There is an increasing appreciation for the role of secreted extracellular vesicles (EVs) and nonvesicular nanoparticles in both physiologic and pathophysiologic conditions. Small EVs (sEVs) are 40- to 200-nm lipid-bilayer enclosed membrane vesicles, whereas exomeres are small (<50 nm), nonmembranous, extracellular nanoparticles.1 , 2 The coronavirus disease 2019 (COVID-19) pandemic is caused by the recent emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).3 For infection, SARS-CoV-2 must bind to the host cell receptor angiotensin-converting enzyme 2 (ACE2) to gain entry.4 ACE2 is also the critical receptor used by SARS-CoV-1, responsible for the 2002–2004 SARS epidemic. Middle East respiratory syndrome (MERS) is a highly lethal respiratory disease initiated by binding of MERS-CoV to its entry receptor dipeptidyl peptidase 4 (DPP4).5 ACE2 and DPP4 are highly expressed in gastrointestinal tissues, and a significant number of COVID-19, SARS, and MERS patients present with gastrointestinal symptoms.6 , 7 The ectodomain of ACE2 can be released by the activity of tumor necrosis factor-α converting enzyme (TACE) and the transmembrane serine protease 2 (TMPRSS2).4 The S1 subunit of the spike (S) protein of SARS-CoV-2 is primed by TMPRSS2 or TMPRSS4 for cell entry4 , 7, and TMPRSS2 can also prime the S protein of MERS-CoV.
It has been demonstrated that human recombinant soluble ACE2, but not mouse ACE2, can inhibit SARS-CoV-2 infection,8 raising the possibility that extracellular ACE2 carried by sEVs or extracellular nanoparticles may act as a decoy to bind the virus. Here, we explore the hypothesis that sEVs and exomeres containing ACE2 can bind SARS-CoV-2.
Methods
Detailed methods are available in the Supplementary Methods.
Results
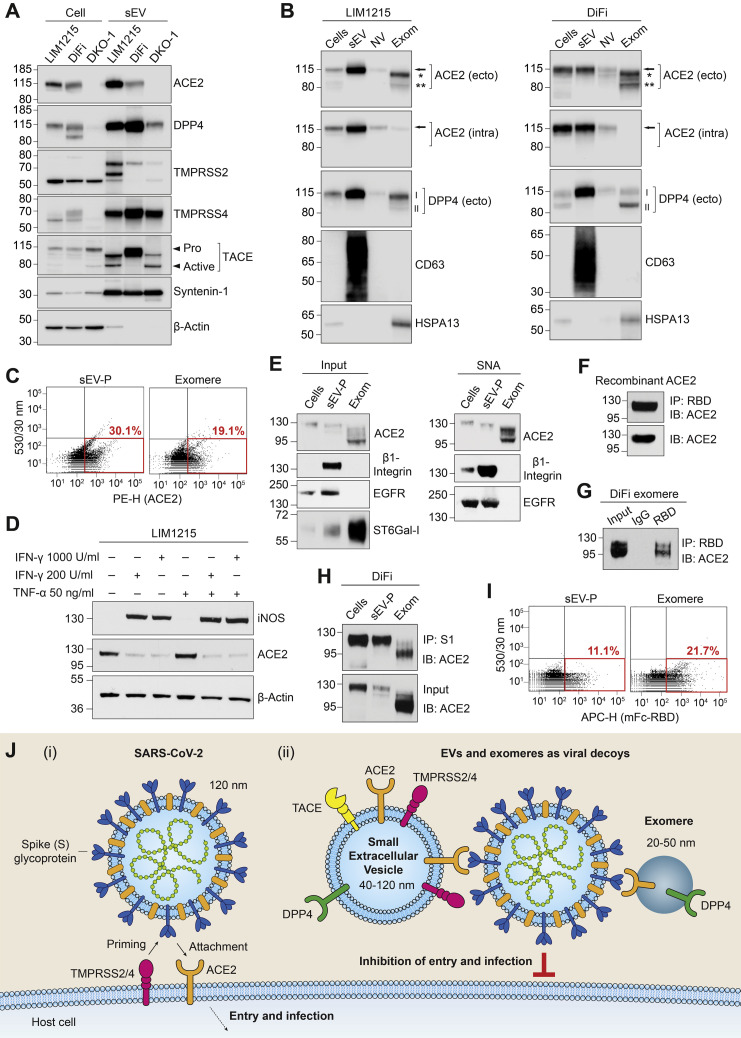
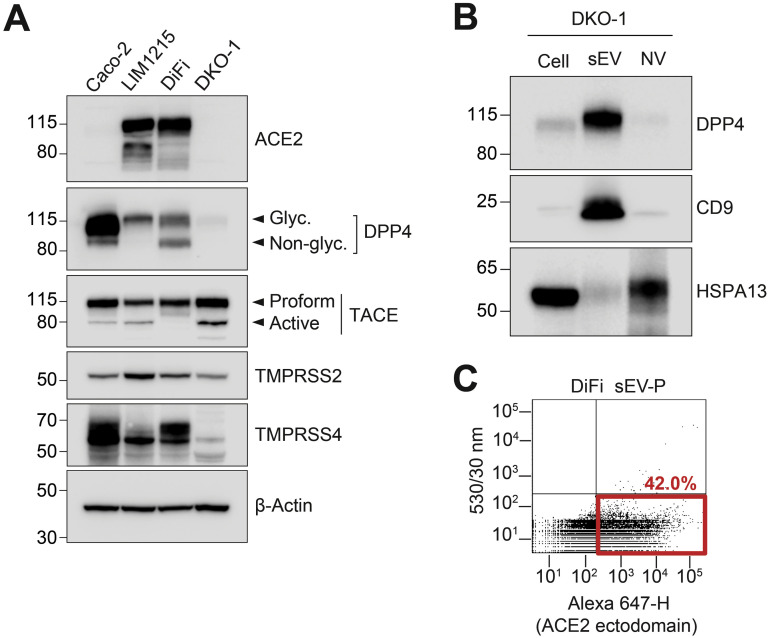
Colorectal cancer cell lines LIM1215 and DiFi express high levels of ACE2, whereas Caco-2 express much less, and ACE2 was undetectable in DKO-1 (Figure 1 A). After high-resolution density gradient purification,1 ACE2 was found to be secreted in LIM1215 and DiFi sEVs, whereas DPP4 was enriched in sEVs from LIM1215, DiFi, and DKO-1 cells (Figure 1 A and Supplementary Figure 1 B). The proteases TACE, TMPRSS2, and TMPRSS4 were all expressed by LIM1215, DiFi, and DKO-1 cells and secreted in sEVs (Figure 1 A and Supplementary Figure 1 A).
