Abstract
An outbreak, caused by an RNA virus, SARS-CoV-2 named COVID-19 has become pandemic with a magnitude which is daunting to all public health institutions in the absence of specific antiviral treatment. Surface glycoprotein and nucleocapsid phosphoprotein are two important proteins of this virus facilitating its entry into host cell and genome replication. Small interfering RNA (siRNA) is a prospective tool of the RNA interference (RNAi) pathway for the control of human viral infections by suppressing viral gene expression through hybridization and neutralization of target complementary mRNA. So, in this study, the power of RNA interference technology was harnessed to develop siRNA molecules against specific target genes namely, nucleocapsid phosphoprotein gene and surface glycoprotein gene. Conserved sequence from 139 SARS-CoV-2 strains from around the globe was collected to construct 78 siRNA that can inactivate nucleocapsid phosphoprotein and surface glycoprotein genes. Finally, based on GC content, free energy of folding, free energy of binding, melting temperature, efficacy prediction and molecular docking analysis, 8 siRNA molecules were selected which are proposed to exert the best action. These predicted siRNAs should effectively silence the genes of SARS-CoV-2 during siRNA mediated treatment assisting in the response against SARS-CoV-2.
Keywords: SARS-CoV-2, Nucleocapsid phosphoprotein, Surface glycoprotein, siRNA, siDirect
1. Introduction
COVID-19, A pandemic affecting lives of billions of people worldwide, has confronted humanity in the commencement of 2020, is caused by a viral pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or 2019-nCoV. Initial symptoms of this disease mainly include fever, cough, fatigue, dyspnea & headache [1,2] or it may be asymptomatic [3]. The spike glycoprotein of SARS-CoV-2 binds directly with the surface cell angiotensin converting enzyme II (ACE2) receptor present on alveolar epithelial cells of lung facilitating virus entry, replication and triggers cytokine cascade mechanism [4]. In severe cases, patient may die due to massive alveolar damage and progressive respiratory failure [1,5]. The current detection process of SARS-CoV-2 carried out by most countries is using real-time RT-PCR, although several other methods are also being developed [[6], [7], [8]]. Incubation period for the virus ranges between 2 and 14 days [9] and in some cases, transmission is also reported during asymptomatic period [10]. Some recent studies suggest that bats are likely reservoir hosts for SARS-Cov-2 but the identity of the intermediate host that might have facilitated transfer to human still remain elusive with some studies indicating pangolins [11]. SARS-CoV-2 is assumed to spread mainly from person-to-person through respiratory droplets produced when an infected person sneezes and coughs or between people who are in close contact [5].
Coronaviruses are genetically classified into four main genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus [12]. The first two genera generally infect mammals, while the last two mostly cause disease in birds. The genome size of coronaviruses ranges between approximately 26–32 kb and includes about 6 to 11 open reading frames (ORFs) [13]. Nucleocapsid protein (N), small envelope protein (E), spike surface glycoprotein (S) and matrix protein (M) are the four major structural proteins of coronavirus and all of which are essential to produce a structurally complete virus [14,15]. The nucleocapsid protein (N) is a multifunctional protein comprising three distinct and highly conserved domains: two structural and independently folded structural regions, namely the N terminal domain and C-terminal domain, which are separated by an intrinsically disordered RNA-binding domain [16]. The primary role of CoV N protein is to package the genomic viral genome into flexible, long, helical ribonucleoprotein (RNP) complexes called nucleocapsids [17]. Apart from these, N protein is essential for viral assembly, envelope formation, genomic RNA synthesis, cell cycle regulation and viral pathogenesis [[18], [19], [20]]. Spike glycoprotein (S) is a viral fusion protein which forms homotrimers protruding from the viral surface [21] and mediates virus entry to cell [22]. S contains two functional subunits: S1 & S2 subunits. The S1 subunit includes the receptor-binding domain(s) and contributes to stabilization of the membrane-anchored S2 subunit that contains the fusion machinery [23]. As the coronavirus S glycoprotein is surface-exposed and mediates entry into host cells and N nucleocapsid protein are essential for genome replication, these could be the main targets for designing therapeutics [24].
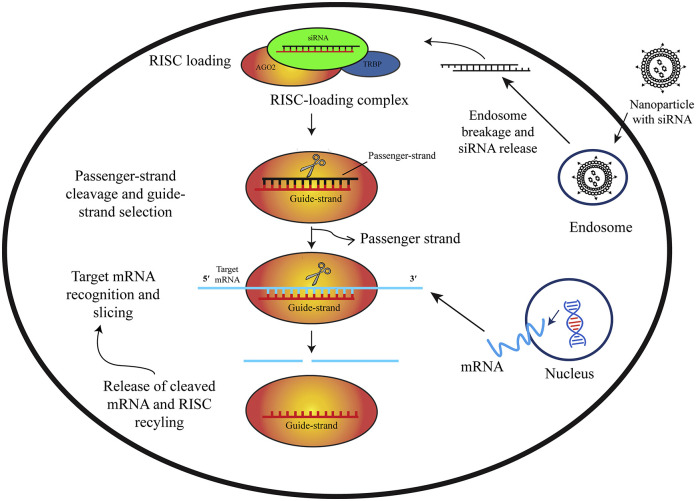
Silencing of mRNA or post-transcriptional gene silencing by RNA interference (RNAi) is a regulatory cellular mechanism. RNAi is a prospective tool for the control of human viral infections [[25], [26], [27]]. Small interfering RNAs (siRNAs) and micro RNAs (miRNAs) are involved in the RNA interference (RNAi) pathway, where they hybridize to complementary mRNA molecules and neutralizes mRNA causing suppression of gene expression or translation [28]. Studies show that, combinations of chemically synthesized siRNA duplexes targeting genomic RNA of SARS-CoV results in therapeutic activity of up to 80% inhibition [29]. siRNAs directed against Spike sequences and the 3′-UTR can inhibit the replication of SARS-CoV(Fig. 1)
Fig. 1.
Graphical representation of the siRNA-mediated gene silencing mechanism. Nanoparticles loaded with siRNA are taken up through endocytosis by the cells. These particles are then trapped into the endosomes. siRNA escape endosomes and release siRNA into the cytoplasm due to pH responsive mechanism or proton sponge effect. Once generated, siRNA is loaded into RNA-indiced silencing complex comprising of RNA-binding protein TRBP and Argonaute (Ago2). Ago2 opts the siRNA guide strand, then excises and ejects the passenger strand. After that, the guide strand pairs with its complementary target mRNA and Ago2 slices the target. After slicing, the cleaved target mRNA is released and RISC is recycled for another few rounds of slicing using the same guide strand [74].
As of November 14, 2020, total infected case is 53,521,552 and among these patients 1,306,135 people have died which means case fatality rate (CFR) is approximately 2.44%. The alarming phenomenon is the exponential growth of total infection case and death number (https://www.worldometers.info/coronavirus/). Treatment of this increased number of people is not possible as no antiviral drug is still available specifically for SARS-CoV-2 and there is a lack on appropriate medical response. In-silico approaches are a general trend to discover novel therapeutic approaches [[30], [31], [32], [33]] and for the viruses there is no exception to this [34]. Therefore, in this study, we have designed siRNAs specific to various conserved region of nucleocapsid phosphoprotein & surface glycoprotein genes of SARS-CoV-2 and finally predicted 8 universal siRNA molecules against nucleoprotein and glycoprotein genes which will inhibit the translations of these proteins and allow the host to discard this infection. siRNAs are designed against both nucleoprotein and glycoprotein as both are important for the survival of virus [22,35] and targeting these proteins may cause viral inhibition [29,36]. Currently, ONPATTRO® and GIVLAARI™ RNAi therapeutics are commercially approved on the market for the treatment of polyneuropathy and acute hepatic porphyria respectively [37]. We hope this study will help to develop a similar treatment strategy for SARS-CoV-2.
