Graphical abstract
Keywords: COVID-19, Deep learning, Syndromic surveillance
Abstract
Coronavirus Disease 2019 has emerged as a significant global concern, triggering harsh public health restrictions in a successful bid to curb its exponential growth. As discussion shifts towards relaxation of these restrictions, there is significant concern of second-wave resurgence. The key to managing these outbreaks is early detection and intervention, and yet there is a significant lag time associated with usage of laboratory confirmed cases for surveillance purposes. To address this, syndromic surveillance can be considered to provide a timelier alternative for first-line screening. Existing syndromic surveillance solutions are however typically focused around a known disease and have limited capability to distinguish between outbreaks of individual diseases sharing similar syndromes. This poses a challenge for surveillance of COVID-19 as its active periods tend to overlap temporally with other influenza-like illnesses. In this study we explore performing sentinel syndromic surveillance for COVID-19 and other influenza-like illnesses using a deep learning-based approach. Our methods are based on aberration detection utilizing autoencoders that leverages symptom prevalence distributions to distinguish outbreaks of two ongoing diseases that share similar syndromes, even if they occur concurrently. We first demonstrate that this approach works for detection of outbreaks of influenza, which has known temporal boundaries. We then demonstrate that the autoencoder can be trained to not alert on known and well-managed influenza-like illnesses such as the common cold and influenza. Finally, we applied our approach to 2019–2020 data in the context of a COVID-19 syndromic surveillance task to demonstrate how implementation of such a system could have provided early warning of an outbreak of a novel influenza-like illness that did not match the symptom prevalence profile of influenza and other known influenza-like illnesses.
1. Introduction
The fast spread of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS CoV-2), has resulted in a worldwide pandemic with high morbidity and mortality rates [1], [2], [3]. To limit the spread of the disease, various public health restrictions have been deployed to great effect, but as of May 2020, international discussion has begun shifting towards relaxation of these restrictions. A key concern is, however, any subsequent resurgence of the disease [4], [5], [6], particularly given that the disease has already become endemic within localized regions of the world [7]. This issue is further exacerbated by significant undertesting, where estimates have found that more than 65% of infections were undocumented [8], [9]. Additionally, increasing levels of resistance and non-adherence to these restrictions has greatly increased resurgence risk.
A key motivation behind the initial implementation of public health restrictions was to sufficiently curb the case growth rate so as to prevent overwhelming hospital capacities [10], [11]. While the situation has been substantially improved, a resurgent outbreak will present much the same threat [11]. Indeed, second-wave resurgence has already been observed in Hokkaido Japan after public health restrictions were relaxed, and these restrictions were re-imposed a mere month after being lifted [12]. Additionally, from a healthcare provider perspective, significant nosocomial transmission rates for the disease have been found despite precautions [13], [14], [15], a significant concern as many of the risk factors in terms of severity and mortality for COVID-19 [2], [16] can be commonly found within an in-hospital population. To avoid placing an even greater burden on already strained hospital resources, it is important that healthcare institutions respond promptly to any outbreaks and modify admission criteria for non-emergency cases appropriately. For both reasons, it is critical to detect outbreaks as early as possible so as to contain them prior to requiring reinstitution of these extensive public health restrictions. Early detection is, however, no mean feat. Reliance on laboratory confirmed COVID-19 cases to perform surveillance introduces significant lag time after the beginning of the potential shedding period as symptoms must first present themselves [17], [18] and be sufficiently severe to warrant further investigation, before test results are received. This is further complicated by limited test reliability, with RT-PCR tests having an estimated sensitivity of 71% [19], and serological tests, despite having high reported specificity, having significant false positive rates. Moreover, asymptomatic carriers, which in some studies have been found to reach as much as 50–75% of the actual case population [20], [21], [22], present significant risk, particularly amongst the healthcare provider population.
It is therefore evident that any surveillance solution relying purely on laboratory-confirmed cases will suffer from a significant temporal delay as compared to when the transmission event actually occurs, suggesting that a syndromic surveillance solution may be necessary [23]. In this study, we aim to perform computational syndromic surveillance for novel influenza-like illnesses such as COVID-19 amongst a hospital’s patient population (comprising both inpatient and outpatient settings) to detect outbreaks and prompt investigation in advance of actual confirmation of cases.
In the following sections, we will first briefly discuss the history of digital syndromic surveillance approaches and provide an introduction to our proposed approach in the background section, expand in further detail and provide dataset procurement and evaluation procedure in the methods section, show the results of our evaluation in the results section, and discuss interpretation of our results and potential pitfalls in the discussion section.
2. Background and related work
Digital syndromic surveillance systems came to the forefront of national scientific attention for bioterrorism preparedness purposes [24], particularly in the wake of the anthrax attacks in the fall of 2001 [25]. Such systems, however, were quickly noted to also be of use in clinical and public health settings [26]. In this section, we will first present an introduction to existing disease surveillance approaches, and then discuss the theoretical justifications behind our proposed approach.
2.1. Syndromic Surveillance for COVID-19 and Other Novel Influenza-Like Illnesses
Approaches that have been explored for disease surveillance [27] include usage of simple statistical thresholds on raw frequency or prevalence data, to statistical modeling and visualization approaches such as Cumulative Sums (CUSUM), Exponentially Weighted Moving Averages (EWMA), and autoregressive modeling [28], [29], [30], [31], [32], [33].
For example, the United States Centers for Disease Control and Prevention (CDC) applies CUSUM modeling to monitor for Salmonella outbreaks [34], while Stern et al. proposed a compound smoothing technique for the same task [35]. Akhtar et al.[36] employed a dynamic neural network model based on the NARX neural network model [37], [38] for Zika surveillance. Anno et al. applied convolutional neural networks on spatiotemporal data for dengue fever hotspot detection [39]. For general disease surveillance, the US CDC operates the EARS system [29], [40] which utilizes an aggregation of historical limits (mean + 2 standard deviations), log-linear regression [41], CUSUM, compound smoothing [35], and autoregressive models (ARIMA)[42] while the US Department of Defence operates the ESSENCE system [43], which uses a variety of statistical modeling techniques including a modification of the Kulldorff scan statistic, CUSUM, EWMA, and autoregressive modeling. A similar work by Reis et al. utilizes much the same methods, implementing CUSUM, EWMA, and SatScan (from which ESSENCE derived its Kulldorff scan statistic) for its detection modules [44]. Lake et al. utilized an ensemble of Bayesian classifiers [45] for general disease surveillance.
More specifically to the syndromic surveillance of influenza-like illnesses (ILI), at a national level, the United States Centers for Disease Control and Prevention operates the ILInet, a national statistical syndromic surveillance solution deriving its data from reports of fever, cough, and/or sore throat without a known non-influenza cause within outpatient settings [28]. ILInet’s detection component implemented statistical cutoffs based on historical data, specifically the mean percentage of patient visits for ILI + 2 standard deviations during off-season weeks over the previous three influenza seasons as a detection threshold [28]. Prior work by Sebastiani et al. [30] and Chan et al. [46] both explored using Bayesian modeling variants to perform the surveillance task. Cheng et al.[47] explored using an aggregation of statistical modeling and machine learning techniques including ARIMA, random forests, support vector regression, and extreme gradient boosting to perform an influenza prediction task, that could then be used for early warning purposes.