Figure 1.
sEVs and exomeres containing ACE2 can bind SARS-CoV-2 through the virus spike (S) protein. (A) Immunoblot analysis of cells and sEVs purified by high-resolution density gradients. (B) LIM1215 (left) and DiFi (right) samples were fractionated followed by immunoblotting. Arrow, full-length ACE2; ∗Ectodomain glycoform 1 of ACE2; ∗∗Ectodomain glycoform 2 of ACE2; ACE2 (ecto), ectodomain-specific monoclonal antibody; ACE2 (intra), intracellular-specific monoclonal antibody; I, glycosylated DPP4; II, nonglycosylated DPP4; anti-CD63 and anti-HSPA13 are used as exosomal and exomere markers, respectively; NV, nonvesicular; Exom, exomere. (C) Fluorescence-activated vesicle sorting analysis for ACE2 expression in sEV pellets (P) and exomeres from DiFi cells using an ectodomain-specific polyclonal antibody directly conjugated to PE. (D) Immunoblot analysis of inducible nitric oxide synthase (iNOS) and ACE2 expression in LIM1215 cells treated with indicated doses of interferon gamma (INF-γ) and TNF-α. (E) Sample inputs from DiFi cells (left) and agarose-conjugated Sambucus nigra agglutinin (SNA) lectin pull-downs (right) were analyzed by immunoblotting. β1-Integrin and epidermal growth factor receptor (EGFR) are known to be α2,6-sialylated and were used as positive controls. (F) Binding of human recombinant soluble ACE2 to the RBD of SARS-CoV-2 S protein. IP, immunoprecipitation; IB, immunoblotting. (G) Binding of DiFi exomeres with RBD of SARS-CoV-2 S protein. (H) Binding of DiFi cells, sEV-P, and exomeres with the S1 subunit of SARS-CoV-2 S protein. (I) Fluorescence-activated vesicle sorting analysis for binding of DiFi sEV-P and exomere to a mouse Fc-tagged RBD of SARS-CoV-2 S protein. (J) Schematic illustration of (i) SARS-CoV-2 viral infection of ACE2-positive cells and (ii) inhibition of infection due to hypothetical viral decoy binding by ACE2-positive sEVs and exomeres. Similarly, DPP4-positive sEVs and exomeres may act as decoys for MERS-CoV.
