Graphical abstract
Keywords: COVID-19, Antiviral, Drug discovery, Docking, Pharmacophore, Therapeutic, C30 endopeptidase
Highlights
-
•
Virtual screening of FDA drugs yields potential therapeutics against COVID19.
-
•
Hit compounds are supposed to inhibit SARS-CoV-2 main protease 3CLpro.
-
•
Best hits include oxytetracycline and other antibiotics.
Abstract
3CL proteases (3CLpro) are only found in RNA viruses and have a central role in polyprotein processing during replication. Therefore, 3CLpro has emerged as promising drug target for therapeutic treatment of infections caused by Coronaviruses. In the light of the recent major outbreak of the SARS-CoV-2 virus and the continuously rising numbers of infections and casualties, there is an urgent need for quickly available drugs or vaccines to stop the current COVID-19 pandemic. Repurposing of approved drugs as 3CLpro inhibitors could dramatically shorten the period up to approval as therapeutic against SARS-CoV-2, since pharmacokinetics and toxicity is already known. Several known drugs, e.g. oxytetracycline, doxorubicin, kanamycin, cefpiramide, teniposide, proanthocyanidin and salvianolic acid B, but also not-approved active compounds from the ZINC15 library were identified as new potential inhibitors of 3CLpro by using different complementary virtual screening and docking approaches. These compounds have the potential to be further optimized using structure based drug design as demonstrated for oxytetracycline.
1. Introduction
The recent worldwide outbreak of SARS-CoV-2 with almost 15 million infected individuals and more than 600000 casualties (Dong et al., 2020) (accessed on 20th July 2020) is a massive challenge for all countries and societies, since at this time no vaccine or small molecule therapeutic is approved and available on the market. In the light of the urgent need for a causal therapy, almost 1500 studies are actively recruiting or enrolling patients by invitation for clinical studies against COVID-19 (https://clinicaltrials.gov/ct2/results?cond = COVID-19&Search=Apply&recrs=a&recrs=f&age_v=&gndr=&type=&rslt=, accessed on 20th Jly 2020).
The main protease of Coronaviruses, chymotrypsin-like protease (3CLpro), processes the large polyprotein 1ab releasing several further enzymes that are crucial for viral replication. Moreover, 3CLpro is unique for Coronaviruses and not found in higher organisms. This predestines 3CLpro as most attractive target for the development of anti-infective agents against SARS-CoV-2 and related Coronaviruses. Consequently, several inhibitors of 3CLpro were developed mostly during the last 17 years right after the first wave of infection caused by the SARS-CoV-1. However, no experimental compound was developed further and reached the market. Most compounds were designed as covalent inactivators that react with the catalytic Cys145. This is reflected by the majority of x-ray structures of 3CLpro enzymes from different Coronaviruses. However, small reversible inhibitors are preferred in many cases, because of less side effects and toxicity, which often arise with covalent inhibitors. Most recently two crystal structures of 3CLpro of SARS-CoV-2 were reported, both of them bound to a covalent inhibitor (Jin et al., 2020; Zhang et al., 2020). Both crystal structures of 3CLpro-complexes (PDB-ID’s 6LU7 and 6Y2F) show perfect overlap with an RMSD-value of 0.48 Å over 300 amino acids. The most prominent crystal structure of 3CLpro of SARS-CoV-1 with a non-covalent inhibitor is PDB-ID: 3V3M (Jacobs et al., 2013). Interestingly, some approved drugs for other indication areas proved to be active against Coronaviruses as well. One of the active substances is the old malaria therapeutic hydroxychloroquine that seemed to be effective in limiting the replication of SARS-CoV-2 in vitro (Cortegiani et al., 2020) and has shown efficacy in clinical studies (Gao et al., 2020). These findings stimulated an intensive debate about the potential benefit of hydroxylchloroquine for COVID-19 patients. However, a large retroperspective study revealed serious dysrhythmias in patients treated with chloroquine or hydroxychloroquine and showed no evidence of benefit in patients with COVID-19 (Geleris et al., 2020). Moreover, a very recent FDA review of safety issues with the use of these chloroquine derivates points out that treatment is associated with serious heart rhythm problems and other safety issues, including blood and lymph system disorders, kidney injuries and liver failure (https://www.fda.gov/drugs/drug-safety-and-availability/fda-cautions-against-use-hydroxychloroquine-or-chloroquine-covid-19-outside-hospital-setting-or, accessed on 20th July 2020). Consequently, the actual NIH treatment guideline (https://www.covid19treatmentguidelines.nih.gov/, accessed on 20th July 2020) recommends against the use of chloroquine or hydroxychloroquine for the treatment of COVID-19, except in clinical trials. Dexamethasone is a corticosteroid that is used to treat many different inflammatory conditions such as allergic disorders, as well as ulcerative colitis, arthritis, lupus, psoriasis and breathing disorders. The actual COVID-19 treatment guideline of the NIH recommends dexamethasone for the treatment of COVID-19 patients due to lower mortality rates when compared with patients who received standard of care (Horby et al., 2020) (https://www.covid19treatmentguidelines.nih.gov/immune-based-therapy/immunomodulators/corticosteroids/, accessed on 20th July 2020,).
Other drugs like cepharanthine, selamectin and mefloquine showed also promising effects in cell culture (Fan et al., 2020). Furthermore, a combination of ribavirin and interferon-α is recommended for treatment of COVID-19 by the National Health Commission in China because of the effect on MERS-CoV (Du et al., 2020). Recently, a clinical trial started to treat COVID-19 with remdesivir, a broad-spectrum antiviral agent that was previously tested in humans with Ebola virus disease. First results in animal models infected with MERS-CoV proved to be promising. Furthermore, the clinically proven camostat mesilate has shown to prevent cellular entry of SARS-CoV-2 upon inhibition of the serine protease TMPRSS2 (Hoffmann et al., 2020). A recent study found that flavonoids like herbacetin, rhoifolin and pectolinarin (Jo et al., 2020) or derivatives of isatin (Liu et al., 2014) block the enzyme activity of 3CLpro from SARS-CoV-2. The availability of crystal structures of 3CLpro enzymes of different Coronavirus strains stimulated several virtual screening campaigns yielding different and poorly overlapping hit compounds (Chen et al., 2020; Ton et al., 2020; Berry et al., 2015; Mukherjee et al., 2011; Tsai et al., 2006) (see detailed comparison below). Only two virtual screens were based on the structure of 3CLpro from the actual SARS-CoV-2 and only one of these efforts used a crystal structure of this target (Ton et al., 2020), whereas the other virtual screen relied on a homology model of the target protein (Chen et al., 2020).