While generally effective, many of these approaches are limited in granularity to a syndrome level: although they perform surveillance of the frequencies or prevalence of a particular syndrome or illness as a whole, they do not make a distinction amongst individual diseases that share similar syndromes. This is an issue for our task at hand as COVID-19′s syndrome very closely resembles that of many other seasonal diseases such as influenza, the common cold, or even allergic reactions. As such, while an outbreak of a novel influenza-like illness like COVID-19 may be registered in these surveillance systems, they may be difficult to discern if the outbreak temporally overlaps with known seasonal illnesses sharing the same syndrome (e.g. if they begin at the height of the influenza season), and the ongoing outbreak may be misattributed to the more benign seasonal disease. The underlying symptom prevalence amongst positive cases of influenza-like illnesses is, however, perceptibly different. For instance, while the symptom prevalence distribution for positive cases of influenza amongst the hospitalized, vaccinated, sub-50, population is 98%, 88%, 83%, 87%, and 96% for cough, fever, headache, myalgia, and fatigue respectively [48], the distribution for the same symptoms is 59%, 99%, 7%, 35%, and 70% respectively for hospitalized COVID-19 positive cases [13]. As an outbreak of COVID-19 will likely affect the background symptom prevalence distribution in a different manner than an outbreak of influenza, we theorize that an approach incorporating symptom prevalence distributions as part of its input data as opposed to the frequency/prevalence of the syndrome as a whole will be able to perform this differentiation and as such trivialize outbreaks of known, relatively benign, seasonal diseases at the user’s discretion.
2.2. Leveraging autoencoders to perform aberration detection detection
In this study, we adapted an approach to perform aberration detection that is commonly used within the general domain, autoencoders [49], [50], [51], for our syndromic surveillance task. An autoencoder (also commonly termed a “Replicator Neural Network”) is a neural network trained in a self-supervised manner to first encode the input into a lower-dimensional form, and then decode this lower-dimension form to reconstruct the input [52]. In other words, a trained autoencoder learns two functions, an encoding function and a decoding function, such that given an input , , , and . A natural property of autoencoders is that their encoding and decoding functions only function properly for input data that is similar to the data for which it is trained: data that differs in its input features compared to the training data will fail to be successfully reconstructed such that .
There are many variations of autoencoder-based aberration detection methods: the simple model with hidden layers of reduced dimensionality to act as a bottleneck and force non-linear transformation that we present here can be considered a base that can be further modified. Past work has involved replacing individual units with more complex networks to address particular characteristics of the input data. For instance, outside of the clinical domain, units in autoencoder networks have been replaced with recurrent neural networks, particularly long short-term memory blocks (LSTMs) [53] to address time-series dependencies in the input data [54], [55], [56], [57]. Convolutional neural networks have also been used in conjunction with autoencoders for the same purpose [58], [59].
Beyond the substitution of individual units within the autoencoder itself with more complex networks, alternative autoencoder architectures exist. For instance, the autoencoder architecture we presented earlier is an example of an undercomplete autoencoder, where the dimensionality of the hidden layer is smaller than that of the input and output layers to prevent the autoencoder from learning the identity function. One alternative that has also been applied to the anomaly detection task is the denoising autoencoder (DAE)[60], [61], whereby noise is added to the input data (e.g. by randomly zeroing certain input nodes). Instead of constraining the hidden representation, this approach aims to build a more robust representation by providing a hint during the network learning process to capture useful features that are able to accurately denoise the input data [62]. Sparse autoencoder architectures have also been proposed for the anomaly detection task [63], [64], [65], where the dimensionality of the hidden layers is on-par with or larger than the input and output layers, but the number of active hidden layer units is restricted.
For the purposes of syndromic surveillance, we theorize that the autoencoder approach can be adapted: given a distribution of symptom prevalence within the clinic, we would expect that distribution to change significantly should an outbreak occur. This implies that during an actual outbreak, the reconstruction error would increase perceptibly as compared to during normal time periods and can thus be plotted against time to provide a readily interpretable visualization of an outbreak of a novel influenza-like illness. As very limited work has been done to apply autoencoder-based anomaly detection to epidemiologic surveillance tasks, we have opted to use a basic undercomplete autoencoder architecture without any special substitutions in the individual neural unit types to verify the underlying theory in an epidemiologic surveillance context. It is important to note that these variations all intended to bolster representation learning and reconstruction performance, usage of these variations does not fundamentally alter the theory behind why we are applying the autoencoders themselves – in that the reconstruction error will increase as input symptom prevalence distributions become more dissimilar to that of the training data.
In other words, to accomplish the COVID-19 and other novel influenza-like illness syndromic surveillance task, we propose that:
-
(1)
By mining the raw mentions of symptoms within a syndrome of interest through an NLP-based approach, we can estimate the prevalence of individual symptoms amongst the overall patient population in a timely manner
-
(2)
By delineating certain time periods as “normal” (i.e. no outbreaks of surveilled target of interest) for autoencoder training purposes, the resulting model can be used to perform syndromic surveillance by measuring the error score of any given day’s input symptom prevalence distribution. Crucially to the COVID-19 and novel ILI detection task itself, “normal” time periods can also contain outbreaks of seasonal influenza, which should lead the model to learn the appropriate symptom prevalence distributions without increasing false alarms during typical influenza seasons.
3. Materials and methods
The true beginnings of the COVID-19 pandemic within the United States is still a subject of much contention, with the date being pushed earlier as investigation continues [66]. As such, it is difficult to directly validate any conclusions about the viability of autoencoder-based syndromic surveillance for COVID-19. We therefore validated our approach incrementally through a three-phase approach:
-
(1)
Validating the utility and accuracy of autoencoder-based anomaly detection for syndromic surveillance on a disease with known outbreak time periods
-
(2)
Validating that given appropriate training data, our autoencoder model can effectively learn the symptom distributions of other differential outbreaks such as seasonal influenza, allergies and the common cold within its underlying model, i.e. be capable of suppressing outbreaks of these other, known, seasonal illnesses from its resulting signal
-
(3)
Applying an autoencoder based anomaly detection approach to syndromic surveillance for COVID-19 over the past year of data and evaluating against currently known key dates of the COVID-19 pandemic
We present an overview of our experimental procedure in Fig. 1 , and outline each step in detail within the ensuing subsections.
Fig. 1.
Experimental Procedure Overview.
3.1. Sign/symptom extraction
Sign and symptom extraction via natural language processing was accomplished via the MedTagger NLP engine [67], [68]. The signs and symptoms chosen were selected via a literature review conducted in early March 2020 for known COVID-19 and influenza symptoms [13], [69]. Specifically, mentions of Abdominal Pain, Appetite Loss, Diarrhea, Dry/Nonproductive Cough, Dyspnea, Elevated LDH, Fatigue, Fever, Ground-Glass Opacity Pulmonary Infiltrates, Headaches, Lymphopenia, Myalgia, Nasal Congestion, Patchy Pulmonary Infiltrates, Prolonged Prothrombin Time, and Sore Throat were used for all three experiments. Additionally, explicit mentions of influenza were used for phase 2 (establishing baseline/incorporating influenza seasons as part of “normal” symptom prevalence distributions) and phase 3 (COVID-19 surveillance task) of our experiment. Only positive present NLP artifacts with the patient as the subject were retained.
3.2. Symptom prevalence distribution dataset
Clinical documentation generated from January 1st 2011 through May 1st 2020 was utilized as part of this study, with the exclusions detailed within the Data Limitations subsection within our Discussion section (January 1st–July 1st 2016, May 1st–July 7th 2018). For each day within this range, a symptom prevalence feature vector was generated, where each element in the vector corresponds to the symptom prevalence of one of the symptoms of interest for that day. We defined symptom prevalence on any given day as the number of unique patients that had a clinical document generated that day containing a NLP artifact corresponding to that symptom (that was positive, present, and had the patient as the subject) divided by the number of unique patients that had at least one clinical document generated on that day.
This dataset was then subdivided into different training and plotting (for simulated surveillance purposes) definitions for each of the tasks at hand. We have provided a summary of these divisions in Table 1 .
Table 1.
