Abstract
The Coronavirus disease 2019 (COVID-19) pandemic has spread to almost all nooks and corners of the world. There are numerous potential approaches to pharmacologically fight COVID-19: small-molecule drugs, interferon therapies, vaccines, oligonucleotides, peptides, and monoclonal antibodies. Medications are being developed to target the spike, membrane, nucleocapsid or envelope proteins. The spike protein is also a critical target for vaccine development. Immunoinformatic approaches are being used for the identification of B cell and cytotoxic T lymphocyte (CTL) epitopes in the SARS-CoV-2 spike protein. Different vaccine vectors are also being developed. Chemical and physical methods such as formaldehyde, UV light or β-propiolactone are being deployed for the preparation of inactivated virus vaccine. Currently, there are many vaccines undergoing clinical trials. Even though mRNA and DNA vaccines are being designed and moved into clinical trials, these types of vaccines are yet to be approved by regulatory bodies for human use. This review focuses on the drugs and vaccines being developed against the COVID-19.
Key Words: SARS-CoV-2, COVID-19, Spike protein, Coronavirus, Drugs, Vaccine
Graphical abstract
Introduction
Coronavirus disease 2019 (COVID-19) is currently having a damaging impact on almost all countries in the world. The pandemic, which has spread to almost all parts of the globe has a basic reproduction number (R0) of 2–2.5, implying that 2–3 people may acquire the infection from an index patient (1). The causative microorganism for the disease is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The disease is characterized by high mortality rate, the absence of medical countermeasures and a large distribution of reservoir (2). The deleterious economic consequences of this pandemic in incalculable with many countries stretching the resources of their healthcare facilities and job losses recorded across several industries. COVID-19, which was first documented in Wuhan, Hubei Province, China at the end of 2019 (3), is highly transmittable and pathogenic (4). Cough, fever, and dyspnea are symptoms of patients suffering from COVID-19 (5). Severe acute respiratory distress, pneumonia, renal failure and death are associated with severe forms of the infection (5). It is challenging to compute the number of asymptomatic individuals infected with COVID-19 (6). For individuals exhibiting symptoms of the disease, cough, fever, rhinitis, fatigue and other signs typically begin to manifest after few days (6). About 75% of COVID-19 patients exhibit symptoms as detected by computed tomography (6). Pneumonia frequently presents in patients during the second or third week of symptomatic COVID-19 infection and the main symptoms of viral pneumonia include blood gas deviations, patchy consolidation, decreased oxygen saturation, ground glass abnormalities, interlobular involvement and alveolar exudates (6). Lymphopenia is frequently observed together with an increase in proinflammatory cytokines as well as inflammatory markers (C-reactive protein) (6). Oxygen therapy and treatment of symptoms constitute the mainstay of therapy while mechanical ventilators are used for patients with pulmonary failure (7).
In the past twenty years, H5N1 influenza A, SARS-CoV, H1N1 2009, and the Middle East respiratory syndrome coronavirus (MERS-CoV) have crossed from birds or mammals to humans (4). These are mostly respiratory disorders some of which are characterized by acute respiratory distress syndrome (ARDS) as well as acute lung injury (ALI), leading sometimes to respiratory failure and death (4).
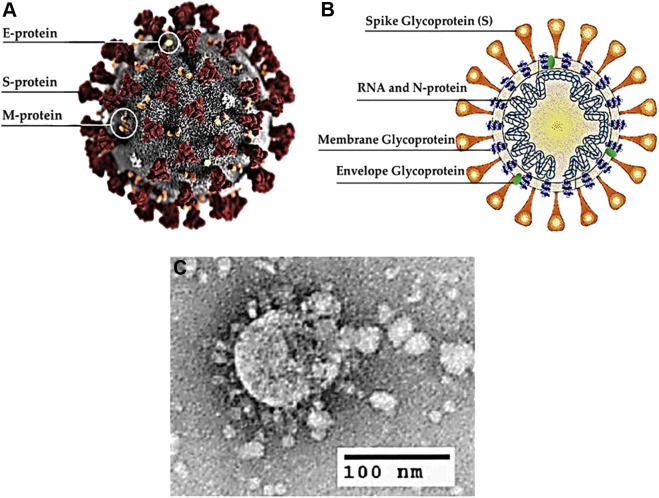
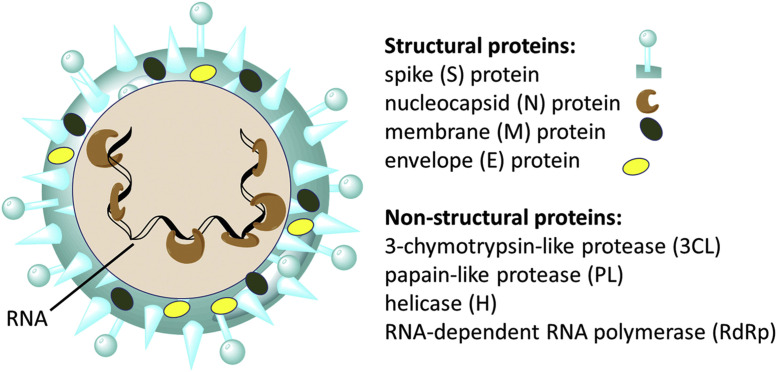
By analyzing the results obtained by sequence alignment and phylogenetic clustering and taking into consideration the common animals found in Wuhan, Qiu et al predicted that pangolin, buffalo, cat, goat, cow, pigeon, sheep swine and/or civet could be the intermediate hosts for SARS-CoV-2 (8). There is no final agreement among researchers on what the specific intermediate host(s) is(are). Some investigators theorize that the likely intermediate host candidate is the pangolin as the genetic sequences of coronaviruses from the mammal is 99% similar compared to those obtained from individuals infected with SARS-CoV-2 (9). The rate of SARS-CoV-2 transmission between individuals is high (10). COVID-19 has become a global pandemic. It is spreading at an exponential rate and there are still no effective drugs with which to treat the disease even though some therapeutic options do exist. According to the Coronavirus Resource Center of John Hopkins University in Baltimore, Maryland, USA, there were 25,349,528 reported cases of COVID-19 and 848,394 deaths worldwide as at August 31, 2020 (11). SARS-CoV-2 encodes four principal structural proteins: the spike (S),membrane (M), envelope (E), and nucleocapsid (N) proteins (12). The S protein is the major trans-membrane glycoprotein that mediates receptor-binding and virion entry (12). An illustration of the SARS-CoV-2 virion and the structure of SARS-CoV-2 (13) are shown in Figure 1 .