Supplementary Figure 1.
(A) Immunoblot analysis of colorectal cancer cells with indicated antibodies. (B) Immunoblot analysis of DKO-1 cells, sEV, and nonvesicular (NV) samples with indicated antibodies. (C) Fluorescence-activated vesicle sorting analysis for ACE2 expression in DiFi sEV-P using an ectodomain-specific monoclonal ACE2 primary antibody followed by an Alexa-647 secondary antibody. ß-actin serves as a loading control.
To investigate further, we fractionated extracellular samples into sEV, nonvesicular, and exomere fractions.1 , 2 Although cells and sEVs contained full-length ACE2, the 2 ectodomain glycoforms of ACE2 were present in exomeres (Figure 1 B). The full-length ACE2 in cells and sEVs was detected with antibodies recognizing ectodomain and intracellular epitopes. However, the 2 shed ectodomain fragments present in exomeres were only detected with the ectodomain-specific ACE2 antibody, confirming identity (Figure 1 B). Additionally, full-length DPP4 was highly enriched in sEVs from both LIM1215 and DIFI cells, whereas shed ectodomain DPP4 was present in exomeres (Figure 1 B). Both DiFi cell-derived ACE2-containing sEVs and exomeres could be analyzed using fluorescence-activated vesicle sorting with a polyclonal antibody recognizing the ectodomain of ACE2 (Figure 1 C). Fluorescence-activated vesicle sorting detection of sEVs was confirmed by using a monoclonal antibody to the ectodomain of ACE2 (Supplementary Figure 1 C).
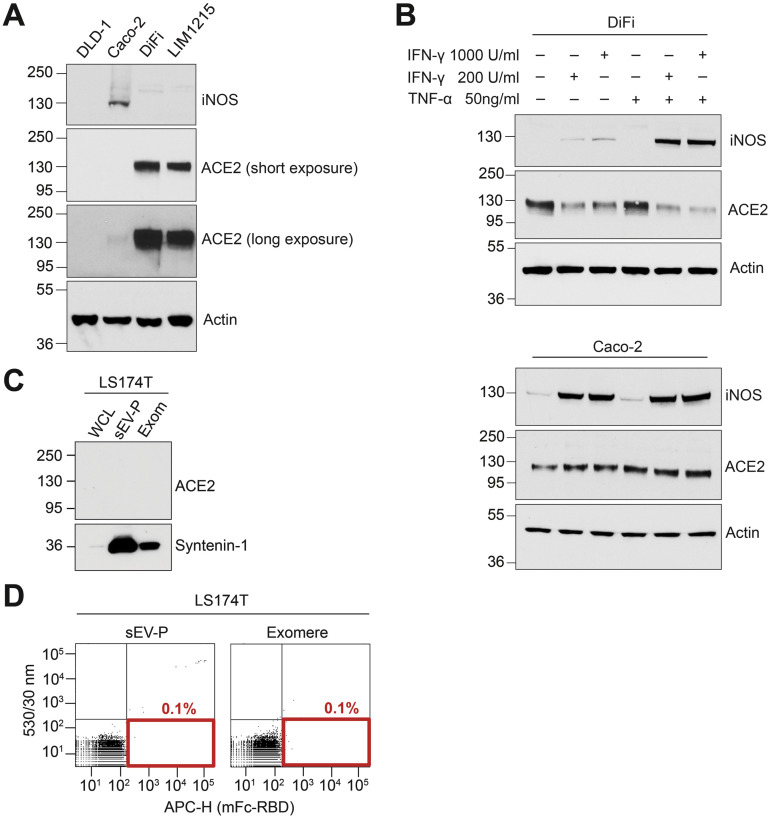
Because inflammatory cytokines modulate expression of ACE29 and inflammation influences ACE2 messenger RNA levels in ileal and colonic tissue,10 we sought to determine the baseline inflammatory state of colorectal cancer cells secreting sEV- and exomere-containing ACE2. Under the growth conditions used for production of sEVs and exomeres, levels of the inflammatory marker, inducible nitric oxide synthase, were clearly detectable in Caco-2 cells but were undetectable in DLD-1, LIM1215, and DiFi cells (Supplementary Figure 2 A). ACE2 levels were high in DiFi and LIM1215 cells but were much lower in Caco-2 and undetectable in DLD-1 cells. However, after treatment with the inflammatory cytokine, interferon gamma, or co-treatments with interferon gamma and TNF-α, the expression of inducible nitric oxide synthase was strongly induced in both LIM1215 and DiFi cells (Figure 1 D and Supplementary Figure 2 B). Furthermore, these inflammatory cytokines significantly reduced ACE2 protein levels in both LIM1215 and DiFi cells, whereas ACE2 levels in Caco-2 cells were relatively unchanged (Figure 1 D and Supplementary Figure 2 B). Thus, under these culture conditions, LIM1215 and DiFi cells were in a noninflammatory state at baseline, but interferon gamma in particular decreased cellular levels of ACE2.