2. Materials and methods
2.1. Sequence retrieval from NCBI
Coding sequences from 139 genomes of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) were retrieved from NCBI Virus [38] portal (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/) (Supplementary Table 1). Nucleocapsid phosphoprotein and surface glycoprotein sequences were manually extracted and curated from the retrieved data using bash scripting in Linux computer platform.
2.2. Multiple sequence alignment & phylogenetic tree construction
ClustalW [39] algorithm was employed to perform multiple sequence alignment (MSA). Maximum likelihood phylogenetic trees were constructed with a bootstrap value of 500. Tamura Nei [40] model of evolution was selected while constructing the phylogenetic tree. MEGA-X [41] and MEGA-CC [42] programs were was used for alignment formation and phylogenetic tree construction respectively. iTOL online tool [43] (https://itol.embl.de/) was used in order to visualize the phylogenetic trees.
2.3. Target recognition & siRNA designing
Target-specific siRNAs were designed with the help of siDirect web server [44]. Rules of Ui-Tei [45], Amarzguioui [46] and Reynolds [47] were used (Table 1 ) and the melting temperature was kept below 21.5 °C as a default parameter for siRNA duplex.
Table 1.
Rules/Algorithm to construct siRNAs.
| Rule name | Description |
|---|---|
| Ui-Tei | A or U present at the 5′ terminus of the sense strand |
| G or C present at the 5′ terminus of the antisense strand | |
| At least 4 A or U residues present in the 5′ terminal 7 bp of sense strand | |
| GC stretch no longer than 9 nt | |
| Amarzguioui | Duplex End A or U differential >0 |
| No U present at position 1 | |
| Strong binding of 5′ sense strand | |
| Presence of A at position 6 | |
| Weak binding in case of 3′ sense strand | |
| Reynolds | 1 point for GC content 30–52% (one point) |
| 1 point for each occurrence of three or more A or U base pair at position 15–19 of sense strand | |
| 1 point for little internal stability at target site (Tm > −20 °C) | |
| 1 point for occupancy of U at position 10 of the sense strand | |
| 1 point for occupancy of A at position 3 of the sense strand | |
| 1 point for occupancy of A at position 19 of the sense strand | |
| 1 point for Absence of G at position 13 of the sense strand | |
| Threshold for efficient siRNAs score ≥ 6 |
2.4. Off target similarity search using BLAST
Blast search was performed against human genome and transcriptome using the standalone blast package [48] to identify the possible off target matches. The e-value was set to 1e-10 to reduce the stringency of the search condition thereby increasing the chances of random matches.
2.5. GC content calculation & secondary structure prediction
OligoCalc [49] was used to calculate the GC content. The secondary structure of siRNAs was predicted along with the respective free energy using MaxExpect [50] program in the RNA structure web server [51]. Here, the higher values of energy indicate better candidates as those molecules are less prone to folding.
2.6. Computation of RNA-RNA interaction through thermodynamics
Higher interaction between the target and the guide strand serves a better predictor for siRNA efficacy. Therefore, the thermodynamic interaction between the target strand and the siRNA guide strand was predicted with the aid of DuplexFold [52] program of the RNA structure web server [51].
2.7. Computation of heat capacity & concentration plot
DINA Melt web server [53] was used to generate heat capacity and concentration plot. The ensemble heat capacity (Cp) is plotted as a function of temperature, with the melting temperature Tm (Cp) (Supplementary Table 6 & Supplementary Table 7). The contributions of each species to the ensemble heat capacity are shown by detailed heat capacity plot. Also, the point at which the concentration of double-stranded molecules of one-half of its maximum value defines the melting temperature Tm (Conc) was shown using the concentration plot- Tm (Conc).
2.8. Predicted siRNA validation
siRNAPred server (http://crdd.osdd.net/raghava/sirnapred/index.html) was used to validate the predicted siRNA species. The predicted siRNAs were evaluated against the Main 21 datasets using support vector machine algorithm and the binary pattern prediction approach. siRNAPred score greater than 1 predicts very high efficacy, score ranging 0.8–0.9 predicts high efficacy and score ranging 0.7–0.8, predicts moderate efficacy. In total, 78 siRNAs were used for efficacy prediction.
2.9. Modelling & docking of argonaute protein 2 and siRNAs
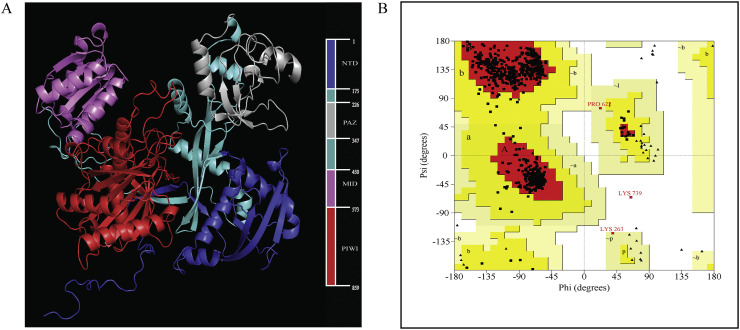
The three dimensional structure of argonaute 2 (Ago2) protein was formed using Modeler 9.25 [54,55] against the crystal structure of RNA bound argonaute protein (pdb id: 4z4d) [56] via homology modelling. The modelled protein was refined in the GalaxyRefine [57,58] server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE). The quality of the model was checked using Ramachandran Plot generated by PROCHECK [59].
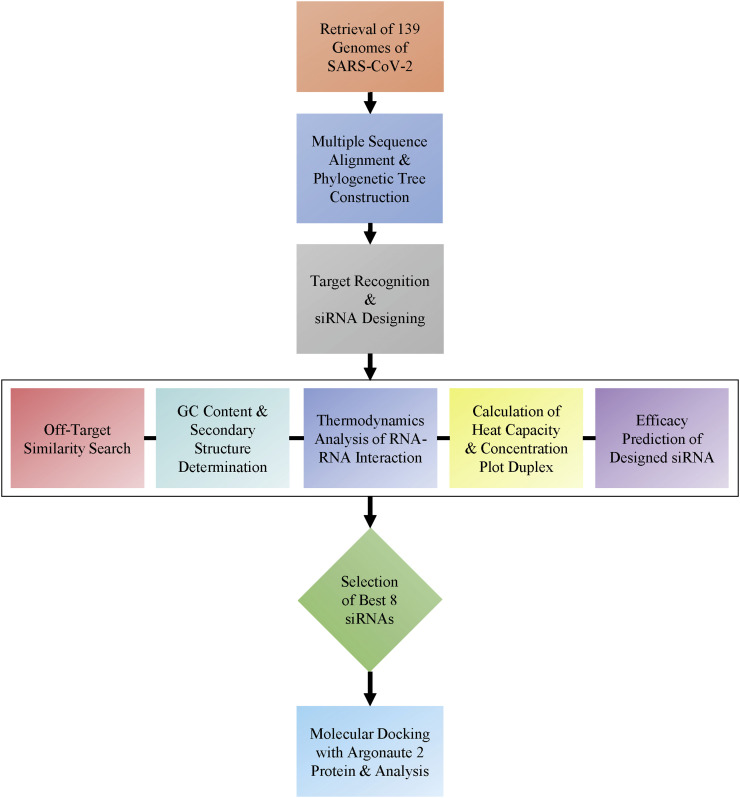
The guide RNA, bound in the structure of 4z4d, was taken as template to model the siRNA. The sequence of the guide RNA structure was changed by mutating and appending the necessary nucleotides in Discovery Studio Visualizer (version 20.1) thereby, obtaining the particular siRNA structure that is reminiscent to the template structure. Thereafter, the siRNAs were optimized in MMFF94 force field using steepest descent algorithm for 1000 steps (convergence at 10e-7) in Avogadro v1.2.0. Finally, molecular docking for the siRNAs against the argonaute protein was performed in HDOCK server (http://hdock.phys.hust.edu.cn/) [60,61]. The best results were taken based on the lowest docking energy score and docked complex where siRNA was bound to the binding cavity of Ago2. The structures were analyzed and the necessary images were generated using the PyMOL Molecular Graphics System (v1.8.4). A graphical workflow of the methodology used in this study was provided as well (Fig. 2 ).