Task-Specific Training and Plotted Data Divisions.
| Task | Training Data (“Normal” Time Periods) | Plotted Data (Surveillance Time Periods) |
|---|---|---|
| Influenza Surveillance | [2011-05-22, 2011-10-02) [2013-05-19,2013-09-29) [2015-05-23, 2015-10-04) [2017-05-20, 2017-10-01) |
[2011-01-01, 2018-05-01*) |
| Seasonal Illness Suppression | [2011-05-22, 2014-01-25†) | [2014-01-25, 2016-01-01) |
| COVID-19 Syndromic Surveillance Task | [2018-08-01§, 2019-06-01) | [2019-06-01, 2020-05-01) |
Mayo Clinic transitioned from its historical EHR to the Epic EHR on this date.
End of moderate or greater ILI activity within the State of Minnesota for 2013–14 influenza season.
Three months after EHR migration began, to allow for clinical workflow changes to be solidified and reduce data volatility.
3.3. Autoencoder architecture and implementation
Our neural network was implemented in Java via the DL4J deep learning framework [70]. For our purposes we used a 5-layer fully-connected stacked autoencoder consisting of [INPUT_DIM, 14, 12, 14, INPUT_DIM] nodes in each respective layer, where INPUT_DIM refers to the dimensionality of the input data. For influenza detection, this was 16 (excluding influenza prevalence), and for all other tasks, this was 17. The activation function used for all layers was the sigmoid activation function, except for the output layer, which used the identity function, with all inputs being rescaled to the [−1, 1] range. The optimization function, their associated learning rate, and the L2 regularization penalty used were selected via five-fold cross-validation, where optimization function was one of AdaDelta [71], AdaGrad [72], or traditional stochastic gradient descent [73] and their respective learning rate was selected from 100 randomly sampled points from the range [0.0001, 0.01], with the exception of AdaDelta, as it is an adaptive learning rate algorithm, and we instead used the recommended default rho and epsilon of 0.95 and 0.000001 respectively. An L2 regularization penalty [74] was selected from 100 random samples in the range [0.00001, 0.001]. The cost function used was mean squared error. For all model training tasks, 30% of the training data from “normal” time periods was withheld for testing purposes, and the aforementioned five-fold cross-validation was then done on the remaining 70% for hyperparameter selection and model training was done using the entire train dataset as one batch, over 1000 epochs utilizing early stopping (5 iterations with score improvement <0.0001) and selecting the model resulting from the epoch that had the best performance against the withheld test dataset.
3.4. Evaluating influenza season detection capabilities
To validate the utility and accuracy of autoencoder-based anomaly detection for syndromic surveillance, we chose syndromic surveillance of influenza seasons as the target task. This task was chosen primarily due to two factors: (1) its relatively well-defined outbreak periods (available both at a national and state level via the CDC Morbidity and Mortality Weekly Reports [75], [76], [77], [78], [79], [80], [81], [82] and the CDC Influenza-Like-Illness (ILI) Activity Tracker [83] respectively) and (2) its similarity in potential input features (due to similar symptom presentations) to our end-goal of performing COVID-19 syndromic surveillance.
For training purposes, we used seasonal date ranges as defined in the US CDC released morbidity and mortality weekly report (MMWR) and selected flu offseason for the odd-numbered years between 2010 and 2018 as our training set [75], [76], [77], [78], [79], [80], [81], [82]. Specifically, the date ranges used for training were [2011-05-22, 2011-10-02), [2013-05-19, 2013-09-29), [2015-05-23, 2015-10-04), and [2017-05-20, 2017-10-01).
For these date ranges, all extracted symptom prevalence information was included for training with the exception of explicit mentions of influenza, as that might provide an unwarranted hint for the task to the underlying trained network.
To evaluate this approach's effectiveness for influenza season detection, we ran the trained autoencoder on all years from 2011 through May of 2018 (when the Epic EHR migration occurred), and plotted the error, as determined by the mean-squared error between the supplied input feature set and the network’s outputs, with a particular focus on detected influenza seasons starting on even years.
The best performing model from training was selected, and the anomaly threshold was determined as the mean + 2 standard deviations of the reported errors derived from the test partition resulting from cross-validation of the normal (training) time periods, with errors higher than this value being deemed anomalous.
The errors were plotted and compared against timespans with elevated influenza activity, both at a national level via the official MMWR defined influenza season and in terms of ILI activity for the state of Minnesota as reported by the CDC ILInet. The distinction is important as while the CDC MMWR reports a national level influenza season, the actual periods of elevated activity differ from state to state, and we would only truly be able to detect anomalies when influenza activity is actually elevated within Minnesota, as that is the source for our data.
3.5. Evaluating autoencoder capability to embed influenza season data as “Normal”
COVID-19 syndromic surveillance is severely complicated by its similar presentation and overlapping timeframe with a variety of seasonal illnesses, such as the common cold, allergies, and influenza. To verify that an autoencoder-based COVID-19 syndromic surveillance solution will be functional, we must first verify that, if supplied as part of its training data, outbreaks of these seasonal illnesses will not be reflected in its resulting error plots. To that end, we again use influenza as the target for evaluation here, due to its relatively well-defined temporal boundaries.
In this phase, we use data from May 22nd 2011 (the end date of the 2010–2011 influenza season) through January 25th 2014 (the end date for observed moderate-or-greater ILI activity in the state of Minnesota for the 2013–2014 influenza season) as our training set.
Unlike in the previous phase, the prevalence of influenza mentions is included within the feature set for training to supply explicit knowledge about the occurrence of and the magnitude of ongoing influenza seasons. Additionally, to ensure a balance in examples, we sampled from the influenza off-seasons such that the number of off-season examples corresponded to the number of in-season examples.
Once training using this dataset was completed, we then ran this new autoencoder model on all data between January 25th 2014 and January 1st 2016 and plotted the mean squared error between the supplied input and the autoencoder’s resultant output, with a focus on even years.
The anomaly threshold was again set to the mean + 2 standard deviations of the test partition error during the training time period and the resultant anomalous spans were used to evaluate the autoencoder’s capability to embed influenza and other seasonal differential data.
3.6. Applying autoencoder-based anomaly detection for COVID-19 syndromic surveillance
At this point in the experiment we will have validated that (a) an autoencoder reconstruction error-based approach to anomaly detection is capable of reflecting both the occurrence and the magnitude of shifts in underlying symptom prevalence distributions, and (b) if included as part of the “normal” training data, autoencoders will successfully reconstruct symptom prevalence distributions occurring during COVID-19’s seasonal differentials. We can thus proceed with the targeted task of this study: syndromic surveillance of the COVID-19 outbreak within the United States, particularly within Olmsted County, Minnesota, the location of the Mayo Clinic Rochester campus.
In this phase, we use data from August of 2018 through June of 2019 (Exclusive) as our “normal” training data. Again, we ensure a 50/50 balance of influenza in-season and off-season examples in our dataset prior to partitioning the data for cross-validation. As with our previous experiments, the anomaly threshold was set to the mean + 2 standard deviations of the test partition error during the training time period.
The resulting model was run on data from June of 2019 through present, and the resulting errors were plotted for further analysis.
4. Results
In this section, we will evaluate and interpret the resulting error plots from our three experiments in order by first verifying that autoencoder-based anomaly detection can be used for syndromic surveillance, then verifying that seasonal illnesses sharing similar syndromes can be suppressed, and then applying our approach to the COVID-19 surveillance problem.