Facing the actual COVID-19 pandemic with exponential growth of infections and without any causative treatment stimulated this study to rapidly identify approved drugs with high potential to inhibit 3CLpro and fight COVID-19. It is anticipated that the development time of a drug with known side effects and toxicity profile to approval could be tremendously shortened. Consequently, this study begins, in step one, with a virtual screen of a collection of approved drugs against the crystal structure of 3CLpro from SARS-CoV-2 using two complementary docking programs, MOE and Autodock Vina. This allowed for consistency checking and increased the confidence in the results. In a second step, the best hits were accurately docked and energy minimized to create the basis for a thorough protein-ligand interaction fingerprints (PLIF) analysis in the third step. The fourth step consisted of the development of a conclusive pharmacophore model based on the most important interactions between the target protein and the ligands. Subsequently, in step five, more than 7 million compounds of the ZINC15 library were screened for compounds fulfilling the requirements of the pharmacophore model. And in the final step six structure based drug design (SBDD) was used to evaluate the perspective and optimization potential of the best compounds (Sander et al., 2015).
2. Methods
2.1. Database preparation
The library of 2683 approved drugs was downloaded as SD-file from the following address: “https://www.selleckchem.com/screening/fda-approved-drug-library.html”. Since the SD-file contained only 2D-structures, at least two 3D-conformers were generated per stereoisomer using free cheminformatics program Osiris Datawarrior (Idorsia Pharmaceuticals. Ltd) (Sander et al., 2015). ZINC15 is a free database of commercially available compounds provided by the Irwin and Shoichet Laboratories at the University of California, San Francisco (UCSF) (https://zinc15.docking.org/) (Sterling and Irwin, 2015). Compounds can be manifold filtered for virtual screening purposes. Here, a collection of 7.2 million 3D protomers was downloaded containing drug-like molecules with molecular weight between 250 and 500 Da, plog < 5.. The compounds of the ZINC15 library were filtered to exclude instable and strongly reactive compounds. Weakly reactive groups such as aldehydes, thiols and Michael acceptors as well as molecules, which contain Pan Assay Interference Compounds (PAINS) patterns, were allowed to pass the filter. The 3D-structure data were transferred in tranches as SD-files using cURL. Subsequently, the SD-files were combined to four larger files to be used in the following pharmacophore search.
2.2. Virtual screening
Two virtual screen were carried out using MOE 2019 software (Chemical Computing Group, Montreal, Canada), as well as Autodock Vina (The Scripps Research Institute, La Jolla, USA) (Trott and Olson, 2010). The latter was implemented in the free version of PyRx environment (available from https://sourceforge.net/projects/pyrx/). For the MOE based virtual screen, AMBER14:EHT force field was used throughout. The crystal structure of 3CLpro of SARS-CoV-2 was obtained from the RCSB Protein Data Bank (PDB-ID: 6LU7). The ligand that is covalently conjugated to the catalytic Cys145 was removed and the Cys145 restored as free thiol. The free target protein was then subjected to the QuickPrep procedure of MOE including corrections for missing atoms, alternate geometries or other crystallographic artifacts, removing water molecules farther than 4.5 Å from any receptor or ligand atom and 3D protonation. The removed covalent ligand was used to define the binding pocket. For virtual screening, the SD-file with 3D structures of approved drugs was loaded into a MOE-specific database and selected as ligand in the docking setup. The triangle matcher and a rigid receptor were selected as further parameters. The docking poses were rated using London dG score for 10 poses per ligand molecule. The screening results were exported as SD-files and multiple results for a particular chemical structure were merged using Osiris Datawarrior to obtain a result file with unique chemical structures preserving multiple docking scores per molecule. For the virtual screen with Autodock Vina, the crystal structure of 3CLpro was also freed from the covalent ligand and then subjected to the Dockprep procedure implemented in the UCSF Chimera program (University of California, USA) (Pettersen et al., 2004). The protocol contains deletion of solvent, replacement of incomplete side chains using a rotamer library, protonation and addition of charges. The prepared protein structure and the
SD-file with 3D structures of the ligand library were loaded and converted to the desired pdbqt-format using PyRx. In the next step, the implemented Vina wizard was exploited to select the compound library as multiple ligands and the protein target. Before starting the virtual screening procedure, a cuboid was used to define the area of the active site pocket. In the last step, the exhaustiveness that gives a measure for the accuracy of docking was set to 8. After that, the Vina wizard started virtual screening. The results were obtained again in pdbqt-format and had to be converted into SD-format for further analysis.
2.3. Molecular docking and energy minimization
Visualization of structural data as well as molecular docking was performed using MOE 2019 software (Chemical Computing Group, Montreal, Canada). The crystal structure of 3CLpro of SARS-CoV-2 (PDB-ID: 6LU7) was obtained from RCSB Protein Data Bank. The structure file was loaded into the program and subjected to structure preparation including 3D protonation for subsequent docking as described before. The partial charges of all protein and ligand atoms were calculated using the implemented Amber14:EHT force field. Molecular docking was performed choosing the triangle matcher for placement of the ligand in the binding site and ranked with the London dG scoring function. The best 50 poses were passed to the refinement and energy minimization in the pocket using the induced fit method and then rescored with the GBVI/WSA dG scoring function. Best poses were further refined by energy minimization of all amino acids in a radius of 10 Å around the ligand.
2.4. Pharmacophore building and search
The complexes structures of 3CLpro with the best scoring ligands were superposed and subjected to the Quickprep procedure implemented in MOE using again the AMBER14:EHT force field in order to enable the analysis of protein-ligand interactions. Protein ligand interaction fingerprints (PLIF) were generated and analyzed on the basis of several docked protein-ligand complexes with optimal binding poses using the corresponding tool of MOE. The PLIFs select the most important interactions for a series of protein-ligand complexes and visualize the result as panel showing the specific contacts of each ligand to adjacent amino acids of the binding pocket. Most importantly, the PLIFs and frequencies of protein-ligand contacts can be used to create a pharmacophore model encoding essential rules that must be fulfilled by a potential ligand of the respective binding pocket. These rules, also called features consist of spheres, where a heavy atom of the ligand with e.g. hydrogen donor or acceptor properties is supposed to be arranged in order to be able to form a particular interaction with an adjacent amino acid. To avoid too high numbers of false positive hits that are branched or bulky molecules, which fulfill the requirements, but would clash with the receptor protein, it is advisable to mimic the binding pocket by a series of overlapping excluded volume spheres. These were generated from selected pocket amino acids using a radius of 2 Å. Molecules with structures that clash with the excluded volume spheres are not considered as hits. A pharmacophore model enables a very rapid search on large 3D structure databases for hits that fulfill all or partial feature requirements. If the pharmacophore model is well defined, the search procedure should very efficiently enrich those chemical entities that produce high docking scores and have increased probability to bind to the respective target protein. The pharmacophore search was started directly from the pharmacophore editor of MOE. To assess the hits of the pharmacophore search regarding binding affinity, all of them were subjected to a follow-up virtual screening procedure using MOE as described above.