Figure 1.
Structure of SARS-CoV-2. (A) Illustration of the SARS-CoV-2 virion created at the Centers for Disease Control and Prevention (CDC). The spikes on the outer edge of the virus particles look like a crown, giving the disease its characteristic name. (B) Schematic representation of the structure of SARS-CoV-2. It has four structural proteins, S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins; the N protein holds the RNA genome, and the S, E, and M proteins together create the viral envelope. (C) An electron microscopic image of a thin section of SARS-CoV-2 within the cytoplasm of an infected cell, showing the spherical particles and cross-sections through the viral nucleocapsid.
(reproduced with permission from reference (13))
Treatment Options
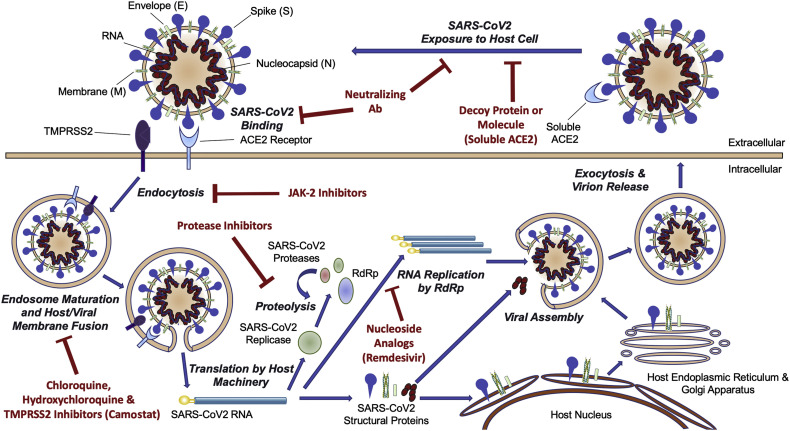
Clinical trials are being carried out in which potential antiviral therapy targets are tested, such as inhibiting viral enzymes that were responsible for genome replication or blocking viral entry into human cells (14). There are numerous potential approaches to pharmacologically fight COVID-19: small-molecule drugs, interferon therapies, vaccines, oligonucleotides, peptides and monoclonal antibodies (15). The medications that can act on a coronavirus can be categorized based on their mechanisms of action (5): (1) those that act on viral proteins and enzymes thus preventing RNA replication and synthesis (2) those that act on the viral structural proteins, inhibiting self-assembly or blocking the virus from tethering to ACE2 (3) those that act on virulence factors and can facilitate the restoration of the host's innate immunity (4) those that can act on human enzymes or receptors thus blocking viral entry (5). The S protein is a critical target for vaccine development (3). Few drugs, however, are being developed to target the membrane, nucleocapsid or envelope proteins. A scheme of SARS-CoV-2 and some of its molecular protein targets (15) are shown in Figure 2 .
Figure 2.
Scheme of SARS-CoV-2 and some of its molecular protein targets.
(reproduced with permission from reference (15))
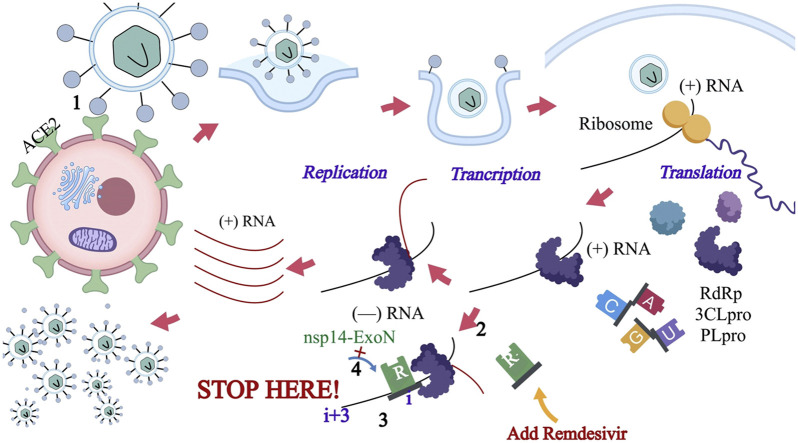
Some of the drugs that are currently being used as investigational therapeutic agents for the management of COVID-19 are repurposed medications that are usually given to patients suffering from other viral infections such as anti-HIV agents or drugs used for the management of influenza. Recently, Yan and colleagues published a report showing the high-resolution structures of full-length ACE2 (16). The authors suggested that the existence at the same time of bonds between the ACE2 dimer and the two S protein trimers (16). Effective pharmacotherapeutic approaches against SARSCoV-2 can either be anchored on the utilization of specific drugs that can inhibit viral attachment and entry or the use of broad-spectrum anti-viral medications (17). Peptidic fusion inhibitors, antiSARS-CoV-2 neutralizing monoclonal antibodies, protease inhibitors, certain antimalarial medications, and anti-ACE2 monoclonal antibodies are possible pharmacotherapeutic options (17). There are conflicting reports in the literature about the clinical efficacy of some investigational medications being used for the management of COVID-19. Because of the current high morbidity and mortality, there is insufficient time to carry out clinical trials and some of the medications are being used on compassionate grounds. Concurrently, clinical trials are being conducted for some of these medications while the trials for others are yet to begin. Putative SARS-CoV-2 Life Cycle and therapeutic targets are shown in Figure 3 (18).