4.1. Autoencoder-based anomaly detection is viable for syndromic surveillance
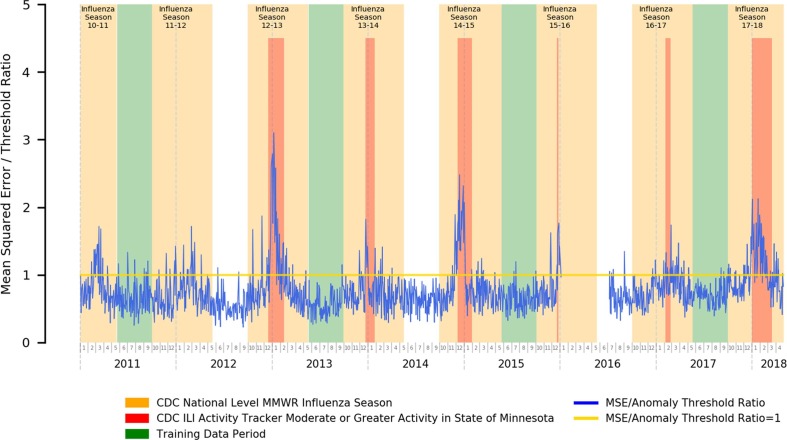
In Fig. 2 , we present the error plot relative to the anomaly threshold of a stacked autoencoder trained using influenza off-season data for the purposes of syndromic surveillance of influenza. We additionally highlight official CDC flu seasons (national level) [75], [76], [77], [78], [79], [80], [81], [82] in orange, and time periods with heightened (moderate or greater) ILI activity [83] within the state of Minnesota (from where our data originates) in red.
Fig. 2.
Mean Squared Error Relative to Anomaly Threshold for an Autoencoder Trained for the Influenza Season Detection Task.
Our error plots and the close congruence between periods of heightened autoencoder reconstruction error and influenza activity does suggest that our approach is fairly successful at performing the influenza syndromic surveillance task. Of particular note, the magnitude of the reconstruction error is also closely tied to the associated severity of the outbreak, as can be seen in the location of our error peaks relative to state-level ILI activity tracking.
As such, our results here suggest that an autoencoder-based anomaly detection approach to syndromic surveillance is capable of picking up and alerting on the underlying changes in the prevalence of influenza-related symptoms in the practice during influenza season as opposed to the off-season, both in terms of identifying that the underlying distribution of symptom prevalence changed and in reflecting the magnitude of the differences in underlying distribution of symptom prevalence compared to normal time periods within its reconstruction error.
These results are promising for our eventual experiment for COVID-19 syndromic surveillance as the underlying assumptions are similar: COVID-19 and influenza share very similar symptoms, but the underlying distribution of the prevalence of individual symptoms within their respective cases will likely differ. It is expected that an autoencoder will be able to pick up on these prevalence distribution differences in a similar manner to the influenza season vs. offseason variation.
4.2. Autoencoders can be trained to suppress alerting on outbreaks of illnesses sharing similar syndromes
In Fig. 3 , we present the mean squared error plot of a stacked autoencoder trained using data covering three influenza seasons and off-seasons, with the aim of verifying that typical influenza seasons can be suppressed from anomalous readings by incorporating their symptom prevalence distributions as part of training data.
Fig. 3.
Mean Squared Error Relative to Anomaly Threshold for an Autoencoder Trained on both Influenza Season and Offseason Data.
Our results demonstrate that our autoencoder has successfully incorporated symptom prevalence data for influenza and other seasonal diseases with similar differentials occurring within the target period, as can be seen by the relatively consistent reconstruction error throughout the year with peaks being dramatically suppressed in magnitude compared to the highly visible peaks in Fig. 2.
4.3. Syndromic surveillance viable for sentinel detection of novel Influenza-Like-Illnesses
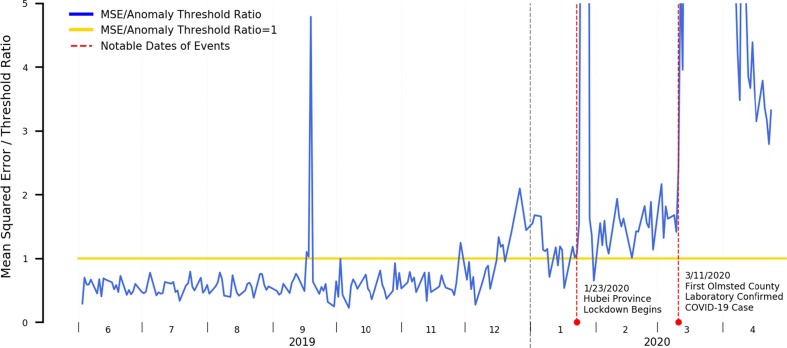
In Fig. 4 , we present the mean error plot of a stacked autoencoder trained using a year of both influenza season and off-season data applied to data from June 1st, 2019 through April 30th, 2020. We additionally annotated the resulting plot with dates pertinent to the COVID-19 epidemic in Minnesota to provide additional context to the detected signals.
Fig. 4.
Mean Squared Error Relative to Anomaly Threshold for an Autoencoder Trained on both Influenza Season and Offseason Data.
Our results suggest the following with respect to the time period prior to the first laboratory confirmed case in the state of Minnesota:
-
(1)
A spike occurring the week of September 15th, 2019. We do not believe this is COVID-19 related and will elaborate more on this in the discussion section.
-
(2)
A persistent, low level of elevated anomalous signals beginning late December through the first laboratory confirmed COVID-19 case within Olmsted County, Minnesota occurring March 11th, 2020. This period is marked by two dramatic spikes occurring January 23rd and March 11th 2020 that we will also discuss in the discussion section. This period of elevated anomalous signals does roughly match the period of heightened state-level ILI activity as reported by the CDC.
When interpreting these results, it is important to note that CDC’s ILI tracker is itself a form of syndromic surveillance and doesn’t explicitly indicate levels of influenza-specific activity, but rather all syndromes with similar symptomatic presentations: specifically, ILInet uses fever, cough, and/or sore throat without a known non-influenza cause as the data through which it performs its tracking [28]. It is therefore expected that our detected anomalous time periods will match, as COVID-19 itself shares many of these symptoms.
The fact that elevated anomalous results appeared in our error plot, however, suggests that the underlying symptom prevalence distributions seen within the clinical practice are atypical of those seen in other influenza seasons: per the second phase of our experiment, we established that “typical” influenza seasons can be suppressed from anomalous readings by incorporating their symptom prevalence distributions as part of training data. We would have thus expected the error rates to have remained largely under the anomaly threshold with no significant peaks, unlike what was observed here.
5. Discussion
While our results are promising, given that autoencoder-based anomaly detection is a relatively black-box method, there are several important points to consider when interpreting the resulting error plots. In this section, we will first discuss potential interpretation pitfalls, then discuss the opportunities our work presents for novel influenza-like illness surveillance in the context of the COVID-19 outbreak, and lastly followed by an outline of limitations of this study.
5.1. Interpreting anomalous signals and potential attribution errors
It is important to note with all our results presented here that the anomaly detection component detects anomalies in the input data, i.e. anomalies in the incoming symptom prevalence distributions. Such anomalies can, however, be caused by a variety of external factors and are not necessarily indicative of an outbreak. As such, while such a system can serve as an early-warning system to alert that an anomaly exists as well as the magnitude of such an anomaly, further human investigation is needed to identify the underlying reasons and confirm whether an outbreak is occurring. Despite our results in Fig. 4 suggesting a sustained elevated anomalous error rate starting around the final week of December through the first laboratory confirmed COVID-19 case, it would still be premature to directly conclude that the anomalous time period is attributable to only COVID-19, such a conclusion would only be possible to achieve had laboratory tests been done during that time period. Instead, it serves only as an indicator of the need for additional investigation.