2.5. Structure based drug design
Once protein-ligand complexes with optimal binding poses are available and additional information are obtained from PLIF analysis, the next step is to dissect those positions of the ligand structure, where beneficial substituents could be introduced to satisfy hydrogen-bond acceptors or donors of pocket amino acids or other types of molecular interaction. Electrostatic maps were calculated to localize such areas, where hydrophobic elements, hydrogen donors or acceptor functions should be arranged. Desired modifications were carried out in silico using the Builder tool of MOE. The effect of the modifications was examined directly afterwards by accurate molecular docking and energy minimization. In the case of a successful optimization, additional contacts are established often leading to higher docking scores.
3. Results and discussion
3.1. Validation of docking procedure
At first, two docking programs, MOE and Autodock vina were evaluated, whether they were able to reproduce the binding pose in x-ray structures of 3CLpro complexes with non-covalent ligands. Redocking ligand ML188, which was also shown to inhibit 3CLpro activity (IC50 = 1.5 μM) (Turlington et al., 2013), into PDB-ID 3V3M yielded excellent overlap with crystallized ligand and RMSD-values of 0.981 Å for MOE and 0.926 Å for Autodock Vina (Fig. S1). The suitability of the docking procedure was also confirmed afterwards, because there was a clear enrichment of known 3CLpro inhibitors compared to all virtually screened compounds and a considerable overlap between the hits of (Chen et al. (2020)) and those of this study (see following sections).
3.2. Virtual screening
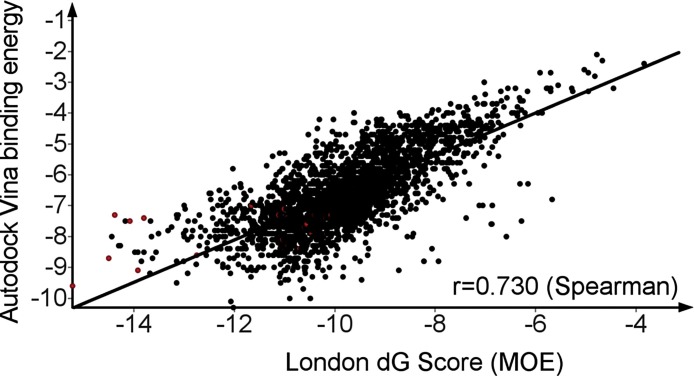
2683 chemical entities from the FDA-approved Drug Library from Selleckchem (https://www.selleckchem.com/screening/fda-approved-drug-library.html) were virtually screened for potential binding to the active site of 3CLpro of SARS-CoV-2 (PDB-ID: 6LU7) using two docking programs, MOE and Autodock vina. The complementary virtual screen allowed to compare both data sets, analyze the correlation of the docking scores of both docking programs and obtain independent confirmation of hits with high scores (Fig. 1 ). Autodock Vina binding energy score and London dG score of MOE showed satisfactory correlation with a Spearman’s rank correlation coefficient of 0.73. As a control, 30 known inhibitors of 3CLpro were included in the virtual screening campaigns as control. The control compounds were clearly enriched by the dual virtual screening approach: 27 out 30 (90 %) known 3CLpro inhibitors were found to have both, Vina score < -7.0 and MOE score < -10, whereas only 782 out of 2683 (29 %) total number of approved drug structures showed this combination of high docking scores. This finding confirmed that the docking protocols for MOE and Autodock Vina were well adjusted for finding inhibitors of 3CLpro.
Fig. 1.
Docking scores of 2683 approved drugs (black dots) and 30 known inhibitors of 3CLpro as control (red dots). Each dot denotes one chemical structure. Autodock Vina (binding energy) is plotted versus the London dG Score of MOE for each chemical entity.
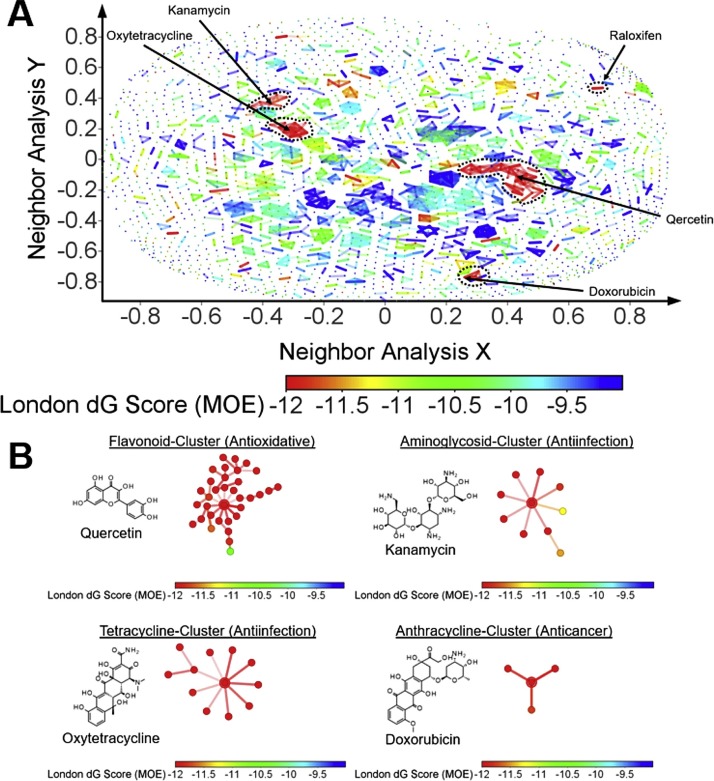
Next, an similarity and Activity Cliff Analysis was performed to visualize the chemical landscape, cluster similar molecules together on a 2D-area and identify clusters and singletons with elevated docking scores (Fig. 2A) (Bajorath et al., 2009). Four major clusters and a few singletons with stand out, including a large flavonoid-, a big tetracycline-, an aminoglycoside- and an anthracycline-cluster (Fig. 2 B). Representative drugs for these clusters are quercetin, oxytetracycline, kanamycin and doxorubicin, respectively. There are also high scoring singletons or clusters of two, e.g. raloxifen. The finding that many flavonoids are among the hits with best docking scores is in excellent agreement with the very recent report of Jo et al., who provide experimental evidence that flavonoids are indeed inhibitors of 3CLpro (Jo et al., 2020). It should be noted, that 4.5 % of the FDA approved drugs and 6 out of 19 hit of the virtual screen PAINS patterns, including the flavonoids quercetin, rutin, homoorientin, all of them flavonoids, eltrombopag and doxorubicin. The concept of PAINS was introduced by Baell and Holloway and addressed the problem of frequent hitters in experimental high throughput screening campaigns, which were often false positive hits (Baell and Holloway, 2010). However, the critical substructural elements of electronic PAINS filters were originally derived from a proprietary library tested in just six assays measuring protein–protein interaction (PPI) inhibition using the AlphaScreen detection technology only. Therefore, Capuzzi et al. “caution against the blind use of PAINS filters to detect and triage compounds with possible PAINS liabilities and recommend that such conclusions should be drawn only by conducting orthogonal experiments” (Capuzzi et al., 2017).Although some of the approved drug molecules contain critical substructures such as labile ester (salvianolic acid B) or possibly redox active groups such as electron rich scaffolds (polyphenols), all compounds were taken further to detailled docking analysis in order to elucidate the potential molecular interactions with 3Clpro, because a wide variety of orthogonal assays have been performed to demonstrate biological activity and safety of the molecules before drug approval.