Figure 3.
Putative SARS-CoV-2 Life Cycle and Therapeutic Targets.
(reproduced with permission from reference (18))
In a different study, Monteil and coworkers provided an in vitro evidence demonstrating that human recombinant soluble ACE2 (hrsACE2) can reduce viral growth (19). The authors also reported that infections of the kidney organoids and human blood vessel organoids can be significantly blocked by hrsACE2 at an early phase (19). Remdesivir, favipiravir and chloroquine are being proposed for the treatment of COVID-19. Other repurposed medications which may be useful include ritonavir/lopinavir alone or used in conjunction with monoclonal antibodies and interferon-β (20). Lopinavir (LPV) has been shown to block coronavirus protease activity in vitro and in animal studies (21). Researchers typically combine ritonavir with lopinavir to increase its plasma half-life by inhibiting cytochrome P450 (14). The targets of protease inhibitors in coronaviruses are 3C-like protease and papain-like protease (22). A randomized controlled trial enrolled COVID-19 patient with dyspnea and desaturation in China and suggested that treatment with lopinavir/ritonavir was comparable to standard care in the time to clinical improvement (14). However, therapy with this drug combination was terminated early because of side effects such as diarrhea, nausea, and hepatotoxicity (14). Leronlimab is a C-C chemokine receptor type 5 antagonist and a humanized monoclonal antibody while galidesivir is a nucleoside RNA polymerase blocker (6). Researchers are investigating the feasibility of using the two medications the management of COVID-19 (6).
SARS-CoV-2 tethers to the alveolar epithelium and subsequently activates both the adaptive immune system and the innate immune system leading to the release of a large amount of cytokines, including interleukin 6 (IL-6) (23). Tocilizumab(TZM) is an anti-IL-6 receptor monoclonal antibody (23). The drug binds to the membrane-bound as well as the soluble IL-6 receptors (mIL-6R and sIL-6R) and blocks mIL-6R and sIL-6R-mediated signal transduction (23). Cytokine release syndrome (CRS) has been documented for many patients with severe COVID-19 and CRS has led to several deaths (23). IL-6 is one of the principal mediators of CRS thus the IL-6R antagonist TZM may be useful for the management of the so called “cytokine storm” observed in COVID-19 patients (23). “Cytokine storm” is characterized by an elevated level on inflammatory markers especially cytokines (24). Indeed, TZM is being used as an investigational agent against SARS-CoV-2.
CR3022 is a monoclonal antibody that was obtained from a convalescent SARS patient and the compound is encoded by the following genes: IGHD3-10, IGHV5-51, IGHJ6 (heavy chain), and IGKV4-1, IGKJ2 (light 56 chain) (25). Even though a high conserved domain in the epitope residues has been reported, CR3022 Fab interacts with SARS-CoV RBD with significantly greater attraction than to SARS-CoV-2 RBD (25). It was postulated that the disparities in the bonding of CR3022 to SARS-CoV-2 or SARS-CoV RBDs may derive from the non-conserved residues found in the epitope (25). CR3022 binds to RBD of the SARS-CoV-2 spike protein (4,26). This is because there is no overlap between the antibody's epitope and the ACE2 receptor-binding motif (4,26). CR3022 may be useful for the management of COVID-19 either alone or in tandem with other neutralizing antibodies (4,26). However, clinical efficacy and safety studies should be carried out before utilizing these drugs for individuals suffering from COVID-19 (20).
Remdesivir (RDV) is a 1′-cyano-substituted adenosine analog, a phosphoramidate prodrug and an RNA-dependent RNA polymerase (RdRp) blocker that acts by inhibiting the synthesis of viral nucleic acid via bond formation with the active site of RdRp (5,22,27). RdRp is a protease that mediates the replication of RNA from an intermediate template (28). Another mechanism of action of RDV involves the avoidance of proofreading by the exoribonuclease of SRS-CoV-2 (22). As a result of these effects, the transcription of the viral RNA is stopped prematurely (22). Remdesevir, originally developed to treat Ebola virus and then dropped (29), is being used as an investigational drug for COVID-19 patients (30). Remdesevir also displays antiviral effect against other types of RNA viruses, such as MERS-CoV and SARS-CoV (31). Even though widespread drug interactions and cardiovascular toxicities have not yet been documented, there were cases of hypotension with ensuing cardiac arrest after a loading dose in one patient (among 175 total) following the use of the remdesivir during the Ebola epidemic (30). Leronlimab (PRO140) is a CC chemokine receptor 5 (CCR5) antagonist and an investigational new medication for the management of COVID-19 (32). CCR5 takes part in diverse biologic processes, such as tumor invasion and metastases, HIV-1 ingress into CD4+ T cells and the pathogenesis of nonalcoholic steatohepatitis (NASH) (32). Favipiravir is another medication used for the management of COVID-19. Although the specific mechanism of action against SARS-CoV-2 is yet to be fully elucidated, the drug is quickly pinpointed as a substrate of the viral RNA polymerase following conversion into an active phosphoribosylated state (22). The drug blocks viral genomic RNA synthesis as a chain terminator (33). An efficient approach to discover drugs against COVID-19 is to determine whether the existing antiviral drugs are effective (34). Favipiravir (6-fluoro-3-hydroxy-2-pyrazinecarboxamide) (FPV) is an oral pyrazinecarboxamide derivative and guanine analogue that potently and selectively blocks the RNA-dependent RNA polymerase (RdRp) of RNA viruses (35). It was recently shown that, as a prodrug, FPV effectively inhibits the SARS-CoV-2 infection in Vero E6 cells (34).