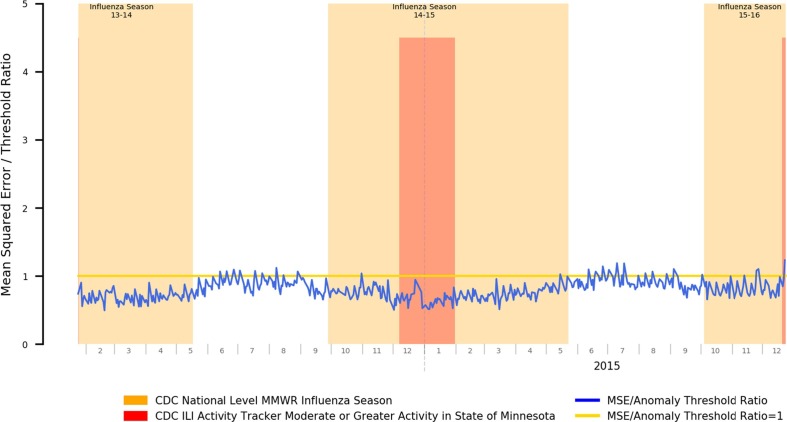
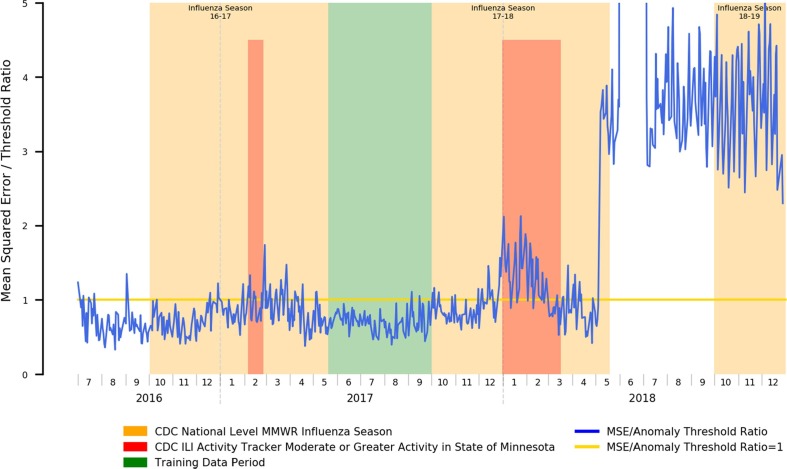
An example of the potential for attribution error can be shown where, in Fig. 5 , we note that while the periods of elevated error rates for the 2017–2018 influenza season do roughly correspond to the official CDC-determined flu season and periods of heightened ILI activity, starting May of 2018, the error rate rises outside the display range of the chart. This anomaly does, in fact, exist in reality, but is not tied to a renewed outbreak of influenza-like illness. Rather, the Mayo Clinic Rochester clinic migrated EHR systems from its historical GE Centricity-based EHR to the Epic EHR, and the go-live date for clinical operations was May 1st. Due to the changes in clinical workflows and associated documentation practices, the underlying distribution of positive symptom prevalence mentions within clinical documentation also dramatically changed, and that anomalous change was appropriately detected.
Fig. 5.
Mean Squared Error Relative to Anomaly Threshold for an Autoencoder Trained for Influenza Season Detection Spanning an EHR Migration Occurring May 2018.
A similar phenomenon is reflected in Fig. 4. A brief spike in the plotted errors occurs mid-September 2019: further investigation leads us to hypothesize that rather than an outbreak of influenza-like illness during this timeframe, this spike was related to media coverage and associated greater patient concern to a local outbreak of E. coli during this same time period originating from a popularly attended state fair [84]. Similarly, two events that triggered greatly increased media coverage and associated public awareness are highlighted in red, the initial lockdown of the city of Wuhan and Hubei province on January 23rd 2020, the event that originally brought the coronavirus outbreak to the public’s attention, and the first laboratory-confirmed COVID-19 case within Olmsted County, Minnesota on March 11th 2020. Instead of directly attributing the spikes to actual [undiagnosed] COVID-19 cases, the news coverage and increased patient concern likely caused a dramatic increase in patient healthcare engagement and a surge of precautionary symptom documentation. Nevertheless, these “public awareness and concern” spikes are typically obvious, as the spike is sudden, relatively large in magnitude, and are temporally co-located with publicly available news sources.
5.2. Autoencoder-based syndromic surveillance: retrospective and prospective opportunities
Had a syndromic surveillance solution similar to what we established in phase 3 of our experiment existed at the time of the Hubei lockdown, anomalous readings would have appeared far in advance of the actual first laboratory-confirmed case even within the United States, and alert on a possible outbreak of a novel influenza-like-illness that did not share similar symptom prevalence distributions as priorly encountered influenza seasons. This information could have been used as an actionable signal for further investigation suggesting a possible spread of COVID-19 within the served community and been a prompt for far more aggressive testing than what was done in practice. From a public health perspective, this could have allowed for earlier intervention and potentially dramatically reduced outbreak magnitude.
From a prospective perspective, such a syndromic surveillance approach can potentially be utilized to provide early warning of future outbreaks, particularly with respect to differentiation from outbreaks of other influenza-like illnesses. As public health restrictions are eased, such capabilities are increasingly critical for detection and early intervention in the case of second-wave outbreaks within the individual hospital’s served communities. It is important to note, however, that clinical workflows for patients presenting with influenza-like illnesses, and by extension documentation practices will have substantially changed in the post COVID-19 era; these changes will cause an artificial surge in the detected anomalous events. Addressing the discrepancy would require model recalibration: with a pretrained model similar to that produced from phase 3 of our experiment, limited retraining of the existing model on a month of “normal” data after resumption of full clinical operations might be sufficient to adapt it to the post COVID-19 data distributions.
Beyond COVID-19 itself, the approach presented here can be adapted to monitor and surveil for any novel ILI that shares similar symptoms, greatly expanding the applicability of our approach beyond the currently ongoing COVID-19 pandemic. Additionally, should the input symptom feature set be expanded beyond symptoms associated with influenza-like illnesses, we theorize that this approach can be applied to syndromic surveillance of other diseases. We have left such explorations to future work.
5.3. Data and study limitations
Our study faced several challenges from a data perspective. Firstly, it must be noted that patient profiles significantly change between normal work-week operations and weekends/holidays, which are far more likely to be acute or emergency care. As such, to prevent these from becoming a confounding factor and unduly influencing our anomaly detection error plots, data points relating to weekends, US federal holidays, Christmas Eve and New Year’s Eve were excluded from our datasets. We do not believe that this has affected the validity of our results, further evidenced by the plot in Fig. 4, showing that the period of elevated ILI activity that occurred from January through mid-March of 2018 was correctly reflected, while December of 2017 did not display anomalous results, indicating that our model is not simply picking up on proximity to holidays. We will, however, work on incorporating weekend and holiday data as part of our models as part of future work.
Additionally, several limitations within our data sources hampered our efforts to evaluate our methods: as previously noted, anomalies may also be caused by problems with the input data unrelated to the syndromic surveillance task. Specifically, in our case, we faced two major EHR/data platform shifts within our source data that led to irregular disruption of clinical documentation within our data warehouse, one occurring throughout the entirety of Q1 2016, and the other occurring beginning May 1st 2018 and lasting through the first week of July 2018 resulting from Mayo Clinic Rochester’s migration to the Epic EHR. The training datasets and results presented thus excluded these time periods (except for illustrative purposes in Fig. 5) as they are known to be anomalous with the reasons for the anomaly being irrelevant to our target tasks (e.g., reasons for anomaly include changes in documentation practices affecting NLP-based prevalence, metadata changes, etc.)
Finally, the fact that an EHR migration did occur significantly hampers the amount of pre-COVID-19 data available for training purposes in phase 3 of our experiment. Due to documentation practice shifts we must use Epic data as part of our training data, and due to the data source disruption as a result of this migration, we were limited to data beginning August of 2018. For future work, we aim to further validate our model on other sites within the Mayo Clinic enterprise that switched EHR systems in 2016, so as to leverage a greater amount of training data.