Fig. 2.
A) Similarity/Activity cliff analysis demonstrating several clusters of similar chemical structures with high docking scores (< -11.5). Representatives of most active clusters are highlighted. A particularly high number of flavonoids has very high scores. B) Most striking clusters with chemical structure of typical reprentatives are shown. Each dot represents a member of this cluster. Pairs of most similar structures are connected by a line. The dots are colored according to reference panels below indicating the respective docking score.
3.3. Molecular docking
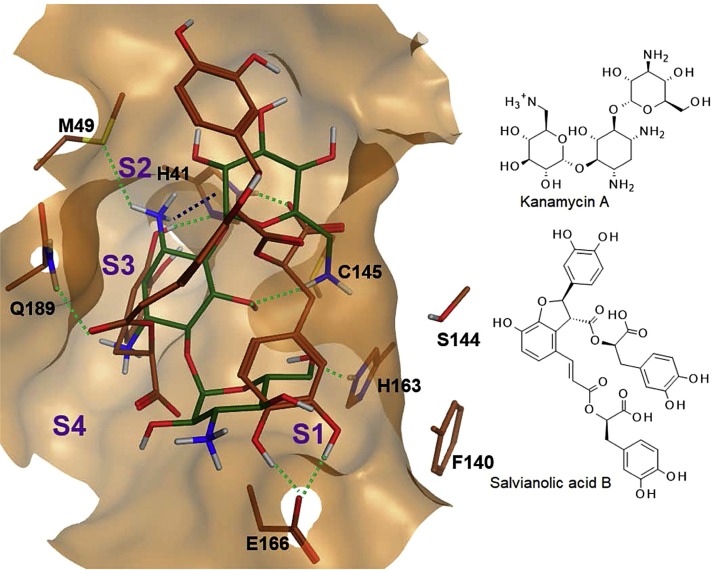
Best hits of the virtual screen were subjected to an accurate more extensive docking procedure involving sampling of 50 docking poses using a secondary GBVI/WSA dG score implemented in MOE. The results of most promising compounds are summarized in Table 1 . Interestingly, most high scoring drugs are antioxidants (flavonoids) or used as anti-cancer or antibiotic therapeutic. The two compounds with best docking scores, kanamycin and salvianolic acid B, show similar binding poses occupying S1- and S2-subpockets (Fig. 3 ). Both ligands form H-bonds with catalytic Cys145, although salvianolic acid B interacts with the backbone nitrogen and kanamycin with the thiolate moiety of Cys145. The particular protein-ligand interactions between the best representative ligands and 3CLpro are shown in Fig. S2 and are analyzed in more detail in the following section.
Table 1.
Docking scores of best hits from virtual screening using MOE.
| Cpd | GBVI/WSA dG | Substance group |
Indication area | Cpd | GBVI/WSA dG | Substance group |
Indication area |
|---|---|---|---|---|---|---|---|
| Naringin | −9.7 | Flavonoid glycosid | Anticancer, antiinflamm., antioxidant | Doxorubicin | −9.1 | Anthracycline | Antibiotic, anticancer |
| Epicatechin | −7.3 | Flavonoid | Antioxidant | Difloxacin | −7.4 | Fluoroquinolone | Antibiotic |
| Homoorientin | −8.8 | Flavonoid glycosid | Anticancer, radical scavenger, antioxidant | Tosufloxacin | −8.8 | Fluoroquinolone | Antibiotic |
| Proantho-cyanidin | −9.2 | Oligomeric flavonoid | Antioxidant, anti-plant pathogens | Etoposide | −9.1 | Lignane glycoside | Anticancer |
| Rutin | −8.9 | Flavonoid, glycoside of quercetin | Antioxidant, anti-haemorrhagic | Teniposide | −9.4 | Liganane glycoside | Anticancer |
| Salvianolic acid B | −10.3 | Benzofuran, polyphenol | Antioxidant, antiinflamm., anticancer | Methotrexate | −8.0 | Anaolog of folic acid | Anticancer, immune suppressive |
| Quercetin | −7.5 | Flavonoid | Anticancer, antiinflamm., antioxidant | Eltrombopag | −8.5 | Other | Chronic idiopathic thrombocytopenic purpura |
| Kanamycin | −10.0 | Aminoglycosid | Antibiotic | Raloxifen | −8.2 | Benzothiophen | Post-menopausal osteoporosis |
| Oxytetracycline | −9.3 | Tetracycline | Antibiotic, antiinflamm. | ML188 (Reference) (Turlington et al., 2013) | −8.8 | ||
| Cefpiramide | −9.6 | Cephalosporin | Antibiotic | ||||
Fig. 3.
Docking pose of kanamycin A (dark green) and salvianolic acid B (brown) within 3CLpro. Conventional hydrogen bonds are shown as dotted green lines. The cation-PI-interaction between kanamycin A and H41 is highlighted by a dotted dark blue line. S1-S4 denotes subpockets. The protein structure is taken from the energy minimized complex with salvianolic acid.