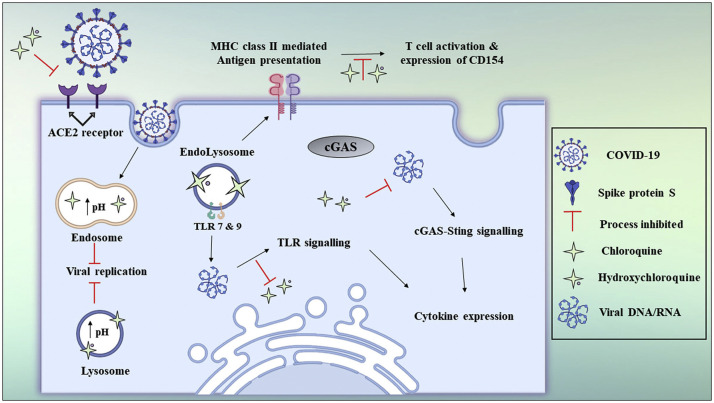
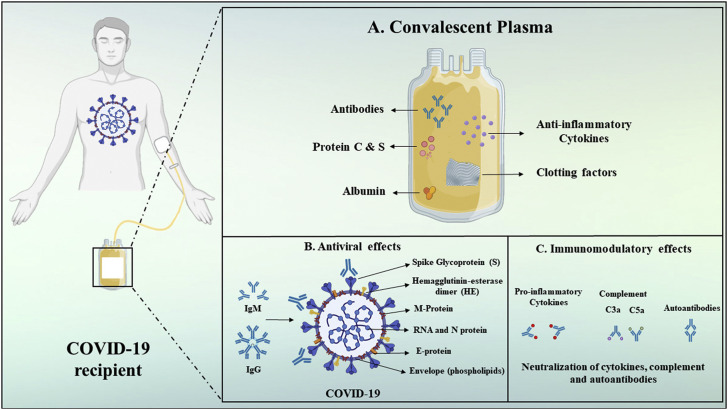
Hydroxychloroquine and chloroquine are utilized, together with antiviral medications, as investigational treatment options for the pharmacotherapy of COVID-19-associated pneumonia (36). It has been postulated that chloroquine inhibits viral particles from tethering to cell surface receptor thus blocking the viral pre-entry phase of COVID-19 (37). The drug acts on quinone reductase 2, which is structurally related to UDP-N-acetylglucosamine 2-epimerases(UNEs) (37). UNEs catalyze sialic acid biosynthesis. Sialic acids are structural constituents of sugar molecules available on cell transmembrane proteins and these are critical factors needed for the recognition of ligands (37). The potent effect of chloroquine against SARS-CoV-1 in vitro were ascribed to impaired glycosylation of ACE2 (37). Chloroquine can also interfere with the pH-dependent endosome-mediated ingress of SARS-CoV viruses (37). Acidic pH is essential for the fusion of the endosomal and viral membranes leading to the cytosolic delivery of the SARS-CoV-1 genome (37). Without an antiviral medication, the virus enters into the lysosome where enzymatic activity as well as the low pH cleaves the viral particle and releases replication enzymes together with the RNA (37). The mechanism of antiviral activity by chloroquine is postulated to entail the swift increase in the endosomal pH, the prevention of endocytosis and the disruption of endosome-virus fusion (37,38). The antiviral mechanisms of chloroquine and hydroxychloroquine (39) are shown in Figure 4 . In another study, blood plasma from convalescent COVID-19 patients was transfused into individuals with SARS-CoV-2 infection with positive and rapid results leading to recovery (4,18). Convalescent plasma components and its mechanisms of action (19) are shown in Figure 5 .
Figure 4.
Chloroquine and hydroxychloroquine antiviral mechanisms.
(reproduced with permission from reference (19))
Figure 5.
Convalescent plasma components and its mechanisms of action.
(reproduced with permission from reference (19))
Ivermectin is a broad spectrum anti-parasitic agent approved by the FDA (14). To test the antiviral activity of ivermectin towards SARS-CoV-2, Caly et al. infected Vero/hSLAM cells with SARS-CoV-2 isolate Australia/VIC01/2020 followed by the addition of ivermectin (40). It was reported that this drug reduced viral RNA up to 5000- fold after 48 h of infection with SARS-CoV-2 (14,40).