Fig. 2.
The graphical workflow of the methodology used in the study.
3. Results
3.1. Evolutionary divergence analysis shows conserved pattern between strains
Phylogenetic tree was constructed using 139 sequences for both nucleocapsid phosphoprotein and surface glycoprotein separately. Only a handful number of sequences showed significant divergence (bootstrap value >60%) (Fig. 3 ). This implied that most of the viral sequences have been conserved and therefore could be used to construct siRNA which should cover a wide range of SARS-CoV-2 strains.
Fig. 3.
Radial phylogenetic tree of A. nucleocapsid phosphoprotein and B. surface glycoprotein using 139 strains of SARS-CoV-2 from around the world. The bootstrap value for tree construction was set to 500 and Tamura-Nei model of evolution was used for both trees.
3.2. siDirect predicted 78 siRNA
siDirect web server predicted 8 siRNA for nucleocapsid phosphoprotein and 70 siRNA for surface glycoprotein (Supplementary Table 4 & Supplementary Table 5) that maintains all the parameters. Seed target duplex stability (Tm) values for all the predicted siRNAs were less than 21.5 °C which suggests the ability of predicted siRNAs to avoid non-target binding.
3.3. Off-target binding exclusion using blast
Standalone blast [48] search against human genome and transcriptome did not reveal any off-target match. This shows that our predicted siRNA would not interact in any places other than the viral target location.
3.4. GC content calculation & secondary structure determination
GC content analysis of the predicted siRNAs was ranged from 31% to 43% for nucleocapsid phosphoprotein (Supplementary Table 6) and 10% to 40% for surface glycoprotein (Supplementary Table 7). Molecules that have GC content below 31.6% were eliminated. Also, the calculated free energy of folding ranged from 1.4 to 2 for nucleocapsid phosphoprotein (Supplementary Table 6) and from 1.3 to 2 for surface glycoprotein (Supplementary Table 7). The associated secondary structures were also determined.
3.5. Thermodynamics of target-guide strand interaction
Free energy of binding between target and guide strand was calculated. The values spanned from −35.8 to −31 for nucleocapsid phosphoprotein (Supplementary Table 6) and −36.6 to −21.6 for surface glycoprotein (Supplementary Table 7).
3.6. Heat capacity & duplex concentration plot determination
The Tm(Cp) and Tm(Conc) were calculated for the predicted siRNAs. The higher values of these two melting temperatures indicate higher effectiveness of the siRNA species. Tm(Conc) values ranged from 71.7 °C to 81.7 °C for nucleocapsid phosphoprotein (Supplementary Table 6) and 66.4 °C to 83.8 °C for surface glycoprotein (Supplementary Table 7). Tm(Cp) values ranged from 72.1 °C to 82.5 °C for nucleocapsid phosphoprotein (Supplementary Table 6) and from 66.3 °C to 85.2 °C for surface glycoprotein (Supplementary Table 7).
3.7. Validation and selection of best 8 siRNAs
siRNAPred [62] checked the effectivity of the predicted siRNAs and values greater than 1 are considered highly effective. 2 siRNAs for nucleocapsid phosphoprotein and 32 siRNAs for surface glycoprotein were found to be highly effective. Based on all the other criteria, 8 siRNAs were selected as best predicted candidates against the nucleocapsid phosphoprotein and the surface glycoprotein genes of SARS CoV-2 (Table 2 ) and advanced for molecular docking.
Table 2.
Best effective siRNA molecules with various parameters.
| Alias | Conserved position | Location of target within mRNA | siRNA target within mRNA | Duplex candidate at 37◦C | Predicted siRNA(guide): siRNA (passenger) | Seed-duplex (Tm) guide | Seed-duplex (Tm) passenger | GC % | Free energy of folding | Free energy of binding | Tm (Conc) | Tm (Cp) | Validity (binary) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n7 | 1031-1253 | 33-55 | AAGCATATTGACGCATACAAAAC | GCAUAUUGACGCAUACAAAAC | UUUGUAUGCGUCAAUAUGCUU | 13.5 | 5.6 | 36 | 1.5 | −31.7 | 75.8 | 77 | 1.047 |
| g15 | 871-1222 | 20-42 | CAGAAACAAAGTGTACGTTGAAA | GAAACAAAGUGUACGUUGAAA | UCAACGUACACUUUGUUUCUG | 21 | 5.6 | 36 | 1.6 | −32.8 | 79.1 | 80 | 1.022 |
| g21 | 871-1222 | 136-158 | TGCCCTTTTGGTGAAGTTTTTAA | CCCUUUUGGUGAAGUUUUUAA | AAAAACUUCACCAAAAGGGCA | 3.2 | 16.1 | 36 | 1.8 | −33.4 | 83.4 | 85 | 1.039 |
| g22 | 871-1222 | 138-160 | CCCTTTTGGTGAAGTTTTTAACG | CUUUUGGUGAAGUUUUUAACG | UUAAAAACUUCACCAAAAGGG | 0 | 18.8 | 33 | 1.9 | −30.4 | 78.6 | 80 | 1.01 |
| g44 | 1842-2389 | 441-463 | CACAATTAAACCGTGCTTTAACT | CAAUUAAACCGUGCUUUAACU | UUAAAGCACGGUUUAAUUGUG | 19.7 | −9.7 | 33 | 1.9 | −30.0 | 79.5 | 80 | 1.102 |
| g46 | 1842-2389 | 482-504 | GACAAAAACACCCAAGAAGTTTT | CAAAAACACCCAAGAAGUUUU | AACUUCUUGGGUGUUUUUGUC | 17.7 | 5.6 | 36 | 1.6 | −32.5 | 80.6 | 82 | 1.076 |
| g59 | 2790-3822 | 304-326 | GTGTGTACTTGGACAATCAAAAA | GUGUACUUGGACAAUCAAAAA | UUUGAUUGUCCAAGUACACAC | 13.8 | 19 | 36 | 1.5 | −33.9 | 80.4 | 81 | 1.007 |
| g70 | 2790-3822 | 1001-1023 | GTGCTCAAAGGAGTCAAATTACA | GCUCAAAGGAGUCAAAUUACA | UAAUUUGACUCCUUUGAGCAC | 7.4 | 16.6 | 38 | 1.9 | −33.7 | 81.7 | 83 | 1.012 |
3.8. Analysis of molecular modelling and docking of best siRNAs & Ago2
According to the Ramachandran plot of Ago2 protein, the percentage of residues resided in core region, allowed region, moderately allowed region, and disallowed region were 95.7%, 4.0%, 0.1%, and 0.1% respectively. The docking score, interaction statistics, interactive residues etc. were provided (Table 3, Supplementary Table 8). Ago2-siRNA complex containing g59 and g15 showed the highest and lowest docking score of −515.5 and − 317.2 respectively, keeping rest of the values in between.