From a methodological perspective, we were constrained in available methods to be unsupervised and/or self-supervised (using “normal” data): given our task to detect novel influenza-like-illnesses of unknown symptom prevalence distributions, it was not feasible to procure labeled “anomalous” data for supervised learning approaches. It is nevertheless important to note that the autoencoder approach is only one of many existing approaches that have been utilized for anomaly detection within the general domain. Other approaches commonly used in this space include k-means clustering [85], [86], [87], one-class SVMs [87], [88], [89], Bayesian networks [90], as well as more traditional statistical approaches such as the chi-square test [91] and principal component analysis [92]. In many systems, such approaches are not taken in isolation, but are rather used in conjunction with others to perform specific sub-components of the anomaly detection task or to provide multiple features for downstream analysis [86], [88], [93], [94]. Our study is not intended to perform a comprehensive benchmarking of available methods, and we have not included comparative metrics here given that we have achieved workable results with only an autoencoder approach as the focus of this work was to test the concept of using aberrations in symptom prevalence distributions to perform syndromic surveillance rather than the model that is used to perform this task. Nevertheless, it is entirely possible that a different model than an autoencoder being used to do this aberration detection task may perform better and as such it may be worth exploring usage and/or integration of many of these other models to improve discriminative power and denoise the signal, and we have left such exploration to future work.
6. Conclusions
Early detection of infectious disease outbreaks is critical to their successful management, but reliance on laboratory confirmation, if even possible for a novel illness, introduces significant temporal delays. For this reason, syndromic surveillance has been utilized so as to provide signals of possible disease outbreaks in advance of signals derived from laboratory confirmed diagnoses. Existing solutions, however, largely focus on known diseases, as well as syndromes as a whole, and may fail to differentiate when syndromes between different illnesses are similar and outbreaks occur co-temporally, as was the case with the initial outbreak of Coronavirus Disease 2019 and seasonal influenza.
To address this, we noted that while syndromes as a whole may be similar, the prevalence of individual symptoms within the syndromes differ between different diseases. We therefore hypothesized that a syndromic surveillance approach incorporating distributions of symptoms as part of its monitoring mechanism as opposed to prevalence of syndromes as a whole may be able to distinguish amongst these diseases, allowing for such an approach to be useful even when outbreaks co-occur with seasonal illnesses sharing similar syndromes.
In this study, we have demonstrated such an approach using autoencoders trained on in-hospital symptom prevalence distributions to perform syndromic surveillance for novel influenza-like-illnesses. We first demonstrated that this approach works on outbreaks with known time boundaries using seasonal influenza as an example use case. We then showed that this approach can be trained to suppress signals for seasonal outbreaks of influenza and similar known and well-managed influenza-like illnesses so as to primarily alert on novel influenza-like-illnesses. We then applied this approach to the initial outbreak of Coronavirus Disease 2019 within the state of Minnesota, and found that the model displayed signals suggesting a possible outbreak more than one month prior to the first laboratory confirmed case.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
Research reported in this publication was supported by the National Center for Advancing Translational Science of the National Institutes of Health under award number U01TR002062 and by the National Library of Medicine under award number R01LM0011934. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Code availability
The NLP engine and associated algorithm used to extract ILI symptoms as described in this study is available within the MedTagger project (https://www.github.com/OHNLP/MedTagger). Please consult the Wiki and README file accessible from the linked page for instructions on how to use for the COVID-19 use case.
The aberration detection/sentinel syndromic surveillance component has been decoupled from institutional data sources and is available at https://github.com/OHNLP/AEGIS. As this is an active project undergoing improvement and new features that may lead to changes in the underlying code inconsistent with what was described in this manuscript, we have tagged the codebase as described in this manuscript with the COVID19 tag.
Data availability
Due to the results of the symptom extraction process being considered protected health information, data is not available as it would be difficult to distribute to anyone not engaged in an IRB-approved collaboration with the Mayo Clinic.
Author contributions
AW: Designed, implemented study, performed experiments. AW, LW, HH, SL, SF, MH, YW, FS: Determined symptom inclusion/exclusion criteria for NLP algorithm and similar contributions to the divisional COVID-19 work group, preparation of NLP algorithm for public distribution, and other miscellaneous project tasks. HH, SL: Generation of graphs and figures as presented in manuscript. AW, SS, JAK, VCK: NLP engine work used for this study, interfacing with institutional data sources. JF, HL: Direction on study design and conceptualization, project leadership. All authors reviewed and contributed expertise to the final manuscript.
References
- 1.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the chinese center for disease control and prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.Phelan A.L., Katz R., Gostin L.O. The novel coronavirus originating in Wuhan, China: challenges for global health governance. JAMA. 2020 doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- 4.Xu S., Li Y. Beware of the second wave of COVID-19. The Lancet. 2020;395:1321–1322. doi: 10.1016/s0140-6736(20)30845-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung K., Wu J.T., Liu D., Leung G.M. First-wave COVID-19 transmissibility and severity in China outside Hubei after control measures, and second-wave scenario planning: a modelling impact assessment. The Lancet. 2020;395:1382–1393. doi: 10.1016/s0140-6736(20)30746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prem K., et al. The effect of control strategies to reduce social mixing on outcomes of the COVID-19 epidemic in Wuhan, China: a modelling study. The Lancet Public Health. 2020;5:e261–e270. doi: 10.1016/s2468-2667(20)30073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., et al. Community transmission of severe acute respiratory syndrome coronavirus 2, Shenzhen, China, 2020. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2606.200239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li R., et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sood N., et al. Seroprevalence of SARS-CoV-2-specific antibodies among adults in Los Angeles county, California, on April 10–11, 2020. JAMA. 2020 doi: 10.1001/jama.2020.8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C.C. Branas et al. Flattening the curve before it flattens us: hospital critical care capacity limits and mortality from novel coronavirus (SARS-CoV2) cases in US counties. medRxiv, 2020. 2020.2004.2001.20049759, 10.1101/2020.04.01.20049759. [DOI]
- 11.Markel H., et al. Nonpharmaceutical interventions implemented by US cities during the 1918–1919 influenza pandemic. JAMA. 2007;298:644–654. doi: 10.1001/jama.298.6.644. [DOI] [PubMed] [Google Scholar]
- 12.S. Neuman, Emergency Declared in Japanese Prefecture Hit by 2nd Wave of Coronavirus Infections, 2020. <http://web.archive.org/web/20200517171614/https://www.npr.org/sections/coronavirus-live-updates/2020/04/13/832981899/emergency-declared-in-japanese-prefecture-hit-by-2nd-wave-of-coronavirus-infecti>.
- 13.Wang D., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected Pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.BBC. Coronavirus: 'Half of A&E team' test positive, 2020. <https://www.bbc.com/news/uk-wales-52263285>.
- 15.Zhou Q., et al. Nosocomial Infections Among Patients with COVID-19, SARS and MERS: a rapid review and meta-analysis. medRxiv. 2020 doi: 10.1101/2020.04.14.20065730. 2020.2004.2014.20065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow N., et al. Preliminary estimates of the prevalence of selected underlying health conditions among patients with coronavirus disease 2019 – United States, February 12–March 28, 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:382–386. doi: 10.15585/mmwr.mm6913e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai Y., et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020 doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong Z.D., et al. Potential presymptomatic transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerg. Infect. Dis. 2020;26:1052–1054. doi: 10.3201/eid2605.200198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang Y., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;200432 doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Day M. Covid-19: identifying and isolating asymptomatic people helped eliminate virus in Italian village. BMJ. 2020;368:m1165. doi: 10.1136/bmj.m1165. [DOI] [PubMed] [Google Scholar]
- 21.Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day M. Covid-19: four fifths of cases are asymptomatic, China figures indicate. BMJ. 2020;369:m1375. doi: 10.1136/bmj.m1375. [DOI] [PubMed] [Google Scholar]
- 23.Henning K.J. Overview of syndromic surveillance: what is syndromic surveillance? MMWR Morb. Mortal. Wkly Rep. 2004;53(Suppl):7–11. [Google Scholar]
- 24.CDC Strategic Planning Workgroup Biological and chemical terrorism: strategic plan for preparedness and response. Recommendations of the CDC Strategic Planning Workgroup. MMWR Recomm Rep. 2000;49:1–14. [PubMed] [Google Scholar]
- 25.Jernigan J.A., et al. Bioterrorism-related inhalational anthrax: the first 10 cases reported in the United States. Emerg. Infect. Dis. 2001;7:933–944. doi: 10.3201/eid0706.010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandl K.D., et al. Implementing syndromic surveillance: a practical guide informed by the early experience. J. Am. Med. Inform. Assoc. 2004;11:141–150. doi: 10.1197/jamia.M1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan P., Chen H., Zeng D. Syndromic surveillance systems: public health and biodefense. Ann. Rev. Inform. Sci. Technol. (ARIST) 2008;42 [Google Scholar]
- 28.United States Centers for Disease Control and Prevention. U.S. Influenza Surveillance System: Purpose and Methods, 2020. <https://web.archive.org/web/20200515174103/https://www.cdc.gov/flu/weekly/overview.htm>.