Vaccine Candidates
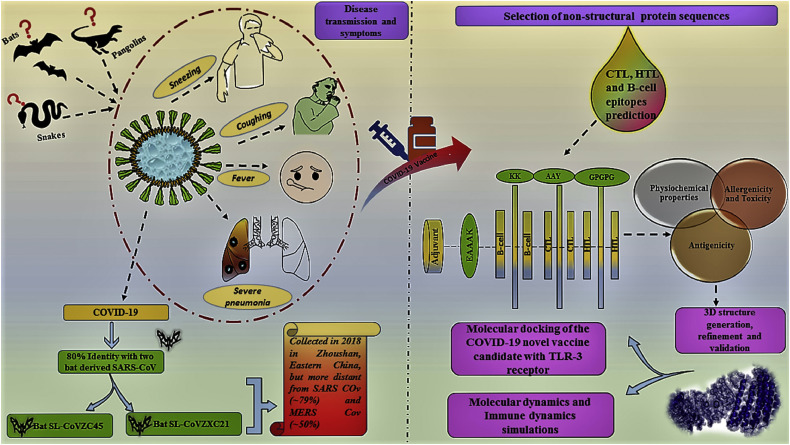
As of 1st June 2020, there were 124 candidate vaccines that were being developed for the prophylaxis of COVID-19 (41). Of these, 10 vaccine candidates had entered phase 1, combined phase 1/2 or phase 2 human clinical trials in adults (41). Multiple strategies are being adopted for the design and production of vaccines against SARS-CoV-2. Schematic flow of the transmission of COVID-19 and work flow used in the designing of vaccine candidate against SARS-CoV-2 (42) are shown in Figure 6 . Chemical and physical methods such as formaldehyde, UV light or β-propiolactonean can be utilized for the preparation of inactivated virus vaccine (43). In contrast, attenuated-virus vaccines can be formulated by using a virus with diminished pathogenesis such as increased anti-inflammatory cytokine levels, reduced neutrophil influx, less lung injury in comparison with the wild-type SARS-CoV-2 (43). Most vaccines are directed against the surface-exposed spike (S) glycoprotein (20). Numerous investigators have utilized vaccine design strategies based on the use of S1-receptor-binding domain (RBD), full-length S protein or expression in virus-like particles (VLP), DNA, or viral vectors (20). It is speculated that the use of spike protein-based vaccines can lead to the production of antibodies that block viral genome uncoating and receptor binding (20). The development of a universal CoV vaccine feasible because the T-cell epitopes of SARS and MERS-CoVs are similar and may induce cross-reactivity (20). SARS-CoV-2 possesses a high genetic homology compared to the SARS-CoV thus SARS-CoV vaccine may display cross-reactivity to SARS-CoV-2 (20). Analysis of S protein sequences of both types of viruses showed highly variable amino acid residues in the S1 subunit (20). That variability implies that vaccines that lead to prophylactically relevant immune response against SARS-CoV might not act effectively on SARS-CoV-2 (20).
Figure 6.
Schematic flow of the transmission of recent outbreak COVID-19 and work flow used in the designing of vaccine candidate against SARS-CoV-2.
(reproduced with permission from reference (43))
The native S protein exists as a trimer on the surface of SARS-CoV-2 (2). In eukaryotes, its ectodomain or S1 subunit is expressed mostly in a monomeric form (2). To synthesize trimeric recombinant codon optimized subunit proteins, Kim et al. fused SARS-CoV-2-S1 and MERS-CoV-S1 sequences to a 27 amino acid foldon segment (2). The foldon was obtained from the C-terminal domain of the T4 fibritin bacteriophage and it can form trimers (2). RS09 or flagellin are known TLR4 or TLR5 agonists respectively. The authors added these immune stimulants to the protein vaccine (2). A 6 histidine tag and a sequence used for the cleavage of the Tobacco Etch Virus (TEV) protease was also incorporated to facilitate metal chelating affinity purification (2). A shuttle carrier (pAd/MERS-S1f) was used in the system by the investigators (1). It was previously demonstrated that SARS-CoV-S1 and MERS-S1 subunit vaccine delivered with an adenoviral vector was more effective than full-length S1, implying that the subunit immunogen may be an optimal vaccine candidate (2). The authors used carboxymethyl cellulose to prepare dissolvable microneedles loaded with the proteins MERS-S1f, MERS-S1fRS09, MERS-S1ffliC, to SARS-CoV-2-S1, or SARSCoV-2-S1fRS09) (2). Micromolding was used to prepare 10 x10 obelisk-shaped microneedles from polydimethylsiloxane (PDMS) templates (2). Next, the authors prepared CMC-based MNArMERS-S1f, MNA-rMERS-S1fRS09, MNA-rMERS-S1ffliC, MNA-rSARSCoV-2-S1, or MNA-rSARS-CoV-2-S1fRS09 vaccines using a two-step spin-drying technique (2). The pre-clinical immunogenicity of the MERS-CoV vaccines administered subcutaneously with conventional hypodermic needles was compared to intracutaneous administration with the prepared dissolving microneedles (2). Virus neutralization assays were carried out and immunoglobulin G antibodies measured (2). Significantly, the SARS-CoV-2 S1 subunit vaccines delivered with microneedles provided effective immune responses that were observed 14 days following vaccination (2). Microneedles are beneficial due to their noninvasiveness and painlessness (44,45).
The main aim of using peptide vaccines is to synthesize T-cell and B-cell epitopes that can induce specific immune responses and are immunodominant (46). Immunogens can be formed by linking a T-cell epitope to the B-cell epitope of a target molecule (46). T-cell epitopes are short peptide fragments (8–20 amino acids), while B-cell epitopes are longer and can be proteins (46). Peptide-based vaccines can also be administered indirectly. Ji et al. used SARS-CoV2 non-replicating, S antigen-expressing cells as presenters and vectors of immunogenic antigens (these are the so called “Icells”) (47). By using irradiated cells as the presenting carriers of SARS-CoV-2 antigens(s), the immune system can recognize viral proteins resulting in an efficient immunity. Generex Biotechnology company uses the direct peptide approach. The company has developed a peptide vaccine against SARS-CoV-2 using synthetic viral peptides as the immunogen(s) and leveraging a patented and proprietary Ii-Key™ immune system activation platform (48). Another company (Novavax) has also developed NVX-CoV2373 which is a vaccine candidate directed against SARS-CoV-2 (49). This is a stable, prefusion protein incorporated into the company's proprietary nanoparticle platform(Matrix-M™) to boost immune responses and stimulate a higher blood concentration of neutralizing antibodies (49).