Supplementary Figure 2.
(A) Immunoblot analysis of inducible nitric oxide synthase (iNOS) and ACE2 expression in cells cultured under standard conditions for EV and exomere isolation. (B) Immunoblot analysis of iNOS and ACE2 expression in cells treated with indicated concentrations of interferon gamma (INF-γ) and TNF-α. (C) Immunoblot analysis for ACE2 from LS174T cells, sEV-Ps, and Exom. (D) Fluorescence-activated vesicle sorting analysis of LS174T, sEV-Ps, and exomeres binding to a mouse Fc-tagged RBD of SARS-CoV-2 spike protein S1. Actin serves as a loading control.
ACE2 and DPP4 are type I and type II transmembrane receptors, respectively, and both are known to be extensively glycosylated in their respective N-terminal or C-terminal ectodomains. The enzyme responsible for α2,6-sialylation, β-galactoside α2,6-sialyltransferase 1, was enriched in DiFi exomeres (Figure 1 E, left). We isolated total α2,6-sialylated proteins by Sambucus nigra agglutinin lectin pull-down and probed for ACE2 by immunoblotting. Full-length ACE2 in cells and sEVs was modified by α2,6-sialylation, as were the 2 ectodomain glycoforms secreted in exomeres (Figure 1 E, right).
Virus-receptor recognition occurs through the receptor-binding domain (RBD) of the S glycoprotein of SARS-CoV-2 with the extracellular peptidase of the ACE2 ectodomain.4 Because the RBD of the SARS-CoV-2 S protein subunit S1 binds the ectodomain of ACE2, the presence of the ectodomain of ACE2 in exomeres suggested these extracellular nanoparticles might bind the virus. We first confirmed a mouse Fc-tagged RBD of the S1 subunit could bind human recombinant soluble ACE2 (Figure 1 F). We next demonstrated that DiFi exomeres containing ACE2 could be bound and pulled down with the mouse Fc-tagged RBD (Figure 1 G). Moreover, S1 subunit could bind DiFi cells, sEVs, and exomeres that all contain ACE2 (Figure 1 H). Fluorescence-activated vesicle sorting analysis further confirmed that ACE2-positive DiFi sEVs and exomeres could bind to the RBD (Figure 1 I), whereas sEVs and exomeres from the ACE2-negative LS174T cell line could not (Supplementary Figure 2 C and D). It has been demonstrated that human soluble recombinant ACE2 can act as a decoy for SARS-CoV-2 and thereby inhibit infection.8 Based on our characterization of extracellular carriers of ACE2 and their binding to SARS-CoV-2 S protein, we propose that ACE2-containing sEVs and exomeres may also act as decoys to inhibit infection (Figure 1 J).