3.4. Protein Ligand Interaction Fingerprint (PLIF) Analysis and comparison with crystallographic fragment screening
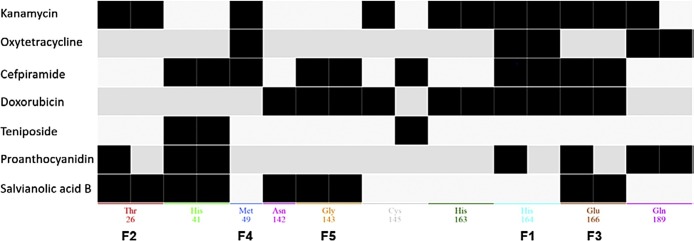
Seven structurally diverse ligands with highest docking scores were selected for the determination of the most important non-covalent contacts between the amino acids of the active site pocket and ligands. The energy minimized complexes between 3CLpro (PDB-ID: 6LU7) and ligands were superposed using MOE showing perfect overlap of the protein structure with slight deviations around the active site, which is expected, because the docking procedure allowed for induced fit around the binding pocket and the final energy minimization within a radius of 10 Å around the bound ligand (Fig. S3). All ligands formed multiple contacts with different sets of acite site flanking amino acids. The PLIF analysis was carried out using MOE resulting in a characteristic pattern of interaction for each of the ligands (Fig. 4 ). The ligands can be roughly grouped together in three blocks. The compounds of the first block (kanamycin, oxytetracycline, and doxorubicin) do not interact with His41 but bind to His164. Two of three compounds interact also with Met64, His163, Glu166 and Gln189. In contrast, all representatives of the second block (teniposide, proanthocyanidin and salvianolic acid B) interact with His41 and two of three compounds of the second block interact also with Thr26, Glu166 and not with His164 or Gln189. The pattern of cefpiramide combines different features of both blocks. Therefore, there are relatively many amino acids of the active site that are able to form significant interactions with structurally diverse chemical structures. This implies that there are also many opportunities to accommodate diverse ligands in the active site pocket of 3CLpro.
Fig. 4.
Different patterns of protein-ligand interaction fingerprints (PLIFs) within the active site of 3CLpro. The F-annotations denote the corresponding feature in the pharmacophore model generated on the basis of this PLIF-analysis.
Moreover, there should be room for the optimization of weak or moderate screening hits by medicinal chemistry methods due to multiple potential interactions with active site amino acids.
A preliminary, but highly relevant crystallographic fragment screening study has been posted on the bioRxiv preprint server at about the time, when this publication has been submitted (Douangamath et al., 2020). The fragment screen identified 71 hits spanning the entire active site. The binding poses of representative non-covalent fragments are compared with the docking pose of kanamycin (Fig. S8). The fragments bind preferably to S1- and S2-subpockets. Non-covalent fragments, which bind into the S1-subpocket, form hydrogen bonds with Gly143, Cys154, His163 and Glu166, while many fragments containing an aromatic ring bind into the S2-subpocket, thereby forming a distinct T-shaped PI-stacking interaction with catalytic His41. One of the S2-binding fragment forms an additional hydrogen bond to Gln189. None of the hits from the virtual screen shows such a deep insertion into the S2-subpocket. Only cefpiramide forms a remotely similar, but parallel PI-stacking between its tetrazole ring and His41. Furthermore, every hydrogen bond between 3Clpro and fragments was also found with virtual screening hits (Fig. 4). However, the hits in this study reveal several additional possible hydrogen bonds involving for example Asn142 and His164 in subpocket S1, as well as Thr26 and Met49 in subpocket S2 (Fig. 4, Fig. S9). Together, the unique T-shaped PI-interaction of non-covalent fragments in subpocket S2 and the additional potential hydrogen bonds identified by docking provide new opportunities to merge fragments and approved drug molecules or parts thereof to optimize inhibitors of 3Clpro.
3.5. Pharmacophore model building
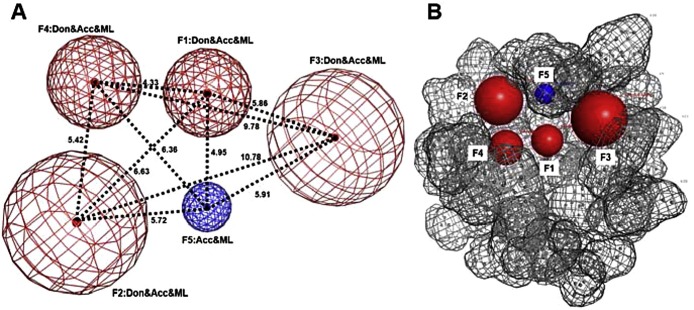
The detailed dissection of the most important molecular interactions between ligands and amino acids, flanking the active site pocket enabled the creation of a pharmacophore model. Such a model is a simplified picture that determines the chemical features in terms of molecular interactions, e.g. H-bond donator or acceptor, and their 3D arrangement within the active site pocket of the receptor protein. The pharmacophore model is completed by overlapping excluded volume spheres that mimic the protein pocket shape. A ligand will not match, if any of its atom centers intersects with an excluded volume. For 3CLpro, a pharmacophore model was generated on the basis of the PLIF-analysis above. The model contains 5 essential features, F1-F5, including the allowed location of H-bond donor, acceptor or metal ligator heavy atoms (Fig. 5 , refer also to Fig. 4). The pharmacophore model is ideally suited to screen larger compound libraries for chemical entities that fulfill the requirements of the chemically and spatially defined model features substantially faster than conventional virtual screens that rely on fast rigid model docking approaches.
Fig. 5.
A) Features (F1-F5) of the pharmacophore model. Red spheres denote areas with preferred presence of an H-bond donor (Don), acceptor (Acc) or metal ligator (ML) heavy atom and the blue sphere the area for H-bond acceptor (Acc) or metal ligator (ML) heavy atom. Black numbers denote the distances in Å between the center of the spheres. B) The same 5 features (solid spheres) of A) embedded in grey overlapping meshed spheres of excluded volumes that mimic the binding pocket of 3CLpro.