Table 3.
siRNA clusters, docking score and interaction statistics.
| Cluster no. | siRNA in Ago2-siRNA complex | HDock docking score | Interaction statistics |
||
|---|---|---|---|---|---|
| Hydrogen bonds | Electrostatic bonds | Hydrophobic bonds | |||
| Cluster 1 | g15 | −317.2 | 45 | 40 | 12 |
| g70 | −341.1 | 27 | 28 | 5 | |
| g46 | −357.8 | 50 | 31 | 11 | |
| g59 | −515.5 | 48 | 32 | 8 | |
| Cluster 2 | g21 | −328.2 | 17 | 13 | 4 |
| g22 | −336.8 | 18 | 11 | 1 | |
| n7 | −356.5 | 18 | 16 | 2 | |
| g44 | −356.9 | 21 | 16 | 3 | |
4. Discussion
COVID-19 is an emerging disease that lays bare the society we have created and its interdependent infrastructure with a surge in cases and deaths since its initial identification. Having no regard for geography, this pandemic has a global reach, and no continent is out of its clutches. Moreover, there is no vaccine available to prevent this disease and no RNAi based treatment is yet in practice or been proposed. So, the next generation medicine, siRNA might be effective in this case, hence it is the focus of our study.
Here, a total of 34 (15 nucleocapsid phosphoprotein and 19 surface glycoprotein) (Supplementary Table 2 & Supplementary Table 3) conserved regions were identified among 139 strains of SARS-CoV-2 from around the world. Phylogenetic analysis revealed that a small number of sequences form significant clades with a bootstrap value greater than 60%. (Fig. 3). Conserved portions that are shorter than 21 nucleotides were omitted from further analysis. Conserved sequences were put to siDirect web server to identify possible targets and to generate corresponding siRNAs. siDirect performs the task in three distinct steps – highly functional siRNA selection, seed-dependent off-target effects reduction, near-perfect matched genes elimination. siRNA targets were found in 18 conserved regions, 5 nucleocapsid phosphoprotein (Supplementary Table 4) and 13 surface glycoprotein (Supplementary Table 5). U,R,A (Ui-Tei, Amarzguioui and Reynolds) rules (Table 1) were applied while predicting the siRNAs to obtain better results. siRNA bond formation with non-target sequences was eliminated by optimizing the melting temperature (Tm) below 21.5 °C. The equation to calculate melting temperature (Tm) is below,
Here,
-
▪
The sum of the nearest neighbor enthalpy change is represented by ΔH (kcal/ mol)
-
▪
The helix initiation constant (−10.8) is represented by A
-
▪
The sum of the nearest neighbor entropy change is represented by ΔS
-
▪
The gas constant (1.987 cal/deg./mol) is represented by R
-
▪
The total molecular concentration (100 μM) of the strand is represented by CT and
-
▪
Concentration of Sodium, [Na+] was fixed at 100 mM
siRNA's functionality is influenced by the GC-content and there is an inverse relationship of the GC-content with the function of siRNA. High GC content in the siRNA corresponds to high content in the target region. siRNA targets with high GC contents might have a propensity for folding which could potentially reduce target accessibility [63]. Usually a low GC content, approximately from 31.6 to 57.9%, is ideal for a siRNA to be effective [63]. Therefore, we calculated the GC content of the predicted siRNAs. Molecules that have GC content lower than 32% were not kept in the final selection. Here, GC content ranged from 10% to 43% for all the 78 predicted species. GC content of finally selected siRNAs is greater than or equal to 33% (Table 2).
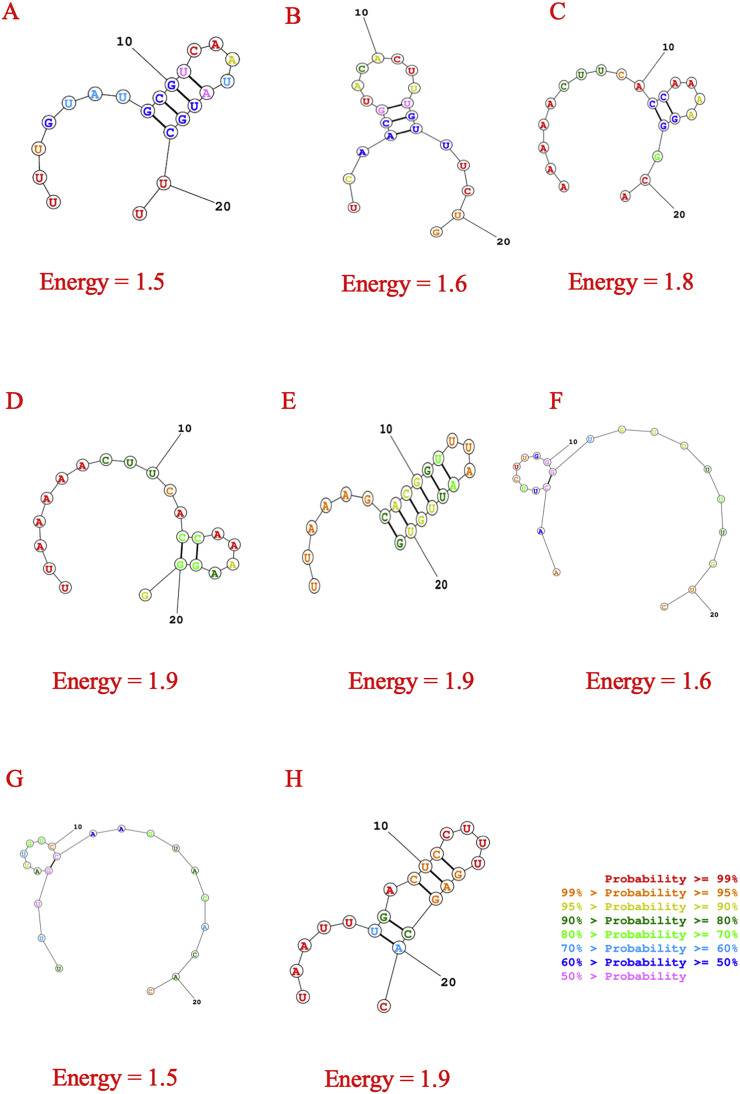
Formation of secondary structure of siRNA may inhibit the RISC mediated cleavage of target. So, the prediction of prospective secondary structure and determination of free energy of corresponding folding is crucial. Here, guide strands of predicted siRNAs were subjected to RNA structure web server in order to predict possible folding structures and corresponding minimum free energies. At 37 °C, finally selected siRNAs have free energy of folding greater than zero (Fig. 4 , Table 2), which suggests the predicted siRNAs are more accessible for efficient binding.
Fig. 4.
Secondary structures of best eight predicted siRNA with probable folding and lowest free energy for consensus sequence. The structures are for A. n7 B.g15 C. g21 D. g22 E. g44 F. g46 G. g59 H. g70 siRNAs.