Discussion
We showed that full-length ACE2 and DPP4, critical receptors for SARS-CoV-1, SARS-CoV-2, and MERS-CoV, are released in sEVs from colorectal cancer cells and that shed ectodomain fragments of ACE2 and DPP4 are enriched in exomeres. To our knowledge, we provide the first evidence that ACE2 in cells, sEVs, and exomeres is α2,6-sialylated. Both sEVs and exomeres released from cells expressing ACE2 can bind to the RBD of the SARS-CoV-2 S protein S1 subunit. Thus, human cells might be used for large-scale production of EVs and exomeres containing fully post-translationally modified ACE2 that can be used as decoys for SARS-CoV-2 to attenuate infection, as has been demonstrated for human soluble recombinant ACE2.8 Studies comparing the efficiency and stability of S protein binding between recombinant soluble ACE2, sEVs containing full-length ACE2, and exomeres containing ectodomain ACE2 may inform the best therapeutic intervention for COVID-19 infections. The inflammatory state of gastrointestinal tissues can influence the expression of ACE2.10 Intriguingly, we find that induction of inflammation can lead to a marked reduction of ACE2 protein expression in LIM1215 and DiFi colorectal cancer cells. Further study is necessary to determine if inflammatory modulation of cellular ACE2 expression results in altered secretion of ACE2-containing sEVs and exomeres. It is possible that inflammation leads to increased secretion of ACE2 in sEVs and exomeres, thereby lowering cellular levels, or that lower cellular levels result in decreased release of ACE2. In either case, the inflammatory state of tissues may influence the proposed decoy effect of extracellular ACE2.
To enable binding to ACE2, TMPRSS2 or TMPRSS4 prime the SARS-CoV-2 S protein. Additionally, the proteases TNF-α converting enzyme and TMPRSS2 cleave ACE2 to release the ectodomain. Thus, the activity of these proteases is critical for infection, and these proteins may represent important clinical targets.4 We found that TMPRSS2, TMPRSS4, and TNF-α converting enzyme are all secreted in sEVs and are thus present in the extracellular environment, allowing them to potentially interact with viral particles. It is currently unknown if the sEV-embedded proteases play any role in priming of coronavirus S proteins or if transfer of these proteases to recipient cells can prime cells for subsequent virus entry into host cells. Our findings have potential important implications for COVID-19, SARS, and MERS.
Acknowledgments
The authors thank Robert H. Carnahan of the Vanderbilt Vaccine Center for useful discussions and providing reagents. The authors acknowledge the support of Vanderbilt Digestive Diseases Research Center and Vanderbilt Ingram Cancer Center.
CRediT Authorship Contributions
Qin Zhang, PhD (Conceptualization: Lead; Investigation: Lead; Methodology: Lead; Writing – original draft: Lead). Dennis K. Jeppesen, PhD (Conceptualization: Lead; Investigation: Lead; Methodology: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead). James N. Higginbotham, PhD (Conceptualization: Lead; Investigation: Lead; Methodology: Lead). Jeffrey L. Franklin, PhD (Resources: Equal). James E. Crowe, Jr, MD (Resources: Supporting; Writing – review & editing: Equal). Robert J. Coffey, MD (Funding acquisition: Lead; Writing – review & editing: Lead).
Footnotes
Conflicts of interest These authors disclose the following: James E. Crowe, Jr has served as a consultant for Takeda Vaccines, Sanofi-Aventis US, Pfizer, Novavax, Lilly, and Luna Biologics; is a member of the Scientific Advisory Boards of CompuVax and Meissa Vaccines; and is Founder of IDBiologics. The Crowe laboratory at Vanderbilt University Medical Center has received sponsored research agreements from Moderna and IDBiologics. The remaining authors disclose no conflicts.
Funding This work was supported by Defense Advanced Research Projects Agency grant HR0011-18-2-0001, National Institutes of Health contract 75N93019C00074, the Dolly Parton COVID-19 Research Fund at Vanderbilt to James E. Crowe, Jr, and National Cancer Institute grants R35 CA197570, UG3 241685, and P50 236733 to Robert J. Coffey.
Prior to acceptance of this manuscript, Penninger and co-workers reported the efficacy of soluble ACE2 in treating a patient with severe COVID-19 infection (Zoufaly A, et al. Lancet Resp Med 2020;8:1154-1158).
Author names in bold designate shared co-first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org and at https://doi.org/10.1053/j.gastro.2020.09.042.
