage was carried out at 95 °C for 20–30 min, followed by detecting the fluorescence intensity of each sample with Bio-rad Real-time fluorescence quantitative PCR instrument (USA).
2.5. Determining LoD of SARS-CoV-2 PAND
To investigate the sensitivity of SARS-CoV-2 PAND, plasmid pUC57-O was extracted with Omega EZNA plasmid mini kit (USA) and the concentration of plasmid pUC57-O was measured with Nanodrop 8000 (ThermoFisher Scientific, USA), followed by serial dilution with dH2O to concentrations of 200, 20, 2, 1.6, 0.8 and 0.2 copy/μl. The serially diluted samples were utilized for SARS-CoV-2 PAND and RT-qPCR detection. RT-qPCR was carried out using SuperScript™ III One-Step RT-qPCR System with Platinum™ Taq High Fidelity DNA Polymerase (ThermoFisher, USA).
2.6. Detecting D614G variant in clinical samples with SARS-CoV-2 PAND
The 23367-23430 nt fragment of the viral genome including nt23403 (D614G mutation) was amplified with primer pairs 614F and 614R (400 nM for each, Table S3). RT-PCR was performed as above mentioned. The PCR product was split equally into two vials for site-directed cleavage of PfAgo with input guide gWT1 and gMT1, respectively. The fluorescence intensity of each sample was measured with Bio-rad Real-time fluorescence quantitative PCR instrument (USA).
2.7. RT-qPCR assay
Clinical nasopharyngeal and oropharyngeal swab samples from patients infected with SARS-CoV-2 were collected and tested by Centers for Disease Control and Prevention (CDC) Wuhan, China. The viral RNA extraction was performed using a KingFisher Flex nucleic acid extraction system (ThermoFisher, USA). Primers for RT-qPCR testing were designed based on the recommendation of the Chinese CDC (Table S4). Conditions for amplification were 45 °C for 10 min, and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min for fluorescence detection by LightCycle 480II fluorescent PCR instrument (Roche, Switzerland). A cycle threshold value (Ct-value) ≤ 37 was defined as a positive test and a Ct-value of 40 or more was defined as a negative test based on the manufacturer's manual. A medium load, defined as a Ct-value of 37 to less than 40, required confirmation by retesting. Both internal controls and negative controls were routinely performed with each batch of tests.
3. Results
3.1. Establishing the workflow of SARS-CoV-2 PAND
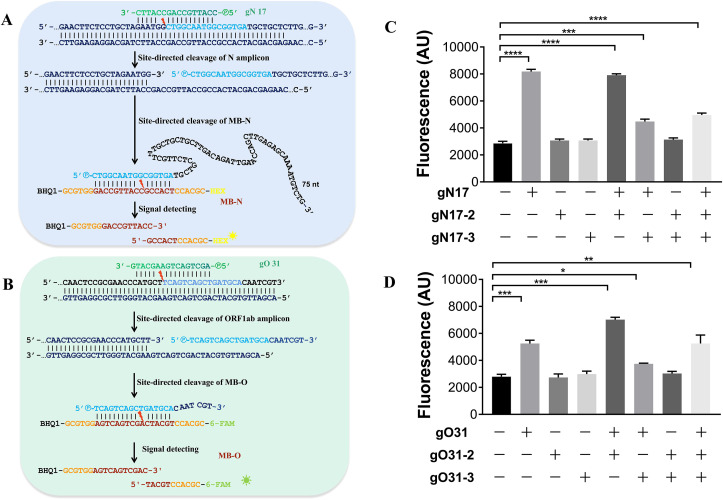
The coding region of N and ORF1ab in the viral genome (GenBank Accession No. MN985325.1) were chosen as the targets based on the guidance of World Health Organization (WHO) (Organization, n.d.). The key point of SARS-CoV-2 PAND is the efficient cleavage of PfAgo/input guide complex to the target DNA. Therefore, we designed 36 guides covering the full amplifying regions of both N and ORF1ab and tested the cleavage activity of corresponding PfAgo/guide complexes to the ssDNA targets (Fig. S2; Fig. S3; Fig. S4). Accordingly, guides gN 17 and gO 31 were chosen for SARS-CoV-2 PAND due to the high activity of the complexes. The cleavage sites and the sequences of guides were indicated in Fig. 2 A and B. To detect N and ORF1ab gene with SARS-CoV-2 PAND, PfAgo protein, guide DNA (gN17 or gO31), molecular beacon (MB-N or MB-O) were added directly to the RT-PCR product amplified using pUC57-ORF1ab and pUC57-N spike-in samples as template since PfAgo demonstrated high activity in RT-PCR buffer supplemented with Mn2+ (Fig. S5). Obvious elevation of fluorescence signals was detected with both targets in comparison with the negative controls (Fig. 2C and D), which proved the feasibility of SARS-CoV-2 PAND. Meanwhile, two more input guides (gN 17-2 and gN 17-3 for N gene; gO 31-2 and gO 31-3 for ORF1ab gene) were designed for each target based on the principle of PAND (Fig. S6). However, the intensity of the fluorescence signal showed no obvious increase. Therefore, we established the workflow for SARS-CoV-2 PAND with only one input guide (Fig. 1). It is worth noting that the gn for N gene is 91 nt, with a 75-nt 3′-unpaired regions to the molecular beacon (Fig. 2A), which is much longer than previous reported guides of Argonaute protein (Dayeh et al., 2018) (Hegge et al., 2019) (Kaya et al., 2016) (Kuzmenko et al., 2019).
