Abstract
There is a strong risk of mutations in the genome sequence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in animals. Consequently, a possibility of zoonotic transfer of a much stronger form of the present virus from animal to human is also very feasible in the near future. Thus, recent incidents of SARS-CoV-2 infection in animals need to be studied very carefully to protect against any future transmissions. Interspecies analysis from animal to human or vice versa is the tool of choice at the present time for understanding zoonotic transfer and improving/accelerated drug discovery for COVID-19.
Main Text
What Is SARS-CoV-2 and COVID-19?
A cluster of pneumonia-type pulmonary diseases was reported from the city of Wuhan in Hubei Province, China.1 The causative virus has been termed SARS-CoV-2/2019-nCoV/HCoV-19 and the disease termed COVID-19 by the World Health Organization (WHO).2 With the announcement of COVID-19 as a global pandemic by the WHO, SARS-CoV-2 is now affecting 213 countries and territories around the world and two international conveyances. As of June 15, 2020, 8,113,679 cases have been confirmed with 439,085 deaths and 4,213,602 recovered; among the active cases, 3,406,424 patients have had a mild condition and 54,568 are in serious or critical condition (https://www.worldometers.info/coronavirus/). This single-strand enveloped RNA virus belongs to the family Coronaviridae, genus Betacoronavirus and subgenus Sarbecovirus; and termed SARS-CoV-2 due to 79.6% similarity with the RNA genome of the SARS coronavirus (SARS-CoV).1,2 Two prominent genomic features of SARS-CoV-2 are its ability to bind to the human receptor angiotensin-converting enzyme 2 gene (ACE2) and the spike protein with a functional polybasic cleavage site at the S1–S2 boundary through the insertion of 12 nucleotides.3 Phylodynamic analysis of 176 full-length genomes and most recent common ancestor data supported a decisive indication of human to human transmission of SARS-CoV-2, denying the fact of a non-human animal reservoir for spreading COVID-19.4 Is current knowledge conclusive? We guess no, because extensive and in-depth studies are required to understand the possible progenitor of the SARS-CoV2.
Why Animals Need to Be Tested for SARS-CoV-2?
There is a strong possibility of zoonotic transfer of the virus from Malayan pangolins (Manis javanica) and/or Rhinolophus affinis bats to a human patient zero due to their genome sequence similarity with SARS-CoV2.5 There is 96% overall similarity of RaTG13 of a Rhinolophus affinis with SARS-CoV-2; this is the closest to SARS-CoV-2 across the genome, supporting the theory of bats as reservoir hosts for its progenitor. The only difference is that the spike diverges in the receptor-binding domain (RBD) of bat SARS-CoV-like coronaviruses, and has weak binding affinity to human ACE2.5 Malayan pangolin coronaviruses display strong similarity to SARS-CoV-2 in the RBD, including all six key RBD residues (L455, F486, Q493, S494, N501, and Y505) shown to be important in human-like ACE2 as a result of nature.6 Interestingly, both animals do not have the polybasic cleavage sites like SAR-CoV2, which raises the chance of mutations occurring near the S1–S2 junction of coronaviruses followed by insertion of a polybasic cleavage site in SARS-CoV-2 naturally.7
Although there is no scientific evidence yet of human to animal transmission of SARS-CoV2, two reports suggested that a Pomeranian and later a German shepherd tested positive for coronavirus in Hong Kong.8 Although both dogs tested weak positive and did not show any symptoms of the disease, the Pomeranian was dead 2 days after it was released from quarantine. In another report, a pet cat has been infected with a novel coronavirus in Belgium and it happened due to close contact with its coronavirus-positive owner.8 The recent literature also claimed that there is a possibility of coronavirus attack on pets such as cats and dogs due to their presence in proximity to humans.9 Due to weak symptoms and lack of testing kits, animals (especially pets) are not tested. It was a bigger surprise when a zoo tiger tested positive in Bronx, New York, due to possible transmission from its caretaker. Later, three more tigers and three African lions in the same zoo showed similar symptoms of dry coughs like the Malayan tiger (https://www.npr.org/2020/04/06/828517076/a-tiger-has-coronavirus-should-you-worry-about-your-pets). Two weeks later, two pet cats in New York state tested positive for COVID-19, making them the first confirmed cases in pet animals in the United States where owners showed symptoms and tested positive for the virus (https://www.theguardian.com/world/2020/apr/22/coronavirus-cats-pets-infected-new-york). In the most recent news, two mink farms have been quarantined after coronavirus infection was confirmed in animals in the Netherlands (https://www.reuters.com/article/us-health-coronavirus-netherlands-mink/dutch-mink-cull-starts-as-coronavirus-spreads-to-10th-farm-idUSKBN23D0KB). As the tiger is also in the cat family (feline), there is a chance of susceptibility to coronavirus in the feline family. As the instances of human to animal transmission are too few to consider, and there are not enough studies, that does not prove that human to animal transmission is impossible. Right now, the priority is saving human lives and, accordingly, the development of drugs. However, there is always the possibility of mutations, insertions, and deletions in the genome sequence of this deadly virus in these animals in the future.
During quarantine, people are almost constantly in proximity of pets (especially emotional support animals), which may be the cause of outbreaks of coronavirus in those animals. It could happen in the near future or maybe it is already happening but with weak symptoms. This could assist zoonotic transfer of much a stronger form of the present SARS-CoV-2 virus from animals to human. Thus, these scattered incidences need to be checked very carefully to avoid any future transmission.