Even though mRNA and DNA vaccines are being designed and moved into clinical trials, these types of vaccines are yet to be approved by regulatory bodies for human use (43). DNA vaccine may be formulated against the SARS-CoV-2 and expressed as an antigen protein within human cells (50). This method is beneficial because it mimics live attenuated vaccines from the standpoint of a coordinated activation of immune responses (50). It is also relatively convenient to prepare DNA vaccines and safety issues are somewhat less (compared to live vaccines) (50). Highly-purified DNA vaccines can be manufactured at a large scale, and they are stable compared to proteins and other biopolymers (51). DNA vaccines have not yet been approved for humans. Some companies are investigating DNA vaccines against SARS-CoV-2 and indeed Inovio Pharmaceuticals is currently carrying out clinical trials on a DNA vaccine. Several vectors are also being investigated for SARS-CoV-2 vaccine candidates. The Oxford Vaccine Group in collaboration with Oxford Jenner Institute is currently conducting clinical trials for an adenoviral (ChAdOx1)vector-based SARS-CoV-2 vaccine (52).
Vaccines based on viral vectors can be constructed and used without an adjuvant but the formulation of such vaccines require antigens with neutralizing epitopes (43). Recombinant adenovirus vectors are relatively safe and their use can engender potent and broad cellular as well as humoral immune response (53). Adenovirus vector construction is quite demanding due to the enormous size of the genome used (36 kilobases) (53). Furthermore, there are sparse restriction sites. Usually, conventional techniques based on homologous recombination are used while some investigators depend on the rare restriction sites, but these approaches are time consuming and challenging to control (53). Some authors have used the Gibson assembly ligation which allows investigators to assemble several overlapping DNA molecules via the combined effect of a DNA polymerase, 5′ exonuclease, and a DNA ligase (53,54). The authors first broke the DNA fragments, obtaining single-stranded DNA overhangs that specifically annealed, and then they were covalently conjugated (54).
There is considerable scientific interest in the use of RNA vaccine for the management of COVID-19. Messenger RNA (mRNA) represents the intermediate phase in the translation of protein-encoding DNA and protein biosynthesis via ribosomes in the cytoplasm (55). Two major types of RNA are presently investigated as vaccines: virally derived, self-amplifying RNA and non-replicating mRNA (55). Self-amplifying RNAs usually the antigen and the required machinery for viral replication while traditional mRNA-based vaccines encode only the antigen of interest with 5′ and 3′ untranslated regions (UTRs) (55). Compared to traditional vaccines, mRNA-based vaccines are highly potent and can be quickly developed, manufactured at a low and safely administered (55). The precision of antigen design necessary to provide both timely and effective responses to emerging threats of epidemics and pandemics (56). The use of mRNA for vaccine formulation has several benefits compared to live attenuated, killed virus, subunit or DNA-based vaccines (55). mRNA is a non-integrating and non-infectious platform, thus is no potential risk of insertional mutagenesis or infection (55). Moreover multiple mRNAs encoding several antigens can be delivered in a single vaccine (57). Moderna® has developed a vaccine candidate (mRNA-1273), which is involved in the synthesis of a prefusion-stabilized conformation of the SARS-CoV-2 S protein (58). The vaccine is currently undergoing clinical trials (58).
Richner and coworkers recently developed a modified non self-amplifying mRNA vaccine that contains an mRNA containing an open reading frame(ORF) (59). The ORF encodes the antigen (59). The authors prepared the mRNA in vitro via T7 polymerase-mediated DNA-dependent RNA transcription where the Uridine-5′-triphosphate (UTP) was substituted with 1-methylpseudoUTP. A linearized DNA template, which contains the 5′ and 3′ untranslated regions (UTRs) with a poly-A tail was used (59). The authors added the donor methyl group S-adenosylmethionine (SAM) to the methylated capped RNA (cap 0), leading to the formation of a cap 1 structure for enhanced efficiency of mRNA translation (59).
There are two primary routes for the development of COVID-19 vaccines: the choice of antigens and the selection of an efficient carrier. Lipid nanoparticles are currently being investigated for the development of COVID-19 vaccines. Lipid nanoparticle (LNP) delivery of modified mRNA has been highlighted in the literature (57). In an interesting study, Geall and coworkers reported that the LNP delivery of a 9 kb self-amplifying RNA significantly enhanced immunogenicity compared with the administration of naked RNA (60). First, the authors constructed DNA plasmids encoding the self-amplifying RNAs. The plasmids were then amplified Restriction digest was used to linearize the DNA (60). The MEGAscript T7 kit was used to transcribe the linearized DNA templates into RNA and the purification was through lithium chloride (LiCl) precipitation (60). The RNA was subsequently capped with a Vaccinia Capping system and purified via LiCl precipitation (60). A modulated ethanol dilution method was used to prepare the LNPs containing these lipid constituents: 1, PEG-DMG 2000, N,N-Dimethyl-2,3-bis([9Z,12Z]-octadeca-9,12-dienyloxy)propan-1-amine [DLinDMA], 2-Diastearoyl-sn-glycero-3-phosphocholine) and cholesterol (60).