Supplementary Methods
Cell Culture
Human colon cancer cell lines, DiFi, DLD-1, DKO-1, LS174T, and LIM1215, were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% bovine growth serum, 1% glutamine, 1% nonessential amino acids, and 1% penicillin/streptomycin at 37°C in a 5% CO2 humidified incubator. Cells were maintained by passage every 3–4 days at 70%–80% confluence and were routinely tested for mycoplasma contamination (Universal Mycoplasma Detection Kit; ATCC, Manassas, VA). All cell culture media were purchased from Corning Cellgro (Manassas, VA), and all cell culture supplements were from Hyclone (Logan, UT) unless stated otherwise.
EV and Nanoparticle Isolation From Cells Cultured in Plastic Cell Culture Dishes
sEV pellets (sEV-Ps) and exomeres were isolated from cell-conditioned medium as previously described.1 The protein concentrations of sEV-Ps and exomere were determined with a Direct Detect (Millipore, Burlington, MA). At no time during the process were samples subjected to temperatures below 4°C.
EV and Nanoparticle Isolation From Cells Cultured in Bioreactors
DKO-1 cells were maintained in CELLine Adhere 1000 (CLAD1000) bioreactors (INTEGRA Biosciences AG, Zizers, Switzerland) at 37°C in a 5% CO2 humidified incubator, as previously described.2, 3, 4 Cell-conditioned medium was harvested from bioreactors every 48 hours, starting from 1 week after inoculation of the bioreactor and continuing for a period of 4 weeks. sEV-P and exomere samples were isolated as described above. At no time during the process were samples subjected to temperatures below 4°C.
High-resolution (12%–36%) Iodixanol Density Gradient Fractionation
Fractionation of sEV-Ps to generate gradient-purified sEV and nonvesicular samples was performed as previously described.2
Protein Isolation From Cells and Extracellular Fractions
To isolate proteins, cultured cells were washed twice with ice-cold phosphate-buffered saline (PBS) and solubilized in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM phenylmethylsulfonyl fluoride) containing a complete protease inhibitor tablet and PhosSTOP tablet (Roche, Indianapolis IN). Lysed samples were incubated on ice for 10 minutes, sonicated 3 times, and cleared by centrifugation at 14,000 rpm for 5 minutes. Supernatant fractions were quantified using direct Detect Infrared Spectrometer (Millipore) and used for immunoblotting. After the final wash step of ultracentrifugation in PBS, sEV-P, nonvesicular, and exomere samples were lysed in RIPA buffer and proteins extracted as described above for cell samples.
Immunoblot Analysis
Immunoblot analysis was carried out as previously described.1 Primary antibodies were as follows: anti-β-Actin (Sigma, A5316), anti-β1-Integrin (BD, 610467), anti-Syntenin-1 (Abcam, ab133267), anti-ACE2 (ab108252, N-terminal), anti-ACE2 (Abcam, ab108209, C-terminal), anti-TMPRSS2 (Abcam, ab92323), anti-TMPRSS4 (Proteintech, 11283-1-AP), anti-TACE (Abcam, ab2051), anti-EGFR (Millipore, 06-847), anti-CD9 (Santa Cruz, SC13118), anti-DPP4 (Cell Signaling Technology, 67138), anti-CD63 (BD Biosciences, 556019), anti-HSPA-13 (Santa Cruz, sc-398297), anti-iNOS (Cell Signaling Technology, 39898) and anti-ST6-Gal1 (R&D, AF5924).
Cytokine Treatment
DiFi, LIM1215, and Caco-2 cells were plated in 6-well plates in Dulbecco’s modified Eagle’s medium containing 10% bovine growth serum at 5 × 105 cells per well for DiFi cells and 2.5 × 105 cells per well for LIM1215 and Caco-2 cells. Treatment with cytokines was carried out as previously described.5 Briefly, 24 hours after seeding, cells were treated with 200 U/mL or 1000 U/mL recombinant human interferon gamma (R&D Systems) in Dulbecco’s modified Eagle’s medium containing 5% bovine growth serum. After 24 hours, cells were treated with 50 ng/mL recombinant human TNF-α (R&D Systems). Twenty-four hours later, cells were washed twice with PBS and lysed in RIPA buffer for protein analysis as described above. Expression of inducible nitric oxide synthase and ACE2 (C-terminal) in cells was examined by immunoblotting.