- 29.Hutwagner L., Thompson W., Seeman G.M., Treadwell T. The bioterrorism preparedness and response Early Aberration Reporting System (EARS) J Urban Health. 2003;80:i89–i96. doi: 10.1007/pl00022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sebastiani P., Mandl K.D., Szolovits P., Kohane I.S., Ramoni M.F. A Bayesian dynamic model for influenza surveillance. Stat Med. 2006;25:1803–1816. doi: 10.1002/sim.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroder C., et al. Lean back and wait for the alarm? Testing an automated alarm system for nosocomial outbreaks to provide support for infection control professionals. PLoS ONE. 2020;15:e0227955. doi: 10.1371/journal.pone.0227955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsui F.C., et al. Technical description of RODS: a real-time public health surveillance system. J. Am. Med. Inform. Assoc. 2003;10:399–408. doi: 10.1197/jamia.M1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lombardo J.S., Buckeridge D.L. John Wiley & Sons; 2012. Disease Surveillance: A Public Health Informatics Approach. [Google Scholar]
- 34.Hutwagner L.C., Maloney E.K., Bean N.H., Slutsker L., Martin S.M. Using laboratory-based surveillance data for prevention: an algorithm for detecting Salmonella outbreaks. Emerg. Infect. Dis. 1997;3:395–400. doi: 10.3201/eid0303.970322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stern L., Lightfoot D. Automated outbreak detection: a quantitative retrospective analysis. Epidemiol. Infect. 1999;122:103–110. doi: 10.1017/s0950268898001939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akhtar M., Kraemer M.U.G., Gardner L.M. A dynamic neural network model for predicting risk of Zika in real time. BMC Med. 2019;17:171. doi: 10.1186/s12916-019-1389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S., Billings S., Grant P. Non-linear system identification using neural networks. Int. J. Control. 1990;51:1191–1214. [Google Scholar]
- 38.Leontaritis I., Billings S.A. Input-output parametric models for non-linear systems part I: deterministic non-linear systems. Int. J. Control. 1985;41:303–328. [Google Scholar]
- 39.Anno S., et al. Spatiotemporal dengue fever hotspots associated with climatic factors in Taiwan including outbreak predictions based on machine-learning. Geospat. Health. 2019;14 doi: 10.4081/gh.2019.771. [DOI] [PubMed] [Google Scholar]
- 40.Hutwagner L., Browne T., Seeman G.M., Fleischauer A.T. Comparing aberration detection methods with simulated data. Emerg. Infect. Dis. 2005;11:314–316. doi: 10.3201/eid1102.040587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farrington C.P., Andrews N.J., Beale A.D., Catchpole M.A. A statistical algorithm for the early detection of outbreaks of infectious disease. J. Royal Statist. Soc. Ser. A (Statist. Soc.) 1996;159:547–563. doi: 10.2307/2983331. [DOI] [Google Scholar]
- 42.Simonsen L., et al. A method for timely assessment of influenza-associated mortality in the United States. Epidemiology. 1997;8:390–395. doi: 10.1097/00001648-199707000-00007. [DOI] [PubMed] [Google Scholar]
- 43.Lombardo J., et al. A systems overview of the Electronic Surveillance System for the Early Notification of Community-Based Epidemics (ESSENCE II) J. Urban Health. 2003;80:i32–i42. doi: 10.1007/pl00022313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reis B.Y., et al. AEGIS: a robust and scalable real-time public health surveillance system. J. Am. Med. Inform. Assoc. 2007;14:581–588. doi: 10.1197/jamia.M2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lake I.R., et al. Machine learning to refine decision making within a syndromic surveillance service. BMC Publ. Health. 2019;19:559. doi: 10.1186/s12889-019-6916-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan T.C., et al. Probabilistic daily ILI syndromic surveillance with a spatio-temporal Bayesian hierarchical model. PLoS ONE. 2010;5:e11626. doi: 10.1371/journal.pone.0011626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng H.Y., et al. Applying machine learning models with an ensemble approach for accurate real-time influenza forecasting in Taiwan: development and validation study. J. Med. Internet. Res. 2020;22:e15394. doi: 10.2196/15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.VanWormer J.J., Sundaram M.E., Meece J.K., Belongia E.A. A cross-sectional analysis of symptom severity in adults with influenza and other acute respiratory illness in the outpatient setting. BMC Infect. Dis. 2014;14:231. doi: 10.1186/1471-2334-14-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hawkins S., He H., Williams G., Baxter R. Proceedings of the International Conference on Data Warehousing and Knowledge Discovery. 2002. Outlier detection using replicator neural networks; pp. 170–180. [Google Scholar]
- 50.Williams G., Baxter R., He H., Hawkins S., Gu L. Proceedings of the 2002 IEEE International Conference on Data Mining. 2002. A comparative study of RNN for outlier detection in data mining; pp. 709–712. [Google Scholar]
- 51.Zhou C., Paffenroth R.C. Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2017. Anomaly detection with robust deep autoencoders; pp. 665–674. [Google Scholar]
- 52.Kramer M.A. Nonlinear principal component analysis using autoassociative neural networks. AIChE J. 1991;37:233–243. doi: 10.1002/aic.690370209. [DOI] [Google Scholar]
- 53.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 54.P. Malhotra et al., Multi-sensor prognostics using an unsupervised health index based on LSTM encoder-decoder. arXiv preprint arXiv:1608.061(2016).
- 55.W. Luo, W. Liu, S. Gao, in: 2017 IEEE International Conference on Multimedia and Expo (ICME), pp. 439–444.
- 56.P. Malhotra et al., LSTM-based encoder-decoder for multi-sensor anomaly detection, 2016, arXiv preprint arXiv:1607.00148.
- 57.M. Du, F. Li, G. Zheng, V. Srikumar, in: Proceedings of the 2017 ACM SIGSAC Conference on Computer and Communications Security 1285–1298 (Association for Computing Machinery, Dallas, Texas, USA, 2017).
- 58.Yin C., Zhang S., Wang J., Xiong N.N. Anomaly detection based on convolutional recurrent autoencoder for IoT time series. IEEE Trans. Syst. Man Cybernet.: Syst. 2020:1–11. doi: 10.1109/TSMC.2020.2968516. [DOI] [Google Scholar]
- 59.Z. Chen, C.K. Yeo, B.S. Lee, C.T. Lau, in: 2018 Wireless Telecommunications Symposium (WTS), pp. 1–5.
- 60.E. Marchi et al., in: 2015 International Joint Conference on Neural Networks (IJCNN). IEEE, pp. 1–7.
- 61.T. Nolle, A. Seeliger, M. Mühlhäuser, in: International Conference on Discovery Science, Springer, pp. 442–456.