Recently, Baruah et al. used an immunoinformatic approach to pinpoint B cell and cytotoxic T lymphocyte (CTL) epitopes in the SARS-CoV-2 spike protein (61). The authors also used molecular dynamics to investigate the bonding between major histocompatibility complex (MHC) class I supertype and the CTL epitopes (61). The authors found out three sequential B cell epitopes, five CTL epitopes, and five discontinuous B cell epitopes in the S protein (61). It was revealed that the CTL epitopes bind to MHC class I channels through several mechanisms such as salt bridge anchors and continuous hydrogen bonds demonstrating the feasibility of using these epitopes to mount an immune response (61). In another study, Ahmed and coworkers identified T cell and B cell epitopes that were similar in both SARS-CoV-2 and SARS-CoV (62). Out of 229 epitopes, approximately 82% were MHC Class I restricted epitopes. Significantly, 102 of the 229 epitopes were prepared from either the N (36) or the S (66) protein (62). Three of the sequences (QPYRVVVLSF, GYQPYRVVVL and PYRVVVLSF) were located entirely in the SARS-CoV receptor-binding motif that is regarded as important for viral ingress into the host cell (62). For the T cell epitopes, the authors analyzed the affiliated MHC alleles and recommended several epitopes that can confer a broad immune response both in China and globally (62).
In another study, Bhattacharya et al. characterized the spike protein of the SARS-CoV-2 for immunogenic epitope design (63). The authors selected 13 epitopes that bind to MHC I and 3 antigenic epitopes that bind to MHC-II (63). The authors also utilized the Immune Epitope Database server to analyze the S protein and found 34 linear B-cell epitopes (63) Analysis of the SARS-CoV-2 sequence T-cell epitopes capable of interacting with the MHC-I and MHC-II molecules was carried out (63). The authors found 8 epitopes against MHC-II and 29 epitopes against MHC-I (63).
Repurposed vaccines are also being studied (41). An oral polio vaccine is under investigation in the United States of America while three multi-centered randomized controlled trials on BCG vaccine administration are being carried out in Netherlands, Australia and South Africa (41). A measles vaccine trial for the prophylaxis of COVID-19 has been registered in Egypt (41).
Conclusion
This review is focused on the global and damaging impact of the COVID-19. A significant quantum of research has been carried out over a relatively brief period of time to gain a better understanding of the structure of SARS-CoV-2 and the effect of this virus on human health with the aim of developing effective countermeasures. Using viral protein information, several research groups are developing vaccines and drugs. Some of these therapeutic and prophylactic agents are presently undergoing clinical trials. It is anticipated that vaccines and drugs will be found to reduce the global public health damage unleashed by this virus.
(ARCMED_2020_1057)
Conflict of Interest
The author declares that there are no conflicts of interest.
Supplementary Data
References
- 1.Dashraath P., Jing Lin Jeslyn W., Mei Xian Karen L., et al. Coronavirus Disease 2019 (COVID-19) Pandemic and Pregnancy. Am J Obstet Gynecol. 2020;222:521–531. doi: 10.1016/j.ajog.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim E., Erdos G., Huang S., et al. Microneedle array delivered recombinant coronavirus vaccines: Immunogenicity and rapid translational development. EBioMedicine. 2020:102743. doi: 10.1016/j.ebiom.2020.102743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poland G.A. Another coronavirus, another epidemic, another warning. Vaccine. 2020;38(10):v–vi. doi: 10.1016/j.vaccine.2020.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adnan Shereen M., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velavan T.P., Meyer C.G. The COVID-19 epidemic. Trop Med Int Health. 2020;25:278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu Y., Zhao Y.B., Wang Q., et al. Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microbes Infect. 2020;22:221–225. doi: 10.1016/j.micinf.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 10.Lai C.-C., Shih T.-P., Ko W.-C., et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coronavirus Resource Center, John Hopkins University, Baltimore, Maryland, USA. 2020. https://coronavirus.jhu.edu/map.html
- 12.Su Q.-D., Yi Y., Zou Y.-N., et al. The biological characteristics of SARS-CoV-2 Spike protein Pro330-Leu650. Vaccine. 2020;38:5071–5075. doi: 10.1016/j.vaccine.2020.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abd El-Aziz T.M., Stockand J.D. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2) - an update on the status. Infect Genet Evol. 2020;83:104327. doi: 10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih H.-I., Wu C.-J., Tu Y.-F., et al. Fighting COVID-19: A quick review of diagnoses, therapies, and vaccines. Biomed J. 2020 doi: 10.1016/j.bj.2020.05.021. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dömling A., Gao L. Chemistry and Biology of SARS-CoV-2. Chem. 2020;6:1283–1295. doi: 10.1016/j.chempr.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan R., Zhang Y., Li Y., et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shanmugaraj B., Siriwattananon K., Wangkanont K., et al. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19) Asian Pac J Allergy Immunol. 2020;38:10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- 18.Atri D., Siddiqi H.K., Lang J.P., et al. COVID-19 for the Cardiologist: Basic Virology, Epidemiology, Cardiac Manifestations, and Potential Therapeutic Strategies. JACC Basic Transl Sci. 2020;5:518–536. doi: 10.1016/j.jacbts.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanessa Monteil H.K., Patricia Prado A.H., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhama K., Sharun K., Tiwari R., et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother. 2020;16:1232–1238. doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhai P., Ding Y., Wu X., et al. The epidemiology, diagnosis and treatment of COVID-19. Int J Antimicrob Agents. 2020;55(5):105955. doi: 10.1016/j.ijantimicag.2020.105955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jean S.-S., Lee P.-I., Hsueh P.-R. Treatment options for COVID-19: the reality and challenges. J Microbiol Immunol Infect. 2020;53:436–443. doi: 10.1016/j.jmii.2020.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C., Wu Z., Li J.-W., et al. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int J Antimicrob Agents. 