Fig. 2.
Establishing SARS-CoV-2 PAND with single input guide. A. SARS-CoV-2 PAND with N gene as the target; B. SARS-CoV-2 PAND with ORF1ab gene as the target. Sequences of 5′-phosphorylated input guides (gN 17, gO 31) are labeled in green. Newly generated single-stranded DNAs (gns) are indicated in blue, while the complementary sequences of gns in the molecular beacons are highlighted in brown; C. the signal intensity of detecting N gene with one, two or three input guides; D. the signal intensity of detecting ORF1ab gene with one, two or three input guides.
3.2. Optimization of SARS-CoV2-PAND
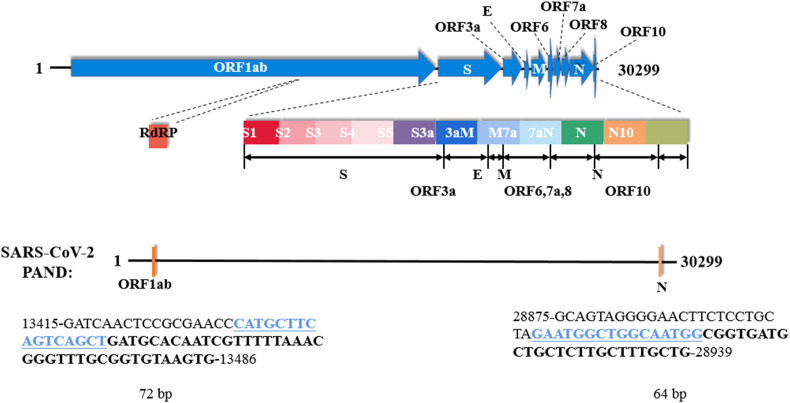
The effect of RT-PCR reagent to the sensitivity of SARS-CoV-2 PAND was investigated. RT-qPCR mix from three different companies all gave high readouts, which proved SARS-CoV-2 PAND has high compatibility with commercial RT-qPCR reagents (Fig. S7). Meanwhile, the boundary of ORF1ab and N amplicons were adjusted slightly to gain higher yield and specificity. The full sequences of the amplicons and the positions of input guides were indicated in Fig. 3 .
Fig. 3.
Schematic of the genome of SARS-CoV-2 and the optimized amplicons of ORF1ab and N gene. The sequences of the input guide gN-17 and gO-31 are highlighted in blue. The 3′-end of both gns are indicated in bold black letters.
We also investigated the correlation between the ratio of guide to the molecular beacon and the readouts of SARS-CoV-2 PAND. The fluorescence signal elevated with increasing ratio of guide to the molecular beacon in the reaction mixture (Fig. S8). Since the signal intensity with a ratio of 1:2 (1 μM of molecular beacon in the reaction mixture) reached approximately 4000 AU and enough for distinguishing positive and negative signals, we chose this ratio for the following assay.
3.3. The sensitivity of SARS-CoV-2 PAND
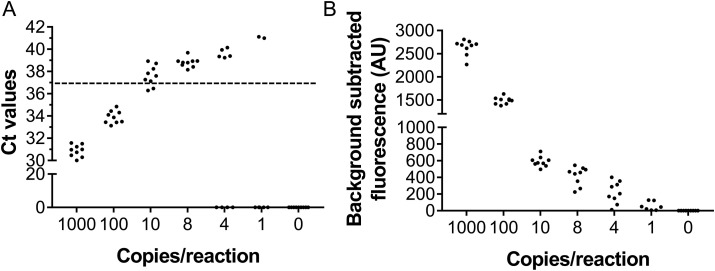
The analytic limit of detection (LoD) of SARS-CoV-2 PAND was compared with SARS-CoV-2 nucleic acid RT-qPCR detection kit from Thermofisher Scientific (MA, USA) with ORF1ab gene as the target. The LoD of the kit was 10 copies per reaction, with 3 out of 9 replicates obtaining positive results (Ct < 37) (Fig. 4 A). When the concentration decreased to 8 or 4 copies per reaction, Ct-values with all nine replicates of each titer were zero or >37 and RT-qPCR failed to detect the target (Fig. 4A). In comparison with RT-qPCR assay, the sensitivity of SARS-CoV-2 PAND was much higher. All replicates except of one gave positive readouts at a titer of 8 or 4 copies per reaction. When the target was further diluted to 1 copy per reaction, four replicates out of nine were still capable of detecting the viral target (Fig. 4B). This result indicated LoD of SARS-CoV-2 PAND was 1 copy per reaction.