3.6. Pharmacophore search
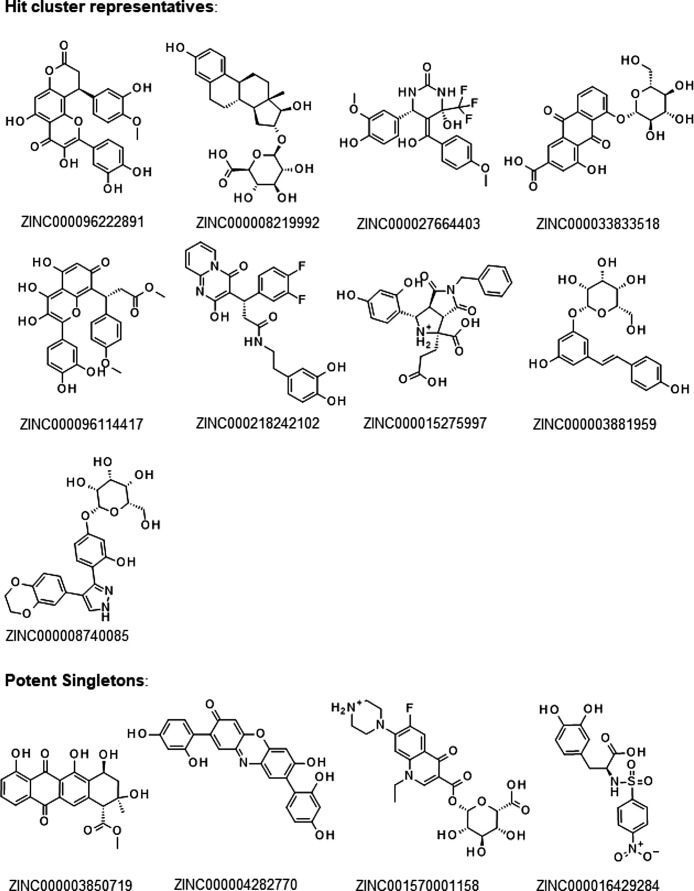
Having identified promising approved drugs as potential inhibitors of 3CLpro, it appeared tempting to exploit the above generated pharmacophore model to extend the chemical space and search for novel chemical entities with possibly better docking scores and activity against 3CLpro. Therefore, 7.2 Million protomers of drug-like molecules were loaded as 3D-structures from the publicly available ZINC15 library (Sterling and Irwin, 2015). A pharmacophore search was performed using MOE program. The search resulted in 5517 hits that fulfilled the requirements of at least 3 out of 5 features defined in the pharmacophore model (hit rate: 0.08 %). This number was good to handle with a classic virtual screen using MOE and London dG score. The result of a subsequent virtual screening of all pharmacophore search hits against 3CLpro was analyzed by Activity Cliff Analysis that was already applied to the virtual screening result of the approved drug library above (Fig. S4). The pharmacophore screen resulted in several clusters with distinct scaffolds. The chemical structure of representatives of the hit clusters and potent singletons are shown in Fig. 6 . Again, flavonoids showed up as the cluster with most members (442 out of 5517 hits) and highest docking scores (e.g. ZINC000096222891). There were also a high scoring cyclopentaphenanthrene cluster with 243 members (e.g. ZINC000008219992), 38 members of a cluster with cyclic ureas with >80 % similarity to ZINC000027664403, a anthrachinone cluster with 37 members some of them glycosylated (e.g. ZINC000033833518), a phenyl-chromen-one cluster with 30 members with a similarity of >80 % to ZINC000096114417, a pyrido-pyrimidinone cluster with 18 members and >80 % similarity to ZINC000218242102 and some smaller clusters with 5–9 members of glycosylated stilbenes (e.g. ZINC000003881959), glycosylated phenyldiazoles (e.g. ZINC000008740085) and octahydropyrrolo-pyrrole-diones (e.g. ZINC000015275997) (Fig. 6).
Fig. 6.
Representative hit structures of pharmacophore search on 7.2 million protomers of ZINC15 library against 3CLpro.
The 232 hits with highest London dG Score (< -13) included 110 compounds with >85 % structural similarity to substances of the approved drug library, mostly flavonoids and other drugs like oxytetracycline, and zidovudine. But the remaining 122 hits are novel chemical entities with high potential to inhibit 3CLpro. These novel compounds include ZINC000096222891, ZINC000008219992, ZINC000218242102, ZINC000015275997, the fluoroquinolone ZINC001570001158 and the sulfonamide ZINC000016429284 (Fig. 6). Best representative hits were subjected to highly accurate flexible docking yielding GBVI/WSA dG scores between -9.5 and -8.8 (Tab. S2). In order to gain more information about the molecular interactions with active site amino acids, a PLIF analysis was performed using the energy minimized complexes between 3CLpro (PDB-ID: 6LU7) and the four ligands with highest docking score < -8.7 (Fig. S5). The PLIF analysis revealed new opportunities for aliphatic-aromatic interactions with Gln189 and H-bond-interactions with Thr190 that were not seen in the PLIF-analysis of approved drugs above (Fig. 4, S6). Most promising compounds without PAINS alert include ZINC000008219992, ZINC000027664403, ZINC000003881959 and ZINC000008740085, representing large to medium sized clusters of analogs, as well as the singletons ZINC001570001158 and ZINC000004282770 (Fig. 6). 7 out of the 13 cluster representitives or singletons (Fig. 6), contain PAINS patterns: ZINC000096114417, ZINC000096222891, ZINC000218242102, and ZINC000016429284 contain a catechol group, ZINC000003850719 and ZINC000033833518 are quinones, and ZINC000015275997contains a mannich substructure. Catechols and quinones are discussed as problematic screening compounds, as they can undergo redox reactions and potentially react with proteins. However, as shown before, such compounds and particularly catechols can still be developed to drugs as 57 catechols and 4 quinones are found in the FDA approved drug library. The mannich bases represented by ZINC000015275997 are in principle able to form highly reactive quinone methides but, also in this case, 20 compounds of the FDA approved drug library contain a mannich base substructure. This demonstrates that compounds with PAINS patterns do not necessarily have poor pharmacokinetic properties or that toxicophores must be PAINS as stated by Capuzzi and others (Capuzzi et al., 2017). Having the PAINS alerts in mind, the biological activity, the mode of action (reversible vs. irreversible) and/or chemical reactivity of these compounds have to be carefully tested by suitable assays with orthogonal readout to decide, which compound should be further developed.