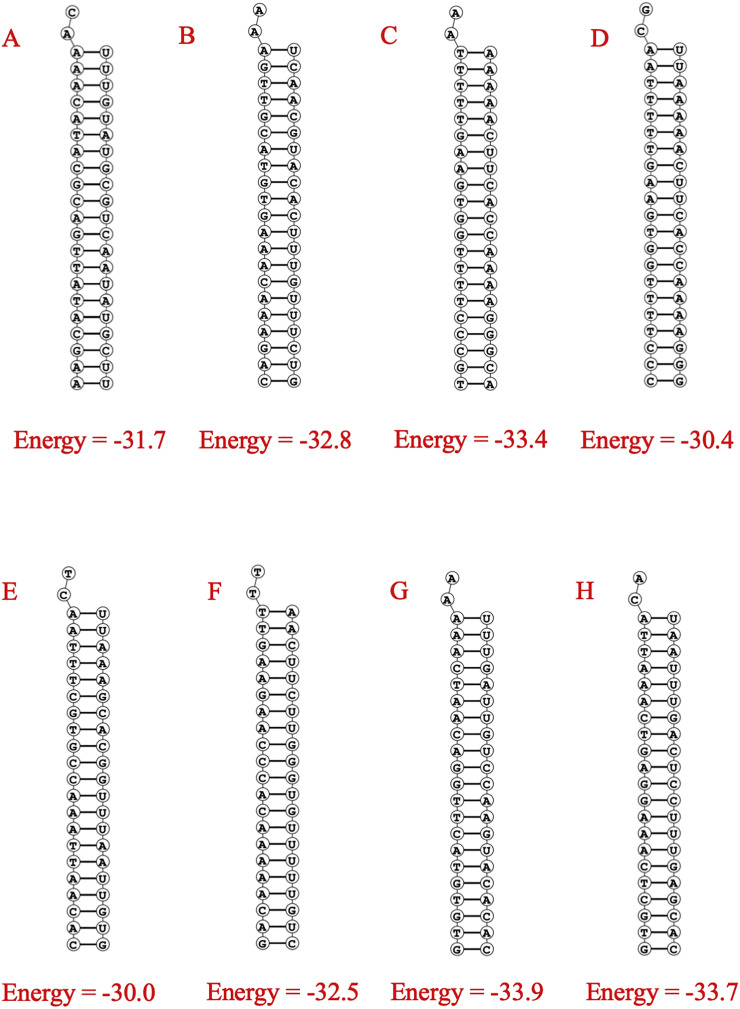
DuplexFold [52] was used to determine the target and guide siRNA interaction and their corresponding binding energy. Lower binding energy indicates better interaction therefore better chance of target inhibition. The values of free energy of binding of all the 78 predicted siRNAs spanned from −36.6 to −21.6 (Supplementary Table 6 & Supplementary Table 7). Finally selected siRNAs have free energy of binding equal or below −30.0 (Fig. 5, Table 2), which suggests the predicted siRNAs are more interactive with their corresponding targets.
Fig. 5.
Structure of binding of siRNA (guide strand) and target RNA with corresponding predicted minimum free energy. The structures are for A. n7 B.g15 C. g21 D. g22 E. g44 F. g46 G. g59 H. g70 siRNAs and their corresponding targets.
The collective heat capacity, denoted as Cp, is plotted as a function of temperature and the melting temperature, denoted as Tm (Cp), was determined. The contribution of each molecule to the collective heat capacity was demonstrated using the inclusive heat capacity plot where melting temperature Tm (Conc), indicates the temperature at which the concentration of double-stranded molecules becomes one-half of its maximum value. DINA Melt web server [53] was used to obtain the melting temperatures. All the selected siRNAs have high Tm value (>75 °C) (Table 2).
siRNAPred [62] was used to determine the inhibition efficacy of the predicted siRNAs. siRNAPred uses Main21 dataset which consist of 2182 siRNAs (21mer) derived from a homogeneous experimental condition to predict the actual efficacy of 21mer siRNAs with high accuracy using the support vector machine based method. Here, siRNA candidates that have validity score greater than one were chosen for the final selection. Considering optimal parameters from the results of GC content calculation, DuplexFold, DINA Melt, and siRNAPred webservers 8 best siRNA candidates were selected (Table 2). These siRNAs are expected to perform better than all the other predicted siRNAs in the study. Afterwards, siRNAs were considered for molecular docking analysis for better understanding their interaction with Ago2 protein which is the key enzyme in RISC.
The complete structure of Ago2 was predicted in Modeler 9.25 standalone package based on homology modelling which is essentially built on target template alignment. The process further comprises steps such as fold designation, model construction and quality control [54]. The missing residues of the protein in the crystal structure were successfully predicted, thus providing the opportunity of covering the maximum interactions between protein and siRNA complex. Afterwards, the model was refined in GalaxyRefine web server to improve the global as well as the local structure quality. The method initially reconstructs side chains, executes side-chain repacking and thereafter performs an overall relaxation by small molecular dynamics simulation in two steps namely, mild and aggressive relaxations. The resultant model had 95.7% residues inside the core region Ramachandran plot thereby surpassing the 90% cut-off and qualifying as a good quality structure.
In the cellular environment, there are multiple factors that are involved in the mechanism of siRNA mediated gene silencing, and one needs to account for all the factor to effectively simulate the interaction patterns and the structural status of all the stakeholders involved in the process which is virtually impossible to this date. De-novo prediction of the siRNA structure will try to obtain a conformation with the lowest energy which may not necessarily be the exact confirmation that will take place while it interacts with Ago2. Molecular docking will also try out other possible conformations of the siRNA, which will further reduce the chance to have a clearer look at the actual event. Therefore considering aforementioned statements, unlike protein structure prediction, a different approach was taken for the prediction of siRNA structure that had similarity with other research groups. The guide RNA bound to 4z4d structure was taken as template and mutated in Discovery Studio Visualizer (version 20.1) to model the siRNA structures. This approach ensured a better conformational reminiscence and a more appropriate depiction of the actual event.
After modelling, molecular docking of the Ago2 and siRNAs were done with the aid of HDOCK server (http://hdock.phys.hust.edu.cn/) [60,61]. The server uses a hybrid-docking algorithm of template-based modelling and free docking and a hierarchical FFT-based docking program, HDOCKlite, to execute traditional global docking. The resultant docking complexes were downloaded from the server and inspected manually to identify best docked complex based on docking score, visual similarity of the complex with 4z4d structural composite and placement of siRNA inside PAZ domain and MID domain of the Ago2. Such criteria were enforced to reduce the spurious results and identify the complex that is most similar to experimental evidence [64].
In order to exert the silencing function, the 5′ and 3′ ends of the siRNA need to be anchored in the MID and PAZ domain of the Argonaute protein respectively [64], although there is a room for flexibility in PAZ domain due to its dynamic nature [65,66]. On the other hand, strong binding of siRNA with Ago2, especially PAZ domain, corresponds to weaker nuclease/RNAi activity [67]. This is expected as 3′ end of siRNA needs to dynamically toggle in between bound and unbound states during the nuclease activity therefore a strong binding with PAZ domain may interrupt this process [67].