Fig. 4.
LoD for RT-qPCR test with Thermofisher nucleic acid detection kit (A) and SARS-CoV-2 PAND assays (B).
3.4. Analysis of clinical samples with SARS-CoV-2 PAND
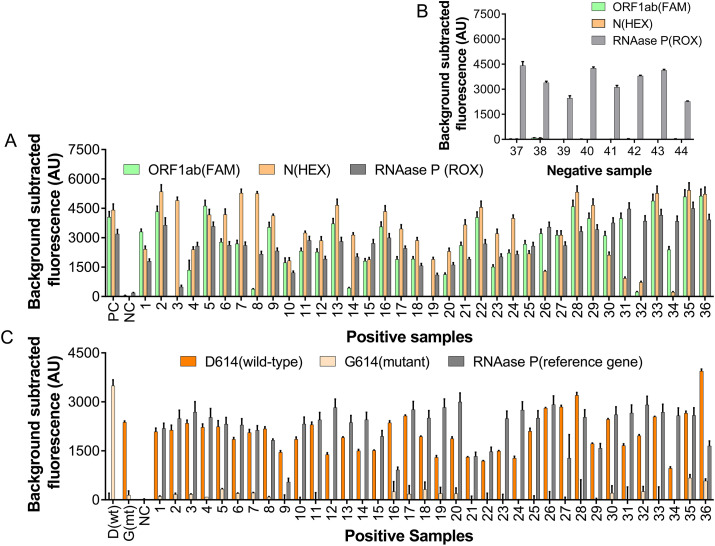
Thirty-six specimens (nasopharyngeal and oropharyngeal swabs) were collected in February 2020 in Wuhan, China from suspected COVID-19 infected individuals and tested positive for COVID-19 infection using RT-qPCR method (Fig. S9). The result was confirmed by SARS-CoV-2 PAND (Fig. 5 A). All samples gave positive readouts for all three channels except of No.3 and 19, with undetectable ORF1ab signals. Since the signal for N gene showed obvious elevation in comparison with the negative control, they were still considered as positive for COVID-19 infection. We deduced the low readout for ORF1ab was caused by the low RNA concentration in these samples because the readouts of the reference gene for these samples were also relatively lower than others. We also performed SARS-CoV-2 PAND with 8 specimens collected in June 2020 and all the results were negative for COVID-19 infection (Fig. 5B), which is consistent with the result of RT-qPCR test carried out in June 2020 (Ct > 40).
Fig. 5.
Identifying SARS-CoV-2 and its D614G mutants with PAND in clinical samples. A. testing results of clinical samples collected in February 2020 with SARS-CoV-2 PAND; B. testing results of clinical samples collected in June 2020 with SARS-CoV-2 PAND; C. detection of nt23403 SNP in clinical samples with PAND.
3.5. Analysis of viral variants with SARS-CoV-2 PAND
SARS-CoV-2 PAND was also applied for large-scale identification of SARS-CoV-2 variants. The D614G point mutation in spike (S) protein caused by a single-based mutation at nt23403 was chosen as the target (Pachetti et al., 2020). Input guides targeting the wild-type and mutant locus were screened and the result indicated gWT1 and gMT1 were able to guide the site-directed cleavage to wild-type and the mutant with high specificity, respectively (Fig. S10A, B). Next, SARS-CoV-2 PAND was performed with the mock samples and the result indicated gWT1 and gMT1 were able to distinguish mutant and wild-type distinctly through comparing the difference between the readouts (Fig. S10C). Therefore, SARS-CoV-2 PAND was designed based on these two input guides (Fig. S11) and applied to identify D614G point mutation of SARS-CoV-2 in clinical samples which were identified as positive in the last session. The readouts of wild-type for all 36 samples were significantly higher than mutant, which indicated that all the samples were wild-type (Fig. 5C). This result is consistent with previous reports that this mutant was discovered mainly in Europe after February 2020 (Korber et al., 2020). In rare cases, two or more alleles may co-exist in one sample, and the alleles in low concentration are hardly detectable. Under this circumstance, NAVIGATER (Nucleic Acid enrichment Via DNA Guided Argonaute from Thermus thermophilus), a TtAgo mediated nucleic acid enrichment method (Song et al., 2020), can be coupled with our method to detect trace amount of rare alleles.