3.7. Comparison with other virtual screening efforts
As mentioned before, several virtual screens have been carried out against proteins of the 3CLpro protein family, but only two against 3CLpro of SARS-CoV-2 (Chen et al., 2020; Ton et al., 2020). From both studies screening results were available for the top 1000 compounds. This enabled the analysis of the overlap between the hits of the different virtual screening approaches. Older virtual screening campaigns were exclusively based on crystal structures of 3CLpro from SARS-CoV-1 and did only disclose data for most promising hits, therefore not allowing comprehensive analysis of hit overlap with other virtual screen (Berry et al., 2015; Mukherjee et al., 2011; Tsai et al., 2006). Generally, it would be expected, that there is a considerable overlap of hits, if the screened compound libraries share a similar chemical space. A comparison of the top 200 hits of the virtual screen applied to the approved drugs with top 200 hits of the pharmacophore search on the ZINC15 library, both this study, revealed 57 shared structures with at least 85 % similarity. The recovery of similar structures was not unexpected, since the binding poses of the best hits from the virtual screen were used to create the pharmacophore model that was used to screen the ZINC15 library. As discussed above, both hit lists share particularly many flavonoids. More interesting was the comparison of virtual screens between different laboratories. Chen and coworkers screened a library of 7173 purchasable drugs against a homology model of 3CLpro of the actual Coronavirus, which was created on the basis of 3CLpro of SARS-CoV-1 (PCB-ID: 2DUC), using Autodock Vina (Chen et al., 2020). The best scoring 4500 molecules screened against chain A or B of the enzyme structure were available for analysis. This number was reduced to 1547 (ca. 22 %) after removing duplicates. These compounds share 41 % structures with exact identity or >85 % similarity with the library of approved drugs that was screened in this study. Comparing the top 200 hits of the virtual screening in this study with the top 200 hits of (Chen et al., 2020) revealed 17 similar compounds (>85 %). All three approaches, that of Chen et al. as well as the virtual screen and pharmacophore search in this study, were able to identify similar compounds as potential inhibitors of 3CLpro, particularly many flavonoids. In an alternative approach, Ton et al. conducted a very large virtual screen on all 1.3 billion compounds from the ZINC15 library against the crystal structure of 3CLpro of the current SARS-CoV-2 using Glide SP Ton et al., 2020). Despite this brute force effort, the top 1000 hits of Ton et al. show no overlap with the top 200 hits of Chen et al. or the virtual screen and pharmacophore screen of this study. This lack of overlap could be at least partly explained by the fact that, in contrast to the virtual screens of Chen et al. and this study, all compounds with PAINS, particularly flavonoids, were removed from the hit list of Ton et al... In order to evaluate the reliability of the virtual screening approach in the current study further, the recovery rate of the already mentioned set of control substances with confirmed experimental inhibitory activity against 3CLpro was analyzed. The recovery rate of the virtual screening and pharmacophore search protocols applied in the current study was clearly better, 7 and 6 out of 30 3CLpro inhibitors, respectively, than for the approach of Chen et al. (2 out of 30) and Ton et al. (0 out of 30). The confirmation of experimental 3CLpro inhibitors provided confidence in the methods used.
3.8. Structure based drug discovery (SBDD)
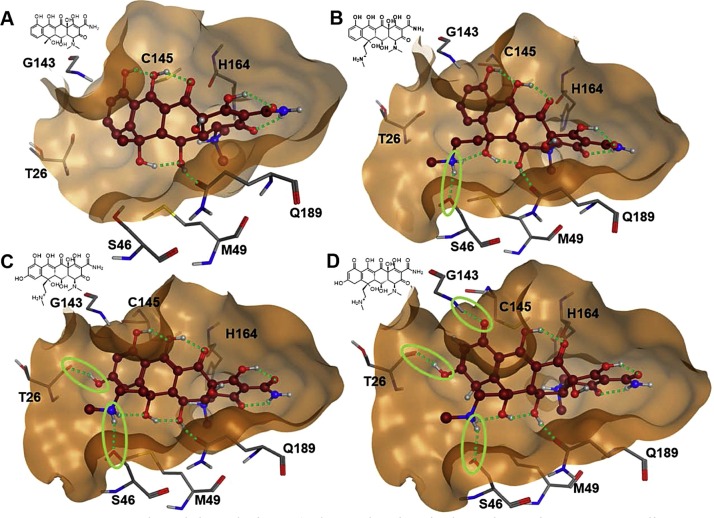
Looking at the PLIF-analysis of approved drugs with highest docking scores shows that every ligand has a specific pattern of molecular interaction with adjacent amino acids within the active site pocket (Fig. 4). None of the ligands exploits all possible contacts to these amino acids. Consequently, there should be room for optimizing the affinity to the binding pocket. Oxytetracycline is a relatively well-tolerated antibiotic that shows one of the highest docking scores. The drug is deeply buried in the active site pocket of 3CLpro, but comes only into contact with His41, Met49, His164 and Gln189, when docked to 3CLpro, whereas other contacts to eligible surrounding amino acids are missing (Fig. 4). Therefore, it was hypothesized that oxytetracycline would be a good candidate to exploit SBDD to optimize the potential affinity of the starting molecule to the target protein. The C-ring of oxytetracycline shows only one contact with Met49 and the D-ring has no contact to adjacent pocket-amino acids Ser46, Thr26, Gly143 or Cys145 (Fig. 7 A). In a first step, the 6-methyl group was changed to a 6-methyl-aminoethyl group, in order to explore a possible hydrogen bond with the side chain of Ser46. After energy minimization, this hydrogen bond was indeed established (Fig. 7B). Since the backbone carbonyl oxygen of Thr26 was in beneficial proximity to the aromatic D-ring, a hydroxyl-function was introduced in 8-position leading to the desired additional hydrogen bond (Fig. 7C). The electrostatic map of the binding pocket revealed an unsatisfied acceptor patch at the 10-position of the D-ring. Therefore, the 10-hydroxygroup was changed to a carbonyl group, which required a modification of the aromatic D-ring into a hydroxy-cyclohexadiene-one (Fig. 7D). The resulting new analog of oxytetracycline formed the anticipated hydrogen bond with the backbone nitrogen of Gly143. Altogether the SBDD optimization of oxytetracycline afforded 4 additional hydrogen bonds with active site pocket flanking amino acids Ser46, Thr26, Gly143 and Cys145, which was accompanied by a significantly improvement of the GBVI/WSA dG docking score from -9.3 to -9.9 (Fig. S7). The introduction of the methylamino-modification into oxytetracycline was by far the most important step for improving binding affinity.
Fig. 7.
Structure based drug design: A) shows the chemical starting point oxytetracycline docked into the active site of 3CLpro. B) 6-Methylaminoethyl-analog of oxytetracycline with an additional hydrogen bond to Ser46. C) 6-Methylaminoethyl-8-hydroxy-analog of oxytetracycline forms a further hydrogen bond with Thr26. D) The 6-Methylaminoethyl-8-hydroxy-analog of oxytetracycline was further modified. The former aromatic D-ring was changed into a hydroxy-cyclohexadien-one enabling two further hydrogen bonds with Gly143 and Cys145. Hydrogen bonds are indicated by dotted green lines.