Lectin Precipitation Assays
Lectin precipitation was carried out as described.6 Briefly, total lysate (approximately 500 μg) from cells or isolated fractions were incubated overnight with 50 μL agarose-conjugated SNA-1 lectin with rotation at 4°C (EY Laboratories, San Mateo, CA). α2,6-Sialylated proteins were pelleted with the SNA lectin by centrifugation at 3000 rpm for 2 minutes and washed 3 times with lysis buffer, followed by 1 wash with PBS. Sialylated proteins were released from the complexes by boiling for 5 minutes in 2× sodium dodecyl sulfate sample buffer. The glycoproteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then immunoblotted to detect target proteins.
Fluorescence-activated Vesicle Sorting Staining and Analysis
sEV-Ps and exomeres derived from DiFi and LS174T cells were stained and analyzed as previously described.1 , 7 For samples incubated with directly conjugated primary antibodies, the samples were washed 3 times and centrifuged at 304,000 ×g with a S100-AT4 fixed angle rotor (75,000 rpm, effective k factor of 29 with 1.5 mL ultramicrofuge tubes filled to capacity) for 30 minutes unless stated otherwise. For samples stained with unconjugated primary antibodies, after incubation for overnight at 4°C, the samples were washed twice, incubated with secondary antibody for 1 hour at room temperature, and then washed 3 times in PBS-HEPES for single-color analysis. Samples incubated with only the secondary antibody were used as negative controls. Primary antibodies were as follows: anti-ACE2 (Bioss, bs-1004R-PE, PE-conjugated), anti-ACE2 (Abcam, ab108252), anti-ACE2 (R&D, AF933), and SARS-CoV-2 mouse Fc fusion (mouse, Crowe Laboratory). Secondary antibodies were donkey anti-goat (H+L) (Invitrogen, A32814, AF488 conjugated).
Immunoprecipitation of ACE2
Cells, sEV-Ps, and exomeres derived from DiFi cells isolated as described above were resuspended in PBS-H, and proteins were lysed in RIPA buffer for immunoblotting. Protein concentrations were determined with Direct Detect (Millipore). DiFi exomeres were incubated overnight at 4°C with SARS-COV-2-RBD-mFc (Crowe Laboratory) followed by incubation with protein G magnetic beads for 2 hours at 4°C. Mouse IgG was used as a negative control for mouse Fc, and human recombinant soluble ACE2 (Abcam, ab151852) was used as a positive control. Samples were incubated with SARS-CoV-2 spike S1 protein-coupled magnetic beads (MBS-K001, ACRO biosystem) overnight at 4°C. The S1 protein coupled to magnetic beads was made in human HEK293 cells and is not glycosylated per information from the manufacturer. After incubation, the beads were washed 3 times in RIPA buffer. Bound proteins were eluted in 2× reducing sample buffer at 70°C for 10 minutes. Proteins were separated and detected as previously described. An anti-ACE2 antibody (Abcam, ab108252) was used to detect ACE2 that binds with either SARS-COV-2-RBD-mFC or SARS-CoV-2 spike S1.
References
- 1.Jeppesen D.K. Cell. 2019;177:428–445. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Q. Cell Rep. 2019;27:940–954. doi: 10.1016/j.celrep.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M., Kleine-Weber H. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raj V.S., Mou H. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding S., Liang T.J. Gastroenterology. 2020;159:53–61. doi: 10.1053/j.gastro.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zang R., Gomez Castro M.F. Sci Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monteil V. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lang A. Virology. 2006;353:474–481. doi: 10.1016/j.virol.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novak J.K. Gastroenterology. 2020;159:1151–1154. doi: 10.1053/j.gastro.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Zhang Q. Cell Rep. 2019;27:940–954. doi: 10.1016/j.celrep.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeppesen D.K. Cell. 2019;177:428–445. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeppesen D.K. J Extracell Vesicles. 2014;3:25011. doi: 10.3402/jev.v3.25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeppesen D.K. Proteomics. 2014;14:699–712. doi: 10.1002/pmic.201300452. [DOI] [PubMed] [Google Scholar]
- 5.Kreeger P.K. Cancer Res. 2009;69:8191–8199. doi: 10.1158/0008-5472.CAN-09-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaikh F.M. Exp Cell Res. 2008;314:2941–2950. doi: 10.1016/j.yexcr.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higginbotham J.N. J Extracell Vesicles. 2016;5:29254. doi: 10.3402/jev.v5.29254. [DOI] [PMC free article] [PubMed] [Google Scholar]