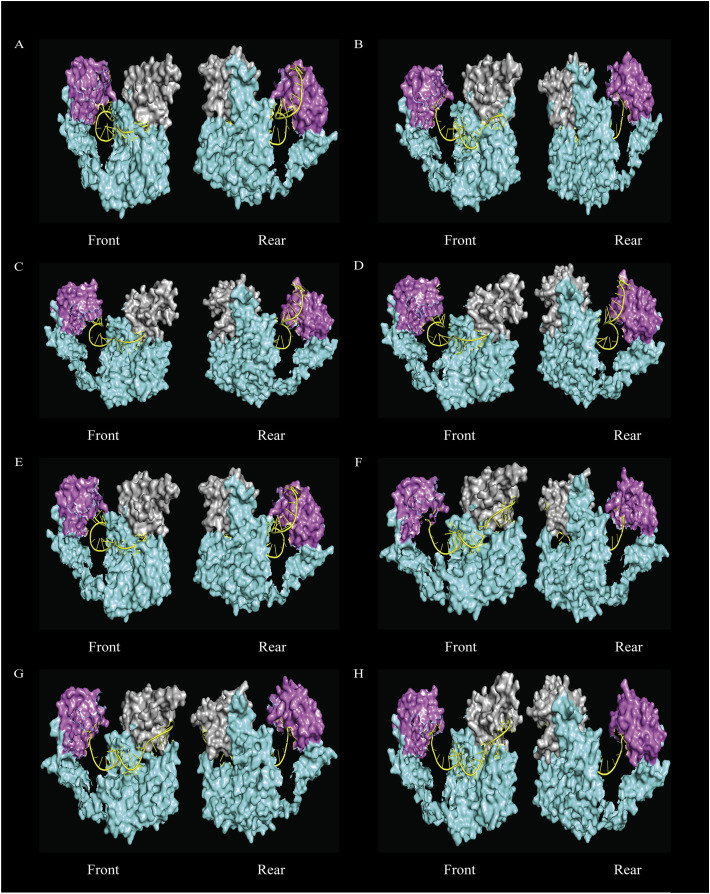
Based on the placement of siRNA inside the Ago2 protein two distinct clusters were found. Four out of the eight siRNA namely, g15, g59, g46, g70, showed anchoring in both MID and PAZ domain forming the Cluster 1 (Fig. 6 ). Although rest of the four siRNAs (n7, g21, g22, g44), constituting Cluster 2, protruded outside of the protein near the 5′ end, they successfully anchored to the PAZ domain of the protein. Cluster 1 showed a consistent bonding pattern with 11 residues of protein where the bonding category and contributing group matches for all the four-member of the group. Ago2 residues such as ARG179, THR526, THR759 formed hydrogen bond, ARG277, LEU522, TYR529 formed hydrophobic interactions, and ARG277 formed an electrostatic interaction with the bases and LYS566 formed a hydrogen bond and ARG68, ARG315, LYS533, ARG812 formed electrostatic with the phosphate backbone of all the Cluster1 siRNAs. TYR529, LYS533 residues of MID domain interacted with first U/A of the siRNA which is also reported by others [68]. Some residues that interacted with all Cluster 1 siRNAs such ARG277, ARG315, THR526, TYR529, LYS533, LYS566, THR759, ARG812 are previously reported to form interactions with other siRNA as well [68,69]. (See Fig. 7.)
Fig. 6.
A. The cartoon representation of the structure of modelled Argonaute 2 (Ago2) protein. The N-terminal domain (NTD), PAZ domain, MID domain and PIWI domain corresponds to blue, gray, pink and red color respectively. Rest of the protein was denoted by Aquamarine color. The sidebar corresponds to range of residues possessed by each domain. B. The Ramachandran plot of modelled Ago2 protein. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Docked structures of siRNA (cartoon view) with Ago2 proteins (surface view). The PAZ domain and the MID domain are colored as gray and pink respectively and rest of the protein denoted by aquamarine color (including N-terminal domain and PIWI domain). The siRNA was designated by yellow color. Front and rear view (180° opposite to Front view along x-axis) were provided and some residues of the N-terminal domains were omitted in order to clearly depict the anchoring of siRNA within Ago2. The structures are for A. n7 B.g15 C. g21 D. g22 E. g44 F. g46 G. g59 H. g70 siRNA complexes respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In contrast to Cluster 1, Cluster 2 showed fewer consistent bonding pattern where only two residues of the protein (MID domain) interacted (ARG460 with base and ARG554 with phosphate backbone) with all four siRNAs of the group. This was not surprising, as siRNAs in Cluster2 were not completely encapsulated inside the Ago2 resulting in fewer interactions. ARG635, ARG792 ubiquitously formed electrostatic interaction with phosphate backbone of all the siRNAs of Cluster 1 and Cluster 2. As both ends placed within the Ago2 protein, Cluster 1 siRNAs more likely to perform better than Cluster 2.
Previously reported interactive residues of Ago2 such as ARG635, ARG710, ARG792 interact with all eight siRNAs [68,69]. Other reported residues do not encapsulate any particular cluster e.g. ARG635, ARG710, ARG792 interacts with n7, g15, g21, g22, g44, g46, g59, g70 whereas ARG761, TYR804 interacts with the aforementioned siRNAs except g21. Table 4 contains previously reported [68,69] protein residues involved in siRNA-Ago2 interactions that also participated in the interaction with siRNAs of the current study.
Table 4.
Previously reported interactive Argonaute protein residues involved in interactions with siRNAs of the current study.
| siRNA | Residues matching the reported residues |
|---|---|
| n7 | ARG351, GLN548, ASN551, ARG635, ARG710, ARG761, ARG792, SER798, TYR804 |
| g15 | ALA221, ARG277, ARG315, ARG351, ILE365, THR526, TYR529, LYS533, GLN548, LYS566, ARG635, LYS709, ARG710, GLN757, THR759, ARG761, ARG792, TYR804, ARG812 |
| g21 | ARG351, ARG635, ARG710, TYR790, ARG792, CYS793 |
| g22 | ARG635, ARG710, ARG761, TYR790, ARG792, CYS793, SER798, TYR804 |
| g44 | ARG351, GLN548, ASN551, ARG635, ARG710, ARG761, ARG792, SER798, TYR804 |
| g46 | ALA221, THR222, HIS271, ARG277, ARG315, HIS316, ARG351, ILE365, THR526, TYR529, LYS533, GLN545, CYS546, GLN548, ASN551, THR559, ASN562, LYS566, ARG635, LYS709, ARG710, GLN757, THR759, ARG761, ARG792, SER798, TYR804, ARG812 |
| g59 | THR222, HIS271, ARG277, ARG315, ARG351, ILE365, GLY524, THR526, TYR529, LYS533, GLN545, CYS546, GLN548, ASN551, THR559, ASN562, LYS566, ARG635, LYS709, ARG710, GLN757, THR759, ARG761, ARG792, TYR804, ARG812 |
| g70 | ASP125, THR222, ARG277, ARG315, ARG351, GLY524, THR526, TYR529, LYS533, ASN551, LYS566, ARG635, ARG710, THR759, ARG761, ARG792, TYR804, ARG812 |
The highest and lowest docking score of Cluster 1 ranged from −317.2 to −515.5 for g15 and g59 respectively. Although Cluster 2 siRNAs did not place effectively in MID domain, the docking scores were in between −328.2 to −356.9 that may confer better efficacy during RNAi. Higher hydrogen bonding is associated with higher RNAi whereas electrostatic interaction shows a moderate correlation with RNAi activity [67]. The total number of hydrogen bonds in the study ranged from 17 to 50 which was, in general, predominant than electrostatic and hydrophobic interactions. Electrostatic and hydrophobic interactions ranged 11–40 and 1–12 respectively.
From the structural perspective, Cluster 1 complexes undoubtedly obtained conformation close to actual event, therefore they are expected to outperform Cluster 2 complexes in RNAi activity. However, from the perspective of docking score the results are inconclusive as g21 & g22 have higher energy than g70 and n7 & g44 have higher docking energy than g46. Therefore, some Cluster 2 siRNAs are expected to show greater RNAi activity than some Cluster 1 siRNAs. Considering the siRNAPred data adds further complexity to the analysis as all of the Cluster 2 siRNAs were predicted to show better efficacy than g15 which is the best candidate from both structural and docking perspective. Finally, due to the inherent difficulty in effective modelling of complex cellular processes such as RISC, molecular docking of siRNA and Ago2 was used a proxy which, although provided some great insights, did not help to further filter out siRNA candidates. Therefore, all the eight siRNAs are proposed to be effective in SARS-CoV-2 treatment and should be further validated in in-vivo for therapeutic purpose.