4. Conclusion
With the sustained development of COVID-19 pandemic, the situation is not optimistic in the coming winter. It is still necessary to develop fast, accurate, low-cost and easy-handled molecular diagnostic methods to identify SARS-CoV-2 and its variants. With its unique merits, SARS-CoV-2 PAND is one of the powerful tools against SARS-CoV-2, and it can also be adopted rapidly to confront the outbreak of other diseases caused by pathogenic microorganisms in the future.
CRediT authorship contribution statement
Fei Wang: Methodology, Data curation, Validation, Visualization. Jun Yang: Methodology, Validation. Ruyi He: Methodology, Data curation, Validation. Xiao Yu: Resources, Validation. Shuliang Chen: Resources. Yang Liu: Investigation. Longyu Wang: Investigation. Aitao Li: Writing - review & editing. Linlin Liu: Resources, Supervision. Chao Zhai: Resources, Investigation, Writing - original draft. Lixin Ma: Conceptualization, Validation, Resources, Funding acquisition, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by a major technological innovation project in Hubei province (2017ACA174).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112932.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- Chen G.-Q., Choi I., Ramachandran B., Gouaux J.E. J. Am. Chem. Soc. 1994;116:8799–8800. [Google Scholar]
- Dayeh D.M., Cantara W.A., Kitzrow J.P., Musier-Forsyth K., Nakanishi K. Nucleic Acids Res. 2018;46:1–13. doi: 10.1093/nar/gky496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Yin K., Li Z., Lalla R.V., Ballesteros E., Sfeir M.M., Liu C. Nat. Commun. 2020;18(1):4711. doi: 10.1038/s41467-020-18575-6. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. RNA. 2020;26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., Chen Y., Feng G., Gu Q., Wang R.R., Zhou Q., Li W. Cell Discov. 2020;19:34. doi: 10.1038/s41421-020-0174-y. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Wang L., Wang F., Li W., Liu Y., Li A., Wang Y., Mao W., Zhai C., Ma L. Chem. Commun. 2019;55:13219–13222. doi: 10.1039/c9cc07339f. [DOI] [PubMed] [Google Scholar]
- Hegge J.W., Swarts D.C., Chandradoss S.D., Cui T.J., Kneppers J., Jinek M., Joo C., Van Der Oost J. Nucleic Acids Res. 2019;47:5809–5821. doi: 10.1093/nar/gkz306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., Wu J., Liao Y., Gou X., Li Y., Wang X., Su H., Gu B., Wang J., Xu T. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Kim N.G., Woolley A.E., Segel M., Rpj Barretto, Ranu A., Macrae R.K., Faure G., Ioannidi E.I., Krajeski R.N., Bruneau R., Huang M.W., Yu X.G., Li J.Z., Walker B.D., Hung D.T., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. N. Engl. J. Med. 2020;8(15):1492–1494. doi: 10.1056/NEJMc2026172. 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4057–4062. doi: 10.1073/pnas.1524385113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield Covid-19 Genomics Group, McDanal C., Perez L.G., Tang H., et al. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmenko A., Yudin D., Ryazansky S., Kulbachinskiy A., Aravin A.A. Nucleic Acids Res. 2019;47:5822–5836. doi: 10.1093/nar/gkz379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li S., Wang J., Liu G. Trends Biotechnol. 2019;37(7):730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Lucia C., Federico P.-B., Alejandra G.C. bioRxiv. 2020 2020.02.29.971127. [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. J. Transl. Med. 2020;22(1):179. doi: 10.1186/s12967-020-02344-6. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She W., Ni J., Shui K., Wang F., He R., Xue J., Reetz M.T., Li A., Ma L. ACS Synth. Biol. 2018;7:2236–2244. doi: 10.1021/acssynbio.8b00245. [DOI] [PubMed] [Google Scholar]
- Shen N., Zhu Y., Wang X., Peng J., Liu W., Wang F., Lu Y., Cheng L., Sun Z. JCI Insight. 2020;21(10) doi: 10.1172/jci.insight.137662. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Hegge J.W., Mauk M.G., Chen J., Till J.E., Bhagwat N., Azink L.T., Peng J., Sen M., Mays J., Carpenter E.L., van der Oost J., Bau H.H. Nucleic Acids Res. 2020;28(4):e19. doi: 10.1093/nar/gkz1165. 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J., Terns R.M., Terns M.P., van der Oost J. Nucleic Acids Res. 2015;26(10):5120–5129. doi: 10.1093/nar/gkv415. 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 2020. Global Surveillance for COVID-19 Caused by Human Infection with COVID-19 Virus: Interim Guidance, 20 March 2020.https://apps.who.int/iris/handle/10665/331506 [Google Scholar]
- Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Viruses. 26. 2020;12(6):582. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.