2020;55:105954. doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chau V.Q., Oliveros E., Mahmood K., et al. The Imperfect Cytokine Storm: Severe COVID-19 with ARDS in Patient on Durable LVAD Support. JACC Case Rep. 2020;2:1315–1320. doi: 10.1016/j.jaccas.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan M., Wu N.C., Zhu X., et al. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. bioRxiv. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian X., Li C., Huang A., et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Y.-C., Deng Q.-X., Dai S.-X. Remdesivir for severe acute respiratory syndrome coronavirus 2 causing COVID-19: An evaluation of the evidence. Travel Med Infect Dis. 2020;35:101647. doi: 10.1016/j.tmaid.2020.101647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lung J., Lin Y.S., Yang Y.H., et al. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J Med Virol. 2020 doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat Biotechnol. Nat Biotechnol. 2020;38:379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 30.Driggin E., Madhavan M.V., Bikdeli B., et al. Cardiovascular Considerations for Patients, Health Care Workers, and Health Systems During the Coronavirus Disease 2019 (COVID-19) Pandemic. 2020;75:2352–2371. doi: 10.1016/j.jacc.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon C.J., Tchesnokov E.P., Feng J.Y., et al. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplon H., Muralidharan M., Schneider Z., et al. Antibodies to watch in 2020. MAbs. 2020;12:1703531. doi: 10.1080/19420862.2019.1703531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiraki K., Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol Ther. 2020;209:107512. doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai Q., Yang M., Liu D., et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering (Beijing) 2020 doi: 10.1016/j.eng.2020.03.007. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKee D.L., Sternberg A., Stange U., et al. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Favalli E.G., Ingegnoli F., De Lucia O., et al. COVID-19 infection and rheumatoid arthritis: Faraway, so close! Autoimmun Rev. 2020;19:102523. doi: 10.1016/j.autrev.2020.102523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Devaux C.A., Rolain J.-M., Colson P., et al. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int J Antimicrob Agents. 2020;55:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colson P., Rolain J.M., Lagier J.C., et al. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents. 2020;55:105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandey A., Nikam A.N., Shreya A.B., et al. Potential therapeutic targets for combating SARS-CoV-2: Drug repurposing, clinical trials and recent advancements. Life Sci. 2020;256:117883. doi: 10.1016/j.lfs.2020.117883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caly L., Druce J.D., Catton M.G., et al. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koirala A., Jin Joo Y., Khatami A., et al. Vaccines for COVID-19: the current state of play. Paediatr Respir Rev. 2020;35:43–49. doi: 10.1016/j.prrv.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ojha R., Gupta N., Naik B., et al. High throughput and comprehensive approach to develop multiepitope vaccine against minacious COVID-19. Eur J Pharm Sci. 2020;151:105375. doi: 10.1016/j.ejps.2020.105375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang W., Yang Y., Rao Y., et al. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vaccines. 2020;5:18. doi: 10.1038/s41541-020-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ita K. Dissolving microneedles for transdermal drug delivery: Advances and challenges. Biomed Pharmacother. 2017;93:1116–1127. doi: 10.1016/j.biopha.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 45.Ita K. Transdermal Delivery of Drugs with Microneedles-Potential and Challenges. Pharmaceutics. 2015;7:90–105. doi: 10.3390/pharmaceutics7030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdelmageed M.I., Abdelmoneim A.H., Mustafa M.I., et al. Design of multi epitope-based peptide vaccine against E protein of human COVID-19: An immunoinformatics approach. BioRxiv. 2020;2020:2683286. doi: 10.1155/2020/2683286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji H., Yan Y., Ding B., et al. Novel decoy cellular vaccine strategy utilizing transgenic antigen-expressing cells as immune presenter and adjuvant in vaccine prototype against SARS-CoV-2 virus. Med Drug Discov. 2020;5:100026. doi: 10.1016/j.medidd.2020.100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Generex Biotechnology Corporation. https://www.generex.com/covid-19
- 49.https://novavax.com/
- 50.Jia R., Yan L., Guo J. Enhancing the immunogenicity of a DNA vaccine against Streptococcus mutans by attenuating the inhibition of endogenous miR-9. Vaccine. 2020;38:1424–1430. doi: 10.1016/j.vaccine.2019.11.083. [DOI] [PubMed] [Google Scholar]
- 51.Bolhassani A., Yazdi S.R. DNA immunization as an efficient strategy for vaccination. Avicenna J Med Biotechnol. 2009 [PMC free article] [PubMed] [Google Scholar]
- 52.Oxford Vaccine Group. https://www.ovg.ox.ac.uk/news/covid-19-vaccine-development
- 53.Luo S., Zhang P., Ma X., et al. A rapid strategy for constructing novel simian adenovirus vectors with high viral titer and expressing highly antigenic proteins applicable for vaccine development. Virus Res. 2019;268:1–10. doi: 10.1016/j.virusres.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 54.Gibson D.G., Young L., Chuang R.-Y., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 55.Pardi N., Hogan M.J., Porter F.W., et al. mRNA vaccines — a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feldman R.A., Fuhr R., Smolenov I., et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 57.John S., Yuzhakov O., Woods A., et al. Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine. 2018;36:1689–1699. doi: 10.1016/j.vaccine.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 58.Moderna. https://www.modernatx.com/
- 59.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell. 2017;168:1114–11125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geall A.J., Verma A., Otten G.R., et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J Med Virol. 2020;92:495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. 2020;12(3):E254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhattacharya M., Ranjan Sharma A., Patra P., et al. Development of epitope-based peptide vaccine against novel Coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. J Med Virol. 2020 doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.