Various research groups also proposed similar siRNA treatment strategy for COVID 19 [70,71]. Chen et al. [72], for example, took a different computational approach to identified 9 siRNAs for 5 different sites of SARS-CoV-2 genome (length 21 – 25nts) using single reference genome obtained from NCBI (https://www.ncbi.nlm.nih.gov/nuccore/MN908947) and analyzing single nucleotide polymorphisms in the target sites. Parikesit & Nurdiansyah [73], alternatively, took 24 sequences of a single gene, performed MSA and used the conserved sequences for in-silico development of siRNA. Unlike aforementioned research groups, this study considered nucleocapsid phosphoprotein and surface glycoprotein genes of 139 genomes of SARS-CoV-2 which provided scope for a more detailed analysis. Interaction between the siRNA and argonaute 2 protein was also taken into consideration in the current study which was previous studies overlooked. Pharmaceutical companies such as Siranomics, Vir Biotechnology & Alnylam Pharmaceuticals (United States), OilX Pharmaceuticals (South Korea) have also identified various RNAi targets and corresponding siRNA agents of SARS-CoV-2 [70]. We believe our study will be an interesting addition to this landscape.
In this study, eight prospective siRNA molecules were proposed to be efficient at binding and cleaving specific mRNA targets of SARS-CoV-2 (Table 2). As the study contain a large array of 139 sequences of SARS-CoV-2 from around the world, the predicted therapeutic agent can be employed to large scale treatment of COVID-19 pandemic.
5. Conclusions
Computational methods can be employed to design and predict siRNA interaction against specific gene target thereby silencing its expression. In this research, eight siRNA molecules were predicted to be effective against nucleocapsid phosphoprotein and surface glycoprotein gene of 139 strains of SARS-CoV-2 virus using computational method considering all maximum parameters in prime conditions and state of art molecular modelling and docking analysis. In order to decelerate the COVID-19 pandemic and recover the affected individuals, the development of siRNA therapeutic approaches could be a promising alternative to traditional vaccine designing.
Accession number, length and location of the SARS-CoV-2 stains used in the study.
Conserved sequence of nucleocapsid phosphoprotein gene.
Conserved sequence of surface glycoprotein gene.
Tm values of predicted siRNA (guide strand and passenger strand) against nucleocapsid phosphoprotein.
Tm values of predicted siRNA (guide strand and passenger strand) against surface glycoprotein.
Effective siRNAs against nucleocapsid phosphoprotein with GC%, free energy of folding and free energy of binding with target.
Effective siRNAs against surface glycoprotein with GC%, free energy of folding and free energy of binding with target.
siRNA and Ago2 interacting residues, bond distance and bond category.
CRediT authorship contribution statement
Umar Faruq Chowdhury: Methodology, Writing - original draft, Visualization. Mohammad Umer Sharif Shohan: Conceptualization, Data curation, Methodology, Writing - original draft, Visualization. Kazi Injamamul Hoque: Visualization, Writing - original draft. Mirza Ashikul Beg: Writing - review & editing. Mohammad Kawsar Sharif Siam: Conceptualization, Conceptualization, Writing - original draft, Visualization, Supervision. Mohammad Ali Moni: Supervision.
Declaration of Competing Interest
The authors declare that they have no competing interests.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song F., Shi N., Shan F., Zhang Z., Shen J., Lu H., Ling Y., Jiang Y., Shi Y. Emerging 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295:210–217. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C.Y., Poon R.W.S., Tsoi H.W., Lo S.K.F., Chan K.H., Poon V.K.M., Chan W.M., Ip J.D., Cai J.P., Cheng V.C.C., Chen H., Hui C.K.M., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan S., Nakajima R., Jain A., de Assis R.R., Jasinskas A., Obiero J.M., Adenaiye O., Tai S., Hong F., Milton D.K., Davies H., Felgner P.L., P.S. Group Analysis of serologic cross-reactivity between common human coronaviruses and SARS-CoV-2 using coronavirus antigen microarray. BioRxiv. 2020 doi: 10.1101/2020.03.24.006544. 2020.03.24.006544. [DOI] [Google Scholar]
- 9.Linton N.M., Kobayashi T., Yang Y., Hayashi K., Akhmetzhanov A.R., Jung S., Yuan B., Kinoshita R., Nishiura H. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J. Clin. Med. 2020;9:538. doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu P., Zhu J., Zhang Z., Han Y. A familial cluster of infection associated with the 2019 novel coronavirus indicating possible person-to-person transmission during the incubation period. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.-G., Ni X.-B., Liao Y.-S., Wei W., Cheung W.Y.-M., Li W.-J., Li L.-F., Leung G.M., Holmes E.C., Hu Y.-L., Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020:1–6. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 12.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11 doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;65:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C., Zheng X., Gai W., Zhao Y., Wang H., Wang H., Feng N., Chi H., Qiu B., Li N., Wang T., Gao Y., Yang S., Xia X. MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget. 2017;8:12686–12694. doi: 10.18632/oncotarget.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 17.de Haan C.A.M., Rottier P.J.M. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H., Gill A., Dove B.K., Emmett S.R., Kemp C.F., Ritchie M.A., Dee M., Hiscox J.A. Mass spectroscopic characterization of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding by using surface plasmon resonance. J. Virol. 2005;79:1164–1179. doi: 10.1128/JVI.79.2.1164-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurst K.R., Ye R., Goebel S.J., Jayaraman P., Masters P.S. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J. Virol. 2010;84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tortorici M.A., Veesler D. Adv. Virus Res. Academic Press Inc.; 2019. Structural insights into coronavirus entry; pp. 93–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard M.W., Travanty E.A., Jeffers S.A., Smith M.K., Wennier S.T., Thackray L.B., Holmes K.V. Aromatic amino acids in the juxtamembrane domain of severe acute respiratory syndrome coronavirus spike glycoprotein are important for receptor-dependent virus entry and cell-cell fusion. J. Virol. 2008;82:2883–2894. doi: 10.1128/jvi.01805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walls A.C., Tortorici M.A., Bosch B.-J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J.E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Gallagher T. Proteolytic processing of middle east respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levanova A., Poranen M.M. RNA interference as a prospective tool for the control of human viral infections. Front. Microbiol. 2018;9:2151. doi: 10.3389/fmicb.2018.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharif Shohan M.U., Paul A., Hossain M. Computational design of potential siRNA molecules for silencing nucleoprotein gene of rabies virus. Futur. Virol. 2018;13:159–170. doi: 10.2217/fvl-2017-0117. [DOI] [Google Scholar]
- 27.Chowdhury F.T., Shohan M.U.S., Islam T., Mimu T.T., Palit P. A therapeutic approach against leishmania donovani by predicting RNAi molecules against the surface protein, gp63. Curr. Bioinforma. 2019;14:541–550. doi: 10.2174/1574893613666180828095737. [DOI] [Google Scholar]
- 28.Hamilton A.J., Baulcombe D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science (80-. ) 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 29.Zheng B., Guan Y., Tang Q., Du C., Xie F.Y., He M.-L., Chan K.-W., Wong K.-L., Lader E., Woodle M.C., Lu P.Y., Li B., Zhong N. Prophylactic and therapeutic effects of small interfering RNA targeting SARS-coronavirus. Antivir. Ther. 2004;9:365–374. [PubMed] [Google Scholar]