4. Conclusions
The current SARS-CoV-2 pandemic motivated this study to rapidly identify already approved drugs that could be repurposed to target 3CLpro, a cysteine protease essential for replication of Coronaviruses. A virtual screening approach yielded several scaffolds with high potential to inhibit 3CLpro including flavonoids, which have very recently been described as experimentally confirmed inhibitors of this enzyme, antibiotics and anticancer agents. Most promising hits were oxytetracycline, naringin, kanamycin, cefpiramide, salvianolic acid b, teniposide, etoposide and doxorubicin. A subsequent pharmacophore search broadened the screened chemical space tremendously by screening 7.2 million compounds from the ZINC15 library. Besides known substance classes like flavonoids several novel scaffolds were discovered including the high scoring molecules ZINC000096222891, ZINC000008219992, ZINC000218242102, ZINC000015275997, the fluoroquinolone ZINC001570001158 and the sulfonamide ZINC000016429284. Moreover, the comprehensive dissection of the essential contacts between ligands and active site pocket amino acids revealed new opportunities to design and optimize inhibitors of 3CLpro. The feasibility of SBDD to optimize hit compounds was demonstrated using already high scoring oxytetracycline. Three substantial modifications at the C- and D-ring of the tetracycline scaffold reinforced binding affinity by four additional hydrogen bonds to adjacent pocket amino acids Ser46, Thr26, Gly143 and Cys145. The results of the study suggest that already approved antibiotics, particularly oxytetracycline, have great potential to inhibit 3CLpro of SARS-CoV-2. It is self-evident, that the efficacy of oxytetracycline and other promising hit compounds has to be experimentally confirmed. But repurposing of e.g. oxytetracycline would tremendously shorten the time period to the clinics, because the safety profile is well known. If the proposed antibiotics turn out to be active against Coronoviruses, they would offer the extra benefit to fight at the same time secondary multi-infections in the lung often observed together with COVID-19.
Author statement
The present study has been conceptualized, performed and written by the sole author.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.compbiolchem.2020.107351.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Baell J.B., Holloway G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53(7):2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Bajorath J., Peltason L., Wawer M., Guha R., Lajiness M.S., Van Drie J.H. Navigating structure-activity landscapes. Drug Discov. Today. 2009;14(13–14):698–705. doi: 10.1016/j.drudis.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Berry M., Fielding B.C., Gamieldien J. Potential broad Spectrum inhibitors of the coronavirus 3CLpro: a virtual screening and structure-based drug design study. Viruses. 2015;7(12):6642–6660. doi: 10.3390/v7122963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuzzi S.J., Muratov E.N., Tropsha A. Phantom PAINS: Problems with the Utility of Alerts for Pan-Assay INterference CompoundS. J. Chem. Inf. Model. 2017;57(3):417–427. doi: 10.1021/acs.jcim.6b00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.W., Yiu C.B., Wong K.Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res. 2020;9:129. doi: 10.12688/f1000research.22457.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care. 2020 doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Ábrányi-Balogh P., Brandaõ-Neto J., Carbery A., Davison G., Dias A., Downes T.D., Dunnett L., Fairhead M., Firth J.D., Jones S.P., Keely A., Keserü G.M., Klein H.F., Martin M.P., Noble M.E.M., O’Brien P., Powell A., Reddi R., Skyner R., Snee M., Waring M.J., Wild C., London N., von Delft F., Walsh M.A. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. bioRxiv. 2020;2020 doi: 10.1038/s41467-020-18709-w. 05.27.118117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du B., Qiu H.B., Zhan X., Wang Y.S., Kang H.Y.J., Li X.Y., Wang F., Sun B., Tong Z.H. [Pharmacotherapeutics for the new coronavirus pneumonia] Zhonghua Jie He He Hu Xi Za Zhi. 2020;43(0):E012. doi: 10.3760/cma.j.issn.1001-0939.2020.0012. [DOI] [PubMed] [Google Scholar]
- Fan H.H., Wang L.Q., Liu W.L., An X.P., Liu Z.D., He X.Q., Song L.H., Tong Y.G. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus (2019-nCoV) related coronavirus model. Chin. Med. J. 2020 doi: 10.1097/CM9.0000000000000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D.K., Kubin C., Barr R.G., Sobieszczyk M.E., Schluger N.W. Observational study of hydroxychloroquine in hospitalized patients with Covid-19. N. Engl. J. Med. 2020;382(25):2411–2418. doi: 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J., Mafham M., Bell J., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., Prudon B., Green C., Felton T., Chadwick D., Rege K., Fegan C., Chappell L.C., Faust S.N., Jaki T., Jeffery K., Montgomery A., Rowan K., Juszczak E., Baillie J.K., Haynes R., Landray M.J. Effect of Dexamethasone in Hospitalized Patients with COVID-19: Preliminary Report. medRxiv. 2020;2020 06.22.20137273. [Google Scholar]
- Jacobs J., Grum-Tokars V., Zhou Y., Turlington M., Saldanha S.A., Chase P., Eggler A., Dawson E.S., Baez-Santos Y.M., Tomar S., Mielech A.M., Baker S.C., Lindsley C.W., Hodder P., Mesecar A., Stauffer S.R. Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J. Med. Chem. 2013;56(2):534–546. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Rcsb Protein Data Bank. 2020 [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35(1):145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Zhu H.M., Niu G.J., Shi E.Z., Chen J., Sun B., Chen W.Q., Zhou H.G., Yang C. Synthesis, modification and docking studies of 5-sulfonyl isatin derivatives as SARS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2014;22(1):292–302. doi: 10.1016/j.bmc.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P., Shah F., Desai P., Avery M. Inhibitors of SARS-3CLpro: virtual screening, biological evaluation, and molecular dynamics simulation studies. J. Chem. Inf. Model. 2011;51(6):1376–1392. doi: 10.1021/ci1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Sander T., Freyss J., von Korff M., Rufener C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015;55(2):460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- Sterling T., Irwin J.J. ZINC 15 – ligand discovery for everyone. J. Chem. Inf. Model. 2015;55(11):2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inform. 2020 doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K.C., Chen S.Y., Liang P.H., Lu I.L., Mahindroo N., Hsieh H.P., Chao Y.S., Liu L., Liu D., Lien W., Lin T.H., Wu S.Y. Discovery of a novel family of SARS-CoV protease inhibitors by virtual screening and 3D-QSAR studies. J. Med. Chem. 2006;49(12):3485–3495. doi: 10.1021/jm050852f. [DOI] [PubMed] [Google Scholar]
- Turlington M., Chun A., Tomar S., Eggler A., Grum-Tokars V., Jacobs J., Daniels J.S., Dawson E., Saldanha A., Chase P. Discovery of N-(benzo [1, 2, 3] triazol-1-yl)-N-(benzyl) acetamido) phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 2013;23(22):6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020 doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.