- 62.Vincent P., et al. Stacked denoising autoencoders: Learning useful representations in a deep network with a local denoising criterion. J. Mach. Learn. Res. 2010;11 [Google Scholar]
- 63.Narasimhan M.G., Kamath S. Dynamic video anomaly detection and localization using sparse denoising autoencoders. Multimedia Tools Appl. 2018;77:13173–13195. [Google Scholar]
- 64.Al-Qatf M., Lasheng Y., Al-Habib M., Al-Sabahi K. Deep learning approach combining sparse autoencoder with SVM for network intrusion detection. IEEE Access. 2018;6:52843–52856. [Google Scholar]
- 65.S. Chang, B. Du, L. Zhang, in: IGARSS 2019-2019 IEEE International Geoscience and Remote Sensing Symposium, IEEE, pp. 5488–5491. [DOI] [PMC free article] [PubMed]
- 66.L. Kamb, When did coronavirus really hit Washington? 2 Snohomish County residents with antibodies were ill in December, 2020. <https://www.seattletimes.com/seattle-news/antibody-test-results-of-2-snohomish-county-residents-throw-into-question-timeline-of-coronaviruss-u-s-arrival/>.
- 67.Liu H., et al. An information extraction framework for cohort identification using electronic health records. AMIA Jt. Summits Transl. Sci. Proc. 2013;2013:149–153. [PMC free article] [PubMed] [Google Scholar]
- 68.Wen A., et al. Desiderata for delivering NLP to accelerate healthcare AI advancement and a Mayo Clinic NLP-as-a-service implementation. NPJ Digit. Med. 2019;2:130. doi: 10.1038/s41746-019-0208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang F.-S., Zhang C. What to do next to control the 2019-nCoV epidemic? The Lancet. 2020;395:391–393. doi: 10.1016/s0140-6736(20)30300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deeplearning4J Development Team. Deeplearning4j: Open-source distributed deep learning for the JVM, 2020. <https://deeplearning4j.konduit.ai>.
- 71.M.D. Zeiler, Adadelta: an adaptive learning rate method, 2012. arXiv preprint arXiv:1212.5701.
- 72.Duchi J., Hazan E., Singer Y. Adaptive subgradient methods for online learning and stochastic optimization. J. Mach. Learn. Res. 2011;12:2121–2159. [Google Scholar]
- 73.L. Bottou, Online Algorithms and Stochastic Approximations. Online Learning and Neural Networks, 1998.
- 74.Ng A.Y. Proceedings of the Twenty-first International Conference on Machine Learning. 2004. Feature selection, L1 vs. L2 regularization, and rotational invariance; p. 78. [Google Scholar]
- 75.United States Centers for Disease Control and Prevention Update: influenza activity – United States, 2010–11 season, and composition of the 2011–12 influenza vaccine. MMWR Morb. Mortal. Wkly. Rep. 2011;60:705–712. [PubMed] [Google Scholar]
- 76.United States Centers for Disease Control and Prevention Update: influenza activity – United States, 2011–12 season and composition of the 2012–13 influenza vaccine. MMWR Morb. Mortal. Wkly Rep. 2012;61:414–420. [PubMed] [Google Scholar]
- 77.United States Centers for Disease Control and Prevention Influenza activity – United States, 2012–13 season and composition of the 2013–14 influenza vaccine. MMWR Morb. Mortal. Wkly Rep. 2013;62:473–479. [PMC free article] [PubMed] [Google Scholar]
- 78.United States Centers for Disease Control and Prevention Influenza activity – United States, 2013–14 season and composition of the 2014–15 influenza vaccines. MMWR Morb. Mortal. Wkly Rep. 2014;63:483–490. [PMC free article] [PubMed] [Google Scholar]
- 79.United States Centers for Disease Control and Prevention Influenza activity – United States, 2014–15 season and composition of the 2015–16 influenza vaccine. MMWR Morb. Mortal. Wkly Rep. 2015;64:583–590. [PMC free article] [PubMed] [Google Scholar]
- 80.United States Centers for Disease Control and Prevention Influenza activity – United States, 2015–16 season and composition of the 2016–17 influenza vaccine. MMWR Morb. Mortal. Wkly Rep. 2016;65:567–575. doi: 10.15585/mmwr.mm6522a3. [DOI] [PubMed] [Google Scholar]
- 81.United States Centers for Disease Control and Prevention Update: influenza activity in the United States during the 2016–17 season and composition of the 2017–18 influenza vaccine. MMWR Morb. Mortal. Wkly Rep. 2017;66:668–676. doi: 10.15585/mmwr.mm6625a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.United States Centers for Disease Control and Prevention Update: influenza activity in the United States during the 2017–18 season and composition of the 2018–19 influenza vaccine. MMWR Morb. Mortal. Wkly Rep. 2018;67:634–642. doi: 10.15585/mmwr.mm6722a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.United States Centers for Disease Control and Prevention. A weekly influenza surveillance report prepared by the influenza division: influenza-like illness (ILI) activity level indicator determined by data reported to ILINet, 2020. <https://gis.cdc.gov/grasp/fluview/main.html>.
- 84.M.D.o. Health, MDH Investigating E. coli O157 Infections Associated with Minnesota State Fair, 2019. <https://www.health.state.mn.us/news/pressrel/2019/ecoli091719.html>.
- 85.Syarif I., Prugel-Bennett A., Wills G. International Conference on Networked Digital Technologies. 2012. Unsupervised clustering approach for network anomaly detection. [Google Scholar]
- 86.Aytekin C., Ni X., Cricri F., Aksu E. 2018 International Joint Conference on Neural Networks (IJCNN) 2018. Clustering and unsupervised anomaly detection with l 2 normalized deep auto-encoder representations; pp. 1–6. [Google Scholar]
- 87.T. Pham, S. Lee, Anomaly detection in bitcoin network using unsupervised learning methods, 2016. arXiv preprint arXiv:1611.03941.
- 88.Erfani S.M., Rajasegarar S., Karunasekera S., Leckie C. High-dimensional and large-scale anomaly detection using a linear one-class SVM with deep learning. Pattern Recogn. 2016;58:121–134. doi: 10.1016/j.patcog.2016.03.028. [DOI] [Google Scholar]
- 89.Amer M., Goldstein M., Abdennadher S. Proceedings of the ACM SIGKDD Workshop on Outlier Detection and Description. 2013. Enhancing one-class support vector machines for unsupervised anomaly detection; pp. 8–15. [Google Scholar]
- 90.C. Kruegel, D. Mutz, W. Robertson, F. Valeur, Bayesian event classification for intrusion detection. In: 19th Annual Computer Security Applications Conference, 2003. Proceedings, 2003, pp. 14–23.
- 91.Ye N., Chen Q. An anomaly detection technique based on a chi-square statistic for detecting intrusions into information systems. Qual. Reliab. Eng. Int. 2001;17:105–112. doi: 10.1002/qre.392. [DOI] [Google Scholar]
- 92.M.-L. Shyu, S.-C. Chen, K. Sarinnapakorn, L. Chang, A novel anomaly detection scheme based on principal component classifier. In: IEEE Foundations and New Direc-tions of Data Mining Workshop, in Conjunction with ICDM'03, 2003, pp. 171–179.
- 93.Zanero S., Savaresi S.M. Proceedings of the 2004 ACM Symposium on Applied Computing. 2004. Unsupervised learning techniques for an intrusion detection system; pp. 412–419. [Google Scholar]
- 94.Zhang Z., Li J., Manikopoulos C., Jorgenson J., Ucles J. Proc. IEEE Workshop on Information Assurance and Security. 2001. HIDE: a hierarchical network intrusion detection system using statistical preprocessing and neural network classification; pp. 85–90. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Due to the results of the symptom extraction process being considered protected health information, data is not available as it would be difficult to distribute to anyone not engaged in an IRB-approved collaboration with the Mayo Clinic.