- 30.Kabir E.R., Siam M.K.S., Mustafa N. ACM Int. Conf. Proceeding Ser. Association for Computing Machinery; New York, NY, USA: 2019. Scaffold of N-(2-(2-(tosylcarbamoyl)hydrazinyl)ethyl)isonicotinamidereveals anticancer effects through selective inhibition of FAP; pp. 1–11. [DOI] [Google Scholar]
- 31.Kabir E.R., Mustafa N., Kawsar M., Siam S., Kabir S.M. ACM Int. Conf. Proceeding Ser. Association for Computing Machinery; New York, New York, USA: 2018. Molecular docking reveals pitavastatin and related molecules antagonize 1DHF and its pseudogene DHFR2 in cancer treatment; pp. 1–9. [DOI] [Google Scholar]
- 32.Kabir E.R., Siami M.K.S., Kabir S.M., Khan A., Rajib S.A. ACM Int. Conf. Proceeding Ser. Association for Computing Machinery; New York, New York, USA: 2017. Drug repurposing: Targeting mTOR inhibitors for anticancer activity; pp. 68–75. [DOI] [Google Scholar]
- 33.Siam M.K.S., Hossain M.S., Kabir E.R., Rajib S.A. ACM Int. Conf. Proceeding Ser. Association for Computing Machinery; New York, New York, USA: 2017. In Silico structure based designing of dihydrofolate reductase enzyme antagonists and potential small molecules that target DHFR protein to inhibit the folic acid biosynthetic pathways; pp. 62–67. [DOI] [Google Scholar]
- 34.Rahman R., Rashid S.M.M., Sayeem M., Sharif I., Sharif K. Surface proteins, potential drug target for antiviral therapy against Nipah virus and in silico drug design. Clin. Biochem. 2011;44:S34. doi: 10.1016/j.clinbiochem.2011.08.1035. [DOI] [Google Scholar]
- 35.Almazan F., Galan C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/jvi.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C.J., Huang H.W., Liu C.Y., Hong C.F., Chan Y.L. Inhibition of SARS-CoV replication by siRNA. Antivir. Res. 2005;65:45–48. doi: 10.1016/j.antiviral.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., Liang X.-J. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., Schäffer A.A., Brister J.R. Virus variation resource – improved response to emergent viral outbreaks. Nucleic Acids Res. 2016;45:D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S., Stecher G., Peterson D., Tamura K. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics. 2012;28:2685–2686. doi: 10.1093/bioinformatics/bts507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naito Y., Yoshimura J., Morishita S., Ui-Tei K. SiDirect 2.0: Updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics. 2009;10:392. doi: 10.1186/1471-2105-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ui-Tei K., Naito Y., Takahashi F., Haraguchi T., Ohki-Hamazaki H., Juni A., Ueda R., Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amarzguioui M., Prydz H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 48.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Z.J., Gloor J.W., Mathews D.H. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA. 2009;15:1805–1813. doi: 10.1261/rna.1643609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bellaousov S., Reuter J.S., Seetin M.G., Mathews D.H. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013;41:W471–W474. doi: 10.1093/nar/gkt290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piekna-Przybylska D., DiChiacchio L., Mathews D.H., Bambara R.A. A sequence similar to tRNA3Lys gene is embedded in HIV-1 U3–R and promotes minus-strand transfer. Nat. Struct. Mol. Biol. 2010;17:83–90. doi: 10.1038/nsmb.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Markham N.R., Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. 2016;2016:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F., Šali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 56.Schirle N.T., Sheu-Gruttadauria J., Chandradoss S.D., Joo C., MacRae I.J. Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. Elife. 2015;4 doi: 10.7554/eLife.07646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heo L., Park H., Seok C. GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee G.R., Heo L., Seok C. Effective protein model structure refinement by loop modeling and overall relaxation. Proteins Struct. Funct. Bioinforma. 2016;84:293–301. doi: 10.1002/prot.24858. [DOI] [PubMed] [Google Scholar]
- 59.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. IUCr, PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 60.Yan Y., Zhang D., Zhou P., Li B., Huang S.-Y. HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45:W365–W373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang S.-Y., Zou X. A knowledge-based scoring function for protein-RNA interactions derived from a statistical mechanics-based iterative method. Nucleic Acids Res. 2014;42:e55. doi: 10.1093/nar/gku077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar M., Lata S., G.R.-P. of the First, U . CSIR-IMTECH; 2009. siRNApred: SVM Based Method for Predicting Efficacy Value of siRNA. 2009. [Google Scholar]
- 63.Chan C.Y., Carmack C.S., Long D.D., Maliyekkel A., Shao Y., Roninson I.B., Ding Y. A structural interpretation of the effect of GC-content on efficiency of RNA interference. BMC Bioinforma. 2009;101(10):1–7. doi: 10.1186/1471-2105-10-s1-s33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Müller M., Fazi F., Ciaudo C. Argonaute proteins: from structure to function in development and pathological cell fate determination. Front. Cell Dev. Biol. 2020;7:360. doi: 10.3389/fcell.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boland A., Tritschler F., Heimstädt S., Izaurralde E., Weichenrieder O. Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep. 2010;11:522–527. doi: 10.1038/embor.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boland A., Huntzinger E., Schmidt S., Izaurralde E., Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic argonaute protein. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10466–10471. doi: 10.1073/pnas.1103946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kandeel M., Kitade Y. Computational analysis of siRNA recognition by the Ago2 PAZ domain and identification of the determinants of RNA-induced gene silencing. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhandare V., Ramaswamy A. Structural dynamics of human argonaute2 and its interaction with siRNAs designed to target mutant tdp43. Adv. Bioinforma. 2016;2016 doi: 10.1155/2016/8792814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uludağ H., Parent K., Aliabadi H.M., Haddadi A. Prospects for RNAi therapy of COVID-19. Front. Bioeng. Biotechnol. 2020;8:916. doi: 10.3389/fbioe.2020.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Habtemariam S., Berindan-Neagoe I., Cismaru C.A., Schaafsma D., Nabavi S.F., Ghavami S., Banach M., Nabavi S.M. Lessons from SARS and MERS remind us of the possible therapeutic effects of implementing a siRNA strategy to target COVID-19: shoot the messenger! J. Cell. Mol. Med. 2020;24:10267–10269. doi: 10.1111/jcmm.15652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen W., Feng P., Liu K., Wu M., Lin H. Computational identification of small interfering RNA targets in SARS-CoV-2. Virol. Sin. 2020;35:359–361. doi: 10.1007/s12250-020-00221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parikesit A.A., Nurdiansyah R. The predicted structure for the anti-sense siRNA of the RNA polymerase enzyme (RdRp) gene of the SARS-CoV-2. Ber. Biol. 2020;19:97–108. doi: 10.14203/beritabiologi.v19i1.3849. [DOI] [Google Scholar]
- 74.Jinek M., Doudna J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession number, length and location of the SARS-CoV-2 stains used in the study.
Conserved sequence of nucleocapsid phosphoprotein gene.
Conserved sequence of surface glycoprotein gene.
Tm values of predicted siRNA (guide strand and passenger strand) against nucleocapsid phosphoprotein.
Tm values of predicted siRNA (guide strand and passenger strand) against surface glycoprotein.
Effective siRNAs against nucleocapsid phosphoprotein with GC%, free energy of folding and free energy of binding with target.
Effective siRNAs against surface glycoprotein with GC%, free energy of folding and free energy of binding with target.
siRNA and Ago2 interacting residues, bond distance and bond category.