Implication of Interspecies Analysis
We would like to bring to your attention a novel technique that allows analysis of data related to various species, providing a broad picture of the action of the virus. Thus, our proposed interspecies-quantitative structure-activity relationship (i-QSAR) analysis from animal to human or vice versa is a useful tool at the present time for drug discovery.10 There is a high chance that the virus may exist in one or multiple animal species. The idea is to confirm the source from patient zero, analyzing much earlier data before November 2019. Exploration of in silico i-QSAR analysis may highlight the issue in the future by correlating and extrapolating the response (activity/toxicity) data from an animal source to human. This could open avenues for drug discovery. Considering bat or pangolin as a possible reservoir source of SARS-CoV-2, we are proposing to exploit these (and possibly other animals) for drug discovery using multiple drug candidates through in vivo/in vitro analysis to generate initial data. Later, these experimental data can be used for in silico modeling using the i-QSAR model, which is a proven tool when there is a gap in the data.10 An important aspect is that interspecies analysis is already an established method, so in future, researchers need to use this approach without hesitation after validation of the developed model. This analysis will incorporate the species-specific mechanism of action followed by extrapolation of response data in the form of drug activity and/or toxicity from bat/pangolin to human. The only limitation of this method is that researchers need to have experimental data for at least 15–20 chemicals (drug molecules) for at least two test species (preferably human and species close to human regarding the genome sequence) for the initial development of the i-QSAR model. Once the model is ready, extrapolation of data can be performed for future chemicals.
With the race against COVID-19, in silico studies have a huge impact to speed up the drug and vaccine discovery process. Along with techniques such as docking, molecular dynamics, homology modeling, we must focus on the less known and entirely unexplored interspecies QSAR analysis. Presently, the amount of data is not enough, but over time, with data resources and understanding of SARS-CoV-2 and COVID-19, interspecies analysis will be possible. Considering the similar genome sequence of SARS-CoV-2 with pangolins and bats, experimental data on drug candidates for pangolins/bats along with the structural and physicochemical properties of the drugs can be correlated with the human response endpoint initially. Later extrapolation of animal data to human data can lead to the inspiring possibility of no human testing through the i-QSAR model developed. The complete scheme of the proposed i-QSAR analysis to combat COVID-19 is reported in Figure 1.
Figure 1.
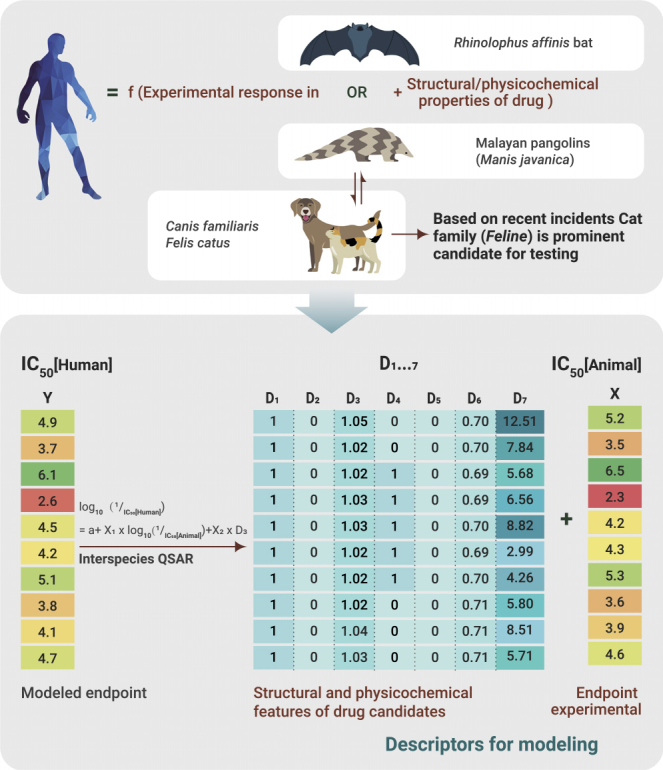
Schematic Protocol of i-QSAR Analysis to Study the Response of Drug Candidates for Humans through Extrapolation of Bats/Pangolins and If Possible, through Cats/Dogs or Common Experimental Mammals.
a is a constant term, X1 and X2 are coefficients of log10(1/IC50[Animal]) and D3 modeled descriptors, respectively, in the interspecies equation.
Once the predictive interspecies QSAR model is established, there will be less requirement for animal testing. Researchers need to test only selected drug candidates for pre-clinical trials instead of too many candidates, which not only maintain the 3R rule but also save many animal lives. There are already reports on coronavirus attacks on dogs and cat. So, future introspection of pets related to coronavirus disease may lead to better accessibility of experimental data as bats and pangolins are endangered species and they are difficult to study compared with dogs/cats. The study involving the interspecies analysis tool is currently being carried out in our laboratory and the results will be published elsewhere.
Acknowledgments
S.K. and J.L. are thankful to the National Science Foundation (NSF/CREST HRD-1547754 and NSF/RISE HRD-1547836) for financial support.
Author Contributions
S.K. and J.L. contributed to the design and writing of this paper.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rambaut, A. (2020). Phylogenetic analysis of nCoV-2019 genomes. http://virological.org/t/356.
- 5.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. e00127–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351.e2. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamada Y., Liu D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009;83:8744. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimm D. Should pets be tested for coronavirus? Science. 2020 doi: 10.1126/science.abc0029. [DOI] [Google Scholar]
- 9.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., et al. Susceptibility of ferrets, cats, dogs, and different domestic animals to SARS-coronavirus-2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kar S., Gajewicz A., Roy K., Leszczynski J., Puzyn T. Extrapolating between toxicity endpoints of metal oxide nanoparticles: predicting toxicity to Escherichia coli and human keratinocyte cell line (HaCaT) with Nano-QTTR. Ecotox. Environ. Saf. 2016;126:238–244. doi: 10.1016/j.ecoenv.2015.12.033. [DOI] [PubMed] [Google Scholar]


